Abstract
Most membrane and secretory proteins are delivered co-translationally to protein translocation channels in their destination membrane by the signal recognition particle (SRP) and its receptor. This co-translational molecular machinery is conserved across all kingdoms of life, though it varies in composition and function. Here we report recent progress towards understanding the mechanism of SRP function, focusing on findings about E. coli SRP’s conformational dynamics throughout the targeting process. These insights shed light on a key checkpoint in the targeting cycle: how SRP regulates engagement of an actively translating ribosome with the translocation machinery at the membrane.
The SRP targeting cycle
About one third of all proteins function in membranes or traverse membranes for secretion. The signal recognition particle (SRP) is an ancient, universally conserved ribonucleoprotein complex that mediates the delivery of ribosome-nascent chain complexes (RNCs) from the cytosol to protein translocation channels (translocons) in the endoplasmic reticulum membrane in eukaryotes (Sec61αβγ complex) or plasma membrane in prokaryotes (SecYEG complex) [1,2]. SRP specifically recognizes and targets RNCs translating membrane or secretory proteins based on the presence of a hydrophobic topogenic signal sequence, typically embedded in an N-terminal cleavable signal peptide or a transmembrane segment, as the nascent chains become exposed after exiting the ribosomal exit tunnel [3]. In eukaryotes, SRP is composed of six proteins [4] and one 7S RNA molecule [5], while in most bacteria SRP has a single protein, Ffh, bound to either a 4.5S (gram-negative bacteria) or 6S (gram-positive bacteria) RNA [6,7]. Across all species, the SRP RNA minimally has an elongated hairpin terminated in a tetraloop (also known as “helix 8”), with additional features, such as a translation-elongation-arrest Alu domain, being appended in eukaryotes and some prokaryotes [8]. Originally thought to be a passive scaffold for SRP protein organization, it is now becoming apparent that the SRP RNA actively drives key steps in SRP activation and membrane targeting [9–11], nicely rationalizing its phylogenetic conservation.
To identify a signal sequence-containing nascent chain and perform its targeting function, SRP first binds to the large subunit of the ribosome via SRP54’s (Ffh in bacteria) N-terminal four-helix bundle and GTPase domains (NG domain), and to both the ribosome and the hydrophobic signal sequence with its methionine-rich M domain [12,13]. Ffh’s NG and M domains are connected by a 30 amino acid flexible linker, which enables relative mobility of these domains within SRP [14–17]. After selection of an RNC, SRP delivers this cargo to a translocon via an interaction with the SRP receptor (SR, or FtsY in bacteria) [18,19]. SRP and SR have homologous NG domains that together form a reciprocally activating, Ras-like GTPase complex [20,21]. At the membrane, SRP releases the RNC to a translocon in a GTP-dependent manner [22], and SRP and its receptor dissociate from one another and are recycled for subsequent rounds of targeting after GTP hydrolysis [23,24]. The ribosome completes synthesis of the protein while remaining bound to the translocon, thus protecting the nascent chain from cytosolic misfolding and harnessing the energy of translation to both synthesize and translocate the nascent chain.
Over three decades of research since the discovery of SRP in mammals [4] have uncovered a wealth of information about the inner workings of this fascinating molecular machine [25]. However, even as new findings are made at increasing speed, some longstanding questions remain. In this brief review, we discuss exciting recent developments in the field, especially as they relate to the conformational dynamics of the SRP and the light that these findings have shed on the mechanism of co-translational protein targeting. We also discuss new questions that have arisen from these recent findings and intriguing puzzles that still await satisfying explanations.
