Abstract
The unambiguous imaging of transplanted cells remains a major challenge to understand their biological function and therapeutic efficacy. In vivo imaging of implanted cells is reliant on tagging these to differentiate them from host tissue, such as the brain. We here characterize a gold nanoparticle conjugate that is functionalized with modified deoxythymidine oligonucleotides bearing Gd(III) chelates and a red fluorescent Cy3 moiety to visualize in vivo transplanted human neural stem cells. This DNA-Gd@Au nanoparticle (DNA-Gd@AuNP) exhibits an improved T1 relaxivity and excellent cell uptake. No significant effects of cell uptake have been found on essential cell functions. Although T1 relaxivity is attenuated within cells, it is sufficiently preserved to afford the in vivo detection of transplanted cells using an optimized voxel size. In vivo MR images were corroborated by a post-mortem histological verification of DNA-Gd@AuNPs in transplanted cells. With 70% of cells being correctly identified using the DNA-Gd-AuNPs indicates an overall reliable detection. Less than 1% of cells were false positive for DNA-Gd@AuNPs, but a significant number 30% of false negatives reveals a dramatic underestimation of transplanted cells using this approach. DNA-Gd@AuNPs therefore offer new opportunities to visualize transplanted cells unequivocally using T1 contrast and use cellular MRI as a tool to derive biologically relevant information that allows us to understand how the survival and location of implanted cells determines therapeutic efficacy.
Keywords: MRI, Gadolinium, nanoparticles, gold, contrast agent, neural stem cells, cell transplantation, Gd-HPDO3A
Graphical Abstract
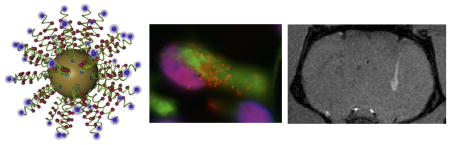
1. Introduction
The regional distribution of transplanted neural stem cells (NSCs) influences their sphere of activity and correlates with the degree of therapeutic efficacy [1]. A greater understanding of the positioning of transplanted NSCs could hence improve our targeting of cell injections to areas crucial to their efficacy. However, mapping the in vivo distribution of transplanted cells remains a major challenge [2, 3]. In the context of intracerebral transplants, a range of 1,000–400,000 cells needs to be detected at a high in vivo spatial resolution (<64 nL voxel) using an unequivocal multi-voxel signal that predominantly derives from transplanted cells with a low risk of false positives (<5%, type I error) and negatives (<20%, type II error) [2, 4]. The selective visualization of transplanted cells by magnetic resonance imaging, nevertheless, requires contrast-inducing particles [3].
Iron oxide (magnetite/maghemite) particles yield a high relaxivity that affords single cell tracking [5]. However, potential blooming artifacts due to air bubbles and small hemorrhage on T2- and T2*-weighted magnetic resonance images (MRI), in addition to a potential for nanotoxicity in neurons [6], complicates an unequivocal interpretation of in vivo cell distribution in the brain [2]. An unequivocal signal can potentially be produced by T1 agents, such as Mn(II) and Gd(III). Mn(II) agents are easily taken-up into cells by substituting for Ca2+ ions. Although this affords the T1 detection of labeled cells [7–10], unchelated manganese which is required for cellular uptake is known to exert cytotoxic effects [11].
Cellular labeling with monomeric gadolinium agents taken-up through endocytosis typically quenches the T1 contrast enhancement due to endosomal sequestration, but electroporation of the agent into the cytoplasm preserves the T1 signal [12]. Several reports indicate the possibility to visualize cells using MRI using this approach [13–18]. However, positive identification of a T1 signal in vivo requires evidence that the agent is localized intracellularly in the transplanted cells (as assessed by an independent marker). This is necessary to prevent false identification of contrast agent inadvertently injected, bound to the outside of the cell membrane or exocytosed from the transplanted cells.
The thermodynamic and kinetic stability of chelated Gd(III) compounds is also essential to avoid cytotoxicity, which can be delayed or avoided if Gd(III) can be contained within the chelate. Macrocylic ligands based on 1,4,7,10-tetraazacyclododecane-1,4,7,10-teraacetic acid (DOTA) exhibit similar thermodynamic stabilities compared to those of the linear diethylene triamine (DTPA) ligand [19], but are more kinetically stable, and are thus a more favorable chelate for Gd(III)-based agents [20, 21]. Immobilization of Gd(III) complexes onto macromolecules or proteins that restrict the motion of Gd(III) chelates can improve the relaxivity compared to monomeric Gd(III)-agents [22]. Intracellular concentration (and hence cellular relaxivity) could be further improved by using this strategy [23].
Achieving a high cellular relaxivity requires an optimized nanoconjugate Gd(III) contrast agent with a high thermostability that affords an efficient cell uptake. In recent work by Song et al [24], Gd(III) labeled DNA gold nanoparticle conjugates (DNA-Gd@AuNP) were shown to be a biocompatible and highly effective platform for cell labeling of HeLa and NIH 3T3 cancer cells [25]. Importantly, the DNA-Gd@AuNP showed an excellent pH stability and inertness towards transmetallation [26], with attached oligonucleotides affording an efficient cell uptake through the CXCR7 scavenger receptor and peri-nuclear localization [27]. Especially in serum-free cultures (as used for human neural stem cells), these oligonucleotides are efficient trans-membrane localization tools. There is further evidence that AuNP in the proximity of Gd(III)-agents serve as “nanoamplifiers” to increase MR relaxivity [28].
Multimodal Gd(III)-conjugated AuNP have been used to visualize pancreatic islets [29] and for targeting tumors [30–32] using MRI, but also show potential as theranostic agents for photothermal therapy to ablate cancers [33]. We here characterize the DNA-Gd@AuNPs to visualize the distribution of human neural stem cells using MRI by performing in vitro assays to determine cell uptake, potential cytotoxicity and cellular relaxivity. Optimal imaging parameters are defined in vitro and applied in vivo to visualize transplanted cells by T1-weighted MRI with validation using fluorescent immunohistochemistry.
2. Methods
2.1. Synthesis and quality assurance of DNA-Gd@AuNP nanoparticles
Gold nanoparticle synthesis
Citrate stabilized AuNPs were synthesized by reduction of HAuCl4 by sodium citrate in aqueous media. Specifically, in an acid washed two necked round bottom flask 0.197 g of gold (III) chloride was added to 0.496 L of water while stirring. The solution was brought to reflux and 0.574 g of trisodium citrate dihydrade pre-dissolved in 0.004 L of water was added. The solution is left to reflux for 30 min, cooled to room temperature, and filtered [24, 34].
Gd(III) labeled oligonucleotide synthesis
Oligonucleotides were synthesized on solid phase CPGs by standard techniques on a MerMade automated synthesizer. All oligonucleotide synthesis reagents and protected 3′-Thiol modifier CPGs were purchased from Glen Research (Sterling, Va). Specifically, the synthesized oligonucleotide consisted of the sequence 3′ – S-S-TTT-TTT-TTT-T*TT-T*TT-T*TT-T*TT-T*TT–Cy3 5′, where C6 amino modifier dT modified bases that are indicated by T*. Cy3 is added as the final base by the use of a Cy3 phosphoramidite coupled under standard conditions. Oligonucelotides were deprotected from the solid phase using AMA conditions [(1:1 methylamine:ammonium hydroxide (sat.)] for one hour. Deprotected DNA was purified from failed sequences by reversed phase high pressure liquid chromatography (HPLC) using a C18 column with 30 mM triethylammonium acetate buffer and a linear gradient of acetonitrile on a Varian ProStar 500 instrument, and eluted product is lyophilized and characterized by matrix assisted laser desorption ionization mass spectrometry (MS-MALDI) using a Bruker AutoFlex III spectrometer. Successful DNA synthesis was confirmed, (m/z) observed: 8762.1, calculated: 8760.7 [M − H]−. Azide functionality was subsequently added by the solution phase coupling of azidobutyrate NHS ester (Glen Research) in the presence of aqueous carbonate. Complete conversion of pendant amines to azides was again confirmed by MALDI, and 5x azide DNA was purified by HPLC and lyophilized, (m/z) observed: 9318.2, calculated: 9326.3 [M − H]−[34].
The inorganic Gd(III) complex was synthesised using standard organic chemistry techniques using a previously reported procedure [34]. Briefly, a primary alkyne bearing linker arm was designed to bear an α,β-unsaturated ketone. This small molecule was refluxed in acetonitrile and potassium carbonate for four days to complete the 1,4-conjugate addition to the tert-butyl protected DO3A macrocyclic ligand. After conjugation, the chelate is deprotected in triflouroacetic acid, metalated and purified by HPLC. Lyophilized product was characterized by electrospray ionization mass spectrometry (ESI-MS) on a Varian 1200L single-quadrupole mass spectrometer, (m/z) observed: 609.2, calculated: 610.7 [M + H]+.
Conjugation of the Gd(III) to the azide modified DNA was performed using Cu(I)-catalyzed 1,3 dipolar cycloaddition (click) chemistry (Figure 1A). Specifically, azide modified DNA and alkyne-bearing Gd(III) complex was dissolved in 1.5 M triethylammonium acetate buffer, to which was added Cu(II) Sulfate, tris-hydroxypropyl triazolylamine (used for stabilization of the Cu(I) oxidation state, and to act as a sacrificial reagent against oxidative cleavage of the DNA backbone), and sodium ascorbate. Upon complete reaction product, DNA was purified by HPLC as prior and final characterization was performed using MALDI (m/z) observed: 12374.3, calculated: 12372.3 [M − H]−
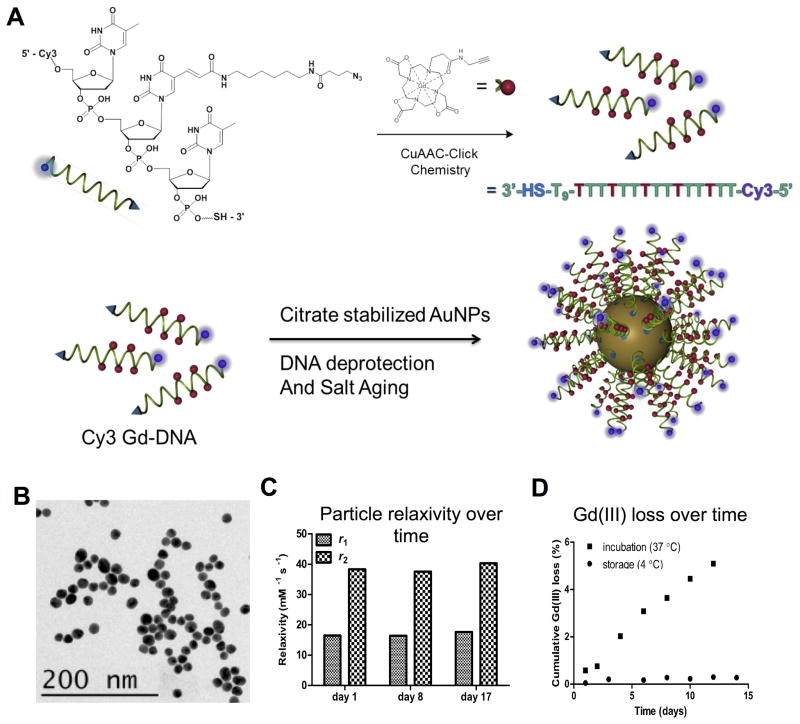
Figure 1. DNA-Gd@AuNP synthesis and stability.
