Abstract
Analysis of a Selected Set of Antimicrobial Peptides
The rapid emergence of resistance to classical antibiotics has increased the interest in novel antimicrobial compounds. Antimicrobial peptides (AMPs) represent an attractive alternative to classical antibiotics and a number of different studies have reported antimicrobial activity data of various AMPs, but there is only limited comparative data available. The mode of action for many AMPs is largely unknown even though several models have suggested that the lipopolysaccharides (LPS) play a crucial role in the attraction and attachment of the AMP to the bacterial membrane in Gram-negative bacteria. We compared the potency of Cap18, Cap11, Cap11-1-18m2, Cecropin P1, Cecropin B, Bac2A, Bac2A-NH2, Sub5-NH2, Indolicidin, Melittin, Myxinidin, Myxinidin-NH2, Pyrrhocoricin, Apidaecin and Metalnikowin I towards Staphylococcus aureus, Enterococcus faecalis, Pseudomonas aeruginosa, Escherichia coli, Aeromonas salmonicida, Listeria monocytogenes, Campylobacter jejuni, Flavobacterium psychrophilum, Salmonella typhimurium and Yersinia ruckeri by minimal inhibitory concentration (MIC) determinations. Additional characteristics such as cytotoxicity, thermo and protease stability were measured and compared among the different peptides. Further, the antimicrobial activity of a selection of cationic AMPs was investigated in various E. coli LPS mutants.
Cap18 Shows a High Broad Spectrum Antimicrobial Activity
Of all the tested AMPs, Cap18 showed the most efficient antimicrobial activity, in particular against Gram-negative bacteria. In addition, Cap18 is highly thermostable and showed no cytotoxic effect in a hemolytic assay, measured at the concentration used. However, Cap18 is, as most of the tested AMPs, sensitive to proteolytic digestion in vitro. Thus, Cap18 is an excellent candidate for further development into practical use; however, modifications that should reduce the protease sensitivity would be needed. In addition, our findings from analyzing LPS mutant strains suggest that the core oligosaccharide of the LPS molecule is not essential for the antimicrobial activity of cationic AMPs, but in fact has a protective role against AMPs.
Introduction
The extensive use of classical antibiotics not only in human medicine, but also in animal farming for treatment and growth promotion has resulted in the development and spread of antibiotic resistance in bacteria. There is now an increasing awareness of the problems for human health caused by antibiotic resistant bacteria among food producing animals, especially since many of the same classes of antibiotics are used in both reservoirs [1]. This has emphasized the need for new solutions to battle infections in farmed animals which are not based on antibiotics that are considered critically important for human health [2].
Antimicrobial peptides (AMPs) represent an attractive alternative to classical antibiotics. AMPs are present in all kingdoms of life and are ancient components of the innate immunity and represent the first line of defense in an infection [3,4]. Despite their diversity in sequence, they generally have an overall positive charge (+2 to +9) and have a substantial proportion of hydrophobic amino acids (= >30%). The length normally varies between 10–50 amino acids. Based on these properties, AMPs are able to fold into amphiphilic three-dimensional structures, which are divided into 4 major groups: α-helical peptides (e.g. Cecropin B, Cecropin P1, Melittin, Cap11, Cap18 and magainins); β-sheet peptides with 2–4 disulfide bridges (e.g. human defensins, plectasin or protegrins); extended peptides which are rich in glycin, proline, tryptophan, and/or histidine (e.g. Indolicidin, Apidaecin); loop peptides with one disulfide bridge (e.g. bactenecin). The majority of the so far characterized AMPs belong to the group of the α-helical peptides and the β-sheet peptides [3][5][6]. AMPs isolated from prokaryotes are called bacteriocins. One of the best characterized bacteriocins is Nisin, which is originally isolated from Lactococcus lactis. Nisin is active against various major Gram-positive food-borne pathogens, including Listeria and Clostridium. It is used as food additive and preservative since 1969 in processed cheese, pasteurized milk and milk products and cooked sausages [7].
Classically, the mechanisms of AMP action are thought of as an interaction with the bacterial cell membrane. Most often, the interaction of an AMP with the membrane will lead to destabilization of the membrane by formation of transient channels, micellarization, and dissolution of the membrane or translocation across the membrane which results in increased membrane permeability. As a result of increased permeability, nutrients and electrolytes will flow out which lead to killing of the bacteria. However, killing might also happen in a more specific or targeted manner by recognition of cell surface proteins as a first step followed by insertion in the membrane. Finally, the mode of action might also be targeting metabolic processes in the bacteria including cell wall synthesis, nucleic acid or protein synthesis, which are vital to the organism [8].
Several studies have reported activity of different peptides useful for food preservation and safety purposes [7][9][10][11]. However, most of these studies have only focused on a single or a few peptides and a single or a few bacterial targets. Thus, a major bottleneck in identifying which new AMPs to choose for further development into practical use is the lack of comprehensive overviews comparing the potency of known AMPs from different sources to a broader range of pathogens. In addition, cytotoxicity and stability is most often ignored, although being crucial parameters in developing successful AMP alternatives.
In this study, we focused on finding AMPs to inhibit zoonotic and fish pathogens. Aquaculture is a high density animal production system characterized by a high use of antimicrobial agents. For this study we have selected a handful of peptides reported to have high antibacterial activity against Gram-negative bacteria. In addition we required the peptides to be composed of ordinary L-amino acids, devoid of posttranslational modifications, and to be shorter than 40 aa. These selection criteria were chosen to allow for subsequent development of recombinant production procedures for peptides with potential applications in food or feed. We compared the potency of the selected AMPs towards a broad range of pathogens under standardized and comparable conditions with respect to antimicrobial activity, hemolytic activity, and stability. In addition, the mode of action of the most potent AMPs has been addressed in the E. coli ATCC25922 which was chosen as model organism. The present work will facilitate the evaluation and identification of AMPs for further development.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains used in this study are listed in Table 1. The Yersinia ruckeri strain was kindly provided by Prof. Kurt Buchmann, University of Copenhagen, Faculty of Health and Medical Sciences, and the Flavobacterium psychrophilum strain was kindly provided by Prof. Inger Dalsgaard, DTU, Denmark. BW25113 is the Escherichia coli wild-type strain, a derivative of the F-,λ- E. coli K12 strain BD792 which was used in generating the KEIO collection [12][13]. E. coli ATCC25922 is a clinical isolate, serotype O6 and is often used as control strain in antimicrobial susceptibility testing. All strains were grown in Mueller-Hinton-II medium, except L. monocytogenes which was grown in BHI medium and F. psychrophilum which was grown in TYES medium (Tryptone yeast extract plus salts [14]). Incubation took place aerobically at 37°C, except for Y. ruckeri and A. salmonicida, which were grown aerobically at RT (20°C), C. jejuni NCTC11168 which was grown under microaerophilic conditions at 42°C and F. psychrophilum 1947 which was grown under aerobic conditions at 15°C. All plates were incubated for 18–20 hours, except the F. psychrophilum plates, which were incubated for 72 hours.
