Abstract
Febrile seizures are the most prevalent type of seizures among children up to 5 years of age (2–4% of Western-European children). Complex febrile seizures are associated with an increased risk to develop temporal lobe epilepsy. To investigate short- and long-term effects of experimental febrile seizures (eFS), we induced eFS in highly febrile convulsion-susceptible C57BL/6J mice at post-natal day 10 by exposure to hyperthermia (HT) and compared them to normotherm-exposed (NT) mice. We detected structural re-organization in the hippocampus 14 days after eFS. To identify molecular candidates, which entrain this structural re-organization, we investigated temporal changes in mRNA expression profiles eFS 1 hour to 56 days after eFS. We identified 931 regulated genes and profiled several candidates using in situ hybridization and histology at 3 and 14 days after eFS. This is the first study to report genome-wide transcriptome analysis after eFS in mice. We identify temporal regulation of multiple processes, such as stress-, immune- and inflammatory responses, glia activation, glutamate-glutamine cycle and myelination. Identification of the short- and long-term changes after eFS is important to elucidate the mechanisms contributing to epileptogenesis.
Introduction
Febrile seizures are the most prevalent type of seizures among children up to 5 years of age (2–4% of Western-European children) [1]. Although simple febrile seizures are generally benign, complex and especially prolonged febrile seizures, lasting longer than 10 minutes with recurring episodes, are thought to contribute to the development of epilepsy (epileptogenesis). Retrospective studies amongst temporal lobe epilepsy (TLE) patients show a higher incidence of complex or prolonged febrile seizures during childhood compared to other types of epilepsy, and hence have been suggested to be a precipitating or aggravating factor in the development of TLE [2–5]. In addition, a series of prospective studies indicate that children with prolonged febrile seizures are at risk of hippocampal injury and development of hippocampal sclerosis, a common pathology among TLE patients [6,7]. Prolonged febrile seizures together with risk factors such as a family history of epilepsy, age of onset and underlying neurological abnormality, increase the risk of epilepsy later in life [8,9]. Genetic predisposition to febrile seizure susceptibility in human and mice shows that the molecular make-up of the brain contributes to the occurrence and impact of febrile seizures [10–12]. Animal models are highly suitable to study the relation between febrile seizures and epilepsy, and provide opportunity to monitor the underlying process of epileptogenesis.
Several animal models for febrile seizures have been developed, in which a temperature controlled chamber is used to trigger hyperthermia and subsequently the development of febrile seizures in rats and mice [13–15]. The development and onset of febrile seizures can be determined by behavioral and by electroencephalographic (EEG) criteria [13,14]. Studies on the effects of experimentally induced prolonged febrile seizures (eFS) in rat and mouse indicate structural and functional changes in temporal lobe brain areas such as the hippocampus and amygdala [15–20]. Hallmarks of such changes in the hippocampus after eFS are mild mossy fiber sprouting and ectopic migration of neonatally generated granule cells in the dentate gyrus, as well as CA1 neuronal hyper-excitability and a shift in activity-dependent synaptic plasticity [15,17,18,21]. Control experiments elegantly showed that these hippocampal changes are eFS-induced and not merely a consequence of the induced hyperthermia [13,15,21,22]. The molecular processes mediating these eFS-induced structural and functional changes are still largely unresolved. In resemblance with humans, eFS elicited in rodents are followed by a latent period until the first signs of spontaneous epileptiform activity as observed among 35–68% of the exposed animals [15,22,23]. The molecular changes that occur during the latent phase which eventually lead to spontaneous epileptiform activity are still largely unknown.
To investigate short- and long-term changes after eFS, we induced eFS in highly febrile convulsion susceptible C57BL/6J mice (post-natal age 10, P10) by warm-air induced hyperthermia (HT)[14]. We compared hippocampal mRNA expression profiles of eFS and normothermic control mice at various times (up to 56 days) after eFS induction. Subsequently we validated several regulated genes by qPCR and immunocytochemistry.
Materials and Methods
Ethics statement
All experiments conformed to institutional guidelines of the University Medical Center Utrecht, approved by the Animal Ethics Committee of Utrecht University, and were conducted in agreement with Dutch laws (Wet op de Dierproeven, 1996) and European regulations (Guideline 86/609/EEC).
Animals
Mice were bred from pairs of C57BL/6J mice (Jackson Laboratories, ME, USA) and raised by their mothers. Birth of new litter was daily checked and the date of birth was defined as postnatal day zero (P0). Between P0-P2, large litters (>6 pups) were reduced to a maximum of six pups with a balanced male/female ratio where possible. Pups were weaned on P21 and housed maximum four per cage. Animals were kept in a controlled 12h light-dark cycle with a temperature of 22 ± 1°C and were given unrestricted access to food (211 RMH-TM diet; Hope Farms, Woerden, NL) and water. Animals were housed in transparent Plexiglas cages with wood-chip bedding and tissue for nest building. Animal reaching the human end-point, defined by < 20% weight loss over 2 days, do not receive care from mother, or loss of grooming before the end of the experiment were deceased from the study. All animals at the end of the experiment or reaching the human endpoint were sacrificed by cervical decapitation.
Warm-air induced prolonged febrile seizures
To induce eFS by hyperthermia (HT), mice were exposed to a temperature-regulated laminar stream of warm air (41–48°C) to quickly raise the core body temperature as previously described [14]. Briefly, body weight was determined in P10 C57BL/6J mice. At least 30 minutes before the HT procedure, mice were injected subcutaneously with temperature sensitive transponders (IPTT-300, PLexx, Elst, NL). Two temperature chambers were used, one for age-, weight-, and litter-matched normothermic control mice (NT; core body temperature maintained at 35°C) and one for HT (defined as core body temperature > 39°C) mice. Chamber temperature was maintained at 32°C for NT mice and at 47°C for HT mice. Mice were placed one by one on the floor of the preheated chambers. Core body temperature was measured every 20 seconds by wireless transporter readout (WRS-6006/6007, Plexx). HT was typically reached within 2.5 minutes. After core temperature of the HT mice had reached 42°C, air temperature was adjusted to maintain a core body temperature between 41.5°C and 42°C. Time and temperature data was logged automatically on a computer (DASHost v1.0, Plexx). HT was maintained for 30 minutes. Then mice were partly submerged in water of room temperature to quickly normalize core body temperature and returned to their mother. NT littermates were simultaneous treated as HT mice, except that their core body temperature was kept at 35°C. The observer scored the stages of behavior and seizures during the experiment, as previously described [14]. Only mice showing continued febrile seizures during at least 17.5 minutes (97.3% of all HT-exposed mice) were used in the experiment.
Tissue collection and sectioning
To explore the temporal relationship of structural changes after eFS, mice were sacrificed by decapitation and brains were removed at three days and fourteen days after eFS. The brains were fresh frozen and stored at -80°C for radioactive in situ hybridization or incubated overnight (ON) in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at 4°C for immunohistochemistry.
Cryo-sectioning for radioactive in situ hybridization
Frozen brains were sectioned on a cryostat (Leica CM3050, Rijswijk, NL). Coronal sections (16 μm) containing the hippocampus were collected on Superfrost slides (Merzel, Germany) by 3 sequential sections per slide until a whole cross section was made of the hippocampus. Sections for protein and mRNA quantification were sampled between Bregma -1.46 mm and -2.54 mm. Slides were stored at -80°C until used for in situ hybridization.
Paraffin-sectioning for immunohistochemistry
After overnight incubation with 4% PFA in PBS, brains were transferred to 70% Ethanol and subsequently embedded in paraffin. Paraffin-embedded brains were sectioned on a microtome (Leica). Coronal sections (7 μm) containing the hippocampus were collected on Superfrost slides (Merzel) by 3 sequential sections per slide until a whole cross section was made of the hippocampus. Slides were stored at room temperature until used for immunohistochemistry.
Immunohistochemistry
To determine eFS-induced changes in protein expression, immunohistochemistry (IHC) was performed on coronal sections (7 μm) from animals fourteen days after eFS (NT n = 10, HT n = 10) using antibodies raised against CNP (mouse, 1:24,000, Sigma Aldrich, USA) and NF (medium polypeptide, mouse, 1:600, DSHB, USA). Using serial dilutions, the optimal working concentration of each antibody was determined. After dehydration, all sections were subjected to antigen retrieval using microwave treatment (7 minutes 650W, 5 minutes 350W) in 0.01 M Na+Citrate pH 6.0. Peroxidase activity was blocked by treatment with 0.3% H2O2 in PBS-0.2% triton-X (PBS-t) for 30 minutes, followed by blocking of non-specific antigen binding with 4% horse normal serum in PBS-t at 37°C for 30 minutes. Sections were incubated with primary antibodies in PBS-t overnight at 4°C. Next, sections were incubated with secondary antibody biotinylated horse anti-mouse (1:400, Brunschwig Chemia, Amsterdam, NL) for 1 hour at room temperature, followed by antigen visualization using avidin-biotin method (Vectastain ABC Elite Kit, Vector Laboratories, USA) with 3,3’-diaminobenzidinetetrachloride (DAB) as the chromogen (Sigma Aldrich). No immunostaining was observed when the protocol was completed without primary antibodies (data not shown). All sections stained for NF or CNP were imaged in one session using a light microscope (Axiophot, Zeiss, Germany), with exposure and light intensity kept constant. Photographs were analyzed using the image-processing program ImageJ (version 1.43, US National Institute of Health, MD, USA). Regions of interest were analyzed bilaterally in the stratum lucidum by an observer blind for treatment group. Expression levels were calculated by gray-scale value inversion and normalization [24]. Mean intensity values of both hemispheres at 3 sections per animal were calculated and used for statistical analysis. Background intensities were measured in the stratum lacunosum. Average intensities per animal were grouped per condition and plotted in Tukey-style boxplots. Pair-wise comparison of average DAB intensity values was tested using the Wilcoxon signed-rank test.
Hippocampal dissection and RNA isolation
One hour, three, fourteen, and fifty-six days after HT animals were sacrificed by decapitation. Brains were quickly removed and hippocampal tissue was dissected, collected and stored at -80°C. Total RNA was isolated, purified and checked for quality as described [25].