Particles consist of a gold nanoparticle core loaded with DNA to which the Gd-HPDO3A and Cy3 moieties are attached (A). Transmission electron microscopy (TEM) of conjugated DNA-Gd@Au nanoparticles for size determination (B). Particles retain their relaxivity properties at 1.41 T over more than 2 weeks (C). There is a small amount of Gd(III) loss at 37 °C, amounting to <6% of total Gd(III) over 2 weeks. However, loss is negligible (<0.4%) when particles are stored at 4 °C (D).
DNA-Gd@AuNP synthesis
Particle conjugation was carried out by deprotection of the 3′ thiol protecting group by chemical exchange using dithiothreitol, and purified from this mixture using G25 sephadex (NAP 5, GE Life Sciences). Purified DNA was mixed with citrate capped gold nanoparticles in a ratio of 1 OD DNA/mL AuNP, followed by salt-aging (Figure 1A), a technique which adds successive increases in salt conditions. The salt concentration was incrementally increased up to 600 mM using five aliquots of 5M NaCl over six hours, at which time the particles were left to stir for 48 hours. Upon completion of this process, particles were centrifuged three successive times to purify excess Gd-DNA and concentrate the particles for subsequent studies. Upon completion of this process, particles were centrifuged three successive times to purify excess Gd-DNA and concentrate the particles for subsequent studies. Particle loading was assessed by inductively coupled plasma-mass spectroscopy (ICP-MS) analysis of fully synthesized particles by observation of the ratio of Gd(III) to Au(III) ratio This was done using either a computer-controlled (Plasmalab software) Thermo (Thermo Fisher Scientific) PQ ExCell ICP-MS equipped with a CETAC 500 autosampler or a computer-controlled (Plasmalab software) Thermo X series II ICP-MS equipped with an ESI (Omaha) SC-2 autosampler. Each sample was acquired using 1 survey run (10 sweeps) and 3 main (peak jumping) runs (100 sweeps). The isotopes selected were 197Au and 156,157Gd. 115In, 165Ho, and 209Bi served as internal standards for data interpolation and instrument stability.
Dynamic light scattering (DLS)
DLS was performed using a Malvern Zetasizer using the number average analysis of multiple consecutive measurements. Specifically, data was collected using an Au refractive index of 0.470, absorbance of 0.100, and a viscosity of 0.8872. All measurements were performed in H2O at 25 °C. Using this instrument, measurements acquired are concentration independent. Herein we report the number of average measurements of particle size, because intensity measurements showed a slightly larger hydrodynamic size and broader distribution (skewed non-linearly by increased scattering of a few larger particles, whose presence were not even observed during TEM analysis). Each analysis consisted of three separate measurements, each of which utilized 16 individual samplings in order to estimate hydrodynamic radius and standard deviation. Mark-Houwink parameters were used in the mathematical model for determination of size and standard deviation. These measurements were primarily used to confirm DNA conjugation and potential aggregation of individual particles. Further confirmation was achieved using the plasmon resonance peak by UV/Vis spectroscopy (see below).
Transmission electron microscopy (TEM)
Nanoparticle characterization was performed on JEOL 1230 and Hitachi HD7700 TEMs using particles dried onto copper grids (Ted Pella, Inc.). Particle size analysis was performed using TEM images and ImageJ software for measurement and statistical analysis of 100 individual particles. Error reported represents the standard deviation of the mean.
Ultraviolet/Visible (UV/Vis) light spectrophotometry
UV/Vis spectra were acquired on a spectrophotometer (Agilent). Colloidal stability of prefunctionalized citrate-capped gold nanoparticles and post-functionalized DNA-Gd@AuNP was confirmed by measuring the plasmon resonance absorbance at 523 nm for citrate stabilized particles and 519 nm for DNA conjugates [35].
Shelf-life stability
Solutions of concentrated DNA-Gd@AuNPs were diluted into duplicate tubes of Hank’s Balanced Saline Solution at buffer strengths of 0.1x, 0.5x and 1x. The particles were stored at 4 °C for two weeks. Every other day, DNA-Gd@AuNPs were centrifuged and sampled for supernatant Gd(III) concentration. Data was processed such that the Gd(III) observed in the supernatant was quantified as a percentage of the original Gd(III) concentrated by ICP-MS (see 2.4). A subsequent trial was performed in 0.1 x Hank’s Balanced Salt Solution (HBSS) at 55 nM of nanoconjugates per tube and stored either at 4 °C, and a second tube of the same concentration was incubated for the same period at 37 °C. ICP-MS was used to quantify the initial particle concentrations, including total Gd(III) content per tube. Particles were stored for 24 hours, at which time they were vortexed and centrifuged (30 minutes at 4°C, 21,000 g). A sample of the supernatant was removed (containing no AuNPs), which was analyzed for Gd(III) content. The Gd(III) content measured was normalized to the initial Gd(III) concentration of each tube, and quantified as percentage of Gd(III) lost from the particle surface into the surrounding solution, as above. This process was repeated in 24 hr increments for a period of two weeks. In parallel with shelf life stability experiments, a separate aliquot of particles was measured for r1 and r2 relaxivities (see below) on the first day after conjugation. Other than the particles used for ICP analysis, particles were recovered and re-concentrated after the measurement of relaxivity and then placed back into storage at 4 °C. This process was repeated on days 8 and 17.
Nanoparticles Relaxivity in solution (Northwestern)
A stock of DNA-Gd@AuNP conjugates was serially diluted for a concentrated stock for a total of five solutions. 250 μL of each concentration was warmed to 37 °C and placed into a Bruker minispec mq60 NMR spectrometer (60 MHz, 1.41 T) for measurement of T1 and T2 relaxation times (in s). Data was collected using an inversion recovery pulse sequence, with a 20s repetition time and 10 data points (average of 4 measurements per point). The remaining volumes of each solution were utilized for ICP analysis of Gd(III). The inverse of the longitudinal relaxation time was calculated to obtain the relaxation rate (R1= 1/T1, in s−1) which was plotted against the Gd(III) concentration (mM). By applying a linear fit to this data, the slope generated is reported as the relaxivity (r1) of the agent (in mM−1 s−1). Linear regressions were fitted with R2 of >0.99. Relaxivity indicates changes in relaxation rate as a function of concentration [36].
Nanoparticles Relaxivity in solution (Pittsburgh)
To ensure that the MR properties of the DNA-Gd@AuNP NP were not compromised during shipping, their relaxivity was measured again within 2 days of their arrival. To evaluate a dose-dependent effect on the MR signal (R1 and R2) different concentrations (0.01, 0.02, 0.05, 0.09, 0.18 mM) were arrayed in phosphate buffered saline (PBS) in 0.1 ml PCR tubes (Axygen) placed in 6% gelatin and contrasted with a commercially available standard of monomeric Gd-HPDO3A (ProHance, Bracco Imaging). Solution phantoms were placed in a 9.4 T (400 MHz) horizontal bore system equipped with VnmrJ 3.1 software (Varian). For transmission and acquisition, a custom made volumetric birdcage quadrature coil (Virtumed LLC), achieving a radiofrequency (RF) power input of up to 55 μT/5 s with an internal diameter of 36 mm and effective length of 25 mm was used. For R1 (1/T1) measurements, a fast spin echo (FSE) sequence with a global inversion recovery (IR) with inversion time (Ti) values of 0.01, 2, 4, 6, 8, 10 and 10.2 s (Table 1 for details of all imaging acquisition parameters), whereas a multi-slice multi-echo (MSME) sequence provided measurements for R2 (1/T2). All scanning was performed with the phantom at 37 °C. From individual images, T1 and T2 maps were created in VNMRJ with the inverse of these (R1 and R2) being computed in Matlab v8.1. (Mathworks). A linear regression was performed to determine the relaxivity (r1 and r2) of the agent as expressed in mM−1s−1.
Table 1.
MR imaging parameters (relaxation time = TR; echo time = TE, field of view = FOV).
| TR (s) | TE (ms) | Averages | FOV (mm) | |||
|---|---|---|---|---|---|---|
| In vitro | Solution | T1 map | 10.4 | 40 | 1 | 45×30 |
| T1w | 0.5 | 2.02 | 1 | |||
| T2 map | 5 | 15–480 | 1 | |||
| T2w | 2 | 27.87 | 1 | |||
| Cells | T1 map | 12.2 | 40 | 1 | 45×25 | |
| T2 map | 3 | 15–480 | 1 | |||
| T2w | 3 | 32.74 | 1 | |||
| Ex vivo | T1 map | 15.4 | 40 | 1 | 30×30 | |
| T2 map | 6 | 7.2–229 | 2 | |||
| T2w | 6.1 | 70.78 | 8 | |||
| In vivo | T1 map | 10.4 | 40 | 1 | 30×30 | |
| T1w | 0.5 | 2.73 | 6 | |||
2.2. Cell Culturing
The human striatal neural stem cell (NSC) line STROC05 (kindly provided by ReNeuron, UK) were cultured and passaged, as previously described in detail [37, 38]. In brief, STROC05 NSCs are conditionally immortalized using cMyc-ERTAM under the control of 4-hydroxytamoxifen (100 nM; Sigma). In the absence of 4-hydroxytamoxifen NSCs will cease proliferation and undergo differentiation. NSCs were expanded on laminin (10 μg/ml) coated flasks until they reached a 70–80% confluency. Recombinant human basic fibroblast growth factor (bFGF; 10 ng/ml; PeproTech) and epidermal growth factor (EGF; 20 ng/ml; PeproTech) were used as mitogens. All culturing was performed without the addition of antibiotics at 37 °C in 5 % CO2.
2.3. Cell Labeling
STROC05 were labeled with DNA-Gd@AuNP NP in vitro under proliferative conditions. For this, cells were grown to a confluency of ~50% and DNA-Gd@AuNPs were added to fresh medium at different concentrations (0, 0.02, 0.2, 2 or 20 nM). Additional concentrations (50, 100, 200 nM) of DNA-Gd@AuNP concentration were also tested, but there was significant cytotoxicity and these were hence excluded from further analysis (Supplementary Figure 1). Cells were incubated for 24 hours prior to being washed 3x with HBSS (Gibco) and harvested using Accutase (Sigma) before being used for in vitro assays.
