Abstract
Lipid areas (Aℓ), bilayer area compressibilities (KA), bilayer bending constants (KC), and monolayer spontaneous curvatures (c0) from simulations using the CHARMM36 force field are reported for 12 representative homogenous lipid bilayers. Aℓ (or their surrogate, the average deuterium order parameter in the “plateau region” of the chain) agree very well with experiment, as do the KA. Simulated KC are in near quantitative agreement with vesicle flicker experiments, but are somewhat larger than KC from x-ray, pipette aspiration, and neutron spin echo for saturated lipids. Spontaneous curvatures of bilayer leaflets from the simulations are approximately 30% smaller than experimental values of monolayers in the inverse hexagonal phase.
Keywords: Area per lipid, bending modulus, area compressibility modulus, spontaneous curvature, Gaussian curvature modulus, CHARMM 36 force field
1. Introduction
The equilibrium surface area per lipid, Aℓ, bilayer area compressibility, KA, bilayer bending constant, KC, and the monolayer spontaneous curvature, c0, are critical mechanical properties of biological membranes. They determine the thickness of the membrane, its ability to compress, expand, or bend, and its propensity to curve. Perhaps surprisingly, there is considerable uncertainty in the experimental values of these properties even for single component bilayers. This is partly because experiments are often well tuned for certain lipids and not others. For example, x-ray methods (Nagle and Tristram-Nagle, 2000) work well for obtaining surface areas of lipids with phosphatidylcholine (PC) head groups, but not for those with phosphatidylethanolamine (PE) head groups. KA are typically obtained by pipette aspiration methods (Evans and Rawicz, 1990), but these have been subject to revision both to take into account undulations (Rawicz et al., 2000), and more recently, “fast” stretching (Evans et al., 2013); they are also not available for many lipids. KC determined by x-ray and pipette aspiration are comparable, but can differ by over a factor of two from KC of the same lipid determined from vesicle flicker experiments (Marsh, 2006; Nagle et al., 2015); the source of the discrepancy is unclear. Lastly, experimental measurements of c0 are not even obtained from bilayers. Rather, measurements are carried out on the inverse hexagonal (HII) phase (Gruner et al., 1986; Marsh, 2006), and the results are frequently extrapolated to the lamellar phase.
The quality of all-atom molecular dynamics (MD) simulations of lipid bilayers has improved dramatically since their advent in the early 1990s (Pastor, 1994). Due to increases in computer power and algorithms, state-of-the-art trajectory lengths have increased from hundreds of picoseconds to hundreds of nanoseconds on standard laboratory clusters, to tens of microseconds on special purpose computers (Shaw et al., 2008). These computer advances have enabled rigorous refinements of force fields (FF). Lastly, there have been important formalistic developments for evaluating pressure profiles in systems with long range electrostatic interactions (Sonne et al., 2005), pressures in droplets and cylinders (Sodt and Pastor, 2012), and bending constants (Watson et al., 2012). These advances enable the determination of bilayer mechanical properties for all-atom MD models, and make it reasonable to propose that MD simulations may help to resolve some of the uncertainties and disagreements associated with experimental measurements.
This paper focuses on Aℓ, KA, KC, and c0 for the CHARMM (Chemistry at HARvard Macromolecular Mechanics) force field C36 (Klauda et al., 2010) for the lipids listed in Table 1. Table 1 also contains relevant nomenclature and abbreviations. Only fully hydrated single component bilayers are considered given the near absence of data for compressibility and bending moduli for those at low hydration or with more than one lipid type. Though values for some of these quantities have been previously published, simulations for most systems have been rerun or extended for uniformity of analysis. Specifically, relatively small systems (72 or 80 lipids) were all simulated for 420 ns, and larger ones (usually 648 lipids) were simulated for at least 120 ns (see Fig. 1 for representative snapshots of the DPPC bilayers). The set of polyunsaturated lipids, PDPC, PDPE, and SDPE, is entirely new. Additionally, previously published values of c0 for DOPE and DOPC (Sodt and Pastor, 2013) relied on the experimental monolayer bending constant from measurements on HII phases; c0 for PSM (Venable et al., 2014) was estimated using the polymer brush model (Rawicz et al., 2000), a popular model that relates KC to KA and bilayer thickness. Here, all values of c0 are evaluated directly from the simulations alone, without any input of experimental properties or assumption of empirical relations among the various mechanical properties.
Table 1.
Simulation system details.
| Lipid | T °K | Chains | Nwater/lipid | Large Systems | Small Systems | ||
|---|---|---|---|---|---|---|---|
| Nlipid | tRun(ns) | Nlipid | tRun(ns) | ||||
| DPPCa | 323 | 16:0,16:0 | 30.4 | 648 | 120 | 72 | 420 |
| DMPCa | 303 | 14:0,14:0 | 25.7 | 648 | 120 | 72 | 420 |
| DOPCa | 298 | 18:1,18:1 | 38.0 | 648 | 175 | 80 | 420 |
| DNPC | 303 | 24:1,24:1 | 50.1 | 648 | 180 | 72 | 420 |
| DOPE | 298 | 18:1,18:1 | 32.0 | 648 | 140 | 72 | 420 |
| POPCa | 303 | 16:0,18:1 | 31.1 | 648 | 140 | 72 | 420 |
| PDPC | 303 | 16:0,22:6 | 36.0 | 648 | 120 | 72 | 420 |
| POPEa | 310 | 16:0,18:1 | 32.0 | 720 | 120 | 80 | 420 |
| PDPE | 303 | 16:0,22:6 | 40.0 | 648 | 120 | 72 | 420 |
| SDPE | 303 | 18:0,22:6 | 40.0 | 648 | 125 | 72 | 420 |
| PSMb | 321 | Sph,16:0 | 29.1 | 648 | 220 | 72 | 420 |
| POPG | 303 | 16:0,18:1 | 45.0 | 648 | 120 | 72 | 420 |
Models derived from C36 validation simulations (Klauda et al., 2010).
Sph indicates the sphingosine chain; large system model derived from sphingomyelin validation simulation (Venable et al., 2014).
Fig 1.
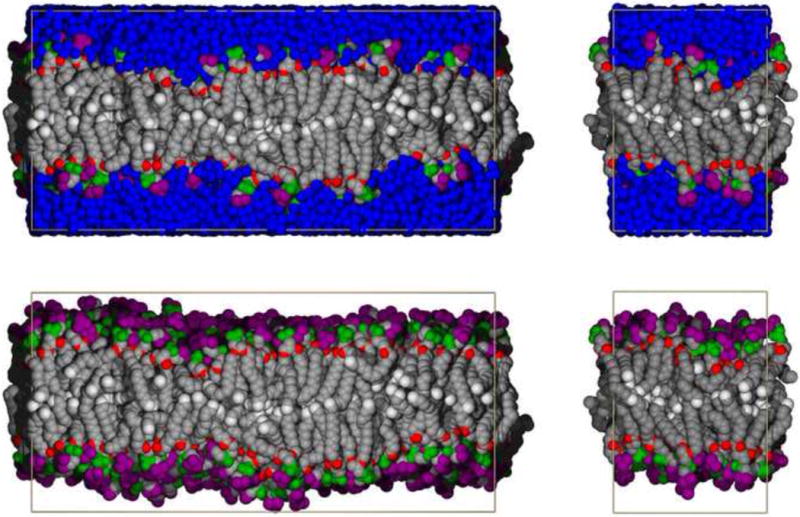
Snapshots of DPPC bilayers containing 648 (left) and 72 lipids (right), with waters shown (top) and omitted for clarity (bottom). Coloring is as follows: nitrogen on head groups (purple); phosphates (green); oxygen (red); methylene groups of lipid chains (grey); terminal methyl groups of acyl chains (white); water (blue).
By way of outline, the Background and Methods Section reviews the strategy used to parameterize C36 (Section 2.1), presents the critical formulae for the calculation of each of the preceding mechanical properties (2.2 to 2.4), and provides the relevant details on the simulations (2.5). The Results and Discussion presents the calculated mechanical values for the lipids in Table 1 (the first part of each section), and compares them with available experiments (the second part). Section 3.1 concerns Aℓ and KA, the relationship of the deuterium order parameter to area (3.1.3), and the effects of time step and smoothing Lennard-Jones interactions (3.1.4). Section 3.2 focuses on KC, and includes a simulation-based test of the polymer brush model (3.2.3), and further evidence that the pressure tensor-based method for calculating the Gaussian curvature modulus may be flawed (3.2.4). Section 3.3 presents values of c0, and comments on the notion of lipid shape (3.3.3).
2. Methods
2.1 Overview of the C36 Lipid Force Field and the problem of surface areas
C36 is a molecular mechanics additive force field consisting of bond, angle, dihedral angle, Lennard-Jones (LJ), and electrostatic terms. It is compatible with the FF for other classes of molecules in CHARMM (Brooks et al., 2009), so there is in principle no difficulty adding a protein or carbohydrate to a lipid bilayer. The quixotic paradigm underlying most molecular mechanics FF is that parameters developed for small model compounds can be combined to form larger ones or assembles like lipid bilayers without further adjustment. That is to say, the atomic parameters are transferrable. The simplicity of the functional form of the FF and the complexity of the membrane/solvent system undermine the paradigm, so C36 is best described as “physics-based as practical”. The original paper (Klauda et al., 2010) and several reviews (Klauda et al., 2008; Pastor and MacKerell, 2011) describe: the underlying functional form; the quantum mechanical calculations used to determine LJ interactions, torsional surfaces, and atomic point charges; simulations on model compounds (e.g., alkanes) to validate the parameters; the fitting to some experimental bilayer data as ultimately was required; and further validation of bilayer simulations against experimental quantities such as NMR 13C T1 relaxation times. C36 has since been extended to ion-lipid interactions (Venable et al., 2013), polyunsaturated lipids (Klauda et al., 2012), cyclic moieties in the acyl chains (Pandit and Klauda, 2012), cholesterol (Lim et al., 2012), sphingomyelin (Venable et al., 2014), cardiolipin (Song et al, in preparation), and numerous other lipids (Wu et al., 2014). Tests of the FF include effects of chain branching (Lim and Klauda, 2011), influence of ester-modified lipids (Villanueva et al., 2013), and the temperature dependence (Zhuang et al., 2014). Deficiencies in C36 include TIP3P water (Jorgensen et al., 1983), which is understructured and overly fluid (Venable et al., 2009; Venable et al., 2010), and the absence of long-range Lennard Jones interactions (discussed briefly in Section 3.1.4). Some of these deficiencies are ameliorated by use of polarizable force fields (Chowdhary et al., 2013; Huang et al., 2014), though the number of available lipid types is presently quite limited.
As implied in the Introduction, the equilibrium area per lipid Aℓ emerged as a critical target in the parameterization. To understand why this is difficult to reproduce one must consider the surface tension γ. For a planar interface such as a locally flat bilayer
| (1) |
where PN and PT are the components of the pressure tensor normal and tangential to the bilayer surface. While PN is a constant value of 1 atm, PT exhibits huge oscillations between negative values at the water/hydrocarbon interface and positive values at the bilayer center (Fig 2). For a stress-free bilayer (the object of most experiments relevant to parameterization) γ = 0 (Jahnig, 1996), or is very close to 0; i.e, the positive and negative contributions of PT(z) must balance. The tuning of a FF is essentially a problem of determining a small difference in large numbers, and any mismatch will lead to expansion or contraction of the bilayer. The difficulty of this task eventually made it necessary to systematically vary the charges on the carbonyl group to obtain agreement with the experimental Aℓ of DPPC. It is fortunate for the simulation field that Nagle and coworkers devoted years to methodically refining the values of surface areas of DPPC and other lipids (Kucerka et al., 2008; Kucerka et al., 2011; Nagle and Tristram-Nagle, 2000). The goal of obtaining agreement with experimental surface areas when simulating in the tensionless ensemble has been reached for a good range of lipids.