Table 1. Strains used in this study.
| Strain | Relevant characteristics /genotype | Reference(s) |
|---|---|---|
| Staphyloccous aureus ATCC29213 | control strain for antimicrobial susceptibility testing | ATCC strain collection |
| Enterococcus faecalis ATCC29212 | control strain for antimicrobial susceptibility testing | ATCC strain collection |
| Pseudomonas aeruginosa ATCC27853 | control strain for antimicrobial susceptibility testing | ATCC strain collection |
| Escherichia coli ATCC25922 | Clinical isolate, Serotype O6, Biotype 1, control strain for antimicrobial susceptibility testing | ATCC strain collection |
| Aeromonas salmonicida ATCC33658 | Type strain | ATCC strain collection |
| Salmonella enterica serovar Typhimurium LT2 | sequenced strain | |
| Listeria monocytogenes N22-2 | Isolate from fish processing industry | [58] |
| Campylobacter jejuni NCTC11168 | Isolate from human feces | NCTC strain collection |
| Flavobacterium psychrophilum 1947 | Prof. Inger Dalsgaard, DTU, Denmark | |
| Yersinia ruckeri 392/2003 | [59] | |
| Escherichia coli BW25113 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514, wild-type strain used in the KEIO collection | [12][13] |
| Escherichia coli JW3596 | BW25113 rfaC::kan | [13] |
| Escherichia coli JW3024 | BW25113 rfaE::kan | [13] |
| Escherichia coli JW3595 | BW25113 rfaF::kan | [13] |
| Escherichia coli JW3606 | BW25113 rfaG::kan | [13] |
| AD120 | Escherichia coli ATCC25922 ΔrfaC | This study |
| AD121 | Escherichia coli ATCC25922 ΔrfaE | This study |
| AD122 | Escherichia coli ATCC25922 ΔrfaF | This study |
| AD123 | Escherichia coli ATCC25922 ΔrfaF | This study |
Antimicrobial peptides
The peptides used in this study are listed in Table 2. Cecropin P1 and Cecropin B were purchased from Sigma-Aldrich. Cecropin P1 with a purity of ≥95% and Cecropin B with a purity of ≥97% and were dissolved in water. Bac2A-NH2 with a purity of ≥95% and Sub5-NH2 with a purity ≥95% were purchased from Anaspec (distributed by BioNordika Denmark A/S). The peptide sequences were confirmed with MS data. Bac2A-NH2 was dissolved in 100% DMSO, whereas Sub5-NH2 was dissolved in water. Cap18 (89.5% purity, 58% net peptide content), Cap11 (94.7% purity, 63% net peptide content), Cap11-1-18m2 (87.9% purity, 57% net peptide content), Bac2A (93.2% purity), Myxinidin (97% purity) and Myxindin-NH2 (97.3% purity, 66% net peptide content) were all synthesized at Genscript. Bac2A, Cap18, Cap11-1-18m2, Myxinidin and Myxinidin-NH2 were dissolved in water and Cap11 was dissolved in 100% DMSO. Melittin (RP10290-1) and Indolicidin (RP11242-0.5) each with a purity of >95% were purchased from Genscript and dissolved in water. The proline rich peptides, Pyrrhocoricin, Apidaecin IA and Metalnikowin I, were purchased from Anaspec each with a purity of ≥95% and dissolved in water. All peptides were dissolved to a stock concentration of 10 mg/ml.
Table 2. Sequence and origin of antimicrobial peptides.
| Peptide | Sequence | Origin | Structure | Reference |
|---|---|---|---|---|
| Cap11 | GLRKKFRKTRKRIQKLGRKIGKTGRKVWKAWREYGQIPYPCRI | mammalian, guinea pig, neutrophils | α-helical | [29] |
| Cap11-1-18m 2 | KLRKLFRKLLKLIRKLLR | truncated derivative of Cap11 | α-helical | [21] |
| Cap18 | GLRKRLRKFRNKIKEKLKKIGQKIQGLLPKLAPRTDY | mammalian, rabbit, neutrophils | α-helical | [28][30] |
| Cecropin P1 | SWLSKTAKKLENSAKKRISEGIAIAIQGGPR | mammalian, pig, small intestine | α-helical | [32][33][34] |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKALG-NH2 | insects, giant silk moth, pupae | α-helical | [33][35] |
| Bac2A | RLARIVVIRVAR | non-amidated version of Bac2A-NH2 | α-helical/β-sheet | |
| Bac2A-NH 2 | RLARIVVIRVAR-NH2 | linear variant of Bactenecin from bovine neutrophils | Linearized version of bactenecin | [36][37] |
| Sub5-NH 2 | RRWKIVVIRWRR-NH2 | synthetic variant of Bac2A-NH2 | Not available | [36] |
| Myxinidin | GIHDILKYGKPS | fish, epidermal mucus of Hagfish | Not available | [38] |
| Myxinidin-NH 2 | GIHDILKYGKPS-NH2 | amidated form of myxinidin | Not available | |
| Pyrrhocoricin | VDKGSYLPRPTPPRPIYNRN | insects, Pyrrhocoris apterus | Not available | [20] |
| Apidaecin IA | GNNRPVYIPQPRPPHPRI | insects, honey bee | Extended, proline rich | [60] |
| Metalnikowin I | VDKPDYRPRPRPPNM | insects, palomena prasina | Not available | [19] |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-NH2 | insects, honey bee | α-helical | [16] |
| Indolicidin | ILPWKWPWWPWR-NH2 | mammalian, bovine neutrophils | Extended | [17] |
Antimicrobial susceptibility testing (MIC testing)
The minimum inhibitory concentrations (MICs) of the AMPs were measured in 96-well microtiter plates according to the Clinical and Laboratory Standards Institute (CLSI, formerly National Committee for Clinical Laboratory Standards [NCCLS]) [15]. Briefly, liquid Mueller-Hinton-II medium containing increasing concentrations of AMPs is inoculated with a defined number of cells (approx. 105 CFUs/ml) in 96-well microtiter plates (polypropylene), whereas each plate also includes a positive growth control and a negative control (sterile control). After incubation, the MIC is determined by the lowest concentration showing no visible growth. All plates were incubated for 18–20 hours, except the F. psychrophilum plates which were incubated for 72 hours.
All the MIC measurements were carried out in duplicate. The MIC of the reference antibiotics was determined by the use of Sensititre panels (Trek Diagnostic Systems Ltd, East Grinstead, UK).
Cytotoxicity assay
The cytotoxicity for each AMP was determined spectrophotometrically by measuring the haemoglobin release from horse erythrocytes. Briefly, fresh defibrinated horse blood was washed three times with PBS, centrifuged for 15 minutes at 1000g and resuspended at 10% (v/v) in PBS. Samples of the washed horse erythrocytes (100 μl) were transferred to a 96 well microtiter plate and mixed with 100 μl AMP solution. PBS was used as a negative control, and 0.2% TritonX-100 was used as a positive control. The microtiter plates were incubated for 60 minutes at 37°C and then centrifuged for 10 minutes at 1300g. The supernatants were transferred to a flat-bottom 96 well polystyrene microtiter plate and the haemoglobin release was monitored by measuring the absorbance at 540 nm. The percentage of hemolysis was calculated as 100 *(Asample−APBS)/(ATritonX-100 –APBS), where Asample is the experimental absorbance of the peptide sample, APBS is the control absorbance of untreated erythrocytes, and ATritonX-100 is the absorbance of 0.2% TritonX-100 lysed cells.
Effect of temperature and proteases on antimicrobial activity
AMPs were heated at 70°C or 90° for 5, 15 or 30 minutes. An untreated control, which was kept at RT, was used as a control. After incubation at 70°C or 90°C, the minimum inhibitory concentrations (MICs) of the peptides were measured in 96-well microtiter plates according the Clinical and Laboratory Standards Institute (CLSI, formerly National Committee for Clinical Laboratory Standards [NCCLS]) [15] (see under Antimicrobial susceptibility testing (MIC testing). E. coli ATCC25922 was used as a test strain for all the AMPs. The effect of proteases, including trypsin (trypsin ultra, NEB, P8101) and proteinase K (NEB, P8107S), on antimicrobial activity of selected AMPs was investigated by incubation with the respective protease at 37°C for either 30 seconds, 2, 5, 15 or 30 minutes. The protease to AMP ratio used in the assay was 1:100 (w/w) and the buffers used were trypsin-ultra reaction buffer (NEB, P8101) for trypsin and 100 mM Tris-HCl pH 7.5 for proteinase K digestion. To ensure that the trypsin or proteinase K themselves have no antimicrobial activity, each of the protease was used alone in the corresponding buffer as a control. After incubation, the samples were cooked for 10 minutes at 90°C to inactivate the protease. Afterwards, the antimicrobial activity was determined by measuring the minimal inhibitory concentration (see Antimicrobial susceptibility testing)). E. coli ATCC25922 was used as a test strain.