2.4. In vitro assays
Agent uptake measurements based on fluorescence
The DNA-Gd@AuNP contains a Cy3 moiety that affords detection of the nanoconjugates using fluorescent microscopy. For the different concentrations, cells were incubated for 24 hours prior to being washed 3x with HBSS and fixed for 15 minutes using 4% paraformaldehyde. Coverslips were stored in 24 well plates with PBS. For analysis, cells were mounted using Vectashield for fluorescence with DAPI (Vector Laboratories). For each of the 3 batches of DNA-Gd@AuNPs a single technical replicate was performed.
Using a M2 microscope (Zeiss) with an X-Cite Series 120Q (Exfo) illumination interfaced with a high sensitivity monochrome camera Axiocam MrM (Zeiss) and StereoInvestigator version 11.07 software (MBF Bioscience), images in 5 fields-of-view per coverslip were acquired in the red channel (range 525–575 nm range), as well as the blue channel (range 335–383 nm) for DAPI. Image acquisition settings were maintained for all images. Cy3 signal intensity in acquired images was quantified using Matlab v8.1. (Mathworks). To obtain a measure of relative fluorescence units (RFU) per cell, the signal intensity was divided by the number of cells (based on manual counting of DAPI+ nuclei) contained within each image.
Survival and proliferation
To determine if cell labeling at different concentrations affects the survival of NSCs or their proliferation, the number of DAPI positive cells present at day 0 (i.e. 24 hrs post-labeling) and day 7 were counted. For this, cells were grown under differentiation conditions (i.e. withdrawal of 4-hydroxytamoxifen, bFGF, and EGF). As STROC05 are adherent cells, once their health is compromised they detach. Hence counting the number of attached cells is a measure of survival.
Nevertheless, survival at 7 days can also be affected by the continued proliferation of NSCs. To measure a potential effect of cell labeling on proliferation, cells were also stained with a Ki67 antibody, which indicates all active phases of the cell cycle (i.e. G1, S, G2 and mitosis) at day 0 and day 7 post-labeling. For immunocytochemistry, fixed cells were washed 3x with PBS prior to the application of the primary rabbit antibody against Ki67 (1:500, Abcam, ab15580). After overnight incubation at room temperature (21 °C), the primary antibody was removed and cells were washed 3x in PBS prior to application of the secondary goat anti-rabbit Alexa 488 (1:500, Molecular Probes, A-11034) for 1 hr. Cells were washed 3x in PBS and mounted with Vectashield for fluorescence with DAPI. The number of Ki67+ cells were counted and expressed as percentage in relation to the total number of DAPI+ cells.
Cell differentiation
To assay potential effects on NSC differentiation into appropriate cell phenotypes, i.e. neurons and astrocytes, or lineage-specific toxicity, cells were allowed to differentiate for 7 days [38]. Only the 20 nM condition was tested, as this was the highest concentration without evidence of detrimental effects. To measure a potential effect on cell differentiation, immunocytochemistry (as described above) was performed on fixed cells to measure the number of neurons, as indicated by staining with a rabbit anti-β-III-Tubulin (Tuj, 1:500, Abcam, ab18207) antibody, and astrocytes, as revealed by mouse anti-Glial Fibrillary Acidic protein (GFAP, 1:3000, Sigma, G3893) antibody on day 0 and day 7. To indicate a % change due to cell labeling, a comparison to unlabeled cells was performed.
2.5. ICP-MS analysis of labeled cells
The Gold and Gadolinium content of cells incubated with different NP concentrations for 24 hours was determined using ICP-MS. Cells were collected in 0.1 mL Accutase. A suspension of 5 μl was digested with 120 μL of nitric acid (70%, Sigma) and concentrated hydrochloric acid (37%, Sigma). To assay for Gd(III), samples were diluted to a total volume of 3 mL with Nanopure water to a final combined acid concentration of 2% (v/v) nitric and 2% (v/v) hydrochloric acids. To probe Au(III) content, samples were diluted to a total of 10 mL with Nanopure water with the same final concentrations of acid with Gd(III) samples.
2.6. Cellular Relaxivity
Due to the potential T1 quenching of Gd(III)-agents’ uptake into cells [12], it is essential to establish if a T1 effect is preserved. For cell pellet phantoms, cells were washed three times in HBSS after cell labeling with 20 nM for 24 hrs prior to being harvested using Accutase and pelleted in a 0.1 mL PCR tube. NSCs labeled with Gd-HPDO3A (20 nM incubation for 24 hrs), as well as unlabeled cells, served as a control. For cell suspension phantoms, cells were re-suspended in DMEM/F12 for counting on a hematocytometer. Counted cells were suspended at 10×103, 50×103, 10×103, 150×104 and 200×104 cells/μL in a final concentration of 6% gelatin.
To calculate a T1 map, images were acquired on a 9.4T horizontal MRI scanner using a sequence employing global Inversion Recovery (IR) with a fast spin echo (FSE) readout (Table 1) with a T1 map being calculated in Matlab. T2 maps were acquired using Multi Slice Multi Echo (MSME) with 32 echoes, where TE was varied between each echo by 15 ms (Table 1). T2-weighted images were acquired using Fast Spin Echo Multi Slice (FSEMS: Table 1). All experiments were conducted at 37 °C.
The Gd(III) concentration of each phantom was calculated from the known cell density and cell uptake of DNA-Gd@AuNP. Plotting of the phantom Gd(III) concentration against R1 (1/T1) using a linear fit specified the relaxivity (r1) in mM−1s−1. Cellular relaxivity (r1 cellular) was measured by calculating the percentage change in relaxivity between unlabeled and labeled cells, as previously described [39]. Based on this, an estimate was made to calculate the minimum number of cells needed for a >5% signal change in T1.
Signal-to-noise (SNR) was calculated by dividing the mean signal from an ROI on the cell pellet by the mean signal outside of the sample, defined as noise. Contrast was defined as the mean signal of the pellet of labeled cells minus the mean signal of the unlabeled pellet [40]. These signal measurements were taken in the center of the pellet to avoid partial volume effect. To determine the influence of partial volume effects on detectability of labeled cells, measurements from the center of the pellet were compared to mean signal measurements that encompassed the whole pellet, i.e. center+corona (defined as the outmost edge visible distinguishable from the gel). As partial volume effects are highly dependent on voxel size, samples were acquired at different voxel volumes (4, 8, 17, 31, 69, 112, 275 nl). The slice thickness was kept constant at 0.5 mm, whilst the matrix size within the 45×25 mm FOV was varied (64×32, 96×48, 128×64, 192×96, 256×128, 384×192 and 512×256).
2.7. Cell Transplantation
All in vivo procedures complied with the Institutional Animal Care and Use Committee (IACUC), as well as NIH guidelines. On the day of implantation, labeled (20 nM of DNA-Gd@AuNP for 24 hrs) and unlabeled cells were suspended at 125×103 cells/μL in 4 mg/ml extracellular matrix hydrogel (ECM, kindly provided by Dr Stephen Badylak, University of Pittsburgh) to promote retention within the brain after transplantation [41]. Viability of cells in this preparation was >85%. Animals were anesthetized using isoflurane (4% induction, 2% maintenance) in medical air and placed in a stereotactic frame (Kopf). A 10 μL Hamilton syringe containing the cell suspension was inserted into the injection site. Using a micro pump (Micro4, WPI) at a speed of 1 μL/min, one animal received 3 different volumes of labeled (left hemisphere) and unlabeled (right hemisphere) to yield a total of 2.50×105, 6.25×105 and 10×105 cells for an ex vivo imaging designed to determine optimal detection parameters. Using this information for an in vivo imaging experiment, a total volume of 5 μL (total of 6.25×105 cells) of cells was injected in a single deposit of unlabeled (right hemisphere) and DNA-Gd@AuNP-labeled cells (left hemisphere) (n=3). After completion, the needle was left in place for two minutes before being retracted. Bone wax was used to fill the skull defect caused by the drill holes. Animals were sutured and given topical analgesic cream (2.5% Lidocaine and 2.5% Prilocaine, Sandoz) and Buprenex (0.05 mg/kg i.p.; Henry Schein).
2.8. Ex- vivo MR Imaging
To determine an appropriate voxel size for in vivo imaging, images for T1 maps were acquired using global Inversion Recovery (IR) with fast spin echo (FSE) readout, whereas T2 maps used a MSME sequence (Table 1) on a 9.4T horizontal MRI scanner. The matrix size was arrayed (48×48, 64×64, 96×96, 128×128, 192×192 and 256×256) within a constant slice thickness of 0.5 mm and FOV of 30×30 mm to yield voxel volumes volumes of 7, 12, 28, 49, 110, and 195 nl.
2.9. In vivo MR imaging
The day following cell implantation, animals underwent MR imaging (9.4 T). For this, animals were anesthetized using isofluorane (4% induction, 1.5% maintenance) in medical air, while their physiology (temperature, respiratory rate) was monitored using a MP150 Biopac system interfaced with AcqKnowledge v4.1 software (Biopac Systems Inc.). Based on the ex vivo voxel size and cell concentration experiment, T1–weighted MRI images were acquired using a Gradient Echo Multi Slice sequence (Table 1).
2.10. Immunohistochemistry
Immediately after imaging, animals were sacrificed using Fatal Plus (Henry Schein) and underwent transcardial perfusion-fixation using 4% Paraformaldehyde (PFA, Electron Microscopy Sciences). Brains were then removed and placed in 4% PFA overnight to ensure complete fixation. Brains were cryoprotected in 30% sucrose for 3–4 days before being sectioned using a cryostat (Leica) at 40 μm thickness. Sections were stored at −20 °C.
For immunohistochemistry, sections were washed 3x with PBS before being incubated in blocking solution (0.5% Triton X100, 10% Normal Goat Serum in PBS) for 1 hr at room temperature. The primary antibody, mouse anti-human nuclei antigen (HNA, 1:1400, Millipore, MAB1281) was diluted in blocking solution and incubated overnight at 4 °C. Sections were washed 3x PBS before exposure to the appropriate anti-mouse AlexaFluor488 secondary antibody (1:500, Life Technologies) for 1hr at room temperature. After 3x PBS washes, 1 μg/mL Hoechst33342 (Sigma) was applied as a nuclear counterstain and coverslipped using Vectashield for fluorescence mounting medium. Images were acquired using the same set-up as described above. Whole slice images were acquired using the Virtual Tissue software module in StereoInvestigator (MBF). The first, last and centre sections containing HNA+ cells were chosen as a representative coverage of the graft with Hoechst+, HNA+ and/or DNA-Gd@AuNPs counted using FIJI image analysis software (NIH). Cells (Hoechst+) were categorized as “correctly identified” if both HNA and the label were present, as a “false negative” cells were defined as HNA+ without DNA-Gd@AuNPs, whereas “false positive” cells contained DNA-Gd@AuNPs but were HNA−.