Microarray analysis
To identify critical and functional mediator genes during epileptogenesis, acute, short-, and long-term effects of prolonged febrile seizures on gene expression were investigated in whole hippocampal samples. Samples were taken from C57BL/6J animals one hour (HT n = 8, NT n = 8), three days (HT n = 6, NT n = 6), fourteen days (HT n = 6, NT n = 6), and fifty-six days (HT n = 6, NT n = 6) after HT. Two-channel oligonucleotide microarray analysis was performed as described [26,27]. Briefly, cDNA from 2 μg total RNA was synthesized using a T7 oligo(dT)24VN primer (Ambion, Cambridgeshire, UK). The T7 Megascript kit (Ambion) was used for cRNA synthesis and its quality analyzed. Cy3 or Cy5 fluorophores (GE Healthcare Europe, Diegem, BE) were coupled to 2μg NT and HT cRNA. The degree of label incorporation was monitored and hybridizations were set up with 1,500 ng, 2–3% Cy5/Cy3 labeled cRNA per channel. Hybridization was performed with a NT and HT sample on the same chip, including a dye-swap (technical replicate). Just before hybridizing, HT and NT sample were mixed and fragmented (AM8740, Austin, TX, USA). The mouse Array-Ready oligo set (version 3, Operon Biotechnologies, Cologne, DE) was printed on UltraGAPS slides (Corning, Schiphol-Rijk, NL). Probe sequences from this array were re-annotated by BLAST-searching against genomebuild version 71_37 at Ensembl. Slides were washed by hand and scanned on an Agilent G2565BA scanner at 100% laser power, 30% PMT. After image analysis using Imagene 5.6.1 (BioDiscovery), Loess normalization was performed [26] per printtip on mean spot-intensities. Gene-specific dye bias was corrected by a within-set estimate [27]. Data were further analyzed by MAANOVA [28], modeling sample, array and dye effects in a fixed effect analysis. P-values were determined by a permutation F2-test, in which residuals were shuffled 10,000 times globally. Gene probes with P < 0.05 after false discovery rate determination (FDR according to Benjamini-Hochberg, BH) were considered significantly changed. In cases of multiple probes per gene, values from the lowest P-value and most regulated probe were used. For all time points a fold change (Fc) cutoff of log2(Fc) = < - 0.125, or > 0.125 was set, all probes with Ensembl annotation for genesymbol and description, excluding annotations for pseudo- and predicted- genes, were used to produce the significant gene list.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [29] and are accessible through GEO Series accession number GSE66762.
Quantitative RT-PCR
cDNA was synthesized from the RNA samples used for the microarray using oligo-dT primers. The qPCR reaction was performed using the LightCycler (Roche, Almere, NL) and the Fast Start DNA Master PLUS SYBRgreen 1 kit (Roche). Primer (Sigma Genosys, Cambridge, UK) sequences are listed in S1 File. Gene expression was calculated as normalized ratio and normalized to the housekeeping gene peptidylprolyl isomerase A (Ppia) [30]. All samples (NT n = 8, HT n = 8) were analyzed in duplicate and reported as mean ± standard error of the mean. To compare qPCR with microarray data, data were analyzed using one-tailed Student’s t-tests, with P < 0.05 considered significant.
Hierarchical clustering analysis
A gene list with all significant differentially expressed genes at one hour, three days and fourteen days after HT was generated based on the microarray analysis. MeV open-source software [31] was used for the generation of heatmap and hierarchical clustering of genes and samples according to Pearson’s correlation of average linkage.
Gene ontology (GO) analysis
For GO analysis, the significant gene list (P < 0.05, Fold change cutoff of log2(Fc) = < -0.125, or > 0.125) and a reference list comprising all unique genes (27,334, excluding pseudo- and predicted- genes) present on the microarray chip were imported into the software tool DAVID [32,33]. In DAVID a total of 11,838 unique genes, among the 27,334 probes, were mapped to the GO Panther database (www.pantherdb.org, version 9.0, release 20 January 2014). These were used to generate a reference gene list. Up- and down- regulated gene lists were mapped against the Panther database. Panther GO classes are greatly abbreviated and simplified to facilitate high-throughput analyses. Significant lists were statistically compared to the reference list to identify significant over-representation of genes (P < 0.05) within the GO classes for biological process. Classes with significant over-representation are shown and marked when, after correcting for multiple testing with BH-correction, P < 0.05.
Radioactive (33P) in situ hybridization
To quantify the expression levels of Nrxn1, Dpysl3, Ptprd, and Slit2 in the hippocampus, 33P in situ hybridization was performed on coronal sections (16 μm) from animals three days after eFS (NT n = 9, HT n = 9). 33P RNA probes were designed against the mRNA for Nrxn1, Dpysl3, Ptprd, and Slit2 (Nrxn1; NM_020252.3 at 7176–7568 bp, Dpysl3; NM_009468.4 position at 1306–1914 bp, Ptprd; NM_011211.3 position at 4865–5415 bp, Slit2; NM_178804.3 position at 2152–2789 bp), and aligned to the region, which included the sequence of the significantly regulated microarray probe. Primer sequences are listed in S1 File. The synthesis of 33P RNA-probes and in situ hybridization were performed as described in previous work [34]. Briefly, sections were hybridized to the 1 x 106 cpm of anti-sense 33P RNA probe in 150 μL hybridization mixture with 50% deionized formamide, 2x standard saline citrate (SSC), 10% dextrane sulphate, 1x Denhardt’s solution, 5 mM ethylenediaminetetraacetic acid (EDTA), and 10 mM phosphate buffer. After overnight hybridization of the probes at 72°C, slides were washed, dried and exposed film (Kodak Bio-Mac, MR). The developed films were digitized using an Epson flatbed scanner (Perfection 4990, Epson America, CA, USA). For each gene, one slide was hybridized to the sense control 33P RNA probe. No signal was observed in presence of the control sense 33P RNA probe (data not shown). Calibration dilutions of 14C micro-scales (Amersham Bioscience, Sweden) and anti-sense probes were used to determine the linear range for image quantification. An observer blind for treatment condition quantified photographs of the film in ImageJ. Regions of interest were placed in the subfields of the hippocampus bilaterally in 3 sections per animal. Expression levels were calculated by gray-scale value inversion, corrected for background gray-scale value and normalized. Mean intensity values of both hemispheres at 3 sections per animal were calculated and used for statistical analysis. Background value was determined within the corpus callosum. Bar graphs depict the average expression levels per condition and subfield, normalized to control (NT) group average with ± standard error of the mean. To compare expression levels of genes in HT versus NT animal couples Student’s t-tests (littermate-paired, two-tailed) were performed.
Statistical analysis
All statistical analyses were performed using R with Stats package (version 3.1.2, http://cran.r-project.org). Thresholds of significance were set at α = 0.05.
Results
Regulation of neurofilament in the hippocampus after eFS
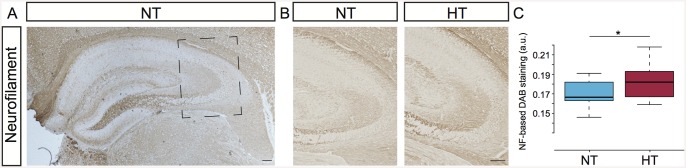
To explore the hypothesis that hyperthermia-induced eFS result in structural reorganization of the hippocampus, we compared neurofilament (NF, medium polypeptide) staining in hyperthermia-exposed mice (HT, n = 10) to normothermia-exposed control littermates (NT, n = 10) (Fig 1). NF is predominantly expressed in the neuropil of the hippocampus, such as the mossy fibers projecting from the DG to the CA3 region. HT exposed mice, compared to NT littermates, showed a significant increase in NF-staining in the stratum lucidum of the CA3 subfield (Wilcoxon signed-rank test P = 0.0049). NF-staining changes were specifically observed in the stratum lucidum and not in other subfields of the hippocampus. In line with published results in rats [15] our data show that eFS in mice trigger structural changes in the hippocampus.
Fig 1. Hyperthermia induces upregulation of neurofilament protein-expression.
(A) Neurofilament (NF) immunostaining in the hippocampus of a normothermic (NT) control animal, 14 days post-hyperthermia exposure. Square marks subfield enlargement; panel B. (B) Immunostaining for NF in the stratum lucidum of NT control animal (left panel) and in hyperthermia-exposed (HT) littermate (right panel). (C) Comparison of NF-based DAB staining in the stratum lucidum between NT and HT littermates. All HT (n = 10) animals showed increased DAB-staining in the stratum lucidum. Pair-wise comparison of DAB-staining intensities showed increased NF expression in HT animals compared to NT littermates (n = 10), Wilcoxon signed-rank test, * indicates a significance of P = 0.0049. Tukey-style box plots. Scalebars: 100 μm.
Regulation of hippocampal gene expression after eFS
To investigate which molecular pathways underlie the structural reorganization after eFS, we compared hippocampal mRNA content from HT and NT mice by microarray hybridization. Hippocampal tissue was sampled one hour, and three, fourteen, and fifty-six days after induction of eFS.
Changes in mRNA expression within the hippocampus after eFS
One hour after eFS (NT n = 8, HT n = 8) 1004 differentially expressed probes were identified. Of these 1004 probes, 579 upregulated and 298 downregulated unique Ensembl annotated genes were identified (S2 File, excluding pseudo- and predicted-genes). The highest upregulated gene (8.04-fold), Cyr61, is an extracellular angiogenic factor. Brain Cyr61 expression is induced by numerous stimuli. Although limited information is available on Cyr61 function, it appears to be involved in vasculature or dendritic ramification of hippocampal neurons [35]. As expected, several highly upregulated genes are immediate early genes (IEGs, for example Cyr61, Jun, Fos, Egr1, and Nr4a1) and heat-shock proteins (HSP110, Hspb1, Hsph1, and Dnajb1) (Table 1). Amongst the highly downregulated genes trophic factor Ptgds, adhesion molecules Pcdh20 and Cldn5, and transcription factors Hes5 and Sox18 were identified.
Table 1. Top 30 differentially expressed genes (up- and down- regulated) 1 hour after eFS.