Construction of LPS mutants of E. coli ATCC25922
Construction of knock-out mutants of E. coli ATCC25922 were constructed using the λ-Red recombinase gene replacement system [12]. Primers for amplification of the npt gene of pKD4 are listed in S1 Table. The correct double-crossover and recombination event was confirmed by primers listed in S1 Table and by sequencing. Finally, the kanamycin cassette was removed [12].
Results
Antimicrobial activity of selected antimicrobial peptides
Fifteen antimicrobial peptides from different classes and origins (Table 2), including the well-characterized AMPs Melittin [16] and Indoclidin [17], were selected and tested for antimicrobial activity. The antimicrobial activity was determined against 10 different bacterial strains from 10 different species (Table 1) as the minimum inhibitory concentration (MIC), summarized in Table 3. In addition, standard antibiotics from different classes were included for the comparison of the antimicrobial activity. No activity against F. psychrophilum could be measured, whereas a varying pattern of activity was found against the other bacterial species tested. Cap18 had the highest antimicrobial activity of all tested AMPs, in particular against Gram-negative pathogens, whereas the other cathelicidin, Cap11, was slightly less active. The antimicrobial activity of both cathelicidins was in general higher against Gram-negative compared to Gram-positive bacteria. In contrast, Cap11-1-18m2, which is a short derivative of Cap11, had only moderate antimicrobial activity against Gram-negative pathogens, but increased antimicrobial activity against Gram-positive bacteria compared to the mother peptide Cap11. Interestingly, Cap11-1-18m2 displayed the same high specific activity against C. jejuni as full length Cap11. Cecropin P1 and Cecropin B showed specific antimicrobial activity against Gram-negative bacteria, whereas no activity was detected against any of the Gram-positive bacteria tested. Cecropin B had moderate antimicrobial activity against all tested Gram-negative pathogens, while Cecropin P1 had specific activity against Y. ruckeri, A. salmonicida and E. coli only. Bac2A and its amidated form Bac2A-NH2 are linear C2A/C11A variants of the naturally occurring bovine peptide bactenecin. Bac2A displayed very low antimicrobial activity against the tested microorganisms except for E. faecalis and L. monocytogenes. Sub5-NH2, a synthetic derivative of Bac2A-NH2 carrying five mutations, had strongly increased antimicrobial activity compared to the mother peptide Bac2A-NH2. No difference in the specificity between gram-negative and gram-positive organisms was observed for the bactenectin-derived peptides. The well-characterized peptide Melittin has good activity against gram-positive bacteria; in particular S. aureus, E. faecalis and L. monocytogenes. Indolicidin shows only moderate activity except for L. monocytogenes. Myxinidin, originally isolated from the epidermal mucus of the hagfish, and it´s amidated form Myxinidin-NH2 displayed no antimicrobial activity under any of the tested conditions. Pyrrhocoricin, Apidaecin IA and Metalnikowin I are all belonging to the family of the glycine-rich peptides [18][19][20]. Only Apidaecin showed specific antimicrobial activity against S. enterica serovar Typhimurium and E. coli, whereas the other AMPs were ineffective (MIC ≥ 256 μg/ml). The potency of the AMPs was compared to the antimicrobial activity of well-characterized antibiotics from different classes either targeting the bacterial cell wall, the protein or nucleic acid synthesis. The MIC values for the reference antibiotics are summarized in Table 3. The solvent DMSO alone had no antimicrobial activity in the concentration range used in the assay (data not shown).
Table 3. Antimicrobial activities of antimicrobial peptides and antibiotics against Gram-negative and Gram-positive bacteria.
| Y. ruckeri 392/2003 | A. salmonicida ATCC33658 | F. psychrophilum 1947 | S. enterica ser. Typhimurium LT2 | C. jejuni NCTC11168 | E. coli ATCC25922 | P. aeruginosa ATCC27853 | S. aureus ATCC29213 | E. faecalis ATCC29212 | L. monocytogenes N22-2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial Peptides | ||||||||||
| Cap18 | 2 | 2 | >256 | 4 | 1 | 4–8 | 4–8 | ≥32 | 8 | 2–4 |
| Cap11 | 4 | 4 | >256 | 8 | 4 | 8–16 | 8 | 16–32 | 16–32 | 32 |
| Cap11-1-18m 2 | 8–16 | 32–64 | >256 | 16 | 2 | 16–32 | 16–32 | 16 | 8–16 | 16 |
| Cecropin P1 | 32 | 64 | >256 | ≥128 | >256 | 16–32 | >256 | >256 | >256 | >256 |
| Cecropin B | 32 | 32–64 | >256 | 32 | 16 | 16–32 | 64 | >256 | >256 | >256 |
| Bac2A-NH 2 | ≥256 | 128 | >256 | 128 | 64–128 | 64 | 128–256 | 128 | 16–32 | 8 |
| Bac2A | 256 | >256 | >256 | >256 | ≥256 | 256 | 256 | >256 | 64 | 32 |
| Sub5-NH 2 | 16–32 | 8 | >256 | 8 | 8 | 4 | 8 | 8 | 4–8 | 2 |
| Myxinidin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Melittin | 16–32 | >64 | n.d. | 32–64 | 2–4 | 16 | ≥64 | 2–4 | 2–4 | 2–4 |
| Indolicidin | ≥64 | ≥64 | n.d. | 64 | 16 | 32 | >64 | 32 | 32 | 4 |
| Myxinidin-NH 2 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Pyrrhocoricin | >256 | >256 | n.d. | >256 | n.d. | >256 | >256 | >256 | >256 | >256 |
| Apidaecin IA | 256 | >256 | n.d. | 64 | n.d. | 32 | >256 | >256 | >256 | >256 |
| Metalnikowin I | >256 | >256 | n.d. | >256 | n.d. | >256 | >256 | >256 | >256 | >256 |
| Antibiotics | ||||||||||
| Ampicillin | 2 | ≤1 | n.d. | ≤1 | 4 | 4 | >64 | ≤1 | ≤1 | ≤1 |
| Gentamicin | 1–2 | 1 | n.d. | 0.5–1 | ≤0.5 | 1 | 1–2 | ≤0.5 | 8–16 | ≤0.5 |
| Colisitin (Polymyxin E) | 1–2 | 2 | n.d. | 2 | 16 | ≤1 | 2–4 | >16 | >16 | >16 |
| Azithromycin | ≤2 | ≤2 | n.d. | 4 | ≤2 | 4 | >64 | ≤2 | 4 | ≤2 |
| Cefotaxime | ≤0.25 | ≤0.25 | n.d. | ≤0.25 | ≥4 | ≤0.25 | >4 | 2 | >4 | >4 |
| Chloramphenicol | ≤8 | ≤8 | n.d. | ≤8 | ≤8 | ≤8 | ≥128 | ≤8 | ≤8 | ≤8 |
| Ciprofloxacin | 0.06 | ≤0.015 | n.d. | ≤0.015 | 0.12 | ≤0.015 | 0.25 | 0.25 | 0.5 | 1 |
| Meropenem | ≤0.03 | ≤0.03 | n.d. | ≤0.03 | ≤0.03 | ≤0.03 | 0.25 | 0.12 | 4 | 0.12 |
| Nalidixic acid | 32 | ≤4 | n.d. | ≤4 | ≤4 | ≤4 | 64 | 64 | ≥128 | ≥128 |
| Ceftaxidime | ≤0.5 | ≤0.5 | n.d. | ≤0.5 | >8 | ≤0.5 | 2 | 8 | >8 | >8 |
| Tetracycline | ≤2 | ≤2 | n.d. | ≤2 | ≤2 | ≤2 | 16 | ≤2 | 16 | ≤2 |
| Tigecycline | ≤0.25 | ≤0.25 | n.d. | ≤0.25 | ≤0.25 | ≤0.25 | 4 | ≤0.25 | ≤0.25 | ≤0.25 |
| Trimethoprim | 1 | ≤0.25 | n.d. | ≤0.25 | >32 | 0.5 | >32 | 2 | ≤0.25 | ≤0.25 |
Data are collected as minimal inhibitory concentrations (MICs) according to the Clinical and Laboratory Standards Institute (CLSI) and expressed in μml. All MIC determinations were carried out in duplicates for AMPs and in triplicate for antibiotics; n.d. = not determined.