2.11. Statistical analyses
Statistical analysis of cell counts and measurements were performed on mean values using one- or two-way analysis of variance (ANOVA) followed by Bonferroni post-hoc analysis using SPSS for Mac v17 (IBM). Percentage change measures for lineage differentiation were compared using two-tailed independent t-tests. A p value of <0.05 was considered significant. Prism v6 (GraphPad) was used to plot data as means ± standard deviation, as well as to calculate the linear regressions for relaxivity measures. Contour maps were drawn in Minitab v17 (Minitab Inc).
3. Results
3.1. Characterization of synthesis and quality control of DNA-Gd@AuNPs
An initial characterization of synthesized gold nanoparticles (AuNPs) by TEM indicates a diameter of these of 15.3±1.4 nm. DNA-Gd(III) strand synthesis were HPLC-purified to 5 chelates per strand. Conjugation of these strands to AuNPs was achieved efficiently using click chemistry. A characterization of DNA-Gd@AuNPs size was performed using dynamic light scattering indicating a hydrodynamic radius of 35±0.7 nm of the nanoconjugate. UV/Vis spectroscopy showed no change in the maximum plasmon resonance wavelength for citrate stabilized particles (523 nm) and DNA-modified particles (519 nm) after functionalization, indicating good colloidal stability (Supplementary Figure 2). Particle loading was assessed by ICP-MS analysis of fully synthesized particles by observation of the ratio of Gd(III) to Au(III) ratio. By calculating the volume of the nanoparticles using a geometric approximation, based on the observed size by TEM (Figure 1B), and applying the density of bulk gold, a particle loading of 326±44 Gd(III) molecules per AuNP was determined. This Gd(III) loading corresponds to a particle loading of 65±9 DNA strands per particle.
3.2. DNA-Gd@AuNP stability and relaxivity
The stability of the nanoparticles is essential to ensure detection of implanted cells. The relaxivity (r1 and r2) of DNA-Gd@AuNP nanoparticles was hence measured in solution for 17 days (Figure 1B). At 1.41 T, r1 was stable at 14.6 mM−1s−1, as was r2 at 38.5 mM−1s−1 (measured at 37 °C). Both measurements indicate a stable particle that produces a robust effect on the MR signal. This was further reflected in the minimal loss of Gd(III) from the nanoparticle construct (Figure 1C). At 4 °C, the cumulative loss of Gd(III) was <0.5%, although a greater loss (<5%) was seen at 37 °C over 17 days. These particles can therefore be safely stored for at least 2 weeks without affecting their stability or relaxivity during shipment at room temperature (21 °C).
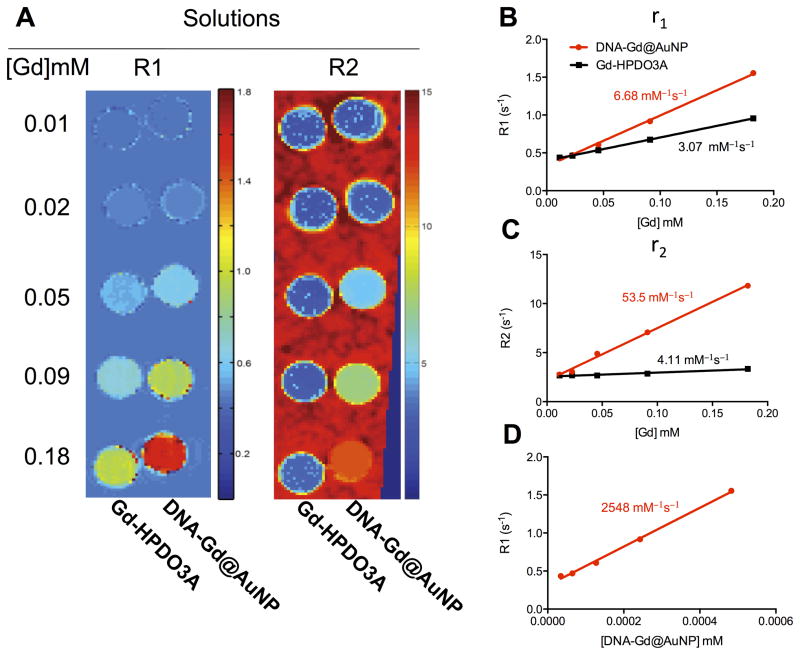
As a further quality control procedure, after shipment of particles, relaxivity of nanoparticles in solution was measured again on a 9.4 T MRI at 21 °C and contrasted with commercially available Gd-HPDO3A (ProHance). Both R1 and R2 were measured for different Gd(III) concentrations to afford a direct comparison as to potential benefits of constructing a gold nanoparticle to improve relaxivity. Indeed, the improvement in relaxivity was clearly evident especially at higher concentrations (Figure 2A). This results in a higher molar relaxivity for the DNA-Gd@AuNPs compared to Gd-HPDO3A, with r1 values of 6.68 and 3.01 mM−1s−1 respectively (Figure 2B). r2 is also higher, at 53.5 compared to 4.11 mM−1s−1 (Figure 2C). It is important to note that this improvement in relaxivity is for each Gd(III) molecule, rather than the polymeric AuNP construct. The r1 for each DNA-Gd@AuNP is 2548 mM−1s−1, a very dramatic ~800 fold improvement compared to monomeric Gd-HPDO3A (Figure 2D). Nanoparticle complexion therefore improves the relaxation properties of Gd-HPDO3A by increasing the relaxivity of each Gd(III) molecule, as well as by complexion into a polymeric construct that can be used to efficiently label cells.
Figure 2. MR relaxivity of DNA-Gd@AuNP nanoparticles.
R1 and R2 maps of DNA-Gd@AuNP and Gd-HPDO3A were generated at 9.4 T (A). DNA-Gd@AuNP was shown to have 2.2x the r1 molar relaxivity of Gd-HPDO3A (B) and 13x the r2 relaxivity (C). Expressing r1 relaxivity on a “per mole of particle” basis suggested that r1 was 830x higher in DNA-Gd@AuNP (D).
3.3. Labeling of neural stem cells
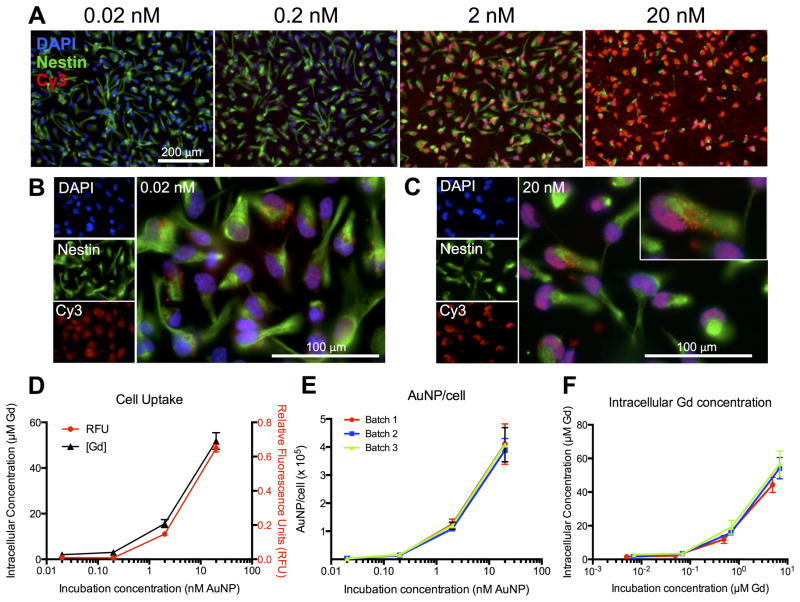
In order to achieve the most sensitive detection of implanted NSCs, the intracellular concentration of Gd(III) must be maximized without any detrimental effects on cells. Oligonucleotides facilitate an efficient cellular uptake with a dose-dependent uptake being evident with increasing amount of NPs. This was evident by DNA-Gd@AuNPs being visible inside NSCs using the Cy3 moiety at 24 hours (Figure 3A). Even at 0.02 nM there was clear evidence of DNA-Gd@AuNPs uptake (Figure 3B), but at 20 nM a far more significant amount of DNA-Gd@AuNPs were present intracellularly (Figure 3C). Quantification of uptake was performed using relative Cy3 fluorescence, as well as an absolute measurement using ICP-MS for Gd(III). Both showed the same pattern of a dose-dependent uptake (Figure 3D). A 20 nM incubation resulted in >3 times the uptake of 2 nM, and ~25 times the uptake of 0.02 nM incubation, yielding an intracellular Gd(III) concentration of ~55 μM. A persistent concern in the use of nanotechnology is the potential variability between batches of nanoparticles. However, three separately synthesized batches of DNA-Gd@AuNPs resulted in a highly reproducible and consistent uptake of NPs into cells (Figure 3E). Nevertheless, the Gd(III) loading of the different batches revealed noticeable variations (Batch 1 = 248 Gd/NP; Batch 2 = 345 Gd/NP; Batch 3 = 350 Gd/NP). As intracellular Gd(III) concentration is determined by DNA-Gd@AuNPs uptake, batches with lower Gd(III) loading result in lower intracellular concentrations (Figure 3F). Quality control of DNA-Gd@AuNPs loading for each batch is hence important to ensure an optimal cellular concentration of Gd(III).
Figure 3. Intracellular uptake of DNA-Gd@AuNP nanoparticles.
Cell Uptake of DNA-Gd@AuNPs at different concentrations is shown by the fluorescent Cy3 moiety (red, A, scale bar = 200 μm). Fluorescence was visible even at the lowest concentration (B, scale bar = 100 μm) and appeared to be localized to the cytoplasm (C). Cell uptake was measured by ICP-MS and RFU per cell, which followed very similar patterns (D). Comparing three separate batches showed that particle uptake was highly consistent between batches (E), but that the variability in Gd(III) loading resulted in variation in intracellular Gd(III) concentration (F).