Fig 2.
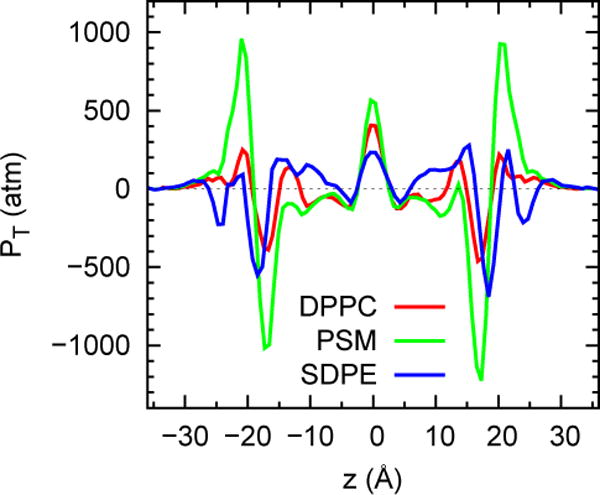
Pressure profiles for DPPC, PSM, SDPE bilayers consisting of 72 lipids from a 420 ns trajectory, with the first 20 ns dropped from the averaging. See Section 2.5 for further details on these and the other simulations presented in the study.
2.2 Calculation of Aℓ and KA, and standard errors
The instantaneous area per lipid Aℓ (t) for pure bilayers was calculated as the area of the simulation cell A divided by the number of lipids per leaflet. This assumes that undulations are small so the difference in projected and local areas is negligible (see Section 3.1.1 for justification). The area compressibility KA is evaluated from:
| (2) |
where 〈A〉 is the average total area, 〈δA2〉 is the mean square fluctuation, kB is Boltzmann’s constant, and T is the temperature. Most simulations (including those presented here) are carried out at only zero surface tension (see section 2.5), so fluctuations are used to obtain KA.
Simulations can also be carried out at series of different fixed areas (e.g., in the NPAT ensemble (Feller et al., 1995; Zhang et al., 1995)), where N is particle number and P is normal pressure). In this case, the surface tension is evaluated at each surface area, and related to KA via the derivative specified in Eq. (2).
The estimation of standard errors (se) for the average of a property X, denoted se[X], demands some attention given the nature of this review. The usual approach is to divide the time series into M equal sized blocks, calculate the average from each block and the standard deviation of the block averages, and divide the standard deviation by . If the blocks are statistically independent, the se calculated in this manner is independent of block size and can be used to specify confidence intervals and carry out hypothesis testing. If the blocks are correlated, the estimated se will decrease with increasing block size, and is not suitable as input for statistical tests. Hence, it should be confirmed that increasingly larger block sizes yield comparable se. A useful consistency check on independent block size involves the normalized correlation function of fluctuations of the time series
| (3) |
where δX (t) = X (t) − 〈X〉 and 〈δX2〉 is the mean squared fluctuation; the root mean squared, fluctuation (rmsf) is 〈δX2〉1/2. The smallest independent block size equals twice the correlation time . It is acceptable to use blocks larger than 2×τX, though the total number of blocks should be at least 5 to 10 to assure precise standard deviations. There may be decay times with low amplitude that are much larger than the estimated τX. In such cases it is reasonable to use blocks larger than the longest decay time to take into account uncertainties in τX. The standard errors in X can also be obtained directly from τX:
| (4) |
The monograph of Allen and Tildesley (Allen and Tildesley, 1987) remains an excellent introduction to these and many other technical topics of MD and Monte Carlo simulation.
2.3 Calculation of KC
2.3.1 Membrane bending energetics
The Helfrich expression (Helfrich, 1973; Safran, 1994) for the bending free energy per unit area of a structureless and homogeneous fluid sheet reads:
| (5) |
Here, KC is the bending modulus, KG is the Gaussian bending modulus (or saddle splay modulus), and c0 is the spontaneous curvature; these parameters characterize the mechanical properties of the surface. The radii of curvature at the particular point on the sheet under consideration are 1/c1 and 1/c2; these quantities specify the local shape of the surface. The Helfrich expression is quite general and applies to both bilayers and to monolayers (monolayer moduli are denoted with superscripts when necessary in what follows). Each leaflet in a symmetric bilayer can have a non-zero spontaneous curvature. The spontaneous curvature of the complete bilayer, however, is zero, because c0 of each leaflet is equal and opposite in sign. From this perspective, the spontaneous curvature of these leaflets might be better called “intrinsic curvature” or “curvature preference”, because the bilayer energy is minimized in the flat geometry. The spontaneous curvatures considered in this review are always for the monolayer leaflets of symmetric bilayers, and are mostly presented in terms of the radius of spontaneous curvature, .
Two different membrane geometries are considered in this review. For membranes that are nominally flat and span the xy plane, small fluctuations away from the perfectly flat state generate c1 and c2 values
| (6) |
Here h(x,y) denotes the “height field” of the sheet – the z-directed displacement away from the minimum energy reference plane at z=0. Lipids in the inverse hexagonal phase (HII) arrange themselves into a densely packed assembly of cylindrical water-filled tubes of radius R. In this case c1 = −1/R and c2 = 0. The preceding sign convention assigns a positive curvature to lipids in a micellar geometry and negative curvature to inverse micelles. It should be noted that the contribution of saddle splay to Eq. 5 is irrelevant for the systems considered in this review. This is obvious for the HII geometry as c2=0. For the quasi-flat geometry, it is easily verified that the total contribution of saddle-splay when integrated over the entire sheet vanishes if the sheet spans a periodic simulation box. In fact, it can be generally shown that the contribution of Gaussian curvature to membrane energetics is important only if the membrane undergoes a change in overall topology (e.g. ripping a hole in the membrane or turning a single vesicle into two vesicles, etc.) (Kamien, 2002).
See (Zimmerberg and Kozlov, 2006) for a succinct biology-focused introduction to membrane curvature, and (Deserno, 2015; Kamien, 2002; Safran, 1994) for reviews of the mathematics.
2.3.2 Calculating KC for “modest sized” bilayers
Because the integrated Gaussian curvature remains constant for a particular surface topology and c0 = 0 for symmetric bilayers, it is evident from Eq. (5) that KC is a particularly important modulus to determine.
When a bilayer fluctuates due to thermal noise its local height h(x,y) varies from that of the minimum energy reference plane; these fluctuations can be related to the bending constant. The analysis is simplest in the Fourier domain where the power spectrum of h(x,y) for a Helfrich sheet takes the form (Safran, 1994):
| (7) |
In principle this expression can be used to extract KC from simulated data, however this approach assumes Eq. (5) (with c0 = 0) is correct and is thus subject to the assumptions inherent to the Helfrich description. In particular, Eq. (7) will be valid only when the molecular structure of the lipids can be neglected and the membrane is behaving as a structureless thin sheet, which is expected to apply only in the limit of small q values (large wavelengths). Deviations from Eq. (7) are observed in finite-sized simulation boxes (Goetz et al., 1999). Over “mesoscopic” length scales on the order of several bilayer thicknesses, these deviations can be attributed to lipid tilting (May et al., 2007; Watson et al., 2011), with lipid protrusions (Goetz et al., 1999) becoming important at length scales below three times the bilayer thickness. For Eq. (7) to faithfully represent simulation data, a simulation box approximately 10 times larger than the bilayer thickness (40 Å), or about 400 Å is required. Even then, only the longest wavelength modes of the box can be analyzed. If Aℓ = 63 Å2 (the value for DPPC), the bilayer amenable to a treatment by Eq. (7) would need to contain about (2 × 400 × 400/63) ≈ 5000 lipids. Unfortunately, this approach is not practical for present-day all-atom simulations, where the number of lipids in a bilayer is typically limited to several hundred.
An alternate approach to determining KC of bilayers was recently developed (Watson et al., 2012), which allows reliable estimates of KC to be extracted from simulations of modestly sized boxes. This method analyzes thermal fluctuations of the lipid director vector field . The lipid director is a vector pointing from lipid head to tail and serves as a means to quantify lipid orientation. The theoretical prediction for the power spectrum of the longitudinal component of reads:
| (8) |
and it should be stressed that this expression follows from a theoretical model that explicitly accounts for lipid tilting (Watson et al., 2012). Lipid tilting adds a correction term to Eq. (7), but the director fluctuations display the simple functional form indicated in Eq. (8) when both bending and tilting are considered theoretically. Deviations from Eq. (8) in simulation data are observed only over wavelengths shorter than 3 bilayer thicknesses (Watson et al., 2012) (Levine et al., 2014). All-atom simulations including 648 lipids were shown to be well converged in our prior study in the sense that Eq. (8) was obeyed over the largest four wavevectors in the simulation box; if convergence is taken for granted, 288 lipid simulations were shown to be sufficient to determine Kc (Levine et al., 2014). A different lipid-orientation-based approach by Khelashvili and coworkers (Khelashvili et al., 2013) has also been proposed to determine Kc. Bending constants obtained via this methodology for pure DPPC, DOPC, and DOPE are accurate to within 20% of those determined via Eq. (8), but these differences are statistically significant (see Supporting Information of (Levine et al., 2014).
An additional benefit of the director spectrum approach (Eq. 8) is that other elastic constants, including the tilt (Kθ) and twist (Ktw) moduli, follow immediately from the analysis (Watson et al., 2012). The transverse component of the lipid directors is predicted to obey:
| (9) |
and comparison to simulation data allows the determination of Kθ Ktw. Kθ is now of practical interest because it may be extracted from x-ray scattering experiments (Jablin et al., 2014).
2.4 Calculation of c0
2.4.2 Monolayers in the HII phase
The spontaneous curvature of lipids is experimentally obtained from the inverse hexagonal (HII) phase. The phase consists of lipid monolayers formed in long hexagonally arranged water-filled tubes, and is preferentially formed by lipids with head groups that appear to be small compared to their tails, such as DOPE, as opposed to DPPC, which contains a larger head group. This qualitative shape-based observation suggests why bilayers composed of certain lipids are more or less susceptible to curvature (see (Brown, 2012) for a recent review), but shape is only one of the driving forces ((Sodt and Pastor, 2014) and Section 3.3.3).
Because c2 = 0, KG = 0 for the lipid tubes in the HII phase, Eq. (5) can be expressed in terms of a single curvature c = c1:
| (10) |
where is the monolayer bending constant, and R = c−1 is the radius of the water filled tube. By convention, the curvature of leaflets in the HII phase is negative. The seminal paper by Gruner, Parsegian, and Rand (Gruner et al., 1986) connected Eq. (10) with experiment. Specifically, their experimental setup consisted of an HII phase in equilibrium with a polyethyleneglycol (PEG) bath. Changing the concentration of PEG changes the osmotic pressure Π in the HII phase, and thereby changes R. and R0 are then obtained from fitting measured values of R and Π to the following relation:
| (11) |
Bending constants and spontaneous curvatures of some other lipids have been obtained using this method (Marsh, 2006), but the list is not especially large: the lipids must either form HII phases, or be easy to dope into the matrix of an HII phase, usually composed of DOPE.
There are nuances in the experiment, some of which pertain to the use of alkanes to stabilize the system, some to assumptions concerning the appropriate bending planes needed to specify the radius of the pore, and others to the additivity of mixtures. These uncertainties and assumptions can now be explicitly assessed by all-atom simulations. For example, Sodt and Pastor (Sodt and Pastor, 2013) simulated the HII phase of DOPE, and estimated and R0 from
| (12) |
where Δp is the pressure difference between the center of the pore and the acyl tail region, and is equivalent to Π in Eq. (11). Very good agreement with experiment was obtained for R0 (−26 ± 2 Å vs. −28.5 Å for experiment), though additional simulations at larger R will be required to reduce the statistical error in to better assess the agreement. The pivotal plane (where the area is constant as the surface bends) was determined to be near the glycerol region of the bilayer, in agreement with the location proposed by Chen and Rand (Chen and Rand, 1997).