SRP’s interactions with actively translating ribosomes
In gram-negative bacteria such as Escherichia coli, SRP does not pause translation upon binding to ribosomes [26]. Instead, due to the absence of an elongation-arrest Alu domain in the short 4.5S SRP RNA, translation continues throughout the targeting cycle, necessitating SRP to interact with a conformationally dynamic, translating ribosome and elongating nascent chain. This nascent chain emerges from the exit tunnel and encounters the scrutiny of multiple protein biogenesis factors, including methionine aminopeptidase (MAP), peptide deformylase (PDF), the nascent polypeptide-associated complex (NAC) (or trigger factor in bacteria), and SRP, all of which sample the neighborhood of the ribosomal exit tunnel and compete with one another for nascent chain binding [27–29]. It follows that SRP, itself a conformationally dynamic molecular machine (see below), must nimbly navigate its interactions with a translating RNC, and do so within a restricted time window, as SRP is unable to target RNCs with nascent chains longer than ~140 amino acids [30,31]. Numerous studies have investigated SRP’s ability to bind RNCs at varying points early in the elongation process and have arrived at a range of conclusions [31–33], with difference likely arising from the use of different species, model substrates, modes of translation stalling, and experimental approaches. A limitation of these experiments was the use of stalled, and most likely conformationally and compositionally heterogeneous RNCs, which may influence SRP’s interactions with the RNCs.
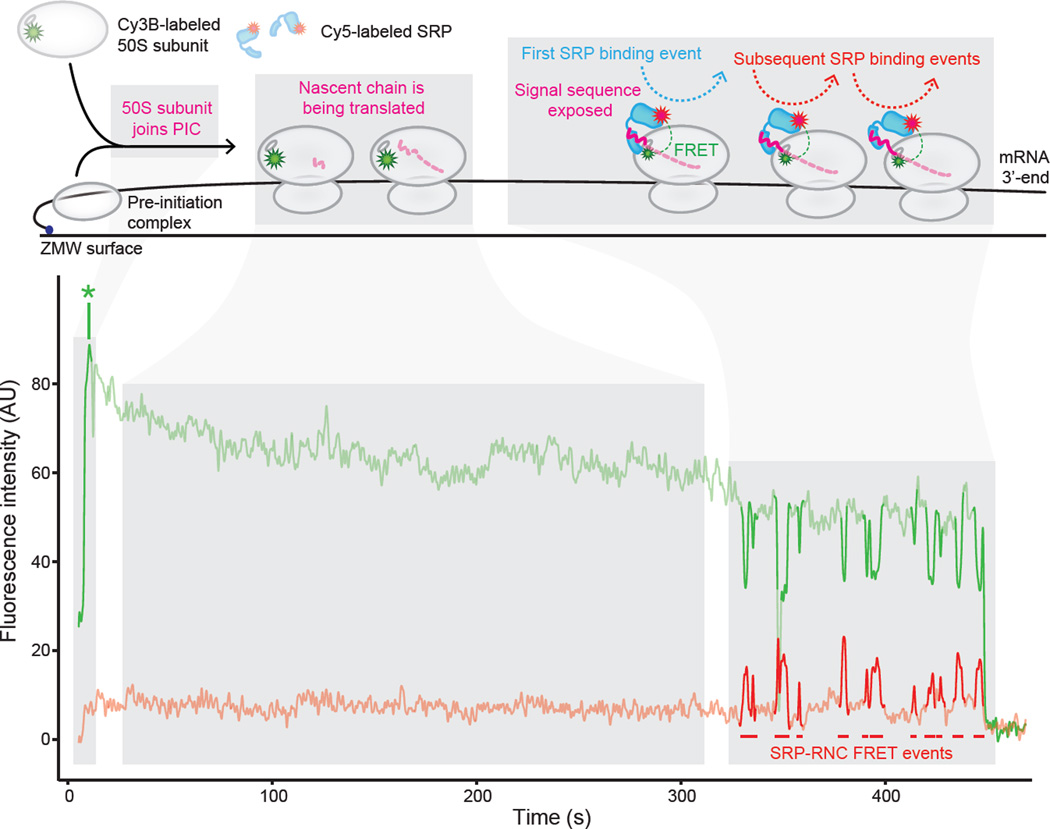
Recently, E. coli SRP’s interactions with individual actively translating ribosomes were studied using a highly-purified translation system and a real-time, single-molecule fluorescence resonance energy transfer-based assay (Figure 1). This study enabled the integration of the key dimension of ribosome dynamics into kinetic measurements of SRP-RNC binding [34]. The approach was pioneered in the Puglisi lab to study real-time transit of tRNAs through actively translating ribosomes [35] and, for this modified assay, employed highly-purified translation factors and site-specifically fluorescently-labeled ribosomes, SRP and tRNAs, as well as zeromode waveguides, to observe binding events with all components present at physiological concentrations [34]. The time series of the fluorescence signal indicated translation initiation (ribosomal subunit association) and elongation along the canonical SRP substrate LepB, and the onset of an anti-correlated donor and acceptor fluorophore signal (i.e. fluorescence resonance energy transfer (FRET)) revealed SRP-binding to actively translating ribosomes and elongating nascent chains. At the temporal resolution of this assay (100 ms), it was demonstrated in agreement with earlier findings [30] that the majority of SRP’s initial binding events (68%) occurred when RNCs have translated between 40 and 55 amino acids (Figure 1), that is, when the signal sequence is first exposed outside the ribosomal exit tunnel [34]. Altering the rate of translation by adjusting the concentration