3.4. Cellular effects of DNA-Gd@AuNPs
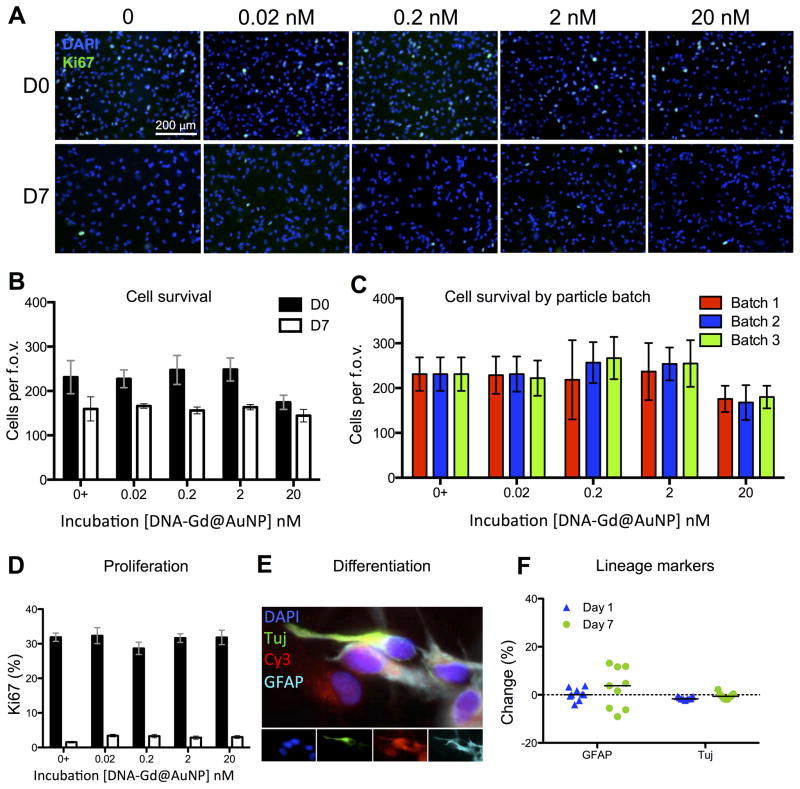
Although uptake of particles was efficient, the effects of labeling on cellular function also require investigation in order to avoid detriment to any potential therapeutic efficacy of the cells. Cell survival and proliferation are key measures to evaluate after cell labeling (Day 0) as well as at Day 7 (Figure 4A). The survival of cells on Day 0 was only significantly reduced with a 20 nM incubation concentration (p<0.05) (Figure 4B). However, this was an acute effect, as by Day 7 all incubation concentration presented a survival equivalent to the control condition where no nanoparticles were present. Batch-to-batch variability was also assessed with respect to its effect on cell survival, and no differences were seen between batches (Figure 4C). No effect of nanoparticles on proliferation was evident at Day 0 or 7 (Figure 4D). A differentiation experiment investigating NSCs potential to differentiation in astrocytes (GFAP+ cells) and neurons (β-III-Tubulin+ cells) (Figure 4E), revealed no significant effect of 20 nM DNA-Gd@AuNP (Figure 4F). Apart from a transient effect on cell survival at day 0, 20 nM DNA-Gd@AuNP, labeling therefore does not affect the cell functions and provides a viable labeling strategy to visualize NSCs using MRI.
Figure 4. Cellular effects of DNA-Gd@AuNPs.
Cells at day 0 and day 7 after labeling were stained with DAPI and Ki67 (A, scale bar = 200 μm). No significant effects were seen on the number of surviving cells at any concentration of DNA-Gd@AuNPs at day 0 or day 7 (B) and this effect remained consistent when three separate batched of particles were tested (C). The percentage of cells expressing Ki67 also remained consistent at all DNA-Gd@AuNP concentrations at day 0 and day 7 (D). Cells were also stained to assess phenotypic changes (E), and no significant differences were seen between those labeled at 20 nM nanoparticles and controls (F).
3.5. Detecting labeled cells using MRI
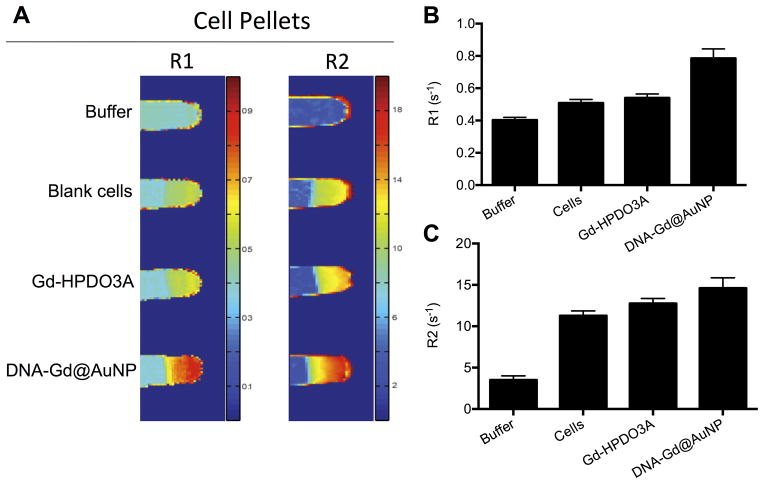
To assess whether the achieved intracellular concentration of Gd(III) was sufficient for visualization of labeled cells using MRI, cells incubated with 20 nM nanoparticles were imaged at 9.4 T and contrasted with cells incubated with an equivalent concentration of Gd-HPDO3A (Figure 5A). The R1 for DNA-Gd@AuNP labeled cells was significantly (1.5x) higher than unlabeled cells (p<0.01) and Gd-HPDO3A-labeled cells (p<0.05) (Figure 5B). Gd-HPDO3A labeling did not result in a significant increase in R1 compared to unlabeled cells. R2 was also significantly (1.3x) higher in DNA-Gd@AuNP labeled cells (p<0.01) compared to unlabeled cells, but did not differ significantly from Gd-HPDO3A-labeled cells (Figure 5C). This is in contrast to the particle characteristics in solution, where they affect R2 more strongly than R1, and shows the importance of the microenvironment to MRI detection.
Figure 5. In vitro imaging of cell pellets.
Blank cells, and cells labeled either with Gd-HPDO3A or DNA-Gd@AuNP were imaged at 9.4 T to generate R1 and R2 maps (A). Cells labeled with DNA-Gd@AuNP increased relaxivity compared to unlabeled cells and those labeled with Gd-HPDO3A on both R1 (B) and R2 (C)
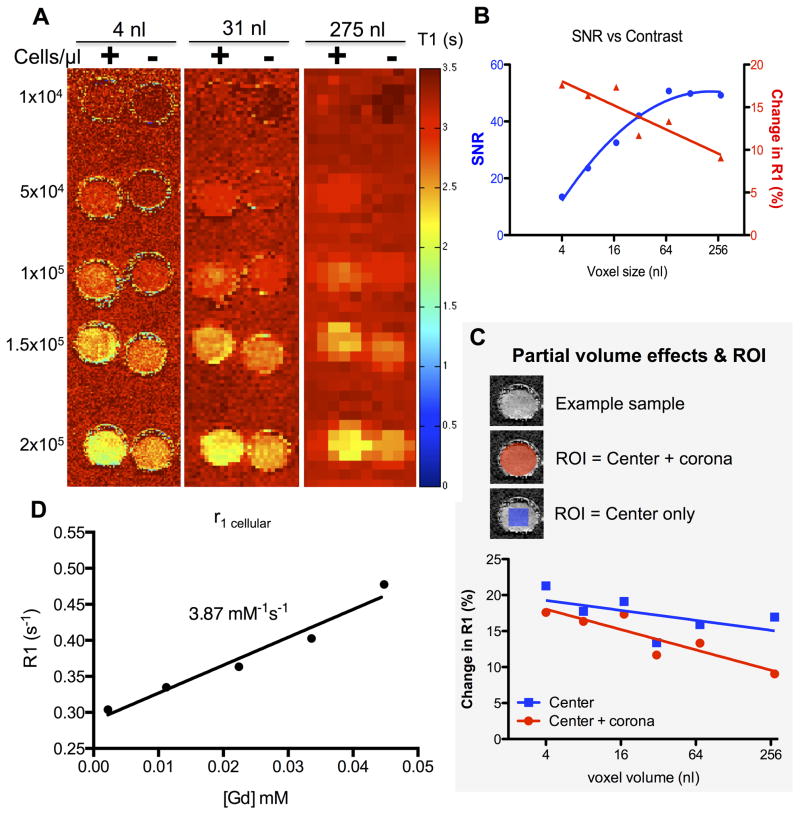
Another important factor to consider is voxel size and the potential for partial volume effects. Suspensions of unlabeled and DNA-Gd@AuNP labeled cells were imaged at different cell densities and voxel sizes (Figure 6A). Larger voxels resulted in a higher signal-to-noise ratio (SNR, i.e. gelatin to non-sample air), but even the smallest voxel size resulted in an SNR of 13 (at 9.4 T). In comparison, the R1 contrast (i.e. labeled versus unlabeled cells) was lower in larger voxels due to a partial volume effect (Figure 6B). An optimal compromise to achieve both a high SNR and CNR can be achieved with a voxel size of 32 nL, whereas maximum 18% increase in R1 in vitro is achieved with a voxel size of 4 nL. A 4 nL voxel size hence affords the detection of ~40 NSCs (= 1×104 cells/μL divided by 250 voxels of 4 nL present in 1 μL volume). A partial volume effect in cell detection can potentially be mitigated by focusing the ROI on the center region, thereby excluding voxels only partially occupied by labeled cells. Partial volume effects are predominantly present in the corona, where voxels cover areas with increasingly smaller fractions of labeled cells (Figure 6C). Nevertheless, in vivo there is no easily defined ROI for detection and hence a smaller voxel volume is the only option to avoid partial volume effects and detect labeled cells. A clear T1 effect was observed here, although some attenuation was evident due to intracellular incorporation with r1 relaxivity of DNA-Gd@AuNPs within cells being reduced to 3.87 mM−1s−1 (Figure 6D) compared to 6.68 mM−1s−1 in solution (i.e. a 42% decrease).
Figure 6. Effect of voxel size and ROI selection on cell detection in vitro.
Labeled and unlabeled cells were suspended in 6% gelatin at different cell densities with T1 maps being generated (A, + = labeled cells, − = blank cells). Increasing voxel size resulted in increased SNR, but less contrast between labeled and unlabeled cells (B). As voxel size increases, peripheral voxels are increasingly susceptible to partial volume effects that attenuate T1 contrast (C). If measurement of T1 is confined to the center of the ROI, there is little attenuation of T1 contrast across voxel sizes, hence abating the partial volume effects. However, measurement of cell distribution will depend on a reliable measurement of peripheral voxels as well as those at the core of the implantation site. The r1 relaxivity of DNA-Gd@AuNPs in cells is 3.87 mM−1s−1 (D), significantly lower than that in solution.