2.4.3 Leaflets in bilayers
Applying monolayer properties to bilayers involves additional assumptions. Eq. (10) must be extrapolated from c ≈ c0 (where monolayer measurements are obtained) to c = 0 (the curvature of a flat bilayer). It is also commonly assumed that the bilayer bending constant is twice that of the extracted . However, alkanes are often added to stabilize the HII phase, which might lead to a “softening” of the bending constant.
While the spontaneous curvature is not directly experimentally accessible for bilayers, it can be obtained from simulation as follows (Safran, 1994):
| (13) |
where is the derivative of the bending free energy per lipid at zero curvature for the upper leaflet, tensor; the limits of the integral extend from the bilayer midplane (z=0) to the upper box length (Lz/2). Note that the right hand side is a product containing . Hence, a value of the bending constant must be provided to extract c0. Here KC for each bilayer is determined from Eq. (8) and divided by 2 to provide the required by Eq. (13).
2.5 Simulation details
2.5.1 General
All simulations were carried out with the CHARMM program (Brooks et al., 2009) in the tensionless NPT ensemble; the extended system barostat and thermostat were employed. In this implementation, the pressure tensor is fully anisotropic and requires a tetragonal prism simulation unit cell, where x = y by constraint, with the bilayer normal vector aligned with the z axis; the cell height and the xy area are coupled to separate pistons. The bulk of the simulations were run with recent (c38b2 and later) versions of the program that include a domain decomposition scheme (Hynninen and Crowley, 2014) for the pairwise non-bond calculation. The electrostatic term was computed via the particle-mesh Ewald method, with a real space cutoff of 12 Å, a κ value of 0.32, and ca. 1 grid point per Å in each direction for the mesh. The van der Waals dispersion term used an LJ functional form with a force-switching function over the interval of 8 to 12 Å for most simulations; the PSM simulations and one DPPC simulation used a 10 to 12 Å interval. Likewise, a 1 fs integration time step was used for nearly all of the simulations, with the exception of one DPPC simulation, which used a 2 fs integration time step.
2.5.2 Models
For small systems with DMPC, DPPC, POPC and POPE, endpoints from previously published simulations were used as the starting point; all others except PSM were constructed de novo using the CHARMM-GUI web interface (Jo et al., 2008). The small PSM system had already been run for 420 ns, and was not extended further. Each large system was created by a 3×3 expansion of a small 72 lipid system in the xy plane, except for the POPE system, where the original system had 80 lipids. For DOPC, an alternate system of 80 lipids was used for the extended small system, rather than the original system with 72 lipids.
3. Results and discussion
3.1 Area per lipid and area compressibility modulus
3.1.1 Simulation
Table 2 lists Aℓ and KA and their standard errors for the large and small systems for the entire set of lipids, and Fig. 3 plots Aℓ (t) for DPPC and PSM over their 420 ns trajectories. Relaxation of area fluctuations is considerably slower for PSM, as evident both from time series (Fig. 3) and the correlation functions (Fig. 4); the relaxation times are approximately 1 ns for DPPC and 10 ns for PSM. Based on this analysis and consistency checks using different block sizes (see Section 2.2), independent block sizes for calculation of standard errors were set to 20 ns for PSM, and to 10 ns for DPPC and the other lipids. The standard errors for Aℓ (se[Aℓ] range from 0.1 to 0.3 Å2.
Table 2.
Area/lipid (Aℓ) and area compressibility (KA) from the simulations compared to each other and to experimental values; standard errors are given in parentheses.
| Lipid | area/lipid (Å2) | KA (dyn/cm) | ||||
|---|---|---|---|---|---|---|
| Large | Small | Expt | Large | Small | Expt | |
| DPPC | 62.9 (0.1) | 63.0 (0.1) | 63.0a | 210 (10) | 230 (20) | 231i |
| DMPC | 61.5 (0.1) | 61.9 (0.2) | 59.9b, 60.6c | 210 (30) | 210 (40) | 234j |
| DOPC | 68.9 (0.1) | 68.9 (0.1) | 67.4a, 72.4d | 290 (20) | 280 (10) | 300k |
| DNPC | 64.1 (0.1) | 63.7 (0.2) | 220 (30) | 240 (20) | ||
| DOPE | 62.3 (0.1) | 62.3 (0.2) | 250 (40) | 260 (20) | ||
| POPC | 65.9 (0.1) | 66.0 (0.2) | 64.3b, 68.3e | 280 (30) | 240 (10) | 180–330l |
| PDPC | 70.8 (0.1) | 71.0 (0.1) | 260 (20) | 220 (10) | ||
| POPE | 58.7 (0.1) | 58.8 (0.2) | 59.8–60.8f | 260 (20) | 280 (20) | 233m |
| PDPE | 64.3 (0.1) | 64.7 (0.1) | 200 (20) | 230 (10) | ||
| SDPE | 63.9 (0.1) | 64.0 (0.1) | 300 (10) | 230 (20) | ||
| PSM | 55.1 (0.1) | 55.4 (0.3) | 310 (40) | 350 (40) | ||
| POPG | 67.5 (0.1) | 68.0 (0.1) | 64.3g, 66.0h | 180 (30) | 220 (20) | |
(Pan et al., 2008)
(Kucerka et al., 2005)
(Rappolt et al., 2003)
(Pan et al., 2014)
(Kucerka et al., 2012)
(Binder and Gawrisch, 2001)
(Rand and Parsegian, 1989).
Fig 3.
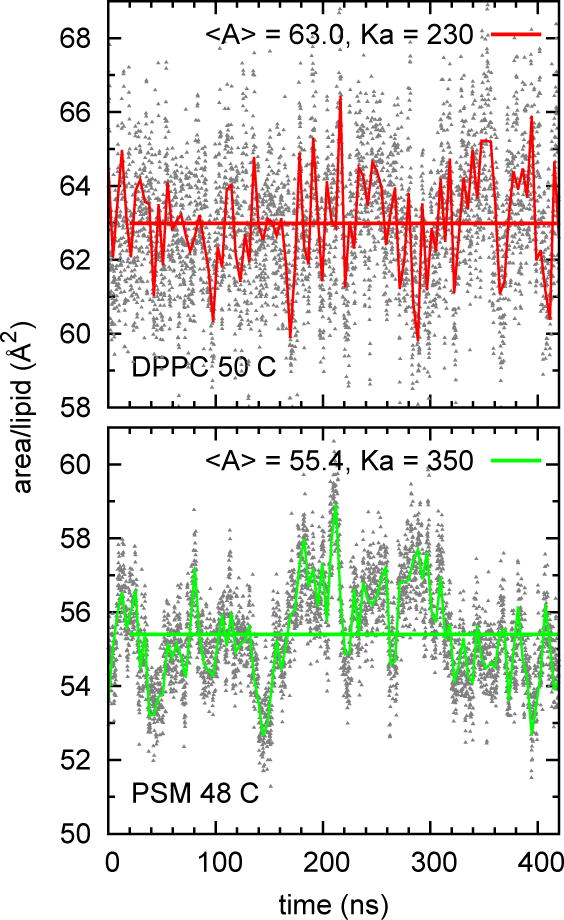
Time series of area for DPPC and PSM (72 lipid, or small, systems). The grey symbols are the instantaneous values at 100 ps intervals; the fluctuating lines are Bezier smoothed 5 ps averages; the horizontal lines are the average over 20–420 ns.
Fig 4.
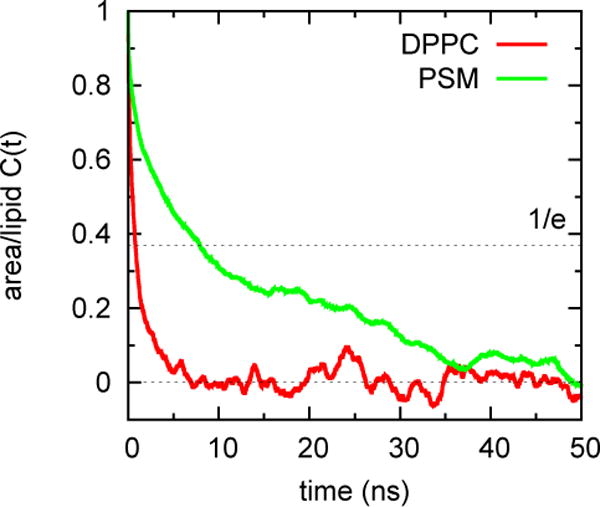
for DPPC and PSM for the time series plotted in Fig. 3.
for DPPC and PSM are 1.83 Å2 and 1.40 Å2, respectively. Inserting these values, the preceding relaxation times, and the time of trajectory used for averaging (Trun = 400 ns) into Eq. (4), yields se[Aℓ] = 0.13 Å2 for DPPC and 0.31 Å2 for DPPC, nearly the same as the estimates from block size in Table 2 for these lipids.
With the exception of DNPC, areas for all of the large systems are slightly smaller than those of the small systems (0.1 to 0.4 Å2). While these differences are comparable to the se for each lipid, on aggregate the trend is statistically significant: p = 0.052 for a paired t-test comparing the differences in area of the big and small systems for all of the lipids, and p = 0.0012 when DNPC is omitted. Hence, as expected, undulations in the large systems only slightly reduce their projected surface areas, in agreement with an earlier prediction (Braun et al., 2011).
KA are mostly between 200 and 300 dyn/cm, with se of 20–40 dyn/cm. The difference in se[Aℓ] (0.2–0.5% of Aℓ) and se[KA] (10–20% of KA) is expected for the area and its fluctuation, and highlights the need for long trajectories when evaluating KA. There is no statistically significant difference in KA for the large and small systems (p = 0.767). PSM has the highest KA (330 ± 30 dyn/cm averaged over the small and large systems), though some of the unsaturated lipids (e.g., DOPC at 285 ± 10 dyn/cm) are close. Saturated lipids DMPC and DPPC (average of 215 ± 5 dyn/cm) are among the lowest. KA for DPPC are approximately twice those obtained with C27r (Klauda et al., 2005), the immediate the precursor of C36. Hence, area compressibilites provide a good and reasonably straightforward test of a force field, though long simulations are required. It is somewhat disconcerting to reread papers from pre-2000 presenting KA from simulations of only a ns (Feller and Pastor, 1999), and it will likely be disconcerting to read present-day papers in 2030.
3.1.2 Comparison with experiment
From Table 2, the surface areas of all but POPG are bracketed by recent experimental values, or are either within 1 Å2 of these values; the simulated Aℓ for POPG is 2.0 Å2 above the higher of two experimental values (a difference of 3%). The chain SCD for PSM agree well with experiment (Venable et al., 2014). The order parameter averaged over the 5 largest values (the so-called “plateau region”) for PDPC is 0.200 from experiment (Leftin and Brown, 2011), and 0.192 from simulation (this study).
In 2000 Evans and coworkers published KA for a number of lipids, primarily unsaturated, using pipette aspiration (Rawicz et al., 2000). After correction for undulations, these lay in a relatively small range, 234–263 dyn/cm, with experimental uncertainties of 10–30 dyn/cm. More recent measurements using higher stretching rates (Evans et al., 2013) led to a revised value of ~300 dyn/cm for DOPC.
The simulated KA for DMPC and DOPC reported here (Table 2) are within the statistical errors of the simulations and experiment, and correctly reproduce the order.