| 1 hour post-eFS | ||||
|---|---|---|---|---|
| Upregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000028195 | Cyr61 | cysteine rich protein 61 | 8.04 | 0.0000 |
| ENSMUSG00000031558 | Slit2 | slit homolog 2 (Drosophila) | 5.68 | 0.0000 |
| ENSMUSG00000052684 | Jun | Jun oncogene | 3.13 | 0.0000 |
| ENSMUSG00000021250 | Fos | FBJ osteosarcoma oncogene | 3.04 | 0.0000 |
| ENSMUSG00000023034 | Nr4a1 | nuclear receptor subfamily 4, group A, member 1 | 2.99 | 0.0000 |
| ENSMUSG00000038418 | Egr1 | early growth response 1 | 2.62 | 0.0000 |
| ENSMUSG00000029657 | Hsph1 | heat shock 105kDa/110kDa protein 1 | 2.46 | 0.0000 |
| ENSMUSG00000004951 | Hspb1 | heat shock protein 1 | 2.40 | 0.0000 |
| ENSMUSG00000024190 | Dusp1 | dual specificity phosphatase 1 | 2.34 | 0.0000 |
| ENSMUSG00000005483 | Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 2.25 | 0.0000 |
| ENSMUSG00000026628 | Atf3 | activating transcription factor 3 | 2.11 | 0.0000 |
| ENSMUSG00000019997 | Ctgf | connective tissue growth factor | 2.08 | 0.0000 |
| ENSMUSG00000022602 | Arc | activity regulated cytoskeletal-associated protein | 1.97 | 0.0000 |
| ENSMUSG00000006057 | Atp5g1 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) | 1.90 | 0.0000 |
| ENSMUSG00000053877 | Srcap | Snf2-related CREBBP activator protein | 1.90 | 0.0000 |
| ENSMUSG00000028214 | Gem | GTP binding protein (gene overexpressed in skeletal muscle) | 1.89 | 0.0000 |
| ENSMUSG00000028907 | Utp11l | UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) | 1.86 | 0.0000 |
| ENSMUSG00000020205 | Phlda1 | pleckstrin homology-like domain, family A, member 1 | 1.75 | 0.0000 |
| ENSMUSG00000020818 | Mfsd11 | major facilitator superfamily domain containing 11 | 1.75 | 0.0000 |
| ENSMUSG00000021810 | Ecd | ecdysoneless homolog (Drosophila) | 1.68 | 0.0000 |
| ENSMUSG00000037266 | D4Wsu53e | DNA segment, Chr 4, Wayne State University 53, expressed | 1.65 | 0.0000 |
| ENSMUSG00000020423 | Btg2 | B cell translocation gene 2, anti-proliferative | 1.64 | 0.0000 |
| ENSMUSG00000030847 | Bag3 | BCL2-associated athanogene 3 | 1.63 | 0.0000 |
| ENSMUSG00000078698 | Mrgpra3 | MAS-related GPR, member A3 | 1.63 | 0.0000 |
| ENSMUSG00000021453 | Gadd45g | growth arrest and DNA-damage-inducible 45 gamma | 1.62 | 0.0000 |
| ENSMUSG00000031765 | Mt1 | metallothionein 1 | 1.61 | 0.0000 |
| ENSMUSG00000021367 | Edn1 | endothelin 1 | 1.59 | 0.0000 |
| ENSMUSG00000073676 | Hspe1 | heat shock protein 1 (chaperonin 10) | 1.56 | 0.0000 |
| ENSMUSG00000032531 | Amotl2 | angiomotin-like 2 | 1.55 | 0.0000 |
| ENSMUSG00000048222 | Mfap1b | microfibrillar-associated protein 1B | 1.54 | 0.0000 |
| Downregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000020863 | Luc7l3 | LUC7-like 3 (S. cerevisiae) | 0.62 | 0.0000 |
| ENSMUSG00000015090 | Ptgds | prostaglandin D2 synthase (brain) | 0.64 | 0.0000 |
| ENSMUSG00000041378 | Cldn5 | claudin 5 | 0.68 | 0.0000 |
| ENSMUSG00000071337 | Tia1 | cytotoxic granule-associated RNA binding protein 1 | 0.71 | 0.0000 |
| ENSMUSG00000030495 | Slc7a10 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 | 0.71 | 0.0000 |
| ENSMUSG00000029570 | Lfng | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | 0.74 | 0.0000 |
| ENSMUSG00000046470 | Sox18 | SRY-box containing gene 18 | 0.74 | 0.0000 |
| ENSMUSG00000028645 | Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | 0.74 | 0.0000 |
| ENSMUSG00000000253 | Gmpr | guanosine monophosphate reductase | 0.77 | 0.0000 |
| ENSMUSG00000030218 | Mgp | matrix Gla protein | 0.78 | 0.0047 |
| ENSMUSG00000026822 | Lcn2 | lipocalin 2 | 0.78 | 0.0009 |
| ENSMUSG00000030711 | Sult1a1 | sulfotransferase family 1A, phenol-preferring, member 1 | 0.79 | 0.0000 |
| ENSMUSG00000040055 | Gjb6 | gap junction protein, beta 6 | 0.80 | 0.0000 |
| ENSMUSG00000048001 | Hes5 | hairy and enhancer of split 5 (Drosophila) | 0.81 | 0.0000 |
| ENSMUSG00000046546 | Fam43a | family with sequence similarity 43, member A | 0.81 | 0.0000 |
| ENSMUSG00000032422 | Snx14 | sorting nexin 14 | 0.81 | 0.0000 |
| ENSMUSG00000029231 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide | 0.81 | 0.0000 |
| ENSMUSG00000050505 | Pcdh20 | protocadherin 20 | 0.81 | 0.0000 |
| ENSMUSG00000022508 | Bcl6 | B cell leukemia/lymphoma 6 | 0.82 | 0.0000 |
| ENSMUSG00000030707 | Coro1a | coronin, actin binding protein 1A | 0.82 | 0.0000 |
| ENSMUSG00000041073 | Nacad | NAC alpha domain containing | 0.82 | 0.0000 |
| ENSMUSG00000016921 | Srsf6 | serine/arginine-rich splicing factor 6 | 0.82 | 0.0000 |
| ENSMUSG00000038175 | Mylip | myosin regulatory light chain interacting protein | 0.82 | 0.0000 |
| ENSMUSG00000017760 | Ctsa | cathepsin A | 0.82 | 0.0000 |
| ENSMUSG00000001240 | Ramp2 | receptor (calcitonin) activity modifying protein 2 | 0.82 | 0.0000 |
| ENSMUSG00000043131 | Mob1a | MOB kinase activator 1A | 0.82 | 0.0000 |
| ENSMUSG00000070167 | SNORA57 | Small nucleolar RNA SNORA57 | 0.83 | 0.0000 |
| ENSMUSG00000075602 | Ly6a | lymphocyte antigen 6 complex, locus A | 0.83 | 0.0000 |
| ENSMUSG00000028655 | Mfsd2a | major facilitator superfamily domain containing 2A | 0.83 | 0.0000 |
| ENSMUSG00000050295 | Foxc1 | forkhead box C1 | 0.84 | 0.0000 |
Fc: Fold change represents expression values of HT animals divided by expression values of NT animals. P-value BH-corrected: Benjamini and Hochberg (BH) error corrected p-values.
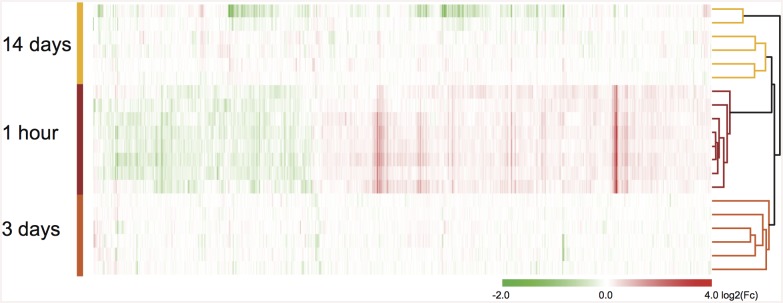
Analysis of the later time points identified 74, 15, and 1 significantly regulated probe(s) three, fourteen, and fifty-six days after eFS, respectively (Table 2, for all time points: NT n = 6, HT n = 6). Hierarchical cluster analysis performed on the total geneset of all differentially expressed genes from one hour, three and fourteen days after eFS (930 Ensembl annotated unique genes, excluding pseudo- and predicted- genes). Fifty-six days after eFS was excluded from the cluster analysis because only one significantly regulated gene was found. Differential expression levels (log2(Fc)) between each HT and NT littermate couple was used for clustering. Based on Pearson correlation of average linkage the HT samples and genes were clustered (Fig 2). Gene expression profiles of all samples showed strong clustering per time-point except for two samples (14 days post-eFS), which showed larger clustering distance to all other samples.
Table 2. Differentially expressed genes (up- and down- regulated) three, fourteen, and fifty-six days post-eFS.
| 3 days post-eFS | ||||
| Upregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000001911 | Nfix | nuclear factor I/X | 1.32 | 0.0414 |
| ENSMUSG00000003410 | Elavl3 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | 1.20 | 0.0319 |
| ENSMUSG00000066551 | Hmgb1 | high mobility group box 1 | 1.16 | 0.0050 |
| ENSMUSG00000071337 | Tia1 | cytotoxic granule-associated RNA binding protein 1 | 1.16 | 0.0195 |
| ENSMUSG00000024501 | Dpysl3 | dihydropyrimidinase-like 3 | 1.16 | 0.0181 |
| ENSMUSG00000024268 | Celf4 | CUGBP, Elav-like family member 4 | 1.14 | 0.0414 |
| ENSMUSG00000001020 | S100a4 | S100 calcium binding protein A4 | 1.14 | 0.0317 |
| ENSMUSG00000028399 | Ptprd | protein tyrosine phosphatase, receptor type, D | 1.14 | 0.0115 |
| ENSMUSG00000024109 | Nrxn1 | neurexin I | 1.13 | 0.0319 |
| ENSMUSG00000038900 | Rpl12 | ribosomal protein L12 | 1.12 | 0.0022 |
| ENSMUSG00000053128 | Rnf26 | ring finger protein 26 | 1.12 | 0.0252 |
| ENSMUSG00000027962 | Vcam1 | vascular cell adhesion molecule 1 | 1.12 | 0.0319 |
| ENSMUSG00000024799 | Tm7sf2 | transmembrane 7 superfamily member 2 | 1.11 | 0.0181 |
| ENSMUSG00000003198 | Zfp959 | zinc finger protein 959 | 1.11 | 0.0312 |
| ENSMUSG00000059325 | Hopx | HOP homeobox | 1.11 | 0.0181 |
| ENSMUSG00000032047 | Acat1 | acetyl-Coenzyme A acetyltransferase 1 | 1.11 | 0.0119 |
| ENSMUSG00000022194 | Pabpn1 | poly(A) binding protein, nuclear 1 | 1.11 | 0.0424 |
| ENSMUSG00000060675 | Pla2g16 | phospholipase A2, group XVI | 1.11 | 0.0331 |
| ENSMUSG00000031451 | Gas6 | growth arrest specific 6 | 1.11 | 0.0414 |
| ENSMUSG00000049932 | H2afx | H2A histone family, member X | 1.11 | 0.0119 |
| ENSMUSG00000024411 | Aqp4 | aquaporin 4 | 1.11 | 0.0119 |
| ENSMUSG00000030605 | Mfge8 | milk fat globule-EGF factor 8 protein | 1.10 | 0.0195 |
| Downregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000052305 | Hbb-b1 | hemoglobin, beta adult major chain | 0.75 | 0.0002 |
| ENSMUSG00000069919 | Hba-a1 | hemoglobin alpha, adult chain 1 | 0.80 | 0.0103 |
| ENSMUSG00000069917 | Hba-a2 | hemoglobin alpha, adult chain 2 | 0.83 | 0.0195 |
| ENSMUSG00000027375 | Mal | myelin and lymphocyte protein, T cell differentiation protein | 0.85 | 0.0012 |
| ENSMUSG00000032517 | Mobp | myelin-associated oligodendrocytic basic protein | 0.86 | 0.0016 |
| ENSMUSG00000006205 | Htra1 | HtrA serine peptidase 1 | 0.88 | 0.0103 |
| ENSMUSG00000040055 | Gjb6 | gap junction protein, beta 6 | 0.88 | 0.0195 |
| ENSMUSG00000006782 | Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | 0.89 | 0.0103 |
| ENSMUSG00000049892 | Rasd1 | RAS, dexamethasone-induced 1 | 0.90 | 0.0119 |
| ENSMUSG00000062591 | Tubb4a | tubulin, beta 4A class IVA | 0.90 | 0.0461 |
| ENSMUSG00000070282 | 3000002C10Rik | RIKEN cDNA 3000002C10 gene | 0.90 | 0.0195 |
| ENSMUSG00000054459 | Vsnl1 | visinin-like 1 | 0.90 | 0.0423 |
| ENSMUSG00000022594 | Lynx1 | Ly6/neurotoxin 1 | 0.90 | 0.0195 |
| ENSMUSG00000036634 | Mag | myelin-associated glycoprotein | 0.90 | 0.0319 |
| ENSMUSG00000079018 | Ly6c1 | lymphocyte antigen 6 complex, locus C1 | 0.91 | 0.0312 |
| ENSMUSG00000090071 | Cdk5r2 | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | 0.91 | 0.0181 |
| ENSMUSG00000017009 | Sdc4 | syndecan 4 | 0.91 | 0.0369 |
| ENSMUSG00000028843 | Sh3bgrl3 | SH3 domain binding glutamic acid-rich protein-like 3 | 0.92 | 0.0119 |
| 14 days post-eFS | ||||
| Upregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000018501 | Ncor1 | nuclear receptor co-repressor 1 | 1.27 | 0.0494 |
| ENSMUSG00000006782 | Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | 1.26 | 0.0295 |
| ENSMUSG00000037625 | Cldn11 | claudin 11 | 1.25 | 0.0078 |
| ENSMUSG00000028385 | Snx30 | sorting nexin family member 30 | 1.21 | 0.0112 |
| ENSMUSG00000063077 | Kif1b | kinesin family member 1B | 1.18 | 0.0475 |
| ENSMUSG00000025038 | Efhc2 | EF-hand domain (C-terminal) containing 2 | 1.17 | 0.0335 |
| ENSMUSG00000029684 | Wasl | Wiskott-Aldrich syndrome-like (human) | 1.15 | 0.0112 |
| ENSMUSG00000054459 | Vsnl1 | visinin-like 1 | 1.15 | 0.0494 |
| ENSMUSG00000036526 | Card11 | caspase recruitment domain family, member 11 | 1.15 | 0.0123 |
| ENSMUSG00000036985 | Zdhhc9 | zinc finger, DHHC domain containing 9 | 1.15 | 0.0475 |
| ENSMUSG00000076439 | Mog | myelin oligodendrocyte glycoprotein | 1.13 | 0.0494 |
| Downregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000023087 | Ccrn4l | CCR4 carbon catabolite repression 4-like (S. cerevisiae) | 0.86 | 0.0295 |
| ENSMUSG00000030218 | Mgp | matrix Gla protein | 0.86 | 0.0037 |
| 56 days post-eFS | ||||
| Downregulated genes | ||||
| Ensembl | genesymbol | Description | Fc | P-value BH-corrected |
| ENSMUSG00000024617 | Camk2a | calcium/calmodulin-dependent protein kinase II alpha | 0.89 | 0.0120 |
Fc: Fold change represents expression values of HT animals divided by expression values of NT animals. P-value BH-corrected: Benjamini and Hochberg (BH) error corrected p-values.