3.6. In vivo detection of transplanted cells
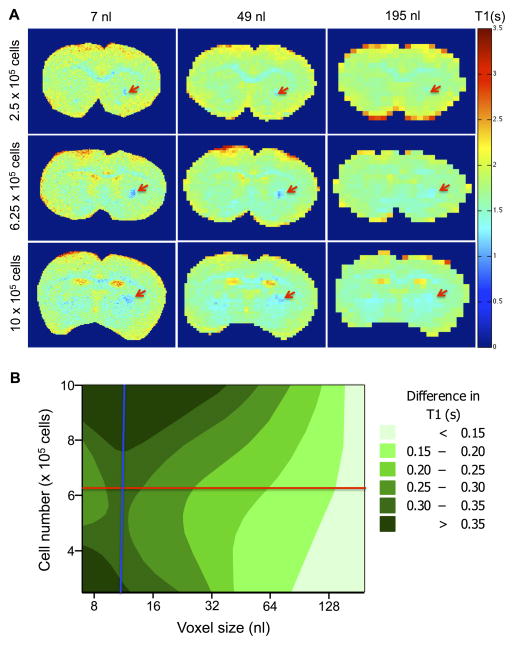
To establish intracerebral detection of transplanted cells, cell number and voxel size were arrayed and T1 maps were generated to measure signal intensity changes due to labeled cells (Figure 7A). Very little change in T1 was evident at a large voxel size (195 nL), whereas deposits of all 3 concentrations were readily identifiable at a voxel size of 49 nL (at 9.4 T). The smaller voxel size of 7 nL provided the greatest detail and contrast for all concentrations. Importantly, the lowest concentration of cells (2.5×105 cells) was more visible at this voxel size. To achieve a robust in vivo detection, ideally a signal change of at least 25% is required. Using the acquired array, a contour map was generated for the experimental space (Figure 7B). It is evident that there is some interaction between voxel size and cell number on the T1 signal change. The aim of a 25% change in T1 for a proof-of-principle study can be targeted with a cell injection of 6.25×105 labeled cells and a voxel volume of 12 nL (229 μm isotropic). This image resolution provides also a sufficient margin of error to achieve the required signal change with lower concentrations of cells.
Figure 7. Effect of voxel size and cell number on ex vivo detection.
T1 maps of 3 different injection sites were acquired at 6 different voxel sizes (A. Red arrows indicate implanted cells). The difference in T1 between labeled and unlabeled cells was compared across cell number and voxel volume, with voxel volume having a more marked effect than cell number. The conditions taken forward for in vivo image acquisition were 12 nL voxel volume (blue line) and 6.25×105 cells (red line, B).
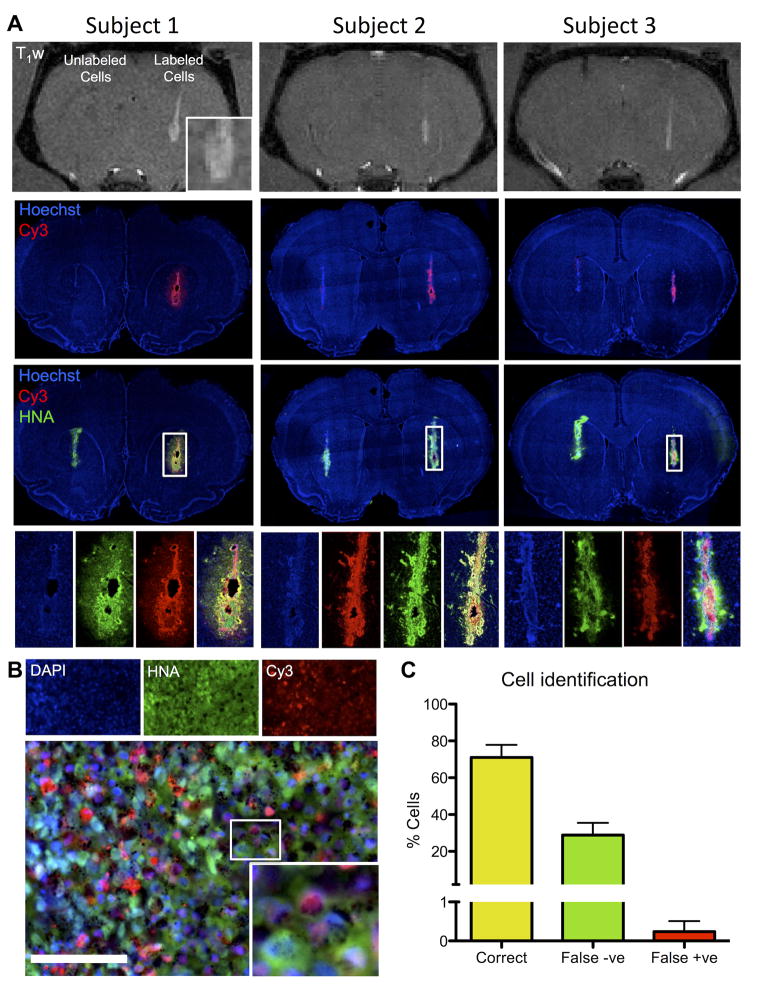
To validate the in vivo T1 detection of transplanted cells in a pilot proof-of-principle study, three animals were transplanted with both labeled (right hemisphere) and unlabeled cells (left hemisphere). The deposit of labeled cells as well as cells within the injection tract were readily distinguishable against the host brain background on T1-weighted images in all animals at 9.4 T (Figure 8A). There was no evidence of a signal change in T1 caused by the unlabeled cells in the homologous region in the opposite hemisphere. A signal change of 17.7% was measured on the T1-weighted images at the center of the deposit compared to non-labeled cells. To verify that the signal change on the T1-weighted images was indeed due to the transplanted cells, immunohistochemical analyses were performed to detect human cells (HNA in green), as well as the Cy3 moiety of the DNA-Gd@AuNPs. Although human NSCs were present in both hemispheres, only those containing DNA-Gd@AuNP were detectable using the T1-weighted MR images. A higher magnification image shows the center of a graft, where co-localization of DNA-Gd@AuNP based on the Cy3 moiety can be seen in human cells (Figure 8B). Aggregates of gold nanoparticles can seen as black spots. In order to assess whether the label may have leaked from transplanted cells, the level of co-localization between HNA and Cy3 was quantified (Figure 8C).
Figure 8. In vivo imaging with histological verification.
Three animals were transplanted with labeled cells in one hemisphere and unlabeled cells in the other. DNA-Gd@AuNP labeled cells were clearly visible on T1-weighted (T1w) MR images (A). The deposit could be seen with some infiltration into the host tissue (inset for subject 1). In all animals with labeled cells, the injection tract was clearly visibile. In contrast, unlabeled cells did not produce a signal in the right hemisphere. Fluorescent histology corroborated these in vivo results indicating that in both hemispheres transplanted cells were present (human nuclei antigen, HNA, in green), but only cells containing DNA-Gd@AuNP nanoparticles (as detected by the red Cy3 moiety) produced a T1 effect on MR images. A higher magnification image shows good colocalisation of HNA and Cy3 (B, scale bar = 100 μm), which is supported by the quantification showing a high level of accurate cell identification with less than 1% of false positives, but a substantial number of false negatives (C).
The majority of cells (71%) were correctly identified based on the presence of DNA-Gd@AuNP, but there was a significant (29%) portion of false negatives (Type II error). However, there was very little transfer of agent with 0.38% host cells containing DNA-Gd@AuNP (Type I error). DNA-Gd@AuNP therefore afford the in vivo reliable detection of transplanted cells using T1-weighted MR imaging with little risk of a Type I error (<5%), but significantly underestimating the total number of transplanted cells (Type II error >20%).
4. Discussion
Achieving an unequivocal in vivo detection of transplanted cells remains a major impediment to understand how the distribution of therapeutic cells influences recovery. We here characterized the use of a new class of MR contrast agents for cellular MRI that uses gold nanoparticles as a platform to assemble multiple components that afford visualization using different modalities, but with added functionality, such as improved cellular uptake. These DNA-Gd@AuNP were efficiently incorporated into cells without the use of a transfection agent or electroporation to produce a high intracellular yield of Gd(III) that preserved T1 relaxivity. At concentrations up to 20 nM nanoparticles, no significant effects on survival, proliferation or differentiation were evident. Batch-to-batch variability was insignificant with good stability and retention of T1 relaxivity. Optimization of MR parameters indicated that voxel size significantly affects the degree of contrast, with smaller voxels providing favorable conditions to detect small numbers of cells. This approach allowed us visualize transplanted cells using T1 contrast in vivo with verification by immunohistochemistry indicating a high level of specificity of the contrast (i.e. low Type I error), although there is a significant underestimation of transplanted cells using this approach (Type II error).
4.1. DNA-Gd@AuNP nanoparticle design facilitates T1 detection
The use of gold nanoparticles as platform to assemble multi-functional and -modal nanoconstructs provides several opportunities that improve on the polymeric conjugation of Gd(III)-complexes [42]. Most notably, gold-based constructs can provide a unique opportunity to tune T1 contrast to achieve optimal relaxivity of each monomer [24, 28]. Grafting Gd(III) onto DNA-Au nanoparticles here almost doubles the relaxivity of each Gd-HPDO3A chelate demonstrating a key improvement in performance compared to merely creating a polymeric compound. It is of note that shape of particles is increasingly emerging as an important design consideration in providing improvements in contrast [34] that potentially will further improve relaxivity yield of each Gd(III)-chelate, but might eventually also allow tuning to multiple independent constructs being visualized using the same contrast mechanism [43]. Conjugation of gadolinium chelates onto the gold nanoparticles further affords an additional capping preventing Gd(III) leaching, hence increasing their stability [44]. High thermostability is essential to avoid ill-effects of Gd(III) ions on cellular health that could compromise long-term detection of transplanted cells [45]. A further key performance characteristic of a putative cellular MRI contrast agent is its reproducibility in manufacturing, as well as relaxivity performance [4]. Indeed, the DNA-Gd@AuNP nanoparticles here provided very consistent performance in physical characteristics, cell uptake, as well as relaxivity further supporting their potential for cellular imaging studies.
Cell uptake into non-phagocytic cells, such as NSCs, remains a major challenge to yield a sufficient intracellular concentration that affords T1 detection, but also to prevent a quenching of the T1 effect. Since the sensitivity of T1 based MR probes is inherently lower than T2 agents [46], a higher concentration is required for detection. Nonspecific endocytosis or pinocytosis are the most commonly used routes of agent uptake due to their simplicity and efficiency. This typically results in localization of the agent to the endosomes/lysosome pathway, leading to a quenching of the T1 effect above 1010 Gd/cell [12] and potentially a rapid clearance through exocytosis [47, 48]. Instead of quenching of T2 relaxivity, lysosome-entrapped agent creates susceptibility effects that favor T2 detection [49] and can be used to track cells [45, 50]. Indeed, the R2 of DNA-Gd@AuNP is 8 times higher than R1 in solution. However, upon incorporation into cells, this ratio is decreased due to the R2 effect of cells. In contrast, there is less change in R1 between solution and cells, hence the ratio of contrast of DNA-Gd@AuNP for R1 improves upon cell internalization. Importantly, preservation of T1 detection (not only increase in R1) is essential to provide an unambiguous detection of labeled cells in contrast to T2-based artefacts, which could increase the in vivo error rate of cell imaging. DNA-Gd@AuNP resulted in 108 Gd/cell, well below the threshold for quenching and consequently resulted in a clear signal change on T1-weighted images. The agent’s microenvironment influences its relaxivity properties, as it affects water diffusion and interaction with the agent’s outer sphere (i.e. chelate). This narrows the range of optimal water exchange (τm) and tumbling rate (τr) for a given field [51]. As these molecular parameters are highly sensitive to protein binding and water access, relaxivity in solution and inside cells is significantly different [49].