3.1.3 Area and Deuterium Order Parameters
Experimentally, the deuterium order parameter, SCD, is measured from the lineshape of the NMR spectrum of deuterated carbons (Seelig, 1977). It is obtained from simulations as
| (14) |
where θ is the angle of the CH vector of the relevant carbon with respect to the bilayer normal. SCD shows characteristic splittings in the head group and for carbon 2 of the acyl chains, is relatively constant in the upper parts of the chains, and then decreases toward the chain ends. See (Klauda et al., 2010) and (Venable et al., 2014) for comparisons of experimental order parameters and those of simulations using C36 for DPPC and PSM bilayers, respectively. The span from carbons 4–8 is often called the “plateau region”, though not all chain types exhibit this exact range (Fig. 5). It has been long recognized that the order parameters of the plateau regions are sensitive to the surface area (Feller et al., 1997; Koenig et al., 1997; Nagle, 1993; Petrache et al., 2000), and these can provide invaluable target data for parameter development when reliable surface areas derived from x-ray and/or neutron data are not available. For example, the C36 parameters for sphingomyelin (Venable et al., 2014) and corrections to the Na+ interactions with charged lipids (Venable et al., 2013) were developed using this approach.
Fig 5.
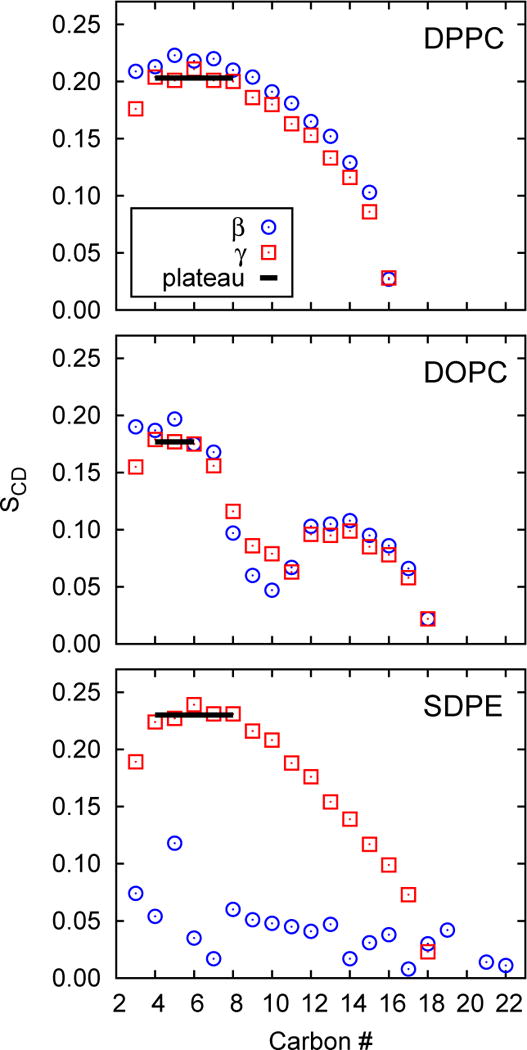
Deuterium order parameters for the γ and β chains of lipids in the large systems; the plateau value 〈S〉 for the γ chain of each system is denoted by a solid line.
Assorted models have been developed to directly relate SCD and surface area (Nagle, 1993; Petrache et al., 2000). This is a difficult endeavor because SCD contains contributions from conformational disorder (gauche-trans states of the acyl chains, and librations within each state), local tilting of the entire lipid (termed wobble), and assorted collective motions (Pastor et al., 1991; Pastor et al., 1988b). The usual approach is to estimate the average projection per methylene group along the bilayer normal, and then relate this to the area by a volume relationship. A nicely-derived and easy to implement model comes from Nagle (Nagle, 1993):
| (15) |
where 〈S〉 is the plateau value (the absolute value is used here as consistent with Eq. 14), VCH2 is the volume of a CH2 group, and bCC is the projected length of the CC bond along the bilayer normal for an all-trans chain oriented along the normal. Setting VCH2 = 27.6 Å3, bCC = 1.27 Å, 〈S〉 = 0.2 (the approximate experimental average of carbons 4–8), Nagle obtained Aℓ = 62.1 Å2 in quite good agreement with experiment for DPPC.
How do areas and deuterium order parameters relate for simulations with C36, where both quantities are known precisely for a wide range of lipids? To begin, Fig. 5 plots the chain order parameters for DPPC, DOPC, and SDPE for the large systems (those from the small systems are nearly identical). The same chains in the other lipids have similar patterns, and the following ranges are used to evaluate the 〈S〉 listed in Table 3: 4–8 for saturated acyl chains (in accord with standard practice); 4–6 for oleyl chains (the presence of the cis double bond at the 9–10 position perturbs the order parameters of carbons 7 and 8); 4–11 for DNPC; and 6–12 for the sphingosine chain of PSM. With the exception of the polyunsaturated lipids, differences between the γ and β chains (often denoted chain 1 and chain 2, respectively) are mostly less than 0.01. SCD for the polyunsaturated chains are close to zero as consistent with the very low conformational barriers between conformers (see Fig 5 for SDPE); these are not appropriate for area estimates using Eq. (15) so 〈S〉 from these chains is not reported or used here. The average 〈S〉 of the γ chains from all systems is 0.216, with a standard deviation of 0.025, in accord with the general notion that the plateau value for fluid phase lipids is approximately 0.2.
Table 3.
Plateau values for the γ and β chains of the large system (see text for specification of ranges)
| Lipid | γ chain | β chain |
|---|---|---|
| DPPC | 0.203 | 0.217 |
| DMPC | 0.221 | 0.221 |
| DOPC | 0.177 | 0.186 |
| DNPC | 0.234 | 0.240 |
| DOPE | 0.211 | 0.215 |
| POPC | 0.198 | 0.197 |
| PDPC | 0.192 | |
| POPE | 0.247 | 0.231 |
| PDPE | 0.225 | |
| SDPE | 0.230 | |
| PSM | 0.276 | 0.261 |
| POPG | 0.188 | 0.184 |
Fig. 6 plots the averages of Aℓ vs. 〈S〉 for the γ chains for the large systems, and includes the prediction from Eq. (15) (with the original parameters). The saturated chains (DPPC, DMPC and PSM), POPE and DOPE and are well described by Eq. (15), which was in fact developed for saturated chains. The polyunsaturated group and lipids with large surface area (low 〈S〉) are not so well described. Perhaps these observations can lead to a refined model.
Fig 6.
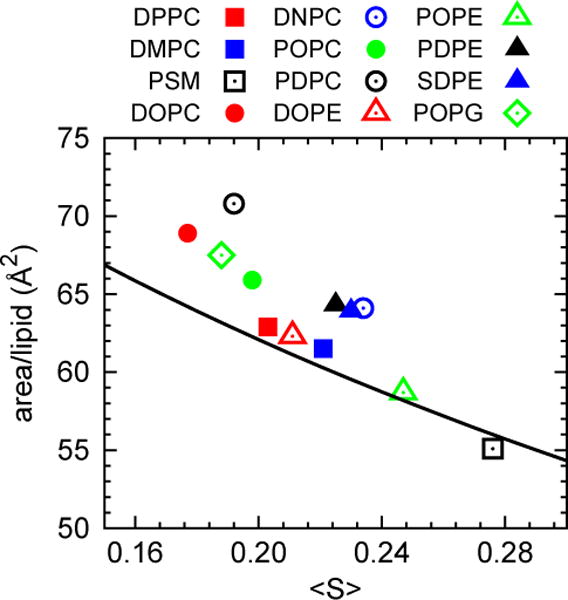
Area per lipid vs. plateau value of the deuterium order parameter 〈S〉 for the γ chains of lipids in the large systems (assorted symbols), and prediction of Eq (15) (solid line).
3.1.4 Time step and Lennard-Jones cutoffs
Increasing the time step Δt is always tempting to simulators. It seems to extend trajectories for free. However, accuracy decreases with increasing Δt, and a trajectory can become unstable (Pastor et al., 1988a). The first entries in Table 4 indicate that increasing Δt from 1 to 2 fs increases Aℓ from 63.0 ± 0.1 to 63.5 ± 0.1 Å2 for a DPPC bilayer, a small but statistically significant change.
Table 4.
Dimensions and KA for small DPPC simulations of 420 ns generated with different time steps and LJ cutoff ranges.a
| Time step | LJ interval (Å) | Area/lipid (Å2) | KA (dyn/cm) | Distances (Å) | |
|---|---|---|---|---|---|
| C2:C2 | P:P | ||||
| 1 fs | 8–12 | 63.0 | 230 | 27.9 | 39.1 |
| 2 fs | 8–12 | 63.5 | 220 | 27.8 | 38.9 |
| 1 fs | 10–12 | 61.6 | 250 | 28.3 | 39.5 |
Standard errors are ca. 0.1 Å2 for area, 0.1 Å for distance, and 20 dyn/cm for KA
Long range electrostatic forces are explicitly included in essentially all modern all-atom FF using various summation methods. In contrast, Lennard-Jones (LJ) interactions are most often cut off, and both the value of the cutoff and the smoothing function should be considered part of the FF. C36 was developed with a force-switching function extending from 8–12 Å. Ideally, all simulations carried out with C36 should use these options. Unfortunately, different programs have slightly different code for smoothing LJ interactions. Furthermore, the CHARMM protein parameters were developed with a force-switching extending from 10–12 Å, and this has become the default in most releases of CHARMM. The last row of Table 4 shows the is decreased by effect of using a 10–12 Å force shifting function on a DPPC bilayer: Aℓ nearly 1.5 Å, a little over 2%.
Lastly, long-range LJ interactions contribute substantially to the surface tension γ of alkane/vapor interfaces. For example, when octane/vapor is simulated with C27r (Klauda et al., 2005), the alkane component of C36, γ = 13.6, 16.0, and 21.1 ± 0.5 dyn/cm for LJ cutoffs of 10, 12 and 30 Å, respectively; the “no-cutoff” result is 20.8 ± 0.5 dyn/cm (Venable et al., 2009). Consequently, C36, which neglects long-range LJ interactions, substantially underestimates the surface tension of lipid monolayers (Klauda et al., 2010), which have an alkane/vapor interface. The next generation of FF will likely be developed with long range LJ interactions, and the issues noted above will disappear. For present studies, this flaw in the parameter set should be recognized.
3.2 Bending, tilt, and twist moduli
3.2.1 Simulation
Table 5 presents KC (large systems only) calculated from Eq. (8). The convergence and precision of the simulations are shown in Figs 7–9 for DPPC and PSM. Constant values for q2S(q) between q = 1 and 0.4 nm−1 (leftmost 4 points of Fig. 7), indicate that the system size (648 lipids) is sufficiently large for these systems. It is evident that KC is substantially larger for PSM. Figs. 8 and 9 plot the time series of S(q) and its correlation function for the smallest q (largest length). Fluctuations are more rapid for DPPC, though both bilayers relax within 5 ns, indicating that a 10 ns block average is acceptable for all of the lipids.
Table 5.
Bending constants KC from simulation (in three different units) and four different experimental method (all in J); standard errors for the simulations are given in parentheses.
| Lipid | Simulation
|
kBT | kcal/mol | Experiment (10−20 J)
|
|||
|---|---|---|---|---|---|---|---|
| 10−20 J | PAa | X-ray | NSEb | Flicker | |||
| DPPC | 15.8 (0.4) | 35.4 | 22.7 | 6.7d | 15.0i | ||
| DMPC | 12.2 (0.5) | 29.3 | 17.6 | 5.6c | 6.6e | 8.2h | 13.0i |
| DOPC | 11.8 (0.4) | 28.8 | 17.0 | 8.5c | 8.3f | 10.9j | |
| DNPC | 22.0 (0.7) | 52.6 | 31.7 | ||||
| DOPE | 11.8 (0.3) | 28.7 | 17.0 | ||||
| POPC | 13.2 (0.4) | 31.7 | 19.1 | 8.5g | 8.6h | ||
| PDPC | 9.6 (0.1) | 23.1 | 13.9 | ||||
| POPE | 13.3 (0.6) | 31.2 | 19.2 | ||||
| PDPE | 9.9 (0.6) | 23.6 | 14.2 | ||||
| SDPE | 10.6 (0.4) | 25.4 | 15.3 | ||||
| PSM | 27.8 (0.8) | 62.7 | 40.0 | ||||
| POPG | 11.3 (0.6) | 26.9 | 16.2 | ||||
pipette aspiration (PA)
neutron spin echo (NSE)
(Guler et al., 2009)
(Kučerka et al., 2005)
(Kucerka et al., 2005)
(Mell et al., 2013)
(Fernandezpuente et al., 1994)
(Gracia et al., 2010).