Fig 2. Hierarchical clustering analysis of mRNA expression after eFS.
Hierarchical clustering analysis of differentially expressed genes (930) in all experimental littermate couples (1 hour, 3 and 14 days post-eFS). Rows depict differential expression levels per HT/NT littermate couple, per time-point after eFS. Clustering was performed based on gene and sample clustering using Pearson correlation on average linkage. Tree cluster, right from heat-map, shows hierarchical distance between samples from the different time points. Differential gene expression (log2(Fc) between HT/NT littermate couples is shown in the heatmap as upregulated (red), downregulated (green), or no change (white) according to colored-scale bar.
To independently validate the one hour after eFS microarray results, we used qPCR to quantify mRNA levels of several regulated genes in the HT/NT couples. Jun, Cyr61, Atf3, Egr1, and Arc were selected for validation (Fig 3). In line with the microarray data, all five genes were found to be upregulated in HT animals compared to NT littermates.
Fig 3. Validation of microarray results by qPCR analysis.
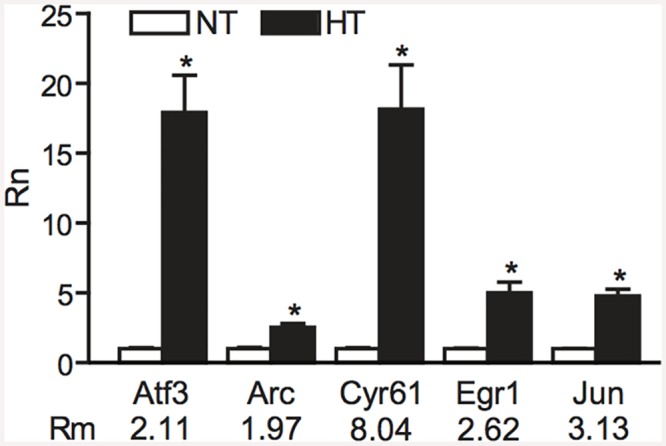
Gene expression levels as determined by qPCR (Rn) in normothermic (NT) and hyperthermic (HT) animals compared to fold-change in expression of 5 genes as detected by microarray analysis (Rm). P < 0.05 was considered significant (one-tailed Student’s t-test). Indicated are mean ± standard error of the mean.
Short- and long-term gene expression profile after eFS
To identify genes related to structural reorganization of the hippocampus we analyzed the gene sets three, fourteen, and fifty-six days after eFS. Compared to one hour after eFS, a limited number of genes was significantly regulated at three, fourteen, and fifty-six days after eFS (Table 2). Three days after eFS 22 upregulated and 18 downregulated unique genes were identified. In the 3 day gene list, several genes implicated in structural reorganization such as cell adhesion molecules (Ptprd, Nrxn1, Dpysl3, and Vcam1) were found to be upregulated, and myelin-sheath related genes (Mobo, Cnp, and Mag) downregulated. Interestingly, the top three downregulated genes were hemoglobin subunits, alpha (Hba-a1 and Hba-a2) and beta (Hbb-b1) chains. These three genes were upregulated at one hour after eFS. Information about the role of hemoglobin expression in the brain is limited [36]. Fourteen days after eFS 11 upregulated and 2 downregulated unique genes were identified. At this time-point, in contrast to the three days after eFS gene set, myelin-sheath related genes (Cnp, Cldn11, Wasl, and Mog) were upregulated. Analysis of fifty-six days after eFS samples revealed only 1 significantly downregulated gene (Camk2a).
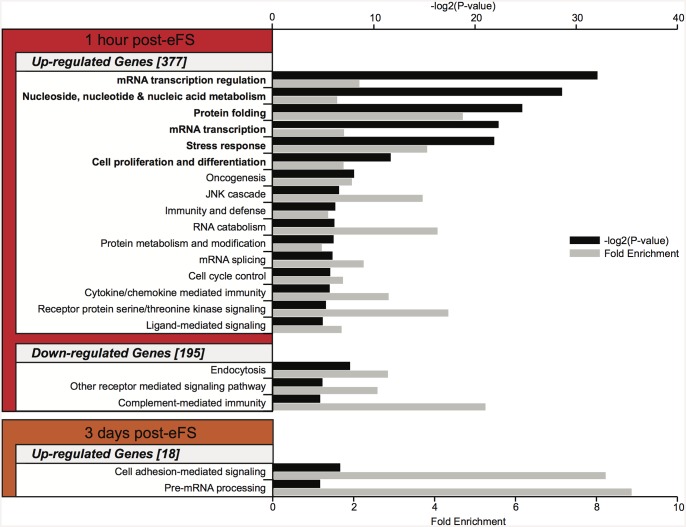
Gene ontology (GO) analysis of regulated genes after eFS
To identify common biological processes underlying changes after eFS we performed GO analysis using the DAVID web tool [32,37] based on GO Panther biological process (BP) annotation database (Mi 2012 Nucleic Acids Research). One hour after eFS upregulated genes significantly enriched GO biological process classifications such as mRNA transcription regulation, protein folding, stress response, and cell proliferation and differentiation (Fig 4, BH error corrected P < 0.05). Downregulated genes one hour after eFS were associated (but not significantly enriched after correction for multiple testing) to GO classification such as endocytosis and complement-mediated immunity. Three days after eFS upregulated genes were associated to GO classifications cell-mediated adhesion signaling and pre-mRNA processing (but did not reach significance after correction for multiple testing). Due to the low number of genes in the geneset no enrichment of GO classification was found within the downregulated genes at three days after eFS, nor within all regulated genes at fourteen and fifty-six days after eFS.
Fig 4. Gene ontology (GO) enrichment analysis on differentially expressed genes after hyperthermia exposure.
GO biological process classifications with a P < 0.05 are depicted (up- and downregulation) per time point. Numbers between brackets represent total Panther GO annotated genes used for GO analysis per geneset. P-value per GO classification is shown (black bar, -log2(P value)), with corresponding fold enrichment (grey bar). Bold denoted GO classifications show significant enrichment (P < 0.05) after Benjamini and Hochberg (BH) error correction.
All together, the GO analysis reveals that within the genelists, specific biological processes can be identified 1 hour and 3 days after eFS. This urges us to identify candidate genes important after hyperthermia-induced eFS.
Characterization of candidate genes implicated in structural reorganization after eFS
Transcriptional regulation at three days after eFS
Microarray gene expression analysis provided a list of significantly regulated genes three and fourteen days after eFS (Table 2). Closer examination of this list by GO analysis indeed showed transcriptional activity of genes related to structural reorganization. Based on literature and fold-change levels a number of candidate genes were selected. To characterize and validate the expression of these candidate genes we performed radioactive in-situ hybridization (RA-ISH) 3 days after eFS. 33P RNA-probes specific against Nrxn1, Dpysl3, Ptprd, and Slit2 were used for analysis of mRNA expression in subfields of the hippocampus.
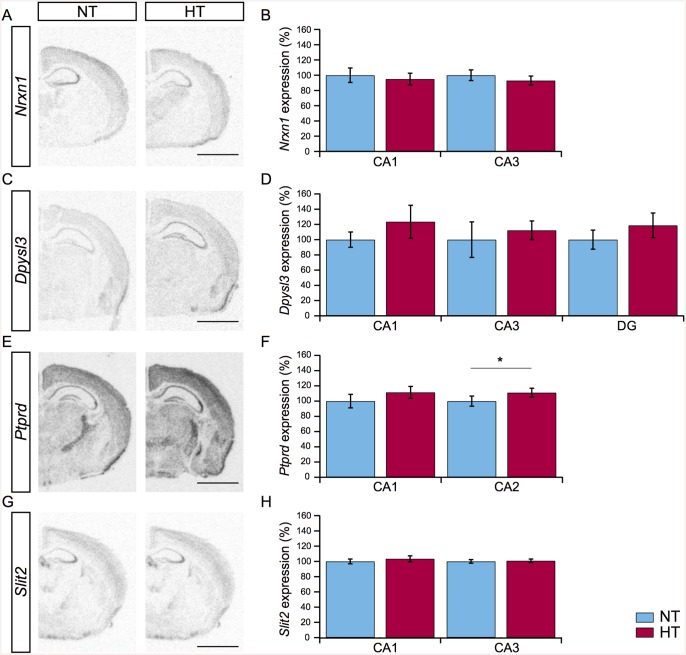
Neurexin1 (Nrxn1) expression was prominent in the CA1 and CA3 subfields of the hippocampus. Nrxn1 is important during the formation of excitatory synapses, synaptogenesis [38] (Fig 5A). Quantification of Nrxn1 showed no significant regulation of expression in HT animals within the CA1 and CA3 subfield (Fig 5B, NT n = 9, HT n = 9, littermate-paired Student’s t-test, P > 0.05). Dpysl3, also known as collapsin response mediator protein 4 (Crmp4), acts as a signaling molecule downstream from Sema3A, and is implicated in the regulation of neuronal dendrite development [39]. Expression of Dpysl3 was quantified in the DG, CA1, and CA3 subfield of the hippocampus. A widespread increase in Dpysl3 expression levels in all three subfields was observed (Fig 5C), but this increase was not statistically different between HT and NT littermates (Fig 5D, NT n = 9, HT n = 9, littermate-paired Student’s t-test, P > 0.05). Ptprd, Receptor-Type Tyrosine-Protein Phosphatase Delta, regulates axon guidance during brain development, and similar to Nrxn1 is important during synaptogenesis [40]. Expression of Ptprd was observed in the CA1 and, remarkably, clearly present in the CA2 subfield known to be resistant to cell death in TLE patients (Fig 5E, [41]). Expression of Ptprd was significantly upregulated in CA2, but not in CA1, in HT animals compared to NT littermates (Fig 5F, NT n = 9, HT n = 9, littermate-paired Student’s t-test, CA2: P = 0.0097 CA1: P > 0.05). Slit2 was included in the analysis, because it was the highest (5.68-fold) upregulated gene involved in structural reorganization, 1 hour after eFS. It regulates axon guidance during development of the hippocampus and is differentially regulated during experimental epileptogenesis and in temporal lobe epilepsy patients [42]. Pronounced expression of Slit2 was observed in the CA3 subfield of the hippocampus (Fig 5G). No significant difference between HT and NT littermates was detected in relative expression levels 3 days after HT (Fig 5H, NT n = 9, HT n = 9, littermate-paired Student’s t-test, P > 0.05).