Here, cell incorporation led to a 42% loss of r1 relaxivity per Gd(III) molecule. Nevertheless, incorporation of Gd(III) into DNA-Gd@AuNP increased the relaxivity of each individual Gd(III) chelate and resulted in an intracellular concentration that permitted in vivo detection. Further improvements in relaxivity, hence might afford a further reduction in required intracellular Gd(III) or afford the detection of fewer cells. Alternatively, modification of the agent to reduce intracellular protein binding, for instance, could preserve the relaxivity of DNA-Gd@AuNP in solution and provide a dramatic improvement in cell detection [34].
4.2. Specificity of cell detection
Apart from the intracellular environment influencing the relaxivity of DNA-Gd@AuNP relaxivity, detection of cells is equally dependent on the imaging volume within which these cells are embedded. Partial volume effects are much discussed in the context of diffusion weighted MRI [52], but little attention has been given to them in the context of cell tracking [2, 4]. To assess efficacy of potential cell transplant therapies and correlate cell presence and migration with clinically relevant outcomes (such as motor coordination) large animal numbers are required per study [53]. This necessitates shorter imaging times per animal in order to increase throughput. Lower resolution images are significantly faster to acquire and have a better SNR, but are likely to result in a partial volume effect that leads to a significant underestimation of the regional distribution of cells. By arraying voxel size and cell number against T1 signal change, a 12 nL voxel size (229 μm isotropic) here provided the optimal intersection of these parameters for in vivo imaging. Although these arrays indicate which parameters provide appropriate imaging conditions, it does not account for what spatial resolution would be required to yield biologically important information. Unfortunately, at present it remains unknown what information would be relevant biologically to determine a relationship between the distribution of transplanted cells and their therapeutic efficacy. Voxel size must therefore be based on a balance between acceptable cell detection thresholds, imaging time constraints, as well as the biological relevance of the information gained from imaging.
Since there is normally very little contrast in a T1-weighted image of the brain, T1 contrast agents are able to generate a very specific signal that is less likely to be confused with naturally occurring phenomena than T2 based agents [4]. However the question of agent specificity to the cells remains an issue. Any agent present outside transplanted cells, whether from the injectate solution or after leakage from labeled cells, will be detected as contrast. Bimodal agents allow validation of MRI contrast using histology, as shown here and elsewhere [41, 54, 55], in order to assess the specificity of cell detection. One area of particular concern is the fate of the label after cell death, as lingering label could produce erroneous contrast [2, 4]. However, there were very few false positives, consistent with reports [16, 56] indicating that at least in the short-term there is little risk to overestimate the implanted cells (i.e. Type I error is <.05%). In contrast, there is a significant number of false negatives, approximately 30% which is higher than what is typically acceptable for a Type II error (20%). Although 100% of cells were labeled prior to injection with a slow proliferation activity in NSCs, the exact source of the underestimation remains unclear. It is conceivable that the fluorescent moiety is detaching and producing an underestimation of labeling using this method of verification. However, there is minimal loss of Gd(III) complexes over two weeks, hence it is unlikely that the fluorescent moiety is detaching this rapidly. Gold nanoparticles are known to eventually undergo a degradation process that leads to their clearance [57], but they have an excellent stability profile [26].
5. Conclusions
We have shown that DNA-Gd@AuNPs have several advantages for the visualization of cell transplants in the brain. Specifically, these nanoconjugates provide an efficient cellular uptake of large quantities of Gd(III) into non-phagocytic NSCs, while preserving a T1 contrast inside cells that affords an robust in vivo detection using T1-weighted MR images. Conjugation of Gd(III) to Au nanoparticles was essential to improve T1 relaxivity of Gd(III) molecules that was attenuated upon intracellular uptake. Further engineering of particles is desirable to potentially further reduce this intracellular attenuation of relaxivity, hence producing an even more efficient contrast agent for cell tracking. Long-term studies with larger group size are nevertheless required to demonstrate the utility of these agents to assess cell survival, as well as graft distribution. Using DNA-Gd@AuNPs offers new opportunities to potentially visualize transplanted cells unequivocally using T1 contrast and use cellular MRI as a tool to derive biologically relevant information that allows us to understand how the survival and location of implanted cells determines therapeutic efficacy.
Supplementary Material
High concentrations of DNA-Gd@AuNPs had a detrimental effect on NSC survival (phase images overlaid with Cy3 moiety of DNA-Gd@AuNPs).
UV/Vis absorbance of citrate-capped and DNA functionalized gold nanoparticles at 25 °C.
Acknowledgments
This study was part funded by the National Institute for Neurological Disease and Stroke (R01NS08226). FJN was supported by a MRC PhD studentship. The authors would like to thank Ms Jessie Liu for her help with quantification of false negatives and positives and Dr Wen Ling for setting up the MR sequences.
Footnotes
7. Author Contributions
FJN performed in vitro and in vivo experiments, post-processed MRI data, and assisted with the first draft of the manuscript. MWR designed and synthesized DNA-Gd@AuNPs, performed shelf life and stability assays and acquired the ICP-MS data. HG performed histology and immunocytochemistry. KWM set-up and acquired MR data on the 1.41 T. TJM conceived of the study, provided funding and oversaw the production and characterization of DNA-Gd@AuNPs. MM conceived of the study, provided funding, oversaw the acquisition and analysis of the cell and in vivo data, and also drafted the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith EJ, Stroemer RP, Gorenkova N, Nakajima M, Crum WR, Tang E, et al. Implantation Site and Lesion Topology Determine Efficacy of a Human Neural Stem Cell Line in a Rat Model of Chronic Stroke. Stem Cells. 2011 doi: 10.1002/stem.1024. [DOI] [PubMed] [Google Scholar]
- 2.Naumova AV, Modo M, Moore A, Murry CE, Frank JA. Clinical imaging in regenerative medicine. Nat Biotechnol. 2014;32:804–18. doi: 10.1038/nbt.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen PK, Riegler J, Wu JC. Stem cell imaging: from bench to bedside. Cell Stem Cell. 2014;14:431–44. doi: 10.1016/j.stem.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modo M, Kolosnjaj-Tabi J, Nicholls F, Ling W, Wilhelm C, Debarge O, et al. Considerations for the clinical use of contrast agents for cellular MRI in regenerative medicine. Contrast Media Mol Imaging. 2013;8:439–55. doi: 10.1002/cmmi.1547. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10901–6. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisanic TR, 2nd, Blackwell JD, Shubayev VI, Finones RR, Jin S. Nanotoxicity of iron oxide nanoparticle internalization in growing neurons. Biomaterials. 2007;28:2572–81. doi: 10.1016/j.biomaterials.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Kim T, Momin E, Choi J, Yuan K, Zaidi H, Kim J, et al. Mesoporous silica-coated hollow manganese oxide nanoparticles as positive T1 contrast agents for labeling and MRI tracking of adipose-derived mesenchymal stem cells. J Am Chem Soc. 2011;133:2955–61. doi: 10.1021/ja1084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillet-Nicolas R, Laprise-Pelletier M, Nair MM, Chevallier P, Lagueux J, Gossuin Y, et al. Manganese-impregnated mesoporous silica nanoparticles for signal enhancement in MRI cell labelling studies. Nanoscale. 2013;5:11499–511. doi: 10.1039/c3nr02969g. [DOI] [PubMed] [Google Scholar]
- 9.Gilad AA, Walczak P, McMahon MT, Na HB, Lee JH, An K, et al. MR tracking of transplanted cells with “positive contrast” using manganese oxide nanoparticles. Magn Reson Med. 2008;60:1–7. doi: 10.1002/mrm.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoki I, Takahashi Y, Chuang KH, Silva AC, Igarashi T, Tanaka C, et al. Cell labeling for magnetic resonance imaging with the T1 agent manganese chloride. NMR Biomed. 2006;19:50–9. doi: 10.1002/nbm.1000. [DOI] [PubMed] [Google Scholar]
- 11.Luo L, Xu H, Li Y, Du Z, Sun X, Ma Z, et al. Manganese-enhanced MRI optic nerve tracking: effect of intravitreal manganese dose on retinal toxicity. NMR Biomed. 2012;25:1360–8. doi: 10.1002/nbm.2808. [DOI] [PubMed] [Google Scholar]
- 12.Terreno E, Geninatti Crich S, Belfiore S, Biancone L, Cabella C, Esposito G, et al. Effect of the intracellular localization of a Gd-based imaging probe on the relaxation enhancement of water protons. Magn Reson Med. 2006;55:491–7. doi: 10.1002/mrm.20793. [DOI] [PubMed] [Google Scholar]
- 13.Cao L, Li B, Yi P, Zhang H, Dai J, Tan B, et al. The interplay of T1- and T2-relaxation on T1-weighted MRI of hMSCs induced by Gd-DOTA-peptides. Biomaterials. 2014;35:4168–74. doi: 10.1016/j.biomaterials.2014.01.073. [DOI] [PubMed] [Google Scholar]
- 14.Shen J, Duan XH, Cheng LN, Zhong XM, Guo RM, Zhang F, et al. In vivo MR imaging tracking of transplanted mesenchymal stem cells in a rabbit model of acute peripheral nerve traction injury. Journal of magnetic resonance imaging : JMRI. 2010;32:1076–85. doi: 10.1002/jmri.22353. [DOI] [PubMed] [Google Scholar]
- 15.Guenoun J, Koning GA, Doeswijk G, Bosman L, Wielopolski PA, Krestin GP, et al. Cationic Gd-DTPA liposomes for highly efficient labeling of mesenchymal stem cells and cell tracking with MRI. Cell Transplant. 2012;21:191–205. doi: 10.3727/096368911X593118. [DOI] [PubMed] [Google Scholar]
- 16.Guenoun J, Ruggiero A, Doeswijk G, Janssens RC, Koning GA, Kotek G, et al. In vivo quantitative assessment of cell viability of gadolinium or iron-labeled cells using MRI and bioluminescence imaging. Contrast Media Mol Imaging. 2013;8:165–74. doi: 10.1002/cmmi.1513. [DOI] [PubMed] [Google Scholar]