Fig 7.
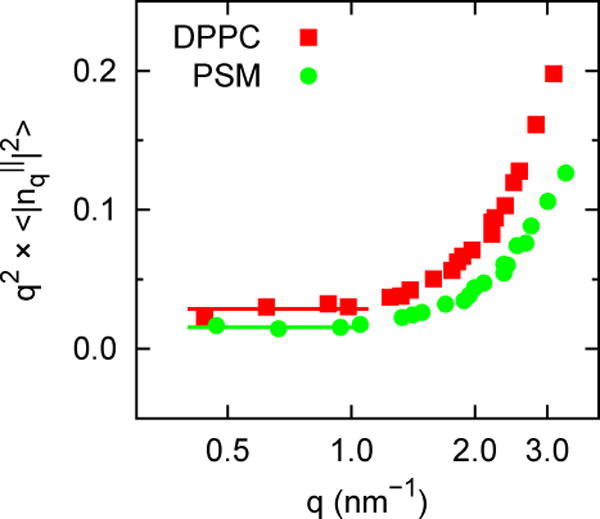
q2S(q) vs q for DPPC and PSM.
Fig 9.
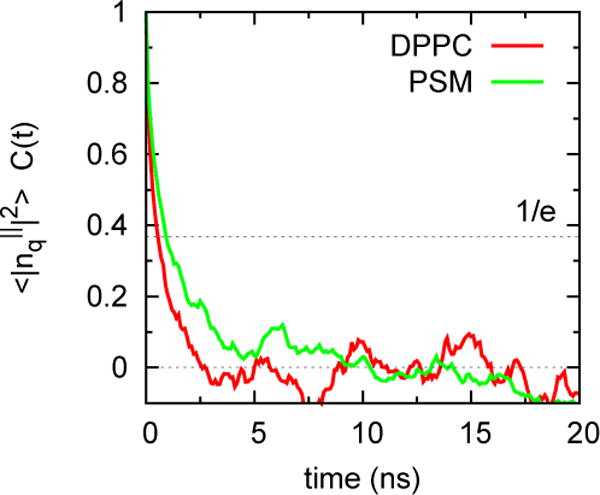
Correlation functions for S(q) for DPPC and PSM for the smallest q (see caption of Fig. 8)
Fig 8.
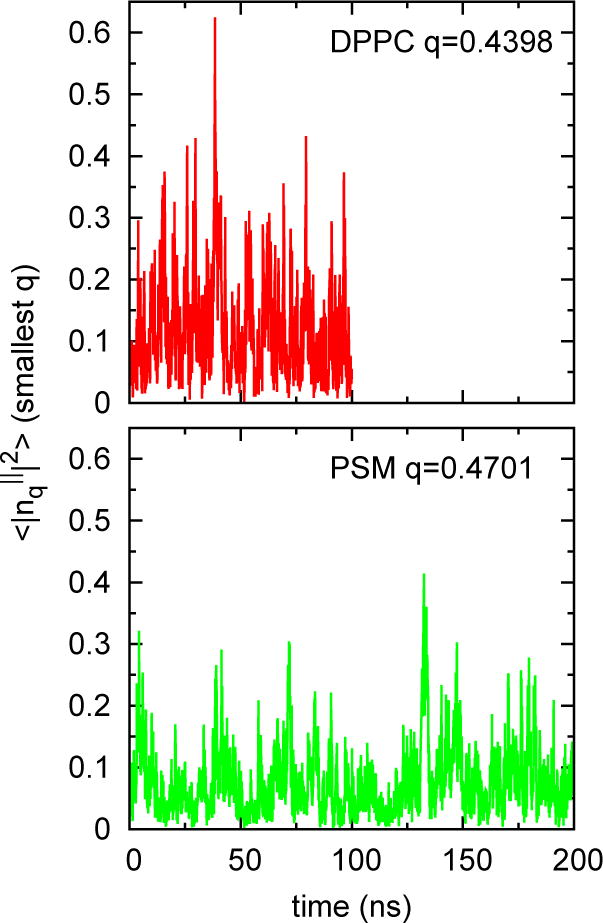
Time series S(q) for DPPC (q = 0.4398 nm−1) and PSM (q = 0.4701 nm−1). These are the smallest q for each system and would be expected to be the slowest to converge.
In contrast to KA, which ranged from 200 dyn/cm (PDPE) to 310 dyn/cm (PSM) for the large systems, the KC vary by nearly a factor of 3. Values for the polyunsaturated lipids cluster at the low end, with PDPC the lowest at 9.6 ± 0.1 × 10−20 J. PSM is the highest at 27.8 ± 0.8 × 10−20 J. Some of the other trends are discussed in Section 3.2.3.
Kθ and Ktw are of mostly theoretical interest, and are presented in Table 6 for completeness. Figure 10 shows results for DPPC and PSM, including the best fit curve for each data set.
Table 6.
Values of tilt (Kθ) and twist (Ktw) moduli obtained from fits to Eq. 9.
| Lipid | Kθ (10−20 J/nm2) | Ktw(10−20 J) | Fit rmsd |
|---|---|---|---|
| DPPC | 4.88 | 2.04 | 0.0039 |
| DPPC | 4.02 | 2.18 | 0.0038 |
| DOPC | 6.40 | 0.99 | 0.0038 |
| DNPC | 4.74 | 2.23 | 0.0054 |
| DOPE | 8.47 | 1.44 | 0.0020 |
| POPC | 5.52 | 1.45 | 0.0040 |
| PDPC | 6.50 | 0.95 | 0.0036 |
| POPE | 8.04 | 2.36 | 0.0019 |
| PDPE | 7.72 | 1.48 | 0.0025 |
| SDPE | 7.48 | 1.65 | 0.0022 |
| PSM | 3.91 | 4.76 | 0.0066 |
| POPG | 6.15 | 1.05 | 0.0032 |
Fig 10.
Fit to Eq. (9) for determination of tilt and twist moduli.
3.2.2 Comparison with experiment
The experimental results listed in Table 5 are grouped by experimental technique: PA, pipette aspiration of giant unilameller vesicles (GUV); X-ray, from x-ray diffraction of lamellar stacks; NSE, from neutron spin echo of lamellar stacks, and Flicker, from thermally excited fluctuations of GUV. The x-ray value of DOPC includes a recent correction accounting for lipid tilt (Jablin et al., 2014); x-ray values for the other lipids do not include this correction. The results for x-ray and PA are close for the lipids studied in this work. KC from flicker experiments are over 100% higher for saturated lipids, and approximately 30% higher for DOPC as compared to x-ray data. The disparity among experimentally determined values for KC is well known and remains poorly understood (Marsh, 2006; Nagle, 2013).
As recently reported (Levine et al., 2014), the simulated KC are nearly identical to those from flicker experiments for DPPC and DOPC for C36. The present study adds the excellent agreement of simulation and flicker experiment for DMPC, and the substantial disagreement of simulation and x-ray for POPC.
Given the uncertain experimental landscape, it is impossible to make any definitive comparison to “experiment”. On the one hand, the close agreement between KC from simulations with those from flicker experiments might be comforting, however the large differences between simulations and other experimental techniques are difficult to rationalize. The periodic boundary conditions, hydration levels and probed length scales of simulation systems are arguably much closer to the lamellar stacks used for x-ray measurements than to the GUVs analyzed in flicker experiments and one might naively expect the best correspondence to be found between x-ray and simulation; the data clearly dispute this hypothesis. One confounding factor that should be emphasized is that the present simulations were carried out with a cut off Lennard Jones interaction (see sec. 3.1.4). Long-ranged van der Waals interactions (acting in aggregate between all molecules) are responsible for stabilizing the lamellar stacks in experiment (Israelachvili, 1992; Safran, 1994), but are absent from the simulations. From this perspective, the simulations might actually be more closely analogous to the GUV geometry than the lamellar stacks observed in x-rays – the simulated bilayers should be behaving essentially as an isolated single membrane. Of course, surface areas from x-ray data were targets of the parameterization of C36, and excellent agreement of simulated and experimental density profiles and scattering functions provided strong support for both simulation methodology (Klauda et al., 2006) and the force field. A reanalysis of x-ray data including the tilt modulus (Jablin et al., 2014) increased the original KC by approximately 10% for DOPC, but the difference still 25% lower than that from the flicker measurement, and 30% lower than simulation.
Recent neutron spin echo results for tetraoleoylcardiolipin (TOCL) at 303 K yielded KC = 10.6 ± 0.6 ×10−20 J, in close agreement 11.4 ± 0.4 ×10−20 J obtained by simulations with the C36 FF at 310 K (Song et al, in preparation). Otherwise, the NSE and x-ray results are close to each other, and lower than simulation. X-ray scattering of tetramyristoylcardiolipin (TMCL) at 323 K yielded KC = 7.5 ± 1.0 ×10−20 J (Boscia et al., 2014) which is comparable with the x-ray determinations of KC for two-chained lipids.
Only two experimental values of tilt modulus appear to be available: 8 ×10−20 J/nm2 for DOPE from an analysis of the Lα to HII transition (Hamm and Kozlov, 2000), and 9.5 ± 0.7 ×10−20 J/nm2 for DOPC from x-ray diffraction of lamellar stacks (Jablin et al., 2014). The simulated values are nearly identical for DOPE, and 30% smaller for DOPC.
3.2.3 A simulation-based test of the polymer brush model
The polymer brush model (PBM) was introduced by Evans and coworkers (Rawicz et al., 2000) to rationalize trends in the bending constants of lipid bilayers and extrapolate them when required. Specifically,
| (16) |
where h is the hydrophobic thickness of the bilayer; it was set to hPP −h0, where hPP is the phosphate to phosphate peaks from x-ray diffraction measurements, and h0 = 10 Å is the head group thickness (the sum from both leaflets).
The results of the present simulations allow an assessment of Eq. (16), starting with the bilayer thickness. Columns 2 and 3 of Table 7 list hPP and hC2C2 for each system. The latter is the separation across the bilayer between the average positions of carbon 2 of the lipid chains, and is often taken to be a measure of hydrophobic thickness. As evident from column 4, hPP − hC2C2 varies between 10.5 and 11.1 among lipids, and averages 10.8 Å. Hence assumptions regarding head group thickness could be refined, although here the original h0 = 10 Å is retained.
Table 7.
Distances across the bilayer, and the resulting polymer brush model values for bending constants compared to simulation
| Lipid | Distances, h (Å)
|
|
% diff | |||
|---|---|---|---|---|---|---|
| C2:C2 | P:P | P-C | ||||
| DPPC | 27.95 | 39.02 | 11.07 | 7.37 | 53% | |
| DMPC | 24.54 | 35.65 | 11.11 | 5.75 | 53% | |
| DOPC | 27.37 | 38.35 | 10.98 | 9.71 | 18% | |
| DNPC | 39.94 | 51.06 | 11.12 | 15.44 | 30% | |
| DOPE | 29.96 | 40.79 | 10.83 | 9.87 | 16% | |
| POPC | 27.52 | 38.54 | 11.02 | 9.50 | 28% | |
| PDPC | 27.15 | 37.91 | 10.76 | 8.44 | 12% | |
| POPE | 30.67 | 41.55 | 10.88 | 10.78 | 19% | |
| PDPE | 29.56 | 40.16 | 10.60 | 7.58 | 23% | |
| SDPE | 31.49 | 42.10 | 10.61 | 12.89 | −21% | |
| PSM | 30.26 | 40.78 | 10.52 | 12.24 | 56% | |
| POPG | 26.88 | 37.49 | 10.61 | 5.67 | 50% | |
The last columns of Table 7 list and the % difference from . The results are similar to the comparison of the and those from pipette aspiration; i.e., for DOPC underestimates by 18%, and the errors for most other unsaturated lipids are all 30% or less (POPG differs by 38%). for DPPC, DMPC, and PSM underestimate by a remarkably precise 53–56%.