Fig 5. Expression of candidate genes mRNA related to structural reorganization.
(A, C, E, G) Representative images of Nrxn1, Dpysl3, Ptprd, and Slit2 gene expression in the hippocampus of NT (left panel) and HT (right panel) animals three days after eFS. (B, D, F, H) Comparison of Nrxn1, Dpysl3, Ptprd, and Slit2 gene expression levels in subfields of the hippocampus in HT (n = 9, red bars) animals compared to NT (n = 9, blue bars) littermates. Ptprd is upregulated in the CA2 subfield of the hippocampus of HT animals, * indicates a significant difference of P = 0.0097, littermate-paired Student’s t-test. Bars represents mean ± standard error of the mean. Scalebars in panel A, C, E, and G: 500μm.
Myelin sheath-associated changes fourteen days after eFS
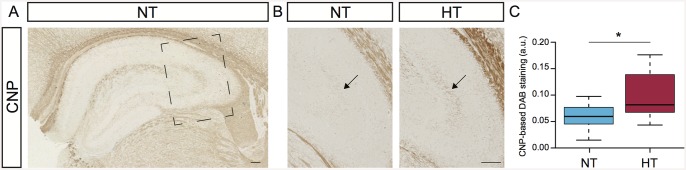
The regulation of myelin-sheath associated genes three and fourteen days after eFS observed by the microarray gene expression analysis prompted us to monitor protein expression of 2',3'-cyclic nucleotide 3' phosphodiesterase (CNP) in hippocampus. This gene was downregulated 3 days after eFS (0.8-fold) and upregulated 14 days after eFS (1.26-fold). CNP is predominantly expressed in myelin sheaths and oligodendrocytes, although it’s physiological function remains ambiguous, loss of CNP affects the stability of axons [43–45]. Immunostaining of CNP protein at fourteen days after eFS revealed expression mostly in the neurophil of the hippocampus (Fig 6A). Interestingly, comparison of CNP expression between HT and NT littermates indicated elevated levels of CNP within the stratum lucidum of the CA3 region, specifically. Quantification and pair-wise comparison of CNP-based DAB staining in the stratum lucidum between HT and NT littermates showed pronounced upregulation of CNP in HT animals (Fig 6B and 6C, Wilcoxon signed-rank test P = 0.0078).
Fig 6. Hyperthermia induced upregulation of 2',3'-cyclic nucleotide 3' phosphodiesterase (CNP) in the stratum lucidum.
(A) Normal expression of CNP in the hippocampus of a normothermic (NT) control animal 14 days post-hyperthermia exposure. Square marks subfield enlargement, panel B and C. (B) Expression of CNP in the stratum lucidum of NT control animal (left panel) and hyperthermia-exposed (HT) littermate (right panel). Arrows indicates CNP-positive staining in the stratum lucidum. (C) Comparison of CNP-based DAB staining in the stratum lucidum between NT and HT littermates. All HT animals showed increased DAB-staining in the stratum lucidum. Increased expression of CNP were observed in HT animals compared to NT littermates, Wilcoxon signed-rank test, * indicates P = 0.0078. Tukey-style box plots. Scalebars: 100μm.
Discussion
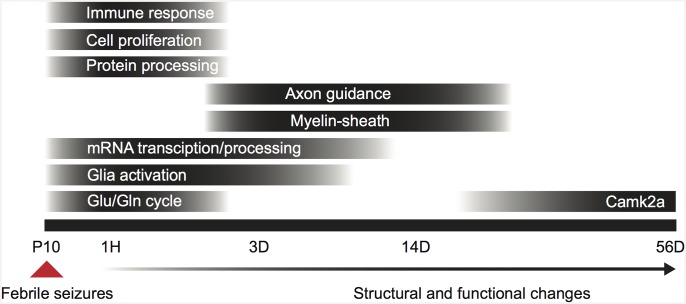
In this study we investigated the short- and long-term changes in hippocampal gene expression following hyperthermia-induced eFS in mice. To pinpoint molecular mechanisms underlying structural reorganization after eFS, we performed a genome-wide transcriptome analysis at 4 time-points: immediately (one hour), shortly (three days) and during the late phase (fourteen and fifty-six days) after induction of eFS. At these time-points we identified 877, 40, 13 and 1 eFS regulated genes, respectively. Here we show that timed regulation of multiple processes, such as heat-, immune- and inflammation response, glutamate-glutamine cycle, myelination, and structural reorganization, are triggered by eFS (Fig 7). Fourteen days after eFS expression of neurofilament, a marker for dentate gyrus mossy fibers, and CNP, a marker for oligodendrocyte activation and myelination, was increased in the stratum lucidum of the hippocampus. This indicates that already 14 days after eFS structural reorganization is taking place (Fig 7). We provide evidence that specific processes and molecular candidates are involved in structural reorganization within the hippocampus at three and fourteen days after eFS. This is the first study to characterize the transcriptional response to eFS in mice throughout a time-course of 56 days.
Fig 7. Timed regulation of biological processes after eFS.
Bar blackness indicates the peak effect of the process. P10: postnatal day 10, 1H: 1 hour post-eFS, 3D: 3 days post-eFS, 14D: 14 days post-eFS, 56D: 56 days post-eFS, Glu/Gln cycle: glutamate/glutamine cycle.
Transcriptional regulation after eFS
Analysis of hippocampal mRNA content identified 877, 40, 13, and 1 statistically significant regulated unique annotated gene(s) one hour, three, fourteen, and fifty-six days after eFS, respectively. Indicating a strong but transient regulation of gene expression during the first hour after eFS. As expected GO-analysis for gene enrichment related to specific biological processes reveals that eFS initially triggers a broad transcriptional response and a cellular response to heat (protein folding, stress response) as a direct response to the hyperthermia (Fig 7). Genes related to heat response included HSP110, Hspb1, Hsph1, and Dnajb1. Expression of IEGs (immediate early genes) such as, Arc, Fos, Egr1, Atf1, Nr4a1, Cyr61, Jun during and directly after eFS is most likely due to the prolonged seizure activity. Expression of these IEGs probably represents profound neuronal activity in the hippocampal network as demonstrated earlier by intra-hippocampal EEG recordings during and after eFS [12,16,46,47]. The lack of regulation of these genes 56 days after eFS is in line with the fact that at this time-point (the majority of) HT mice do not yet suffer from recurrent seizures. Comparing the genelists at the different time-points revealed that 35% of all regulated genes (14 out of 40 genes) three days after eFS were also regulated one hour after eFS. The majority of upregulated genes three days after eFS (5 out of 6 genes) showed downregulation 1 hour after eFS. In contrast, downregulated genes (5 out of 8 genes), with exception of the top-three regulated hemoglobin chain genes, 3 days after eFS were also downregulated 1 hour after eFS. Fourteen days after eFS, with exception of CNP, no overlap was observed with the regulated genes three days after eFS. However, 23% (3 out of 13 genes) of the regulated genes fourteen days after eFS did overlap with those 1 hour after eFS. Due to the low number of differentially expressed genes per time-point it is difficult to make over-time expression wave profiles per GO biological process [48,49]. The observed switch between one hour and three days after eFS from down- to upregulation of several genes indicates that over-time expression wave profiles can be observed after eFS.
Glutamate-glutamine cycle
As a consequence of increased hippocampal neuronal activity tight regulation of glutamate-glutamine cycle is of importance to avoid excitotoxic damage. Regulation of this process has already been described to play a key role in the susceptibility for febrile seizures and epileptogenesis in TLE patients [50–53]. Neuronal expressed glutaminase (Gls) converts glutamine to excitatory neurotransmitter glutamate. Glial glutamate transporters, such as excitatory amino acid transporter 2 (EAAT2, Slc1a2), avoid abnormal levels of extracellular released glutamate, by internalizing the neurotransmitter in the glial cells where it in turn is converted to glutamine by glutamine synthetase (Glul). One hour after eFS Glul is strongly reduced (0.90-fold), whereas Gls and Slc1a2 are upregulated (1.19-fold and 1.21-fold, respectively). The opposing regulation of Gls and Glul expression indicates an increase in glutamate production and less efficient synthesis of glutamine. As a consequence, changes in Glul and Gls expression may therefore contribute to an increase in neuronal excitability. Additionally, the upregulation of Slc1a2 might also reflect compensation to the increased glutamate production. Together with earlier findings of altered HCN-channel expression and function after eFS, the above-stated observations of Glul, Gls, and Slc1a2 expression indicate strong functional changes in neuronal excitability [17,21,54].
Immune and inflammatory response
Inflammatory processes, including regulation of chemokine and cytokine mediated immune response and complement-mediated immunity, are triggered during the first hour after eFS (Fig 7). Genes important in these processes included, CCL-2, CCL-3, ILr10-a/b, C1qa, and Cr1l. Febrile seizures in rats also show a robust immune response directly after exposure to HT [49,55]. Similarly, electrical kindling of the dentate gyrus as a model for TLE, showed overlap in biological processes with our genelists. This initial regulation of genes related to stress- immune- and inflammatory response among several epileptic animal models indicates that seizure activity, independent of the precipitating event, triggers immune response in the hippocampus [48]. A growing body of evidence shows involvement of immune and inflammatory processes in epilepsy and febrile seizures [25,56,57]. For example the role of interleukin-1ß (IL-1ß) in febrile seizures is a prominent topic of debate [5,58]. IL-1ß is a, pyrogenic, fever-promoting proinflammatory cytokine, which promotes the development of febrile seizures. Typically, loss of IL-1ß receptor in mice results in resistance to eFS [59]. We did not observe differential regulation of IL-1ß or its receptor, which suggests that regulation of hippocampal IL-1ß and its receptor is not important after eFS. However this does not exclude function of IL-1ß during eFS.