Hemolytic activity
Not only the antimicrobial activity, but also the ability to differentiate between bacterial and mammalian cells is an important factor for a successful antimicrobial peptide. The cytotoxicity of the selected AMPs was therefore determined using a hemolytic assay based on lysis of washed horse erythrocytes. A minimal peptide concentration of 64 μg/ml, which was above or in the MIC range for the corresponding AMPs, and a maximum peptide concentration of 256 μg/ml, limited by the experimental setup, was used. Melittin showed a very high hemolytic activity at the peptide concentration of 128 μg/ml (110% compared to the 0.1% Trition X-100 control), Cap11-l-18m2 showed a significant hemolytic activity (52% compared to the triton X-100 control) at the peptide concentration of 64 μg/ml and indolicidin was slightly hemolytic (12% compared to the 0.1% trition X-100 control at the peptide concentration of 128 mg/ml), whereas the other tested peptides had no to minimal hemolytic activity indicating that they might be safe to use in the concentrations tested (Table 4). The solvent DMSO alone had no hemolytic activity in the concentration range used in the assay (data not shown).
Table 4. Hemolytic activities of the antimicrobial peptides against horse erythrocytes.
| Peptide | Peptide Concentration [μg/ml] | Hemolytic Activity[%]* |
|---|---|---|
| Cap11 | 64 | 4 ± 0 |
| Cap11-1-18m2 | 64 | 52 ± 6 |
| Cap18 | 64 | 1 ± 0 |
| Melittin | 128 | 110 ± 1 |
| Indolicidin | 128 | 12 ± 0 |
| Cecropin P1 | 256 | 0 ± 0 |
| Cecropin B | 256 | 0 ± 0 |
| Bac2A | 256 | 0 ± 0 |
| Bac2A-NH2 | 256 | 0 ± 0 |
| Sub5-NH2 | 256 | 0 ± 0 |
| Myxinidin | 256 | 0 ± 0 |
| Myxinidin-NH2 | 256 | 0 ± 0 |
| Pyrrhocoricin | 256 | 0 ± 0 |
| Apidaecin IA | 256 | 0 ± 0 |
| Metalnikowin I | 256 | 0 ± 0 |
* The hemolytic activity is measured in duplicates and given as the average ± SD in % relative to full lysis induced by 0.2% Triton X-100.
Thermostability and protease stability
To address the question of stability, the thermostability and protease stability against the commercial available proteases trypsin and proteinase K was investigated. Only AMPs with high antimicrobial activity (MIC ≤ 32 μg/ml for at least two bacterial species) were included in the stability assays. E. coli ATCC25922 was used as test strain for both, the thermostability and protease stability assays.
The thermostability was measured by the determination of the antimicrobial activity of the peptides after incubation for 5, 15 and 30 minutes at 70°C or 90°C. All the tested peptides retained their antimicrobial activity even after incubation at 70°C or 90°C for either 5, 15 or 30 minutes (Table 5). All the tested AMPs are stable at high temperatures.
Table 5. Thermostability of antimicrobial peptides.
| Temperature | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Incubation Time[min] | |||||||||
| Antimicrobial Activity MIC [μg/ml] | |||||||||
| Cap18 | Cap11 | Cap11-1-18m2 | Cecropin B | Cecropin P1 | Melittin | Indolicidin | Sub5 | ||
| 70°C | 0 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 |
| 5 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 | |
| 15 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 | |
| 30 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 8 | |
| 90°C | 0 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 |
| 5 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 | |
| 15 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 | |
| 30 | 8 | 8 | 32 | 16 | 16 | 16 | 32 | 4 | |
Data are collected as minimal inhibitory concentrations (MICs) according to the Clinical and Laboratory Standards Institute (CLSI) and expressed in μml. All MIC values are the average of five independent experiments.
The remaining antimicrobial activity after incubation with either trypsin or proteinase K for different incubation times is summarized in Table 6. The antimicrobial activity of Cecropin B, Cecropin P1and Melittin is completely abolished after a very short incubation of only 30 seconds with proteinase K. Cap18 and Indolicidin show a two-fold decreased antimicrobial activity after incubation with proteinase K for 2 minutes; respectively 4-fold decreased antimicrobial activity after incubation for 30 minutes. Sub5 shows a similar pattern with a 2-fold reduced activity after 30 sec, a 4-fold reduction after 2 minutes, an 8-fold reduction after 5 minutes, and a 16-fold reduction after 15 or 30 minutes. Interestingly, Cap11-1-18m2 showed a 2-fold increased antimicrobial activity after very short incubation of 30 seconds or 2 minutes with proteinase K. However, incubation of more than 15 minutes with proteinase K leads to the complete loss of antimicrobial activity of Cap11-1-18m2.
Table 6. Protease stability of antimicrobial peptides.
| Protease | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Incubation Time[min] | ||||||||||
| Antimicrobial Activity MIC [μg/ml] | ||||||||||
| Cap18 | Cap11 | Cap11-1-18m2 | Cecropin B | Cecropin P1 | Melittin | Indolicidin | Sub5 | Gentamicin | ||
| Proteinase K | 0 | 8 | 16 | 16 | 16 | 16 | 16 | 32 | 4 | 1 |
| 0.5 | 8 | 16 | 8 | >64 | >64 | >64 | 32 | 8 | 1 | |
| 2 | 16 | 16 | 8 | >64 | >64 | >64 | 32 | 16 | 1 | |
| 5 | 16 | 32 | 16 | >64 | >64 | >64 | 64 | 32 | 1 | |
| 15 | 32 | 64 | >64 | >64 | >64 | >64 | 64 | 64 | 1 | |
| 30 | 32 | >64 | >64 | >64 | >64 | >64 | 64 | 64 | 1 | |
| Trypsin | 0 | 8 | 8 | 16 | 16 | 32 | 16 | 32 | 4 | 1 |
| 0.5 | >64 | 16 | 8 | >64 | >64 | >64 | >64 | 4 | 1 | |
| 2 | >64 | 32 | 8 | >64 | >64 | >64 | >64 | 8 | 1 | |
| 5 | >64 | 64 | 8 | >64 | >64 | >64 | >64 | 8 | 1 | |
| 15 | >64 | >64 | 32 | >64 | >64 | >64 | >64 | 16 | 1 | |
| 30 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 64 | 1 | |
Data are collected as minimal inhibitory concentrations (MICs) after incubation with proteinase K or trypsin according to the Clinical and Laboratory Standards Institute (CLSI) and expressed in μml. All MIC determinations were carried out in triplicates.
The incubation of Cap18, Cecropin P1, Cecropin B, Melittin and Indolicidin with trypsin lead to a complete loss of antimicrobial activity after only 30 seconds of incubation. For Cap11, the incubation with trypsin resulted in a 2-fold decrease of antimicrobial activity after 30 seconds, a 4-fold decrease after 2 minutes, 8-fold decrease after 5 minutes and a complete loss of activity after 15, respectively 30 minutes. Sub5 followed a similar pattern; incubation with trypsin reduced the antimicrobial activity by factor 2 after 2 and 5 minutes, by factor 4 after 15 minutes incubation and by factor 16 after 30 minutes incubation. A short incubation with trypsin of up to 5 minutes increased the antimicrobial activity of Cap11-1-18m2 by factor 2. Incubation times longer than 5 minutes lead to a 2-fold reduction, while 15 minutes of incubation decreases the activity by factor 2, incubation times longer than 30 minutes are leading to a complete loss of antimicrobial activity. In contrast to thermostability, the majority of the tested AMPs were highly sensitive towards the proteases trypsin and proteinase K. The antibiotic gentamicin was used as a control, which retained its full antimicrobial activity after incubation with trypsin or proteinase K for 30 minutes.