The preceding results imply that the PBM model can be useful for scaling lipids of similar chains, and perhaps head groups, even though Eq. (16) does not simultaneously model all of the simulation data. Figure 11 plots vs. with similar lipids grouped. The set DPPC, DMPC, and PSM (squares) is clearly offset from the others, and the slope is somewhat different.
Fig 11.
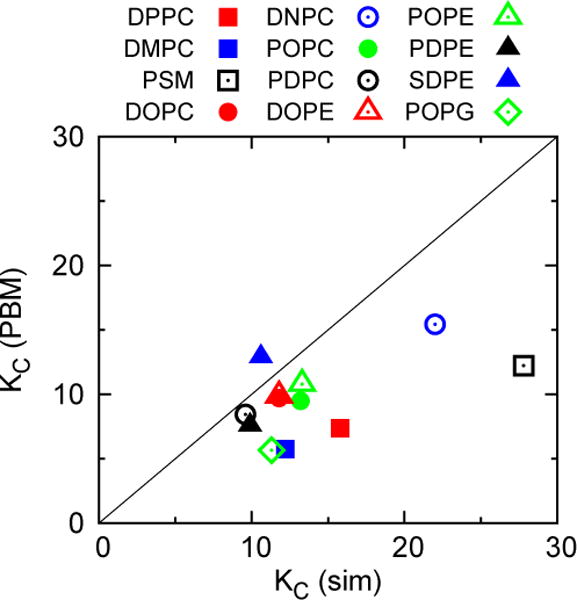
(calculated from KA and P:P distances listed in Tables 2 and 7, respectively) vs. with lipids grouped by type.
3.2.4 Gaussian curvature modulus
The calculation of the Gaussian curvature modulus from simulation is presently controversial, though a method applicable to simple systems (not yet practical for all-atom simulations) is now available (Hu et al., 2012). This method yields for both a very simple coarse-grained model (Hu et al., 2012) and the MARTINI model (Hu et al., 2013), in accord with available experiment results (Marsh, 2006). Hu et al. (2012 and 2013) also considered the following expression for the Gaussian curvature modulus of a leaflet:
| (17) |
where δ is the position of the neutral plane (where the area does not change upon bending) with respect to the bilayer midplane. The Gaussian curvature modulus KG for the bilayer is then evaluated from
| (18) |
Values of ranged from −0.12 to −0.33 for MARTINI DMPC depending on the value of the neutral surface (Hu et al., 2013); a value of −0.9 for their simple CG model was only obtained for any unphysically small value of the neutral surface (Hu et al., 2012). On this basis it was concluded that Eq. (17) is not a reliable formalism for obtaining the Gaussian curvature modulus from simulations. Given the ease of applicability of Eq. (17), it is of interest to evaluate results for the present systems.
To begin, Table 8 lists values of the neutral plane δ evaluated from the 4-parameter fit described in (Watson et al., 2011). Values of δ are mostly between 9.5 and 10 Å, and are not easily related to the dominant features of the pressure profile. In particular, the prominent minima in PT(z) evident in Fig 2 for DPPC, PSM, and SDPE are at |z| = 16.7, 17.3, and 18.4 Å, respectively (these locations are close to the average of the glycerol carbons). The adjacent maxima in PT(z) a smaller |z| are at 13.3, 13.6, and 15.3 Å for the three preceding bilayers. The remaining columns of Table 8 list (from Eq. (17)), , KG (from Eq. (18)), KG/KC for each system. The wide range of unphysically small values, including some with the wrong sign, lends further support to the argument that Eq. (17) may be flawed. Results for other estimates of the neutral plane (not shown) are similarly variable.
Table 8.
Location of the neutral planes δ with respect to the bilayer center, leaflet and bilayer Gaussian curvature moduli, and their ratios with the total curvature moduli.
| Lipid | δ (Å) | (10−20 J) |
|
KG (10−20 J) | KG/KC | |
|---|---|---|---|---|---|---|
| DPPC | 10.05 | −1.33 | −0.17 | −2.78 | −0.18 | |
| DMPC | 9.48 | −1.32 | −0.22 | −3.23 | −0.26 | |
| DOPC | 9.41 | −0.84 | −0.14 | −0.08 | −0.01 | |
| DNPC | 12.66 | −1.38 | −0.13 | −0.97 | −0.04 | |
| DOPE | 9.97 | 0.24 | 0.04 | 6.80 | 0.58 | |
| POPC | 9.64 | −1.09 | −0.17 | −1.38 | −0.10 | |
| PDPC | 9.34 | −0.64 | −0.13 | 0.17 | 0.02 | |
| POPE | 10.12 | 0.06 | 0.01 | 5.85 | 0.44 | |
| PDPE | 9.72 | 0.42 | 0.09 | 5.90 | 0.60 | |
| SDPE | 10.31 | 0.52 | 0.10 | 6.44 | 0.61 | |
| PSM | 9.75 | −4.10 | −0.30 | −13.24 | −0.48 | |
| POPG | 9.57 | −1.43 | −0.25 | −2.76 | −0.24 |
3.3 Spontaneous curvature
3.3.1 Simulation
As specified by Eq. (13), the first moment of the symmetrized pressure tensor is integrated from the bilayer midplane to the upper edge of the simulation cell to obtain the product . Fig. 12 plots this integral as a function of z to illustrate stable estimates for DPPC, PSM, and SDPE. SDPE was added for this figure (and Fig. 2) to include a result for a lipid with substantial negative curvature; i.e., the difference in sign of of PSM and SDPE is clear to see.
Fig 12.
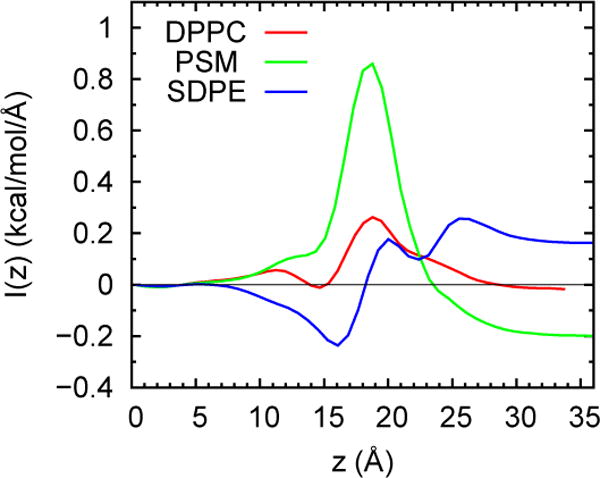
vs z for DPPC, PSM, and SDPE. Value at large z equals for each lipid. The bilayer center is at z = 0.
Table 9 lists and R0 for the small systems. As noted in the Introduction, this Table revises previous estimates for DOPE and DOPC (Sodt and Pastor, 2013), and for PSM (Venable et al., 2014) to reflect the use of KC calculated explicitly from the bilayer simulations (i.e., from Table 5). Spontaneous curvatures for DPPC and POPG are both near zero, though with opposite sign. R0 for the assorted PE’s range from −37 to −47 Å, with standard errors of approximately 1 Å. As expected, R0 increases when the PE head group is substituted for PC: DOPC/DOPE = 3.7, POPC/POPE = 6.7, and PDPC/PDPE = 3.2. PSM shows significant positive curvature, likely resulting from the usual hydrogen bond network.
Table 9.
Spontaneous radius of curvature, R0, calculated from the derivative of the bilayer free energy of bending,
| Lipid | (kcal/mol/Å) | R0 (Å) | R0 Range (Å) | Expt |
|---|---|---|---|---|
| DPPC | −0.0046 (0.0020) | 2465 | 4362, 1718 | |
| DMPC | −0.0225 (0.0022) | 392 | 434, 357 | |
| DOPC | 0.0610 (0.0025) | −140 | −134, −146 | −87.3a |
| DNPC | 0.0511 (0.0053) | −310 | −281, −346 | |
| DOPE | 0.2281 (0.0036) | −37 | −37, −38 | −28.5a |
| POPC | 0.0303 (0.0029) | −315 | −287, −348 | |
| PDPC | 0.0565 (0.0026) | −123 | −118, −129 | |
| POPE | 0.2041 (0.0031) | −47 | −46, −48 | |
| PDPE | 0.1864 (0.0032) | −38 | −37, −39 | |
| SDPE | 0.1889 (0.0029) | −41 | −40, −41 | −27.5b |
| PSM | −0.1858 (0.0032) | 108 | 110, 106 | |
| POPG | 0.0036 (0.0024) | −2220 | −1339, −6485 |
(Teague et al., 2013)
3.3.2 Comparison with experiment
This subsection begins with a direct comparison of simulation and the values reported in the original experimental studies, and then to experimental values transformed from the pivotal to the neutral plane.
From Table 9, the spontaneous curvatures of DOPC, DOPE, and SDPE leaflets from simulation are 37%, 23% and 33% smaller than monolayers from experiment, respectively. Assuming that the simulations are accurate, this result implies that the “curvature frustration” in bilayers is lower than previously assumed. Recall that direct simulations of the HII phase (Sodt and Pastor, 2013) of DOPE yielded good agreement with experiment for the spontaneous curvature (an overestimate of 10%, and within the error of the methods), lending confidence to the present 30% underestimates.
The experimental monolayer bending constants extracted for the preceding HII phases are 3.8 × 10−20 J (DOPC), 4.7 × 10−20 J (DOPE), and 4.9 × 10−20 J (SDPE). After multiplying by 2 to plausibly compare with the simulated bilayer bending constants, the difference between simulation and experiment is appreciable: is higher than by 56% (DOPC), 26% (DOPE) and 9.3% (SDPE). The experimental bilayer bending constants for DOPC are also higher than twice (12% for PA, 9% for x-ray, and 43% for flicker). Hence, it appears that bilayers are stiffer than might be anticipated from HII phase monolayers.
A more rigorous comparison of simulation and experiment entails transforming the experimental values to the neutral plane. The relationships for monolayers are (Leikin et al., 1996):
| (19) |
| (20) |
It is clear from Eqs. (19) and (20) that the corrections lead to an increase in c0 (decrease in R0), and a decrease in KC. This exacerbates the differences between the simulation results for bilayers and experimental results for monolayers. Marsh (Marsh, 2006) estimates that the corrections for DOPE are 5% for c0 and 17% for . Corrections are smaller for DOPC, as expected because c0 is smaller: they are 0% for c0 and 3% for . Hence, the conclusions from the direct comparison are unchanged.
3.3.3 The question of shape
It is useful at this point to reconsider the notion that the spontaneous curvature of a lipid in a bilayer can be intuited by looking at its shape. To this end, Fig. 13 presents the average shapes of the lipids studied here (see Figure caption for how these were generated). It is evident that all of the lipids are basically cylindrical, as would be expected from objects filling a flat bilayer. However, it would be difficult to rationalize that the spontaneous curvatures of POPG and DPPC are both near zero (though opposite in sign), yet the head group of POPG appears to be the smallest of all of the lipids shown. This is because the head group of POPG is charged, and the repulsive interactions in the water/bilayer interface counteract the seemingly wide “cone-shaped” excursions of the chains. The influence of charge on spontaneous curvature in the HII phase has already been recognized (Fuller et al., 2003), so this result extends the conclusion to the lamellar phase. To continue, DNPC (the largest tail of the PC lipids here) might be expected to have the largest negative curvature. However, it is closest to POPC, and less than half of DOPC. It would be difficult to guess that PSM has substantial positive curvature; considerations of inter-lipid hydrogen bonding and the position of the neutral plane are required to understand this (Venable et al., 2014). Hydrogen bonding among head groups is also an important contributor to the spontaneous curvature of DOPE (Sodt and Pastor, 2014).