Based on our data, increased expression of chemokines CCL-2 (1.25-fold) and CCL-3 (1.17-fold) suggests regulation of blood-brain barrier permeability. Interestingly, both chemokines are found to be upregulated in TLE patients at mRNA transcript and protein level [25,57]. Expression of CCL-3 at the apical membrane of vascular endothelial cells triggers adhesion of leucocyte, whereas basal localization of CCL-2 to the membrane facilitates the migration of the leucocytes through the endothelial surface [60–62]. The regulation of chemokines therefore contribute to the permeability of the blood-brain barrier, which is associated with epileptogenesis [56]. In addition, both CCL-2 and CCL-3 trigger Ca2+ signaling in hippocampal neurons resulting in increased excitability and synaptic transmission [63,64].
Cytokine receptor IL-10ra and IL-10rb are differentially regulated 1 hour after eFS. The ligand, IL-10, is generally regarded to have anti-epileptogenic effects [65,66], and is upregulated in hippocampus and cortex of TLE patients [57]. IL-10 and its receptor are both upregulated as a consequence of excitotoxic injury [67]. This suggests that, as a compensatory mechanism, IL-10 expression is induced to avoid exacerbation of the neuronal damage. Interestingly, the alpha and beta subunit of the heterotetrameric IL-10 receptor are up- (1.13-fold) and downregulated (0.90-fold), respectively, after eFS. Unfortunately not much is known about the differences in characteristics of IL-10 receptor subunits, especially regarding the receptor function in the brain. Also little is known about the specific functional implications of the downregulated complement factors C1qa (0.88-fold) and Cr1l (0.91-fold) in epilepsy. Reduced expression of the complement factors may suggest suppression of the complement cascade. However, activation of the complement system, i.e. upregulation of complement mediators including C1qa and C1r, has been reported in both experimental and human temporal lobe epilepsy [68] and is thought to contribute to neuronal hyper-excitability and seizures [56]. In summary, our findings indicate that chemokines, cytokines and complement factors are regulated as a result of eFS.
Genes involved in structural reorganization after eFS
Overrepresentation of genes to GO classification cell-mediated adhesion signaling indicates that structural reorganization seen 14 days after eFS is initiated 3 days after eFS (Fig 7). In this study we found significant upregulation of Ptprd within the CA2 region of the hippocampus 3 days after eFS. Ptprd localizes to the pre-synapse where it interacts with post-synaptic neurotrophin receptor tyrosine kinases (Trks) and bi-directionally activate signal transduction cascades to recruit synaptic proteins important for establishing and maintaining excitatory synapses [40]. Ablation of Ptprd may therefore result in altered CA1 synaptic transmission and learning-deficits [69]. The observed increase of Ptprd within the CA2 may therefore trigger changes at excitatory synapse projecting to the CA1 [70]. Multiple lines of evidence suggest regulation of class 3 semaphorins after kainate acid-induced epilepsy in rodents [71,72]. Dpysl3, a downstream effector of Sema3A, was significantly regulated 3 days after eFS in our microarray and its mRNA expression was found in all subfields of the hippocampus. However, the widespread increase of Dpysl3 expression in HT animals did not reach significance. Validation of the candidate genes from the array, (Dpysl3, Nrxn1, and Slit2,) by in situ hybridization proved to be difficult. Most likely, this is due to heterogeneity in the transcriptional response of HT and NT littermates and detection sensitivity of RA-ISH.
Fourteen days after eFS, upregulation of CNP protein in the stratum lucidum confirmed our hypothesis that structural reorganization occurs after eFS. The upregulation of CNP indicates involvement of oligodendrocyte activation and myelin production. Interestingly, CNP mRNA expression is downregulated (0.89-fold) three days after eFS, similar to Mobp, Mal, and Mag (0.86, 0.85, and 0.90-fold, respectively), which all represent myelination and oligodendrocyte activation. However, fourteen days post HT CNP expression as well as myelin oligodendrocyte glycoprotein (Mog, 1.13-fold) is upregulated (1.26-fold). It would be interesting to learn what this switch in regulation of expression during the early to late phase after eFS means in terms of oligodendrocyte activation and myelination in the hippocampus. Possibly the reversal in expression has to do with different phases in structural reorganization, in which de- and re-myelination are required. It should be noted that mossy fibers to the CA3 are normally non-myelinated, hence the name stratum lucidum [73]. However, myelinated axons can be detected within the stratum lucidum, in particular after lesion of the enthorino-hippocampal perforant pathway [74]. This lesion induced axon sprouting in the stratum lucidum and recruited oligodendrocytes thought to myelinate the newly formed axon sprouts. Thus, our observed changes in CNP levels after eFS may indicate oligodendrocyte recruitment possibly contributing to myelination of newly sprouted axons. What the significance of these processes and oligodendrocytes is during epileptogenesis remains to be determined [75].
All together, our data suggest that proteins related to structural organization of the nervous system, such as cell adhesion molecules, axon guidance proteins, and myelin production are regulated after eFS. Kainate acid-induced experimental mesial temporal lobe epilepsy in rats trigger regulation of proteins involved in structural organization, axon guidance signaling pathways, 14 days post-induction [76]. The observed eFS induced changes in axon guidance and myelin-related genes, as well as the immune response activity mentioned earlier, might therefore resemble processes underlying epileptogenesis independent of the precipitating event.
Glia activation
Interestingly, regulation of glia-associated genes was observed 1 hour and 3 days after eFS. Gjb6 (Connexin 30), a gap junction protein expressed in glial cells, remained downregulated (0.80-fold) until 3 days after eFS. Gap junction proteins are important for cell-to-cell communication and exchange of small signaling molecules (<1000 Da) [77]. Other glia-associated proteins, such as S100a4 and Aqp4, were upregulated 3 days after eFS (1.14- and 1.11-fold, respectively). Aqp4 (aquaporin 4) is a water channel present at the astroglial endfeet contacting capillaries regulating transmembrane water transport [78]. Both Aqp4 and Gjb6 play important roles in regulation of osmolarity and energy metabolite housekeeping, therefore contribute to the epileptogenesis and are related to seizure severity [79]. Genetic ablation of either of the two genes results in prolongation of seizure duration or lowering of threshold for epileptic activity, respectively [79,80]. It remains to be determined by which mechanism these channels/junctions contribute to epileptogenesis. The regulation of these genes, including glia marker S100a4, indicates activation of glial cells shortly after eFS.
Regulation of Camk2a 56 days after eFS
At 56 days after eFS, we found Camk2a to be the sole significantly (down)-regulated gene (Fig 7). Our results are in line with a recent study showing that Camk2a protein localization in the post-synaptic density (PSD) is reduced 25 days after recurrent febrile seizures [81]. In the same study, the phosphorylation state of Camk2a is changed in favor of Thr305 over Thr286 after recurrent febrile seizures. The reduction in Thr286 phosphorylation results in higher dissociation rates of Camk2a from the PSD [82]. Phosphorylation of Camk2a is a major trigger to consolidate synaptic potentiation during the early phase of long-term potentiation (LTP), an important mechanism for synaptic plasticity related to hippocampal learning [82]. The observed reduction in phosphorylation of Thr286 and presence of Camk2a in the spine by Xiong et al. [81] together with our observation of reduced Camk2a expression, may reflect a compensatory mechanism as a consequence to increased neuronal activity. Since lower number of and phosphorylation of Camk2a (Thr286), would result in decreased probability to elicit LTP at the synapse. Transcriptome analysis in the CA3 after electrical kindling of the DG also showed marked reduction of Camk2a during the latent phase, in addition to reduction directly and 1 week after kindling [48]. Our data support the notion that regulation of CaMK2a is involved in epileptogenesis.
Heterogeneity in transcriptional response during late phase after eFS in mice
As mentioned earlier, in rat studies about 35–68% of all eFS animals show spontaneous epileptiform activity [15,22,23], which indicates that there is heterogeneity in response to eFS. All individual samples taken one hour, three days and fourteen days after eFS were used for hierarchical clustering analysis and identified three clusters based on their transcription profiles. All samples within each cluster belonged to the same time-point. This indicates that transcriptional profiles of all individuals per time-point show a high degree of similarity, especially during the early phase after eFS. However, fourteen days after eFS sample cluster behavior shows a certain degree of heterogeneity in transcriptional response. This heterogeneity may reflect the process of epileptogenesis and its relation to susceptibility to develop spontaneous recurrent seizures. Because of the study design we cannot correlate transcriptional profiles with spontaneous recurrent seizure outcome. Further longitudinal studies in mice will be required to determine which animals develop spontaneous recurrent seizures after eFS and whether changes in transcriptome profiles can be used to predict seizure outcome.
Concluding Remarks
The eFS solely may not be responsible for initiation of epileptogenesis but contribute to its process. The observed changes in structural organization and expression of genes related to this process confirm that eFS induce changes in the brain. These changes most likely contribute to or occur simultaneously with functional changes, such as HCN-channel conductance, altered GABAergic function, increased neuronal excitability, and shifted activity-dependent synaptic plasticity, described in other studies related to febrile seizures [14,18,47,54,83]. Other precipitating and susceptibility factors, such as stress [84] or genetic background [10,11], in concert with prolonged eFS, are likely to contribute to the development of TLE. It is of interest to study which factors increase the chance of spontaneous recurrent seizures later in life. The combination of these factors with prolonged eFS, or repetitive exposure to eFS, will be required to validate our findings and provide valuable insight in the molecular mechanism behind the process of epileptogenesis. The observed local changes in NF and CNP expression indicate that hippocampal subfield transcriptome analysis after eFS might contribute to the identification of candidate genes. Additionally, identification of markers for epileptogenesis would greatly facilitate the ability to identify mice that will develop spontaneous recurrent seizures. This is the first study to identify regulated genes during a time-course of 56 days after eFS in mice (Fig 7). Our findings, in support with previous studies, identified initial regulation of immune and inflammation responses and glutamate-glutamine cycle. During the early phase and late phase glial-, myelin-, and axon guidance/cell adhesion related genes are regulated, together with increased expression of NF and CNP. These latter findings point to structural reorganization of the hippocampus after eFS in mice.
Supporting Information
Table lists all primers used for qPCR experiments and the generation of 33P RNA probes for radioactive in situ hybridization.
(XLS)
Fc: Fold change represents expression values of HT animals divided by expression values of NT animals. P-value BH-corrected: Benjamini and Hochberg (BH) error corrected p-values.
(XLS)
Acknowledgments
We thank members of the P.N.E.d.G laboratory for assistance and discussion.
Data Availability
All relevant data are within the paper and its Supporting Information files. In addition microarray raw data are available from the NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE66762.