Lipopolysaccharides are not target for the AMPs
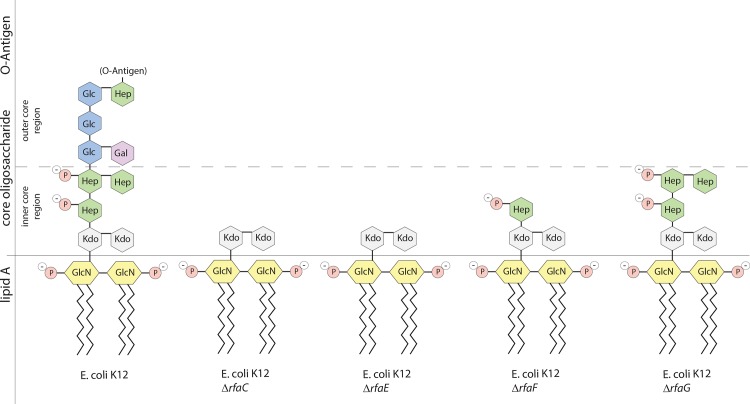
Previous studies have shown that various AMPs including Cap18, Cap11, Melittin and Indolicidin have LPS binding properties [21][22][23][24][25]. To investigate the potential role of LPS involved in the mechanism in more detail, the antimicrobial activity of Cap18, Cap11, Cap11-1-18m2, Cecropin P1, Cecropin B, Indolicidin, Melittin and Sub5 was investigated in a series of LPS mutants compared with their parental strains, BW25113, an E. coli K12 strain, and the reference strain ATCC25922. In this study, the rfaC, rfaE, rfaF and rfaG genes were selected for mutation (Table 7, Fig 1). These genes are involved in the synthesis and assembly of the core oligosaccharide, the middle part of the LPS molecule connecting the lipid A and the O-antigen. Except for Cap11, all the tested AMPs showed higher antimicrobial activity in the BW25113 LPS mutants compared to the parental strain BW25113 (Table 8). The highest increase (4-8x) in antimicrobial activity was detected for the AMP Melittin in all the BW25113 mutant strains. Cecropin P1 and Cecropin B is 4-8x more active in the BW25113ΔrfaF mutant, 2-4x more active in the BW25113ΔrfaC and BW25113ΔrfaF mutants, up to 2x fold more active in the BW25113ΔrfaG mutant compared to the wild-type BW25113. Sub5 showed a 2-4x increased antimicrobial activity, Cap11-1-18m2 and Indolicidin a 2x increased and Cap18 up to 2x increased antimicrobial activity in all BW25113 LPS mutants. Similar results were obtained for the LPS mutants in the E. coli reference strain ATCC25922. The highest increase in antimicrobial activity was observed for Melittin in the ΔrfaC, ΔrfaE and ΔrfaF mutant strains in which the antimicrobial activity is 8x higher compared to the ATCC25922 wild-type. A 4-fold increase in antimicrobial activity was measured for Melittin in the ATCC25922ΔrfaG mutant, for Cap11 and Cecropin P1 in the ΔrfaC, ΔrfaE and ΔrfaF mutant strain, for Cecropin B in the ΔrfaC and ΔrfaE mutant strains and for Sub5 in all tested ATCC25922 LPS mutants. Cap18, Cap11-1-18m2 and Indolicidin showed a 2-fold increase in activity in all the tested ATCC25922 LPS mutants, Cap11, Cecropin P1 and Cecropin B were 2-fold more active in the ATCC25922ΔrfaG background. Summarizing, LPS plays a central role in protecting against the antimicrobial activity of the tested AMPs, which all showed higher antimicrobial activity in LPS defective mutants.
Table 7. Function and Phenotype of the LPS genes selected in this study.
| Gene Name | Alternative Gene Name(s) | Function in core oligosaccharide assembly | Character of the LPS core | Reference |
|---|---|---|---|---|
| waaC | rfaC | LPS heptosyltransferase I (HepI). Adds the first heptose sugar onto the Kdo2 moiety. | Heptoseless | [61],[62] |
| waaE | rfaE, hldE | Heptose 7-phosphate kinase/heptose 1-P adenyltransferase | Heptoseless | [63][64] |
| waaF | rfaF | LPS heptosyltransferase II (HepII). Transfers the second heptose sugar onto the heptosyl-Kdo2 moiety. | Kdo2 with one heptose | [61] |
| waaG | rfaG | LPS glycosyltransferase I. Add the first glucose to the outer core oligosaccharide | Intact 3 heptose, but outer core-less. Reduced phosphorylation of the inner core | [65][57] |
Fig 1. Schematic lipopolysaccharide structures of LPS mutants used in this study.
Schematic LPS structures especially highlighting the core oligosaccharide portion of LPS are illustrated. Structures of the major glycoforms of the core oligosaccharide are based on the structural analysis of an E. coli K12 derivative, W3100 [66]. Each sugar or amino sugar of the core oligosaccharide is shown by a green (Hep), violet (Gal), grey (Kdo) or blue (Glc). Phosphate groups on modified sugars are shown by red circles. Hep: L-glycero-D-manno-heptose, Kdo: 3-deoxy-D-manno-oct-2- ulosonic acid, GlcN: N-acetylglucosamine, Glc: glucose, Gal: galactose.
Table 8. Antimicrobial activity of selected AMPs in different LPS backgrounds.
| Strain | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antimicrobial Activity MIC [μg/ml] | ||||||||
| Cap18 | Cap11 | Cap11-1-18m2 | Cecropin P1 | Cecropin B | Indolicidin | Melittin | Sub5 | |
| BW25113 wild-type | 4–8 | 8 | 16 | 16–32 | 16–32 | 32 | 16–32 | 4–8 |
| BW25113 ΔrfaC | 4 | 8 | 8 | 8 | 8 | 16 | 4 | 2 |
| BW25113 ΔrfaE | 4 | 8 | 8 | 8 | 8 | 16 | 4 | 2 |
| BW25113 ΔrfaF | 4 | 8 | 8 | 4 | 4 | 16 | 4 | 2 |
| BW25113 ΔrfaG | 4 | 8 | 8 | 16 | 16 | 16 | 4 | 2 |
| ATCC25922 wild-type | 4 | 16 | 32 | 32 | 32 | 32 | 16 | 4 |
| ATCC25922 ΔrfaC | 2 | 4 | 16 | 8 | 8 | 16 | 2 | 1 |
| ATCC25922 ΔrfaE | 2 | 4 | 16 | 8 | 8 | 16 | 2 | 1 |
| ATCC25922 ΔrfaF | 2 | 4 | 16 | 8 | 16 | 16 | 2 | 1 |
| ATCC25922 ΔrfaG | 2 | 8 | 16 | 16 | 16 | 16 | 4 | 1 |
Data are collected as minimal inhibitory concentrations (MICs) according to the Clinical and Laboratory Standards Institute (CLSI) and expressed in μml. All MIC determinations were carried out in triplicates.
Discussion
The number of antibiotic resistant pathogens is increasing and the capacity of currently available antimicrobial compounds to control bacterial infections is diminishing. Antimicrobial peptides are an alternative to classical antibiotics to control and fight bacterial infections. However, the lack of standardized protocols and comprehensive overviews comparing the antimicrobial activity, cytotoxicity and stability of known AMPs from different sources is one of the major challenges in identifying AMPs for further development into practical use. Very often direct comparison of the antimicrobial activity of different AMPs from different publications is not possible due to different assay conditions and different peptide purities. This study, comparing AMPs from different origins using the same assay conditions, allows a direct comparison of the antimicrobial activity, hemolytic activity, and stability of a selection of AMPs against a broader range of bacteria. In this study, we demonstrate that Cap18 and Cap11, both members of the cathelidicin family, displayed a potent efficacy in particular against Gram-negative pathogens, including pathogens found in fish and poultry production. Direct comparison of MIC values obtained in this study with previously published results is not feasible due to different assays, different strains, and different growth conditions [26][27][28][29][21][30][31][32][33][34][35][36][37]. In S2 Table we have, however, compared our results to previously reported antimicrobial activities of the selected peptides. Our results confirm the anti-Gram-negative activity of Cap18, Cap11, and Cap11-1-18m2, previously reported against E. coli. Our data extend the range of Gram-negatives investigated for these peptides. For Myxinidin and Pyrrhocoricin our data contradicts previously published results [20,38,39]. One possible explanation for the different results of Pyrrhocoricin could be attributed to the fact that Cudic et al. used the C-terminal amidated form of Pyrrhocoricin and Cocianich et al. used Pyrrhocoricin directly isolated from Pyrrhocoris apterus. For the other peptides we see a mixture of confirmations and contradictions in situations where the combination of peptide and microorganism has been investigated previously. Our analysis allow this set of peptides to be compared under identical conditions against a broad range of microorganisms, and thereby to select the potentially most suited candidate for development into an antibacterial product.