Fig 13.
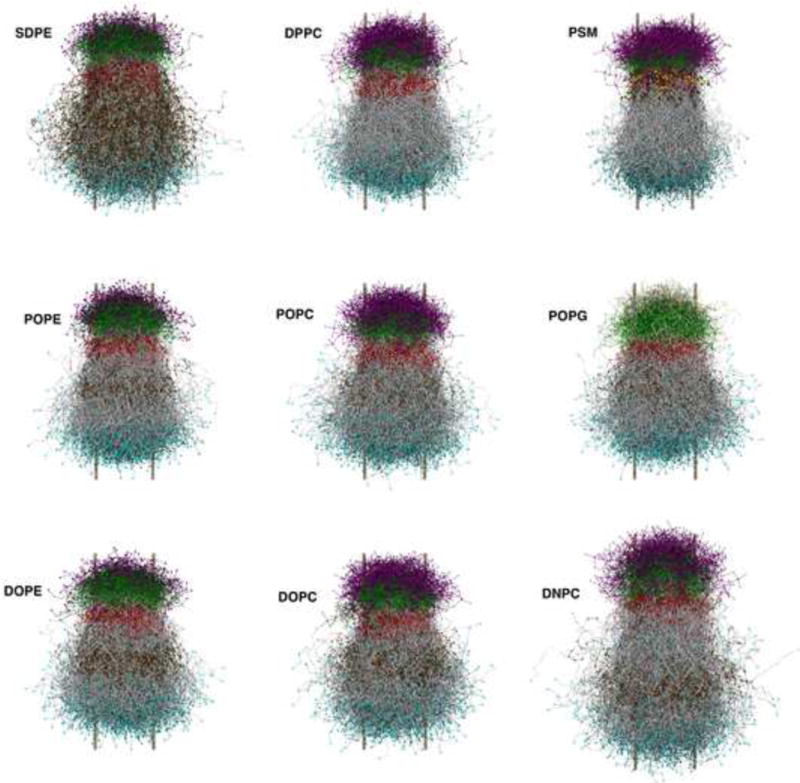
The average shapes of lipids in bilayers. The shapes were generated from the final coordinates of large systems by translating each lipid to place its center of geometry at the origin, with lipids in the bottom leaflet rotated by 180°. Coloring is as follows: nitrogen on head groups (purple); phosphates (green); oxygen (red); methylene groups of lipid chains (grey); terminal methyl groups of acyl chains (cyan); amide oxygen on PSM (yellow). The two metal colored bars on the side of each lipid array specify the diameter of the cylinder whose area equals the average area/lipid.
4. Summary and Conclusions
While exclusively focused on the CHARMM 36 force field, the results presented here provide a snapshot of the broader state of molecular dynamics simulations of lipid bilayers. Simulations with C36 and most of the other FF now yield good to excellent agreement with experimental surface areas Aℓ and deuterium order parameters for most lipids. Caution must be applied regarding the Lennard-Jones cutoffs (Table 4), especially for bilayers near their fluid to gel phase transitions. Nevertheless, when simulation and experiment do not agree, it is not overly brash to question the experiment.
Since Aℓ was a specific target for about half of the lipids presented here (Table 1), agreement with experiment (Table 2) is to be expected, though gratifying. However, area compressibility KA, bending modulus KC, and spontaneous curvature c0 were not targets, so their agreement or disagreement with experiment is more interesting.
KA are in quite good agreement with experiment (Table 2), which was not the case with precursors to C36 (Section 3.1.2). Area compressibilites are easy to calculate (though simulations should be at least several hundred ns), and should be reported with new or revised parameter sets.
The peaceful coexistence of simulation and experiment ends when comparing bending constants. Results from simulation are close to those from flicker experiments, and these are substantially larger (a factor of more than two for DPPC and DMPC) than those from pipette aspiration, x-ray, and neutron spin echo (Table 5). A clue to these discrepancies may lie in the analysis of the polymer brush model (Table 7 and Fig. 11). The unsaturated lipids DPPC, DMPC, and PSM show a qualitatively different dependence on membrane thickness, implying that assumptions reasonable for unsaturated lipids might not extend to this set.
Tilt and twist moduli are included for the entire set of lipids for completeness (Table 6) Agreement with experiment of Kθ is excellent for DOPE, and 30% smaller the experiment for DOPC. Perhaps these Kθ could provide starting values for fitting experimental data to models for extracting KC.
Gaussian curvatures for bilayers obtained from the second moment of the pressure profile appear to be unreliable (Table 8), in agreement with previous analysis using coarse grained models.
Spontaneous curvatures calculated from simulation (Table 9) mostly follow expected trends: lipids with PE head groups have substantial negative c0, those with PC head groups have much lower c0. However, not all are easy to predict from simple considerations of shape (Figure 13), and a detailed analysis of molecular interactions is advised. A direct comparison of simulation and experiment for DOPE, DOPC, and SDPE indicates that c0 from simulation are approximately 30% smaller than those from experiment. Monolayer bending constants from simulation are larger than those from experiment, assuming the simulated bilayer constants can be divided by 2 to correctly compare with experiment. These results should be considered when assuming the correspondence of bilayers and monolayers in the HII phase.
Even if some spontaneous curvatures or bending constants from the present set of simulations with C36 are quantitatively incorrect, this compilation should provide a valuable starting place for a systematic improvement of molecular mechanics force fields for lipids. The comparison of the simulated deuterium order parameter plateau values and surface areas (Section 3.1.3) can be used to refine models predicting surface areas from experimental SCD.
This study has been restricted to single-component bilayers, partially because there is little quantitative experimental data available related to the mechanical properties of composite systems, and partially because the underlying theoretical formalisms used in analyzing the simulations assume (often implicitly) a homogeneous membrane structure devoid of any modulation in elastic properties. In the case of multi-component membranes there exists, at least in principle, a coupling between local membrane composition and elastic behavior that is expected to complicate both the interpretation of simulation data and the comparison between small simulation boxes and the larger systems studied experimentally. For example, a lipid bilayer populated with a collection of non-interacting membrane proteins is expected to behave as a Helfrich sheet (obeying Eq. 5) over sufficiently long wavelengths. The effective bending moduli associated with such a composite system reflect both the elastic properties of the lipid bilayer and of the proteins embedded within it (Netz and Pincus, 1995), however it is infeasible to run detailed simulations on a composite lipid/protein system that are large enough to directly observe this long-wavelength behavior. Further, the known approximations one might hope to use to extrapolate long-wavelength behavior in the composite system from simpler and smaller simulations are valid only in certain limiting regimes, even for vastly simplified toy models (Sigurdsson et al., 2013). In the case of lipid mixtures, the coupling between composition and curvature has recently been implicated as a possible driving force for the formation of lipid rafts/microdomains (Giang et al., 2015). Even in ostensibly “homogeneous” lipid mixtures it seems unlikely that lipids are perfectly ideally mixed and the extent of non-ideality should, in general, be coupled to the local shape and density of the membrane. Such complications are not considered in the theoretical models underlying the analyses presented in this work, but will be pursued in future studies.
Highlights.
Mechanical properties of 12 different bilayers are evaluated from simulation.
The CHARMM 36 force field yields excellent area and compressibility.
The relation of surface area and NMR order parameters does not fit simple models.
Bending constants equal those from flicker experiments, but not others.
Spontaneous curvatures of leaflets in bilayers are 30% lower than in hexagonal phases.
Acknowledgments
We thank Alexander Sodt and Markus Deserno for helpful conversations during the preparation of this manuscript, Max Watson for critical technical advice on obtaining neutral surfaces, and we acknowledge our codevelopers of the CHARMM lipid force field, particularly Jeffery Klauda, Alexander MacKerell, Jr., and Douglas Tobias. The generous advice over many years from Michael Brown, Evan Evans, Klaus Gawrisch, Stuart McLaughlin, John Nagle, Adrian Parsegian, and Steven White regarding experimental data has been invaluable to our efforts. This research was supported by the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute, and by the National Science Foundation (Grant Nos. CHE-1153096 and CHE-1465162), and used the high-performance computational capabilities at the National Institutes of Health, Bethesda, MD (NHLBI LoBoS and Biowulf clusters).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen MP, Tildesley DJ. Computer Simulations of Liquids. Clarendon Press; Oxford: 1987. [Google Scholar]
- Boscia AL, Treece BW, Mohammadyani D, Klein-Seetharaman J, Braun AR, Wassenaar TA, Kloesgen B, Tristram-Nagle S. X-ray structure, thermodynamics, elastic properties and MD simulations of cardiolipin/dimyristoylphosphatidylcholine mixed membranes. Chemistry and Physics of Lipids. 2014;178:1–10. doi: 10.1016/j.chemphyslip.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun AR, Brandt EG, Edholm O, Nagle JF, Sachs JN. Determination of Electron Density Profiles and Area from Simulations of Undulating Membranes. Biophysical Journal. 2011;100:2112–2120. doi: 10.1016/j.bpj.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. Journal of Computational Chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MF. Curvature Forces in Membrane Lipid-Protein Interactions. Biochemistry. 2012;51:9782–9795. doi: 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Rand RP. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophysical Journal. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary J, Harder E, Lopes PEM, Huang L, MacKerell AD, Jr, Roux B. A Polarizable Force Field of Dipalmitoylphosphatidylcholine Based on the Classical Drude Model for Molecular Dynamics Simulations of Lipids. Journal of Physical Chemistry B. 2013;117:9142–9160. doi: 10.1021/jp402860e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno M. Fluid lipid membranes: From differential geometry to curvature stresses. Chemistry and Physics of Lipids. 2015;185:11–45. doi: 10.1016/j.chemphyslip.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Evans E, Rawicz W. Entropy-Driven Tension and Bending Elasticity in Condensed-Fluid Membranes. Physical Review Letters. 1990;64:2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- Evans E, Rawicz W, Smith BA. Back to the future: mechanics and thermodynamics of lipid biomembranes. Faraday Discussions. 2013;161:591–611. doi: 10.1039/c2fd20127e. [DOI] [PubMed] [Google Scholar]
- Feller SE, Pastor RW. Constant surface tension simulations of lipid bilayers: The sensitivity of surface areas and compressibilities. Journal of Chemical Physics. 1999;111:1281–1287. [Google Scholar]
- Feller SE, Venable RM, Pastor RW. Computer simulation of a DPPC phospholipid bilayer: Structural changes as a function of molecular surface area. Langmuir. 1997;13:6555–6561. [Google Scholar]
- Feller SE, Zhang YH, Pastor RW. Computer-Simulation of Liquid/Liquid Interfaces. II. Surface-Tension Area Dependence of a Bilayer and Monolayer. Journal of Chemical Physics. 1995;103:10267–10276. [Google Scholar]
- Fuller N, Benatti CR, Rand RP. Curvature and bending constants for phosphatidylserine-containing membranes. Biophysical Journal. 2003;85:1667–1674. doi: 10.1016/s0006-3495(03)74596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giang H, Shlomovitz R, Schick M. Microemulsions, modulated phases and macroscopic phase separation: a unified picture of rafts. Essays Biochem. 2015;57:21. doi: 10.1042/bse0570021. [DOI] [PubMed] [Google Scholar]
- Goetz R, Gompper G, Lipowsky R. Mobility and elasticity of self-assembled membranes. Physical Review Letters. 1999;82:221–224. [Google Scholar]
- Gruner SM, Parsegian VA, Rand RP. Directly measured deformation energy of phospholipid H-II hexagonal phases. Faraday Discussions. 1986;81:29–37. doi: 10.1039/dc9868100029. [DOI] [PubMed] [Google Scholar]
- Hamm M, Kozlov MM. Elastic energy of tilt and bending of fluid membranes. Eur Phys J E. 2000;3:323–335. [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers – theory and possible experiments. Zeitschrift Fur Naturforschung C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Hu M, Briguglio JJ, Deserno M. Determining the Gaussian Curvature Modulus of Lipid Membranes in Simulations. Biophysical Journal. 2012;102:1403–1410. doi: 10.1016/j.bpj.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, de Jong DH, Marrink SJ, Deserno M. Gaussian curvature elasticity determined from global shape transformations and local stress distributions: a comparative study using the MARTINI model. Faraday Discussions. 2013;161:365–382. doi: 10.1039/c2fd20087b. [DOI] [PubMed] [Google Scholar]