Funding Statement
This work was supported by the National Epilepsy Fund of the Netherlands, grant number: NEF 10-17 and 03-12, (https://www.epilepsie.nl), and the Epilepsies of Childhood Foundation, grant number: N/A, website: N/A. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hauser WA. The prevalence and incidence of convulsive disorders in children. Epilepsia. 1994;35 Suppl 2: S1–6. [DOI] [PubMed] [Google Scholar]
- 2. French JA, Williamson PD, Thadani VM, Darcey TM, Mattson RH, Spencer SS, et al. Characteristics of medial temporal lobe epilepsy: I. Results of history and physical examination. Ann Neurol. 1993;34: 774–780. 10.1002/ana.410340604 [DOI] [PubMed] [Google Scholar]
- 3. Theodore WH, Bhatia S, Hatta J, Fazilat S, DeCarli C, Bookheimer SY, et al. Hippocampal atrophy, epilepsy duration, and febrile seizures in patients with partial seizures. Neurology. 1999;52: 132–136. [DOI] [PubMed] [Google Scholar]
- 4. Engel J. Mesial temporal lobe epilepsy: what have we learned? The Neuroscientist. 2001;7: 340–352. [DOI] [PubMed] [Google Scholar]
- 5. Dubé CM, Brewster AL, Baram TZ. Febrile seizures: mechanisms and relationship to epilepsy. Brain and Development. 2009;31: 366–371. 10.1016/j.braindev.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shinnar S, Bello JA, Chan S, Hesdorffer DC, Lewis DV, MacFall J, et al. MRI abnormalities following febrile status epilepticus in children: the FEBSTAT study. Neurology. 2012;79: 871–877. 10.1212/WNL.0b013e318266fcc5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis DV, Shinnar S, Hesdorffer DC, Bagiella E, Bello JA, Chan S, et al. Hippocampal sclerosis after febrile status epilepticus: the FEBSTAT study. Ann Neurol. 2014;75: 178–185. 10.1002/ana.24081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cross JH. Fever and fever-related epilepsies. Epilepsia. 2012;53 Suppl 4: 3–8. 10.1111/j.1528-1167.2012.03608.x [DOI] [PubMed] [Google Scholar]
- 9. Neligan A, Bell GS, Giavasi C, Johnson AL, Goodridge DM, Shorvon SD, et al. Long-term risk of developing epilepsy after febrile seizures: a prospective cohort study. Neurology. 2012;78: 1166–1170. 10.1212/WNL.0b013e31824f807a [DOI] [PubMed] [Google Scholar]
- 10. Eckhaus J, Lawrence KM, Helbig I, Bui M, Vadlamudi L, Hopper JL, et al. Genetics of febrile seizure subtypes and syndromes: a twin study. Epilepsy Research. 2013;105: 103–109. 10.1016/j.eplepsyres.2013.02.011 [DOI] [PubMed] [Google Scholar]
- 11. Hessel EVS, de Wit M, Wolterink-Donselaar IG, Karst H, de Graaff E, van Lith HA, et al. Identification of Srp9 as a febrile seizure susceptibility gene. Ann Clin Transl Neurol. 2014;1: 239–250. 10.1002/acn3.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hessel EVS, van Gassen KLI, Wolterink-Donselaar IG, Stienen PJ, Fernandes C, Brakkee JH, et al. Phenotyping mouse chromosome substitution strains reveal multiple QTLs for febrile seizure susceptibility. Genes, Brain and Behavior. 2009;8: 248–255. 10.1111/j.1601-183X.2008.00466.x [DOI] [PubMed] [Google Scholar]
- 13. Baram TZ, Gerth A, Schultz L. Febrile seizures: an appropriate-aged model suitable for long-term studies. Developmental Brain Research. 1997;98: 265–270. 10.1016/S0165-3806(96)00190-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Gassen KLI, Hessel EVS, Ramakers GMJ, Notenboom RGE, Wolterink-Donselaar IG, Brakkee JH, et al. Characterization of febrile seizures and febrile seizure susceptibility in mouse inbred strains. Genes, Brain and Behavior. 2008;7: 578–586. 10.1111/j.1601-183X.2008.00393.x [DOI] [PubMed] [Google Scholar]
- 15. Koyama R, Tao K, Sasaki T, Ichikawa J, Miyamoto D, Muramatsu R, et al. GABAergic excitation after febrile seizures induces ectopic granule cells and adult epilepsy. Nature Medicine. 2012;18: 1271–1278. 10.1038/nm.2850 [DOI] [PubMed] [Google Scholar]
- 16. Dube C, Chen K, Eghbal-Ahmadi M, Brunson K, Soltesz I, Baram TZ. Prolonged febrile seizures in the immature rat model enhance hippocampal excitability long term. Ann Neurol. 2000;47: 336–344. [PMC free article] [PubMed] [Google Scholar]
- 17. Kamal A, Notenboom RGE, de Graan PNE, Ramakers GMJ. Persistent changes in action potential broadening and the slow afterhyperpolarization in rat CA1 pyramidal cells after febrile seizures. European Journal of Neuroscience. 2006;23: 2230–2234. 10.1111/j.1460-9568.2006.04732.x [DOI] [PubMed] [Google Scholar]
- 18. Notenboom RGE, Ramakers GMJ, Kamal A, Spruijt BM, de Graan PNE. Long-lasting modulation of synaptic plasticity in rat hippocampus after early-life complex febrile seizures. Eur J Neurosci. 2010;32: 749–758. 10.1111/j.1460-9568.2010.07321.x [DOI] [PubMed] [Google Scholar]
- 19. Jansen JFA, Lemmens EMP, Strijkers GJ, Prompers JJ, Schijns OEMG, Kooi ME, et al. Short- and long-term limbic abnormalities after experimental febrile seizures. Neurobiology of Disease. 2008;32: 293–301. 10.1016/j.nbd.2008.07.010 [DOI] [PubMed] [Google Scholar]
- 20. Toth Z, Yan XX, Haftoglou S, Ribak CE, Baram TZ. Seizure-induced neuronal injury: vulnerability to febrile seizures in an immature rat model. Journal of Neuroscience. 1998;18: 4285–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brewster A, Bender RA, Chen Y, Dubé C, Eghbal-Ahmadi M, Baram TZ. Developmental febrile seizures modulate hippocampal gene expression of hyperpolarization-activated channels in an isoform- and cell-specific manner. J Neurosci. 2002;22: 4591–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubé C, Richichi C, Bender RA, Chung G, Litt B, Baram TZ. Temporal lobe epilepsy after experimental prolonged febrile seizures: prospective analysis. Brain. 2006;129: 911–922. 10.1093/brain/awl018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwak S-E, Kim J-E, Kim SC, Kwon O-S, Choi S-Y, Kang T-C. Hyperthermic seizure induces persistent alteration in excitability of the dentate gyrus in immature rats. Brain Research. 2008;1216: 1–15. 10.1016/j.brainres.2008.04.026 [DOI] [PubMed] [Google Scholar]
- 24. Otte WM, Dijkhuizen RM, van Meer MPA, van der Hel WS, Verlinde SAMW, van Nieuwenhuizen O, et al. Characterization of functional and structural integrity in experimental focal epilepsy: reduced network efficiency coincides with white matter changes. PLoS ONE. 2012;7: e39078 10.1371/journal.pone.0039078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Gassen KLI, de Wit M, Koerkamp MJAG, Rensen MGA, van Rijen PC, Holstege FCP, et al. Possible role of the innate immunity in temporal lobe epilepsy. Epilepsia. 2008;49: 1055–1065. 10.1111/j.1528-1167.2007.01470.x [DOI] [PubMed] [Google Scholar]
- 26. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30: e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, van Hooff SR, et al. Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol Syst Biol. 2009;5: 266 10.1038/msb.2009.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. The analysis of gene expression data. 2003. 10.1007/0-387-21679-0_14 [DOI] [Google Scholar]
- 29. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pernot F, Dorandeu F, Beaup C, Peinnequin A. Selection of reference genes for real-time quantitative reverse transcription-polymerase chain reaction in hippocampal structure in a murine model of temporal lobe epilepsy with focal seizures. J Neurosci Res. 2010;88: 1000–1008. 10.1002/jnr.22282 [DOI] [PubMed] [Google Scholar]
- 31. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34: 374–378. [DOI] [PubMed] [Google Scholar]
- 32. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 34. Boender AJ, Koning NA, van den Heuvel JK, Luijendijk MCM, van Rozen AJ, la Fleur SE, et al. AAV-mediated gene transfer of the obesity-associated gene Etv5 in rat midbrain does not affect energy balance or motivated behavior. PLoS ONE. 2014;9: e94159 10.1371/journal.pone.0094159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malik AR, Urbanska M, Gozdz A, Swiech LJ, Nagalski A, Perycz M, et al. Cyr61, a matricellular protein, is needed for dendritic arborization of hippocampal neurons. J Biol Chem. 2013;288: 8544–8559. 10.1074/jbc.M112.411629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richter F, Meurers BH, Zhu C, Medvedeva VP, Chesselet M-F. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. Journal of Comparative Neurology. 2009;515: 538–547. 10.1002/cne.22062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4: P3 [PubMed] [Google Scholar]
- 38. de Wit J, Sylwestrak E, O'Sullivan ML, Otto S, Tiglio K, Savas JN, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64: 799–806. 10.1016/j.neuron.2009.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Niisato E, Nagai J, Yamashita N, Abe T, Kiyonari H, Goshima Y, et al. CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus. Devel Neurobio. 2012;72: 1447–1457. 10.1002/dneu.22007 [DOI] [PubMed] [Google Scholar]
- 40. Takahashi H, Arstikaitis P, Prasad T, Bartlett TE, Wang YT, Murphy TH, et al. Postsynaptic TrkC and presynaptic PTPσ function as a bidirectional excitatory synaptic organizing complex. Neuron. 2011;69: 287–303. 10.1016/j.neuron.2010.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Babb TL, Brown WJ, Pretorius J, Davenport C, Lieb JP, Crandall PH. Temporal lobe volumetric cell densities in temporal lobe epilepsy. Epilepsia. 1984;25: 729–740. [DOI] [PubMed] [Google Scholar]
- 42. Fang M, Liu G-W, Pan Y-M, Shen L, Li C-S, Xi Z-Q, et al. Abnormal expression and spatiotemporal change of Slit2 in neurons and astrocytes in temporal lobe epileptic foci: A study of epileptic patients and experimental animals. Brain Research. 2010;1324: 14–23. 10.1016/j.brainres.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 43. Edgar JM, McLaughlin M, Werner HB, McCulloch MC, Barrie JA, Brown A, et al. Early ultrastructural defects of axons and axon-glia junctions in mice lacking expression of Cnp1. Glia. 2009;57: 1815–1824. 10.1002/glia.20893 [DOI] [PubMed] [Google Scholar]
- 44. Myllykoski M, Raasakka A, Han H, Kursula P. Myelin 2',3‘-cyclic nucleotide 3’-phosphodiesterase: active-site ligand binding and molecular conformation. PLoS ONE. 2012;7: e32336 10.1371/journal.pone.0032336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33: 366–374. 10.1038/ng1095 [DOI] [PubMed] [Google Scholar]