of elongation factor G (EF-G) present in the system showed that SRP-RNC initial binding occurs mainly after signal sequence emergence from the ribosome exit tunnel, regardless of the time it takes to translate a LepB nascent chain of that length. It remains to be seen how the other protein biogenesis factors that compete for nascent chain binding influence SRP’s interaction with actively translating ribosomes in this assay and whether these results obtained in the bacterial system can be generalized to eukaryotic components.
Figure 1. SRP binds to actively translating RNCs after emergence of a signal sequence-containing nascent chain.
Representative single-molecule fluorescence vs. time trajectory of Cy3B-labeled 50S ribosomal subunits, Cy5-labeled SRP, and unlabeled translation mix delivered at time = 0 to pre-initiation complexes assembled on a truncated LepB mRNA (encoding the first 115 amino acids), immobilized in Zero Mode Waveguides, and imaged by TIRF microscopy. The top panel shows a cartoon representation of the molecular events throughout the time series. The bottom panel shows the fluorescence intensity of the Cy3B (green), and Cy5 (red) signals upon 532 nm excitation. “AU” indicates “arbitrary units”. * denotes 50S ribosomal subunit joining. Figure from (Noriega et al. 2014 eLife).
Towards a mechanism for translocon engagement
Following cargo selection, an SRP-RNC complex needs to locate a vacant translocon at the membrane. This process is aided by the SRP receptor, though the mechanism through which the SRP receptor identifies a vacant translocon is unclear. In bacteria, FtsY can be both soluble and peripherally membrane-associated [36] and is thought to cycle between the cytosol and the membrane [37]. FtsY may initially bind SRP in either location [38]. It has a strong preference for binding anionic phospholipids such as PG and cardiolipin [38,39], though how this preference is harnessed in the membrane targeting cycle remains to be determined. From crosslinking experiments it has been shown that FtsY binds to the translocon through its NG domain at residues on cytosolic loops C4 and C5 of SecYEG at a site that overlaps with a binding site of the ribosome with the translocon [40–42]. Structures of the FtsY-SecYEG complex are lacking, however, so the nature of this interaction is still not well understood. The SRP receptor in eukaryotes presents an even more intriguing puzzle as it is composed of two subunits, SRα and SRβ, both of which are GTPases – the former being soluble and the latter being an integral membrane protein [43]. SRα is highly homologous to FtsY and dimerizes with SRβ at the membrane [44]. In yeast, SRβ directly interacts with the translocon (Ssh1p) as shown by a split ubiquitin assay, and this interaction is critical for both cell growth and cotranslational protein targeting [45]. SRβ is one of the translocon’s many interaction partners, however, and there is evidence that its contacts are dynamic and regulated by substrate length and identity [46], thus raising questions about the timing and regulation of the SRβ-translocon interaction during the SRP targeting cycle and translocation process.