Besides a high antimicrobial activity, low cytotoxicity is a desirable characteristic for AMPs as potential drug candidates. In general, AMPs are binding to the bacterial surface by electrostatic interactions. However, some types of AMPs are able to interact not only with the bacterial surface, but also with the host cells which leads to cell lysis. Very often, the toxicity of AMPs against eukaryotic cells is one of the major obstacles for their clinical application. Melittin has a strong hemolytic activity and is a prototype of AMPs which are inducing pores, which is in agreement with previous findings [40]. Only Melittin, Cap11-1-18m2, Indolicidin, and Cap11 showed hemolytic activity; Cap11 only showed very low hemolytic activity. Using those peptides as drug candidates would need further modifications reducing the hemolytic activity and increasing the specificity for bacterial cells. Several different approaches to improve the specificity are described in literature. Previous studies, show that changing the net charge and reducing the hydrophobicity by introducing hydrophilic amino acids is leading to a decreased hemolytic activity [41][42][43]. Other studies have demonstrated that the introduction of D-amino acids can lower the hemolytic activity of α-helical AMPs [44]. Cap18 displayed no hemolysis in our assay using horse erythrocytes. The absence of cytotoxicity should however be expanded to other relevant species of erythrocytes before Cap18 can safely be developed into products for specific applications.
A challenge with cationic AMPs in various therapeutic contexts is its susceptibility to proteolytic degradation. Many bacteria have developed proteases, e.g. elastase in Pseudomonas aeruginosa, aureolysin and V8 in Staphylococcus aureus, leading to fast degradation of the AMPs. Moreover, gastrointestinal digestive enzymes, such as pepsin and trypsin, and proteases in the serum contribute to the low proteolytic stability of the majority of AMPs. Our data show that Cecropin P1, Cecropin B, Melittin and Indolicidin are highly sensitive to proteolytic degradation by trypsin or proteinase K. Cap18, Cap11, Cap11-1-18m2 and Sub5 are more stable towards proteinase K digestion. However, only Cap11, Cap11-1-18m2 and Sub5 showed partial stability towards trypsin digestion. In summary, these findings corroborate that especially antimicrobial peptides which have a cationic character will show fast degradation. Recent research focused on finding ways to improve proteolytic stability of cationic antimicrobial peptides. Similar strategies which can be used to reduce cytotoxicity are showing potential to reduce protease susceptibility. The incorporation of non-natural amino acids such as D-amino acids or amino acid derivate of arginine, the modification of the terminal regions including acetylation, amidation and hydrophobic tagging and the use of non-peptidic backbones (peptidomimetics) have been shown to improve the protease stability of AMPs [45] [46][47][42][48].
AMPs to be used as feed ingredients should preferably be heat stable, as feed processing usually includes a pelleting process involving temperatures between 50°C and 90°C [49]. In order to reduce the risk of spreading microbial contaminants with the feed the higher end of the temperature range is recommended [50]. Feed enzymes will either have to be applied after the pelleting process or the enzyme need to optimized for thermal stability [51]. In contrast to enzymes all the tested AMPs are highly thermostable.
The mode of action of AMPs largely depends on the bacterial cell surface and the amino acid composition of the peptide itself. According to a previously described model for the mode action, AMPs are initially attracted to the bacterial surface most likely by electrostatic bonding between the cationic peptide and the bacterial surface. Cationic peptides are likely first attracted by negatively charged lipopolysaccharide molecules in Gram-negative bacteria [8]. The negatively charged Lipopolysaccharide molecule (LPS) is one of major molecular components of the outer membrane in Gram-negative bacteria. LPS consist of three distinct components: LipidA acts as hydrophobic anchor in the outer membrane. The core oligosaccharide (core OS) composed of different sugar molecules connects the lipid A with the O-antigen, a structurally variable polysaccharide made up of repeating oligomeric units (Fig 1). In addition, bacterial membranes are primarily composed of negatively charged lipids including phosphatidylglycerol, cardiolipin and the zwitter ionic phosphatidylethanolamine. Even though the negatively charged LPS molecules are important for initial attraction of cationic peptides to the membrane, the outer membrane itself also acts as an effective permeability barrier against various harmful agents, including hydrophobic antibiotics [52][8][53][54]. In particular, mutants in the core oligosaccharide of the LPS have been shown to be more susceptible to hydrophobic agents. E. coli and S. Typhimurium strains that lack heptose in the LPS show a deep rough phenotype which is characterized by a reduction in the outer membrane protein content and increased sensitivity towards detergents and hydrophobic antibiotics [55][56]. An E. coli F540 rfaG mutant which is defective in the inner core of the LPS molecule, has a destabilized outer membrane and exhibits a 80% reduction of heptose phosphorylation which leads to an increased susceptibility towards SDS and Novobiocin [57]. Our data clearly indicates that LPS in E. coli not only acts in the attraction and attachment of antimicrobial peptides to the outer membrane, but also functions as a protection barrier against cationic AMPs, very similar to antibiotics. The mutants defective in the core LPS showed increased susceptibility to the majority of the tested AMPs. The ΔrfaC and ΔrfaE mutants, which are supposed to contain a core without heptose in E. coli K12 BW25113 and the clinical isolate ATCC25922, showed increased susceptibility towards Cap18, Cap11-1-18m2, Cecropin P1, Cecropin B, Indolicidin, Melittin and Sub5. Similarly, both ΔrfaF mutants, having a core oligosaccharide consisting of only one heptose, were more susceptible for Cap18, Cap11-1-18m2, Indolicidin, Melitin and Sub5. A slight difference in susceptibility was measured for Cecropin P1 and Cecropin B in the different E. coli backgrounds. Cecropin P1 and Cecropin B showed increased antimicrobial activity in BW25113ΔrfaF background compared to ATCC25922ΔrfaF and both ΔrfaC and ΔrfaE mutants. More distinct differences in susceptibility were measured in rfaG mutant background, which has an intact inner part of the core oligosaccharide and is only missing a functional outer part of the core oligosaccharide. The biggest differences in susceptibility between the two E. coli strains, the K12 derivative BW25113 and the clinical isolate ATCC25922, were measured for Cap11 in all tested LPS mutants. The antimicrobial susceptibility of Cap11 in the LPS mutants was identical to the wild-type BW25113 strain. In contrast, the antimicrobial activity was increased by factor 4 in the ΔrfaC,ΔrfaE,ΔrfaF and by factor 2 in the ΔrfaG mutants compared to the wild-type ATCC25922. These data suggest that the different degrees of susceptibility are depending on one hand on the character of the LPS mutation and on the other hand on the nature of the AMP. Interestingly, LPS mutants lacking heptose completely and as consequence also lacking all the negatively charged phosphate groups, showed a higher susceptibility to the majority of the tested AMPs. This indicates that LPS is not essential for antimicrobial activity of cationic peptides, even though in previous models LPS are regarded as needed for attraction and attachment via electrostatic bonding. In addition, the O-antigen does not seem to have an obvious function in the mode of action for the tested cationic AMPs since the susceptibility of all tested AMPs in E. coli K12 BW25113, which is missing the O-antigen, and the clinical isolate ATCC25922 belonging to the serotype O6 are the same, except for Cap11 and Cap11-1-18m2 which show only a very minor difference.