- Huang J, Lopes PEM, Roux B, MacKerell AD., Jr Recent Advances in Polarizable Force Fields for Macromolecules: Microsecond Simulations of Proteins Using the Classical Drude Oscillator Model. Journal of Physical Chemistry Letters. 2014;5:3144–3150. doi: 10.1021/jz501315h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynninen AP, Crowley MF. New Faster CHARMM Molecular Dynamics Engine. Journal of Computational Chemistry. 2014;35:406–413. doi: 10.1002/jcc.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelachvili J. Intermolecular and Surface Forces. 2nd. Academic Press; San Diego, CA: 1992. [Google Scholar]
- Jablin MS, Akabori K, Nagle JF. Experimental Support for Tilt-Dependent Theory of Biomembrane Mechanics. Physical Review Letters. 2014;113 doi: 10.1103/PhysRevLett.113.248102. [DOI] [PubMed] [Google Scholar]
- Jahnig F. What is the surface pension of a lipid bilayer membrane? Biophys J. 1996;71:1348–1349. doi: 10.1016/S0006-3495(96)79336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. Software news and updates – CHARNIM-GUI: A web-based grraphical user interface for CHARMM. Journal of Computational Chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Kamien RD. The geometry of soft materials: a primer. Reviews of Modern Physics. 2002;74:953–971. [Google Scholar]
- Khelashvili G, Kollmitzer B, Heftberger P, Pabst G, Harries D. Calculating the Bending Modulus for Multicomponent Lipid Membranes in Different Thermodynamic Phases. Journal of Chemical Theory and Computation. 2013;9:3866–3871. doi: 10.1021/ct400492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Brooks BR, MacKerell AD, Venable RM, Pastor RW. An ab initio study on the torsional surface of alkanes and its effect on molecular simulations of alkanes and a DPPC bilayer. Journal of Physical Chemistry B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- Klauda JB, Kučerka N, Brooks BR, Pastor RW, Nagle JF. Simulation-based Methods for Interpreting X-ray Data from Lipid Bilayers. Biophys J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Monje V, Kim T, Im W. Improving the CHARMM Force Field for Polyunsaturated Fatty Acid Chains. Journal of Physical Chemistry B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. Journal of Physical Chemistry B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, MacKerell AD, Jr, Pastor RW. Considerations for Lipid Force Field Development. In: Feller SE, editor. Current Topics in Membranes: Computer Modeling of Membrane Bilayers. 2008. pp. 1–48. [Google Scholar]
- Koenig BW, Strey HH, Gawrisch K. Membrane lateral compressibility determined by NMR and X-ray diffraction: Effect of acyl chain polyunsaturation. Biophysical Journal. 1997;73:1954–1966. doi: 10.1016/S0006-3495(97)78226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophysical Journal. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucerka N, Nieh MP, Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochimica Et Biophysica Acta-Biomembranes. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Leftin A, Brown MF. An NMR database for simulations of membrane dynamics. Biochimica Et Biophysica Acta-Biomembranes. 2011;1808:818–839. doi: 10.1016/j.bbamem.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikin S, Kozlov MM, Fuller NL, Rand RP. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes. Biophysical Journal. 1996;71:2623–2632. doi: 10.1016/S0006-3495(96)79454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ZA, Venable RM, Watson MC, Lerner MG, Shea JE, Pastor RW, Brown FLH. Determination of Biomembrane Bending Moduli in Fully Atomistic Simulations. Journal of the American Chemical Society. 2014;136:13582–13585. doi: 10.1021/ja507910r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JB, Klauda JB. Lipid chain branching at the iso- and anteiso-positions in complex chlamydia membranes: A molecular dynamics study. Biochimica Et Biophysica Acta-Biomembranes. 2011;1808:323–331. doi: 10.1016/j.bbamem.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Lim JB, Rogaski B, Klauda JB. Update of the Cholesterol Force Field Parameters in CHARMM. Journal of Physical Chemistry B. 2012;116:203–210. doi: 10.1021/jp207925m. [DOI] [PubMed] [Google Scholar]
- Marsh D. Elastic curvature constants of lipid monolayers and bilayers. Chemistry and Physics of Lipids. 2006;144:146–159. doi: 10.1016/j.chemphyslip.2006.08.004. [DOI] [PubMed] [Google Scholar]
- May ER, Narang A, Kopelevich DI. Role of molecular tilt in thermal fluctuations of lipid membranes. Physical Review E. 2007;76 doi: 10.1103/PhysRevE.76.021913. [DOI] [PubMed] [Google Scholar]
- Nagle JF. Area/Lipid of Bilayers from NMR. Biophysical Journal. 1993;64:1476–1481. doi: 10.1016/S0006-3495(93)81514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF. Introductory Lecture: Basic quantities in model biomembranes. Faraday Discussions. 2013;161:11–29. doi: 10.1039/c2fd20121f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF, Jablin MS, Tristram-Nagle S, Akabori K. What are the true values of the bending modulus of simple lipid bilayers? Chemistry and Physics of Lipids. 2015;185:3–10. doi: 10.1016/j.chemphyslip.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochimica et Biophysica Acta-Reviews on Biomembranes. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz RR, Pincus P. Inhomogeneous Fluid Membranes – Segragation, Ordering, and Effective Rigidity. Physical Review E. 1995;52:4114–4128. doi: 10.1103/physreve.52.4114. [DOI] [PubMed] [Google Scholar]
- Pandit KR, Klauda JB. Membrane models of E-coli containing cyclic moieties in the aliphatic lipid chain. Biochimica Et Biophysica Acta-Biomembranes. 2012;1818:1205–1210. doi: 10.1016/j.bbamem.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Pastor RW. Molecular Dynamics and Monte Carlo Simulations of Lipid Bilayers. Current Opinion in Structural Biology. 1994;4:486–492. doi: 10.1016/s0959-440x(97)80115-7. [DOI] [PubMed] [Google Scholar]
- Pastor RW, Brooks BR, Szabo A. An Analysis of the Accuracy of Langevin and Molecular-Dynamics Algorithms. Molecular Physics. 1988a;65:1409–1419. [Google Scholar]
- Pastor RW, MacKerell AD., Jr Development of the CHARMM Force Field for Lipids. J Phys Chem Letters. 2011;2:1526–1532. doi: 10.1021/jz200167q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor RW, Venable RM, Karplus M. Model for the Structure of the Lipid Bilayer. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:892–896. doi: 10.1073/pnas.88.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor RW, Venable RM, Karplus M, Szabo A. A Simulation Based Model of NMR T1 Relaxation in Lipid Bilayer Vesicles. Journal of Chemical Physics. 1988b;89:1128–1140. [Google Scholar]
- Petrache HI, Dodd SW, Brown MF. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by H-2 NMR spectroscopy. Biophysical Journal. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophysical Journal. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran SA. Statistical Thermodynamics of Surfaces, Interfaces and Membranes. Westview Press; Boulder, CO: 1994. [Google Scholar]
- Seelig J. Deuterium Magnetic-Resonance – Theory and Application to Lipid-Membranes. Quarterly Reviews of Biophysics. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, Ho CR, Ierardi DJ, Kolossvary I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan Y, Spengler J, Theobald M, Towles B, Wang SC. Anton, a Special-Purpose Machine for Molecular Dynamics Simulation. Comm of the ACM. 2008;51:91–97. [Google Scholar]
- Sigurdsson JK, Brown FLH, Atzberger PJ. Hybrid continuum-particle method for fluctuating lipid bilayer membranes with diffusing protein inclusions. Journal of Computational Physics. 2013;252:65–85. [Google Scholar]
- Sodt AJ, Pastor RW. The tension of a curved surface from simulation. Journal of Chemical Physics. 2012;137 doi: 10.1063/1.4769880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodt AJ, Pastor RW. Bending free energy from simulation: correspondence of planar and inverse hexagonal lipid phases. Biophysical Journal. 2013;104:2202–2211. doi: 10.1016/j.bpj.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodt AJ, Pastor RW. Molecular Modeling of Lipid Membrane Curvature Induction by a Peptide: More than Simply Shape. Biophysical Journal. 2014;106:1958–1969. doi: 10.1016/j.bpj.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonne J, Hansen FY, Peters GH. Methodological problems in pressure profile calculations for lipid bilayers. J Chem Phys. 2005;122 doi: 10.1063/1.1862624. [DOI] [PubMed] [Google Scholar]
- Venable RM, Chen LE, Pastor RW. Comparison of the Extended Isotropic Periodic Sum and Particle Mesh Ewald Methods for Simulations of Lipid Bilayers and Monolayers. Journal of Physical Chemistry B. 2009;113:5855–5862. doi: 10.1021/jp900843x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable RM, Hatcher E, Guvench O, MacKerell AD, Pastor RW. Comparing Simulated and Experimental Translation and Rotation Constants: Range of Validity for Viscosity Scaling. Journal of Physical Chemistry B. 2010;114:12501–12507. doi: 10.1021/jp105549s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable RM, Luo Y, Gawrisch K, Roux B, Pastor RW. Simulations of Anionic Lipid Membranes: Development of Interaction-Specific Ion Parameters and Validation Using NMR Data. Journal of Physical Chemistry B. 2013;117:10183–10192. doi: 10.1021/jp401512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable RM, Sodt AJ, Rogaski B, Rui H, Hatcher E, MacKerell AD, Jr, Pastor RW, Klauda JB. CHARMM All-Atom Additive Force Field for Sphingomyelin: Elucidation of Hydrogen Bonding and of Positive Curvature. Biophysical Journal. 2014;107:134–145. doi: 10.1016/j.bpj.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva DY, Lim JB, Klauda JB. Influence of Ester-Modified Lipids on Bilayer Structure. Langmuir. 2013;29:14196–14203. doi: 10.1021/la403919h. [DOI] [PubMed] [Google Scholar]
- Watson MC, Brandt EG, Welch PM, Brown FLH. Determining Biomembrane Bending Rigidities from Simulations of Modest Size. Physical Review Letters. 2012;109 doi: 10.1103/PhysRevLett.109.028102. [DOI] [PubMed] [Google Scholar]
- Watson MC, Penev ES, Welch PM, Brown FLH. Thermal fluctuations in shape, thickness, and molecular orientation in lipid bilayers. Journal of Chemical Physics. 2011;135 doi: 10.1063/1.3660673. [DOI] [PubMed] [Google Scholar]
- Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, Im W. CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. Journal of Computational Chemistry. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Feller SE, Brooks BR, Pastor RW. Computer-Simulation of Liquid/Liquid Interfaces. I. Theory and Application to Octane/Water. J Chem Phys. 1995;103:10252–10266. [Google Scholar]
- Zhuang X, Makover JR, Im W, Klauda JB. A systematic molecular dynamics simulation study of temperature dependent bilayer structural properties. Biochimica Et Biophysica Acta-Biomembranes. 2014;1838:2520–2529. doi: 10.1016/j.bbamem.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nature Reviews Molecular Cell Biology. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]