- 17.Hsiao JK, Tsai CP, Chung TH, Hung Y, Yao M, Liu HM, et al. Mesoporous silica nanoparticles as a delivery system of gadolinium for effective human stem cell tracking. Small. 2008;4:1445–52. doi: 10.1002/smll.200701316. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Shao Y, He H, Tan Y, Tian X, Xie F, et al. Gadolinium(3+)-doped mesoporous silica nanoparticles as a potential magnetic resonance tracer for monitoring the migration of stem cells in vivo. Int J Nanomedicine. 2013;8:119–27. doi: 10.2147/IJN.S38213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nwe K, Bernardo M, Regino CA, Williams M, Brechbiel MW. Comparison of MRI properties between derivatized DTPA and DOTA gadolinium-dendrimer conjugates. Bioorg Med Chem. 2010;18:5925–31. doi: 10.1016/j.bmc.2010.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao D, Ai T, Goerner F, Hu X, Runge VM, Tweedle M. MRI contrast agents: basic chemistry and safety. Journal of magnetic resonance imaging : JMRI. 2012;36:1060–71. doi: 10.1002/jmri.23725. [DOI] [PubMed] [Google Scholar]
- 21.De Leon-Rodriguez LM, Martins AF, Pinho MC, Rofsky NM, Sherry AD. Basic MR relaxation mechanisms and contrast agent design. Journal of magnetic resonance imaging : JMRI. 2015 doi: 10.1002/jmri.24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warsi MF, Chechik V. Strategies for increasing relaxivity of gold nanoparticle based MRI contrast agents. Phys Chem Chem Phys. 2011;13:9812–7. doi: 10.1039/c0cp02508a. [DOI] [PubMed] [Google Scholar]
- 23.Gambino G, Engelmann J, Tei L, Botta M, Logothetis NK, Mamedov I. Multimodal contrast agents for in vivo neuroanatomical analysis of monosynaptic connections. Biomaterials. 2013;34:7135–42. doi: 10.1016/j.biomaterials.2013.05.064. [DOI] [PubMed] [Google Scholar]
- 24.Song Y, Xu X, MacRenaris KW, Zhang XQ, Mirkin CA, Meade TJ. Multimodal gadolinium-enriched DNA-gold nanoparticle conjugates for cellular imaging. Angew Chem Int Ed Engl. 2009;48:9143–7. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y, Kohlmeir EK, Meade TJ. Synthesis of multimeric MR contrast agents for cellular imaging. J Am Chem Soc. 2008;130:6662–3. doi: 10.1021/ja0777990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira MF, Mousavi B, Ferreira PM, Martins CI, Helm L, Martins JA, et al. Gold nanoparticles functionalised with stable, fast water exchanging Gd3+ chelates as high relaxivity contrast agents for MRI. Dalton Trans. 2012;41:5472–5. doi: 10.1039/c2dt30388d. [DOI] [PubMed] [Google Scholar]
- 27.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21:2250–6. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian X, Shao Y, He H, Liu H, Shen Y, Huang W, et al. Nanoamplifiers synthesized from gadolinium and gold nanocomposites for magnetic resonance imaging. Nanoscale. 2013;5:3322–9. doi: 10.1039/c3nr00170a. [DOI] [PubMed] [Google Scholar]
- 29.Arifin DR, Long CM, Gilad AA, Alric C, Roux S, Tillement O, et al. Trimodal gadolinium-gold microcapsules containing pancreatic islet cells restore normoglycemia in diabetic mice and can be tracked by using US, CT, and positive-contrast MR imaging. Radiology. 2011;260:790–8. doi: 10.1148/radiol.11101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Q, Li K, Wen S, Liu H, Peng C, Cai H, et al. Targeted CT/MR dual mode imaging of tumors using multifunctional dendrimer-entrapped gold nanoparticles. Biomaterials. 2013;34:5200–9. doi: 10.1016/j.biomaterials.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Sun SK, Dong LX, Cao Y, Sun HR, Yan XP. Fabrication of multifunctional Gd2O3/Au hybrid nanoprobe via a one-step approach for near-infrared fluorescence and magnetic resonance multimodal imaging in vivo. Anal Chem. 2013;85:8436–41. doi: 10.1021/ac401879y. [DOI] [PubMed] [Google Scholar]
- 32.Li K, Wen S, Larson AC, Shen M, Zhang Z, Chen Q, et al. Multifunctional dendrimer-based nanoparticles for in vivo MR/CT dual-modal molecular imaging of breast cancer. Int J Nanomedicine. 2013;8:2589–600. doi: 10.2147/IJN.S46177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Y, Zhang D, Wu M, Liu Y, Zhang X, Li L, et al. Lipid-AuNPs@PDA nanohybrid for MRI/CT imaging and photothermal therapy of hepatocellular carcinoma. ACS Appl Mater Interfaces. 2014;6:14266–77. doi: 10.1021/am503583s. [DOI] [PubMed] [Google Scholar]
- 34.Rotz MW, Culver KS, Parigi G, MacRenaris KW, Luchinat C, Odom TW, et al. High relaxivity Gd(III)-DNA gold nanostars: investigation of shape effects on proton relaxation. ACS Nano. 2015;9:3385–96. doi: 10.1021/nn5070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurst SJ, Lytton-Jean AK, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313–8. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohrer M, Bauer H, Mintorovitch J, Requardt M, Weinmann HJ. Comparison of magnetic properties of MRI contrast media solutions at different magnetic field strengths. Invest Radiol. 2005;40:715–24. doi: 10.1097/01.rli.0000184756.66360.d3. [DOI] [PubMed] [Google Scholar]
- 37.Johansson S, Price J, Modo M. Effect of inflammatory cytokines on major histocompatibility complex expression and differentiation of human neural stem/progenitor cells. Stem Cells. 2008;26:2444–54. doi: 10.1634/stemcells.2008-0116. [DOI] [PubMed] [Google Scholar]
- 38.El-Akabawy G, Medina LM, Jeffries A, Price J, Modo M. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the Hedgehog pathway. Stem Cells Dev. 2011;20:1873–87. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 39.Brekke C, Morgan SC, Lowe AS, Meade TJ, Price J, Williams SC, et al. The in vitro effects of a bimodal contrast agent on cellular functions and relaxometry. NMR Biomed. 2007;20:77–89. doi: 10.1002/nbm.1077. [DOI] [PubMed] [Google Scholar]
- 40.Magnotta VA, Friedman L, First B. Measurement of Signal-to-Noise and Contrast-to-Noise in the fBIRN Multicenter Imaging Study. J Digit Imaging. 2006;19:140–7. doi: 10.1007/s10278-006-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bible E, Dell’Acqua F, Solanky B, Balducci A, Crapo PM, Badylak SF, et al. Non-invasive imaging of transplanted human neural stem cells and ECM scaffold remodeling in the stroke-damaged rat brain by (19)F- and diffusion-MRI. Biomaterials. 2012;33:2858–71. doi: 10.1016/j.biomaterials.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmoudi M, Serpooshan V, Laurent S. Engineered nanoparticles for biomolecular imaging. Nanoscale. 2011;3:3007–26. doi: 10.1039/c1nr10326a. [DOI] [PubMed] [Google Scholar]
- 43.Zabow G, Dodd SJ, Koretsky AP. Shape-changing magnetic assemblies as high-sensitivity NMR-readable nanoprobes. Nature. 2015;520:73–7. doi: 10.1038/nature14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mogilireddy V, Dechamps-Olivier I, Alric C, Laurent G, Laurent S, Vander Elst L, et al. Thermodynamic stability and kinetic inertness of a Gd-DTPA bisamide complex grafted onto gold nanoparticles. Contrast Media Mol Imaging. 2015;10:179–87. doi: 10.1002/cmmi.1616. [DOI] [PubMed] [Google Scholar]
- 45.Modo M, Beech JS, Meade TJ, Williams SC, Price J. A chronic 1 year assessment of MRI contrast agent-labelled neural stem cell transplants in stroke. Neuroimage. 2009;47(Suppl 2):T133–42. doi: 10.1016/j.neuroimage.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helm L. Optimization of gadolinium-based MRI contrast agents for high magnetic-field applications. Future medicinal chemistry. 2010;2:385–96. doi: 10.4155/fmc.09.174. [DOI] [PubMed] [Google Scholar]
- 47.Kim KS, Park W, Na K. Gadolinium-chelate nanoparticle entrapped human mesenchymal stem cell via photochemical internalization for cancer diagnosis. Biomaterials. 2015;36:90–7. doi: 10.1016/j.biomaterials.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Cromer Berman SM, Kshitiz, Wang CJ, Orukari I, Levchenko A, Bulte JW, et al. Cell motility of neural stem cells is reduced after SPIO-labeling, which is mitigated after exocytosis. Magn Reson Med. 2013;69:255–62. doi: 10.1002/mrm.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kok MB, Hak S, Mulder WJ, van der Schaft DW, Strijkers GJ, Nicolay K. Cellular compartmentalization of internalized paramagnetic liposomes strongly influences both T1 and T2 relaxivity. Magn Reson Med. 2009;61:1022–32. doi: 10.1002/mrm.21910. [DOI] [PubMed] [Google Scholar]
- 50.Modo M, Mellodew K, Cash D, Fraser SE, Meade TJ, Price J, et al. Mapping transplanted stem cell migration after a stroke: a serial, in vivo magnetic resonance imaging study. Neuroimage. 2004;21:311–7. doi: 10.1016/j.neuroimage.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 51.Caravan P, Farrar CT, Frullano L, Uppal R. Influence of molecular parameters and increasing magnetic field strength on relaxivity of gadolinium- and manganese-based T1 contrast agents. Contrast Media Mol Imaging. 2009;4:89–100. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roine T, Jeurissen B, Perrone D, Aelterman J, Leemans A, Philips W, et al. Isotropic non-white matter partial volume effects in constrained spherical deconvolution. Frontiers in neuroinformatics. 2014;8:28. doi: 10.3389/fninf.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crum WR, Giampietro VP, Smith EJ, Gorenkova N, Stroemer RP, Modo M. A comparison of automated anatomical-behavioural mapping methods in a rodent model of stroke. J Neurosci Methods. 2013;218:170–83. doi: 10.1016/j.jneumeth.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Detante O, Valable S, de Fraipont F, Grillon E, Barbier EL, Moisan A, et al. Magnetic resonance imaging and fluorescence labeling of clinical-grade mesenchymal stem cells without impacting their phenotype: study in a rat model of stroke. Stem Cells Transl Med. 2012;1:333–41. doi: 10.5966/sctm.2011-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Modo M, Cash D, Mellodew K, Williams SC, Fraser SE, Meade TJ, et al. Tracking transplanted stem cell migration using bifunctional, contrast agent-enhanced, magnetic resonance imaging. Neuroimage. 2002;17:803–11. [PubMed] [Google Scholar]
- 56.Pawelczyk E, Jordan EK, Balakumaran A, Chaudhry A, Gormley N, Smith M, et al. In vivo transfer of intracellular labels from locally implanted bone marrow stromal cells to resident tissue macrophages. PLoS One. 2009;4:e6712. doi: 10.1371/journal.pone.0006712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolosnjaj-Tabi J, Javed Y, Lartigue L, Volatron J, Elgrabli D, Marangon I, et al. The One Year Fate of Iron Oxide Coated Gold Nanoparticles in Mice. ACS Nano. 2015 doi: 10.1021/acsnano.5b00042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
High concentrations of DNA-Gd@AuNPs had a detrimental effect on NSC survival (phase images overlaid with Cy3 moiety of DNA-Gd@AuNPs).
UV/Vis absorbance of citrate-capped and DNA functionalized gold nanoparticles at 25 °C.