In summary, Cap18, isolated from rabbit neutrophils, is of all the tested AMPs the most active and has the highest antimicrobial activity in particular against Gram-negative foodborne pathogens. Cap18 also showed very low toxicity to horse erythrocytes and was stable at high temperatures. All these characteristics indicates that Cap18 has potential for further development as e.g. food and feed ingredient against infections caused by Gram-negative foodborne pathogens. However, Cap18 is sensitive to trypsin and proteinase K in vitro and further improvement addressing protease stability will be needed. In addition, our results indicate that the LPS do not play a central role in the mechanism of cationic AMPs in Gram-negative bacteria. Our findings indicate that LPS is not important in the attraction of cationic peptides to the bacterial surface of Gram-negative bacteria, but in fact acts as a protection barrier. However, other factors than LPS might also be involved in the mode of action, since various cationic AMPs show antimicrobial activity against Gram-positive bacteria which are lacking LPS.
Supporting Information
(DOC)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Financial support for the project was obtained from the the Danish Ministery of Food, Agriculture and Fisheries programme supporting Green Development and Demonstration (GUDP grant nr.: 3405-10-0124) (http://naturerhverv.dk/tvaergaaende/gudp/).
References
- 1. Aarestrup FM, Wegener HC, Collignon P. Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev Anti Infect Ther. 2008;6: 733–750. 10.1586/14787210.6.5.733 [DOI] [PubMed] [Google Scholar]
- 2. Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis. 2009;49: 132–141. 10.1086/599374 [DOI] [PubMed] [Google Scholar]
- 3. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415: 389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 4. Maróti G, Kereszt A, Kondorosi É, Mergaert P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol. 2011;162: 363–374. 10.1016/j.resmic.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 5. Hancock REW, Brown KL, Mookherjee N. Host defence peptides from invertebrates—emerging antimicrobial strategies. Immunobiology. 2006;211: 315–322. 10.1016/j.imbio.2005.10.017 [DOI] [PubMed] [Google Scholar]
- 6. Hancock REW, Lehrer R. Cationic peptides: A new source of antibiotics. Trends in Biotechnology. 1998. pp. 82–88. 10.1016/S0167-7799(97)01156-6 [DOI] [PubMed] [Google Scholar]
- 7. Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek. 1996;69: 193–202. [DOI] [PubMed] [Google Scholar]
- 8. Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3: 238–250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- 9. Hagiwara A, Imai N, Nakashima H, Toda Y, Kawabe M, Furukawa F, et al. A 90-day oral toxicity study of nisin A, an anti-microbial peptide derived from Lactococcus lactis subsp. lactis, in F344 rats. Food Chem Toxicol. Elsevier Ltd; 2010;48: 2421–2428. 10.1016/j.fct.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 10. Tiwari BK, Valdramidis VP, O’ Donnell CP, Muthukumarappan K, Bourke P, Cullen PJ, et al. Application of natural antimicrobials for food preservation. J Agric Food Chem. 2009;57: 5987–6000. 10.1021/jf900668n [DOI] [PubMed] [Google Scholar]
- 11. Weinberg ED. Antibiotic properties and applications of lactoferrin. Curr Pharm Des. 2007;13: 801–811. 10.2174/138161207780363095 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2: 2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michel C, Antonio D, Hedrick RP. Production of viable cultures of Flavobacterium psychrophilum: approach and control. Res Microbiol. 1999;150: 351–358. Available: http://www.ncbi.nlm.nih.gov/pubmed/10422696. [DOI] [PubMed] [Google Scholar]
- 15. Wikler MA, Cockerill FR, Bush K, Dudley MN, Eliopoulos GM, Hardy DJ, et al. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—eighth edition Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 16. Habermann E. Bee and Wasp Venoms. Sci. 1972;177: 314–322. 10.1126/science.177.4046.314 [DOI] [PubMed] [Google Scholar]
- 17. Selsted ME, Novotny MJ, Morris WL, Tang YQ, Smith W, Cullor JS. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J Biol Chem. 1992;267: 4292–4295. [PubMed] [Google Scholar]
- 18. Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. Apidaecins: antibacterial peptides from honeybees. EMBO J. 1989;8: 2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chernysh S, Cociancich S, Briand J-P, Hetru C, Bulet P. The inducible antibacterial peptides of the Hemipteran insect Palomena prasina: Identification of a unique family of prolinerich peptides and of a novel insect defensin. Journal of Insect Physiology. 1996. pp. 81–89. 10.1016/0022-1910(95)00085-2 [DOI] [Google Scholar]
- 20. Cudic M, Condie B a, Weiner DJ, Lysenko ES, Xiang ZQ, Insug O, et al. Development of novel antibacterial peptides that kill resistant isolates. Peptides. 2002;23: 2071–2083. Available: http://www.ncbi.nlm.nih.gov/pubmed/12535685. [DOI] [PubMed] [Google Scholar]
- 21. Okuda D, Yomogida S, Kuwahara-arai K. Augmentation of the antimicrobial activities of guinea pig cathelicidin CAP11-derived peptides by amino acid substitutions. Int J Mol Med. 2009; 501–508. 10.3892/ijmm [DOI] [PubMed] [Google Scholar]
- 22. Nagaoka I, Hirota S, Niyonsaba F, Hirata M, Adachi Y, Tamura H, et al. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-alpha by blocking the binding of LPS to CD14(+) cells. J Immunol. 2001;167: 3329–3338. [DOI] [PubMed] [Google Scholar]
- 23. Larrick JW, Hirata M, Balint RF, Lee J, Zhong J, Wright SC. Human CAP18: A novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63: 1291–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falla TJ, Nedra Karunaratne D, Hancock REW. Mode of action of the antimicrobial peptide indolicidin. J Biol Chem. 1996;271: 19298–19303. 10.1074/jbc.271.32.19298 [DOI] [PubMed] [Google Scholar]
- 25. Bhunia A, Domadia PN, Bhattacharjya S. Structural and thermodynamic analyses of the interaction between melittin and lipopolysaccharide. Biochim Biophys Acta—Biomembr. 2007;1768: 3282–3291. 10.1016/j.bbamem.2007.07.017 [DOI] [PubMed] [Google Scholar]
- 26. Gutsmann T, Hagge SO, Larrick JW, Seydel U, Wiese a. Interaction of CAP18-derived peptides with membranes made from endotoxins or phospholipids. Biophys J. Elsevier; 2001;80: 2935–2945. 10.1016/S0006-3495(01)76259-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larrick JW, Hirata M, Shimomoura Y, Yoshida M, Zheng H, Zhong J, et al. Antimicrobial activity of rabbit CAP18-derived peptides. Antimicrob Agents Chemother. 1993;37: 2534–2539. 10.1128/AAC.37.12.2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mason DJ, Dybowski R, Larrick JW, Gant VA. Antimicrobial Action of Rabbit Leukocyte CAP18. Antimicrob Agents Chemother. 1997;41: 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagaoka I, Hirota S, Yomogida S, Ohwada A, Hirata M. Synergistic actions of antibacterial neutrophil defensins and cathelicidins. Inflamm Res. 2000;49: 73–79. 10.1007/s000110050561 [DOI] [PubMed] [Google Scholar]
- 30. Travis SM, Anderson NN, Forsyth WR, Espiritu C, Conway BD, Greenberg EP, et al. Bactericidal activity of mammalian cathelicidin-derived peptides. Infect Immun. 2000;68: 2748–2755. Available: http://www.ncbi.nlm.nih.gov/pubmed/10768969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yomogida S, Nagaoka I, Yamashita T. Purification of the 11- and 5-kDa antibacterial polypeptides from guinea pig neutrophils. Arch Biochem Biophys. 1996;328: 219–226. 10.1006/abbi.1996.0166 [DOI] [PubMed] [Google Scholar]
- 32. Boman HG, Agerberth B, Boman A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect Immun. 1993;61: 2978–84. Available: http://www.ncbi.nlm.nih.gov/pubmed/8514403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kjuul AK, Büllesbach EE, Espelid S, Dunham R, Jørgensen TÒ, Warr GW, et al. Effects of cecropin peptides on bacteria pathogenic to fish. J Fish Dis. 1999;22: 387–394. 10.1046/j.1365-2761.1999.00191.x [DOI] [Google Scholar]