- 46. Schuchmann S, Schmitz D, Rivera C, Vanhatalo S, Salmen B, Mackie K, et al. Experimental febrile seizures are precipitated by a hyperthermia-induced respiratory alkalosis. Nature Medicine. 2006;12: 817–823. 10.1038/nm1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hamelin S, Pouyatos B, Khalaf-Nazzal R, Chabrol T, Francis F, David O, et al. Long-term modifications of epileptogenesis and hippocampal rhythms after prolonged hyperthermic seizures in the mouse. Neurobiology of Disease. 2014;69: 156–168. 10.1016/j.nbd.2014.05.025 [DOI] [PubMed] [Google Scholar]
- 48. Gorter JA, van Vliet EA, Aronica E, Breit T, Rauwerda H, Lopes da Silva FH, et al. Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J Neurosci. 2006;26: 11083–11110. 10.1523/JNEUROSCI.2766-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung K-H, Chu K, Lee S-T, Park K-I, Kim J-H, Kang K-M, et al. Molecular alterations underlying epileptogenesis after prolonged febrile seizure and modulation by erythropoietin. Epilepsia. 2011;52: 541–550. 10.1111/j.1528-1167.2010.02916.x [DOI] [PubMed] [Google Scholar]
- 50. Eid T, Thomas MJ, Spencer DD, Rundén-Pran E. Loss of glutamine synthetase in the human epileptogenic hippocampus: possible mechanism for raised extracellular glutamate in mesial temporal lobe epilepsy. The Lancet. 2004. 10.1016/S0140-6736(03)15166-5 [DOI] [PubMed] [Google Scholar]
- 51. van Gassen KLI, van der Hel WS, Hakvoort TBM, Lamers WH, de Graan PNE. Haploinsufficiency of glutamine synthetase increases susceptibility to experimental febrile seizures. Genes, Brain and Behavior. 2009;8: 290–295. 10.1111/j.1601-183X.2008.00471.x [DOI] [PubMed] [Google Scholar]
- 52. van der Hel WS, Notenboom RGE, Bos IWM, van Rijen PC, van Veelen CWM, de Graan PNE. Reduced glutamine synthetase in hippocampal areas with neuron loss in temporal lobe epilepsy. Neurology. 2005;64: 326–333. 10.1212/01.WNL.0000149636.44660.99 [DOI] [PubMed] [Google Scholar]
- 53. Proper EA, Oestreicher AB, Jansen GH, Veelen CW, van Rijen PC, Gispen WH, et al. Immunohistochemical characterization of mossy fibre sprouting in the hippocampus of patients with pharmaco-resistant temporal lobe epilepsy. Brain. 2000;123 (Pt 1): 19–30. [DOI] [PubMed] [Google Scholar]
- 54. Richichi C, Brewster AL, Bender RA, Simeone TA, Zha Q, Yin HZ, et al. Mechanisms of seizure-induced “transcriptional channelopathy” of hyperpolarization-activated cyclic nucleotide gated (HCN) channels. Neurobiology of Disease. 2008;29: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z, Fan Y, Xu J, Li L, Heng D, Han S, et al. Transcriptome analysis of the hippocampus in novel rat model of febrile seizures. PLoS ONE. 2014;9: e95237 10.1371/journal.pone.0095237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vezzani A, French J, Bartfai T, Baram TZ. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7: 31–40. 10.1038/nrneurol.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kan AA, de Jager W, de Wit M, Heijnen C, van Zuiden M, Ferrier C, et al. Protein expression profiling of inflammatory mediators in human temporal lobe epilepsy reveals co-activation of multiple chemokines and cytokines. J Neuroinflammation. 2012;9: 207 10.1186/1742-2094-9-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gatti S, Vezzani A, Bartfai T. Mechanisms of fever and febrile seizures: putative role of the interleukin-1 system. Febrile seizures Academic Press; 2002. [Google Scholar]
- 59. Dubé C, Vezzani A, Behrens M, Bartfai T, Baram TZ. Interleukin-1beta contributes to the generation of experimental febrile seizures. Ann Neurol. 2005;57: 152–155. 10.1002/ana.20358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chui R, Dorovini-Zis K. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. J Neuroinflammation. 2010;7: 1 10.1186/1742-2094-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Greenwood J, Heasman SJ, Alvarez JI, Prat A, Lyck R, Engelhardt B. Review: leucocyte-endothelial cell crosstalk at the blood-brain barrier: a prerequisite for successful immune cell entry to the brain. Neuropathol Appl Neurobiol. 2011;37: 24–39. 10.1111/j.1365-2990.2010.01140.x [DOI] [PubMed] [Google Scholar]
- 62. Louboutin J-P, Chekmasova A, Marusich E, Agrawal L, Strayer DS. Role of CCR5 and its ligands in the control of vascular inflammation and leukocyte recruitment required for acute excitotoxic seizure induction and neural damage. FASEB J. 2011;25: 737–753. 10.1096/fj.10-161851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kuijpers M, van Gassen KLI, de Graan PNE, Gruol D. Chronic exposure to the chemokine CCL3 enhances neuronal network activity in rat hippocampal cultures. Journal of Neuroimmunology. 2010;229: 73–80. 10.1016/j.jneuroim.2010.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Y, Tang H, Liu J, Dong J, Xiong H. Chemokine CCL2 modulation of neuronal excitability and synaptic transmission in rat hippocampal slices. J Neurochem. 2011;116: 406–414. 10.1111/j.1471-4159.2010.07121.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ishizaki Y, Kira R, Fukuda M, Torisu H, Sakai Y, Sanefuji M, et al. Interleukin-10 is associated with resistance to febrile seizures: genetic association and experimental animal studies. Epilepsia. 2009;50: 761–767. 10.1111/j.1528-1167.2008.01861.x [DOI] [PubMed] [Google Scholar]
- 66. Levin SG, Godukhin OV. Protective effects of interleukin-10 on the development of epileptiform activity evoked by transient episodes of hypoxia in rat hippocampal slices. Neurosci Behav Physi. 2007;37: 467–470. 10.1007/s11055-007-0036-1 [DOI] [PubMed] [Google Scholar]
- 67. Gonzalez P, Burgaya F, Acarin L, Peluffo H, Castellano B, Gonzalez B. Interleukin-10 and interleukin-10 receptor-I are upregulated in glial cells after an excitotoxic injury to the postnatal rat brain. J Neuropathol Exp Neurol. 2009;68: 391–403. 10.1097/NEN.0b013e31819dca30 [DOI] [PubMed] [Google Scholar]
- 68. Aronica E, Boer K, van Vliet EA, Redeker S, Baayen JC, Spliet WGM, et al. Complement activation in experimental and human temporal lobe epilepsy. Neurobiology of Disease. 2007;26: 497–511. 10.1016/j.nbd.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 69. Uetani N, Kato K, Ogura H, Mizuno K, Kawano K, Mikoshiba K, et al. Impaired learning with enhanced hippocampal long-term potentiation in PTPdelta-deficient mice. The EMBO Journal. 2000;19: 2775–2785. 10.1093/emboj/19.12.2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kohara K, Pignatelli M, Rivest AJ, Jung H-Y, Kitamura T, Suh J, et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nature Neuroscience. 2014;17: 269–279. 10.1038/nn.3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Holtmaat A, Gorter JA, de Wit J, Tolner EA, Spijker SS, Giger RJ, et al. Transient downregulation of Sema3A mRNA in a rat model for temporal lobe epilepsy: a novel molecular event potentially contributing to mossy fiber sprouting. Experimental Neurology. 2003;: 142–150. 10.1016/S0014-4886(03)00035-9 [DOI] [PubMed] [Google Scholar]
- 72. Barnes G, Puranam RS, Luo Y, McNamara JO. Temporal specific patterns of semaphorin gene expression in rat brain after kainic acid-induced status epilepticus. Hippocampus. 2003;13: 1–20. 10.1002/hipo.10041 [DOI] [PubMed] [Google Scholar]
- 73. Blackstad TW, Kjaerheim Å. Special axo-dendritic synapses in the hippocampal cortex: electron and light microscopic studies on the layer of mossy fibers. J Comp Neurol. 1961;117: 133–159. 10.1002/cne.901170202 [DOI] [PubMed] [Google Scholar]
- 74. Drøjdahl N, Nielsen HH, Gardi JE, Wree A, Peterson AC, Nyengaard JR, et al. Axonal plasticity elicits long-term changes in oligodendroglia and myelinated fibers. Glia. 2010;58: 29–42. 10.1002/glia.20897 [DOI] [PubMed] [Google Scholar]
- 75. Parent JM, Bussche von dem N, Lowenstein DH. Prolonged seizures recruit caudal subventricular zone glial progenitors into the injured hippocampus. Hippocampus. 2006;16: 321–328. 10.1002/hipo.20166 [DOI] [PubMed] [Google Scholar]
- 76. Sharma AK, Searfoss GH, Reams RY, Jordan WH, Snyder PW, Chiang AY, et al. Kainic acid-induced F-344 rat model of mesial temporal lobe epilepsy: gene expression and canonical pathways. Toxicol Pathol. 2009;37: 776–789. 10.1177/0192623309344202 [DOI] [PubMed] [Google Scholar]
- 77. Mylvaganam S, Ramani M, Krawczyk M, Carlen PL. Roles of gap junctions, connexins, and pannexins in epilepsy. Front Physiol. 2014;5: 172 10.3389/fphys.2014.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. NSC. 2004;129: 905–913. 10.1016/j.neuroscience.2004.08.053 [DOI] [PubMed] [Google Scholar]
- 79. Steinhäuser C, Seifert G, Bedner P. Astrocyte dysfunction in temporal lobe epilepsy: K+ channels and gap junction coupling. Glia. 2012;60: 1192–1202. 10.1002/glia.22313 [DOI] [PubMed] [Google Scholar]
- 80. Binder DK, Yao X, Zador Z, Sick TJ, Verkman AS, Manley GT. Increased seizure duration and slowed potassium kinetics in mice lacking aquaporin-4 water channels. Glia. 2006;53: 631–636. 10.1002/glia.20318 [DOI] [PubMed] [Google Scholar]
- 81. Xiong Y, Zhou H, Zhang L. Influences of hyperthermia-induced seizures on learning, memory and phosphorylative state of CaMKIIα in rat hippocampus. Brain Research. 2014. [DOI] [PubMed] [Google Scholar]
- 82. Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;: 1–27. 10.1038/nrn3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Swijsen A, Avila A, Brône B, Janssen D, Hoogland G, Rigo J-M. Experimental early-life febrile seizures induce changes in GABA(A) R-mediated neurotransmission in the dentate gyrus. Epilepsia. 2012;53: 1968–1977. 10.1111/j.1528-1167.2012.03694.x [DOI] [PubMed] [Google Scholar]
- 84. van Campen JS, Jansen FE, de Graan PNE, Braun KPJ, Joëls M. Early life stress in epilepsy: a seizure precipitant and risk factor for epileptogenesis. Epilepsy Behav. 2014;38: 160–171. 10.1016/j.yebeh.2013.09.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table lists all primers used for qPCR experiments and the generation of 33P RNA probes for radioactive in situ hybridization.
(XLS)
Fc: Fold change represents expression values of HT animals divided by expression values of NT animals. P-value BH-corrected: Benjamini and Hochberg (BH) error corrected p-values.
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. In addition microarray raw data are available from the NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE66762.