- 34. Lee JY, Boman A, Sun CX, Andersson M, Jörnvall H, Mutt V, et al. Antibacterial peptides from pig intestine: isolation of a mammalian cecropin. Proc Natl Acad Sci U S A. 1989;86: 9159–62. Available: http://www.ncbi.nlm.nih.gov/pubmed/2512577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore a J, Beazley WD, Bibby MC, Devine D a. Antimicrobial activity of cecropins. J Antimicrob Chemother. 1996;37: 1077–1089. Available: http://www.ncbi.nlm.nih.gov/pubmed/8836811. [DOI] [PubMed] [Google Scholar]
- 36. Hilpert K, Volkmer-Engert R, Walter T, Hancock REW. High-throughput generation of small antibacterial peptides with improved activity. Nat Biotechnol. 2005;23: 1008–1012. 10.1038/nbt1113 [DOI] [PubMed] [Google Scholar]
- 37. Wu M, Hancock RE. Improved derivatives of bactenecin, a cyclic dodecameric antimicrobial cationic peptide. Antimicrob Agents Chemother. 1999;43: 1274–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/10223951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian S, Ross NW, MacKinnon SL. Myxinidin, a novel antimicrobial peptide from the epidermal mucus of hagfish, Myxine glutinosa L. Mar Biotechnol (NY). 2009;11: 748–757. 10.1007/s10126-009-9189-y [DOI] [PubMed] [Google Scholar]
- 39. Cociancich S, Dupont A, Hegy G, Lanot R, Holder F, Hetru C, et al. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem J. 1994;300 (Pt 2: 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee M-T, Sun T-L, Hung W-C, Huang HW. Process of inducing pores in membranes by melittin. Proc Natl Acad Sci U S A. 2013;110: 14243–14248. 10.1073/pnas.1307010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang JF, Xu YM, Hao DM, Huang YB, Liu Y, Chen YX. Structure-guided de novo design of a-helical antimicrobial peptide with enhanced specificity. Pure Appl Chem. 2010;82: 243–257. 10.1351/pac-con-09-01-12 [DOI] [Google Scholar]
- 42. Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51: 1398–1406. 10.1128/AAC.00925-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang Z, Vasil AI, Hale JD, Hancock REW, Vasil ML, Hodges RS. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Biopolymers. 2008;90: 369–383. 10.1002/bip.20911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang Y, He L, Li G, Zhai N, Jiang H, Chen Y. Role of helicity of α-helical antimicrobial peptides to improve specificity. Protein Cell. 2014;5: 631–642. 10.1007/s13238-014-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knappe D, Henklein P, Hoffmann R, Hilpert K. Easy strategy to protect antimicrobial peptides from fast degradation in serum. Antimicrob Agents Chemother. 2010;54: 4003–4005. 10.1128/AAC.00300-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malmsten M, Kasetty G, Pasupuleti M, Alenfall J, Schmidtchen A. Highly Selective End-Tagged Antimicrobial Peptides Derived from PRELP. Neyrolles O, editor. PLoS One. 2011;6: e16400 10.1371/journal.pone.0016400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strömstedt AA, Pasupuleti M, Schmidtchen A, Malmsten M. Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob Agents Chemother. 2009;53: 593–602. 10.1128/AAC.00477-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giuliani A, Rinaldi AC. Beyond natural antimicrobial peptides: Multimeric peptides and other peptidomimetic approaches. Cellular and Molecular Life Sciences. 2011. pp. 2255–2266. 10.1007/s00018-011-0717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. EFSA. Scientific Opinion of the Panel on Biological Hazards / Microbiological risk assessment in feedingstuffs for food-producing animals. EFSA J. 2008; 1–84. Available: http://www.elika.net/datos/articulos/Archivo297/BIOHAZ_PiensosSalmonella08.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okelo PO, Wagner DD, Carr LE, Wheaton FW, Douglass LW, Joseph SW. Optimization of extrusion conditions for elimination of mesophilic bacteria during thermal processing of animal feed mash. Anim Feed Sci Technol. 2006;129: 116–137. 10.1016/j.anifeedsci.2005.12.011 [DOI] [Google Scholar]
- 51. Amerah AM, Gilbert C, Simmins PH, Ravindran V. Influence of feed processing on the efficacy of exogenous enzymes in broiler diets. Worlds Poult Sci J. 2011;67: 29–46. 10.1017/S0043933911000031 [DOI] [Google Scholar]
- 52. Lohner K. New strategies for novel antibiotics: Peptides targeting bacterial cell membranes. General Physiology and Biophysics. 2009. pp. 105–116. 10.4149/gpb_2009_02_105 [DOI] [PubMed] [Google Scholar]
- 53. Guilhelmelli F, Vilela N, Albuquerque P, Derengowski L da S, Silva-Pereira I, Kyaw CM. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Frontiers in Microbiology. 2013. 10.3389/fmicb.2013.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vaara M. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrobial Agents and Chemotherapy. 1993. pp. 354–356. 10.1128/AAC.37.2.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vaara M, Nurminen M. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrob Agents Chemother. 1999;43: 1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shlaes DM, Shlaes JH, Davies J, Williamson R. Escherichia coli susceptible to glycopeptide antibiotics. Antimicrob Agents Chemother. 1989;33: 192–197. 10.1128/AAC.33.2.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yethon JA, Vinogradov E, Perry MB, Whitfield C. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J Bacteriol. 2000;182: 5620–5623. 10.1128/JB.182.19.5620-5623.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wulff G, Gram L, Ahrens P, Vogel BF. One group of genetically similar Listeria monocytogenes strains frequently dominates and persists in several fish slaughter- and smokehouses. Appl Environ Microbiol. 2006;72: 4313–4322. 10.1128/AEM.02288-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fouz B, Zarza C, Amaro C. First description of non-motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiss (Walbaum), cultured in Spain. J Fish Dis. 2006;29: 339–346. 10.1111/j.1365-2761.2006.00723.x [DOI] [PubMed] [Google Scholar]
- 60. Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. antibacterial peptides from. 1989;8: 2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gronow S, Brabetz W, Brade H. Comparative functional characterization in vitro of heptosyltransferase I (WaaC) and II (WaaF) from Escherichia coli. Eur J Biochem. 2000;267: 6602–6611. 10.1046/j.1432-1327.2000.01754.x [DOI] [PubMed] [Google Scholar]
- 62. Zamyatina A, Gronow S, Puchberger M, Graziani A, Hofinger A, Kosma P. Efficient chemical synthesis of both anomers of ADP L-glycero- and D-glycero-D-manno-heptopyranose. Carbohydr Res. 2003;338: 2571–2589. 10.1016/S0008-6215(03)00319-7 [DOI] [PubMed] [Google Scholar]
- 63. Valvano MA, Marolda CL, Bittner M, Glaskin-Clay M, Simon TL, Klena JD. The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-L- glycero-D-manno-heptose. J Bacteriol. 2000;182: 488–497. 10.1128/JB.182.2.488-497.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McArthur F, Andersson CE, Loutet S, Mowbray SL, Valvano MA. Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno- heptose biosynthesis. J Bacteriol. 2005;187: 5292–5300. 10.1128/JB.187.15.5292-5300.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Parker CT, Kloser AW, Schnaitman CA, Stein MA, Gottesman S, Gibson BW. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174: 2525–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Müller-Loennies S, Lindner B, Brade H. Structural Analysis of Oligosaccharides from Lipopolysaccharide (LPS) of Escherichia coli K12 Strain W3100 Reveals a Link between Inner and Outer Core LPS Biosynthesis. J Biol Chem. 2003;278: 34090–34101. 10.1074/jbc.M303985200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.