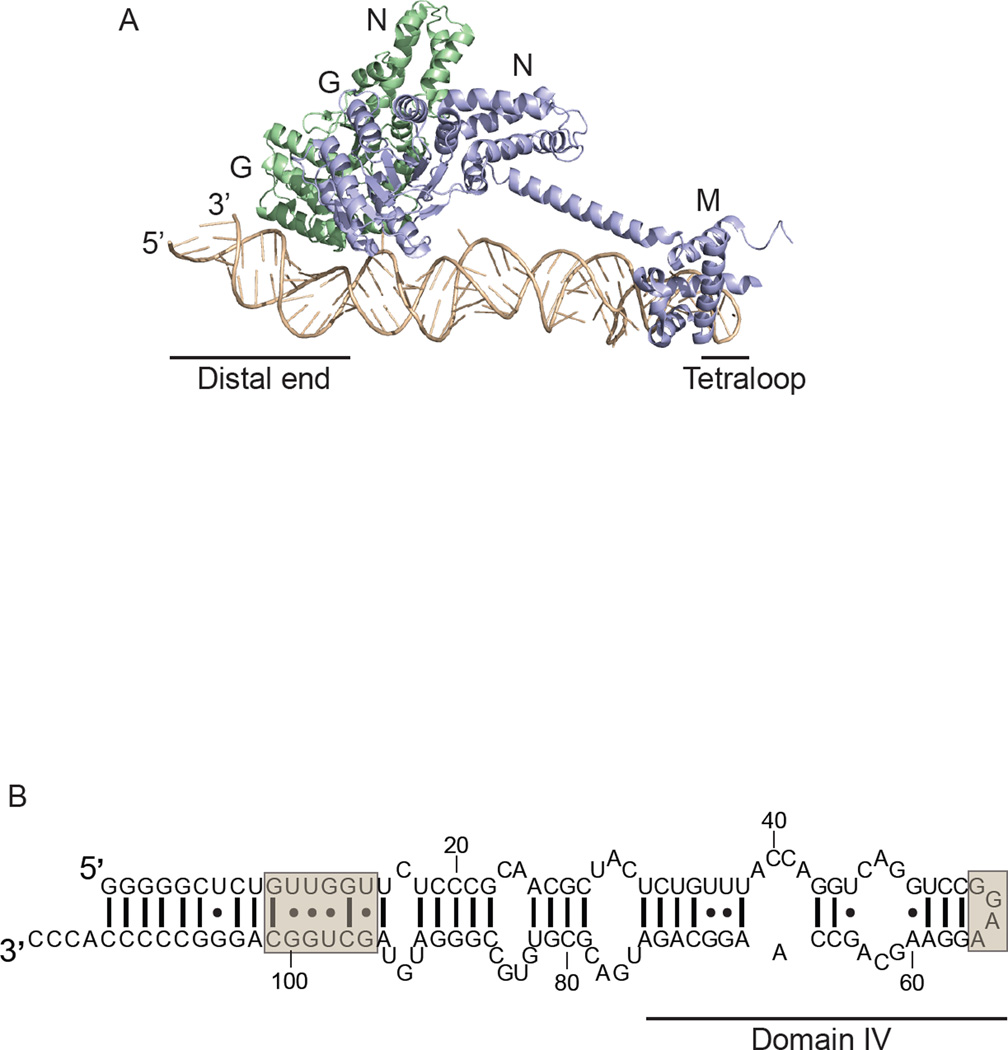
Cryo-EM reconstructions of SRP-RNC complexes [47–49] and RNC-translocon complexes [50–53] show that SRP’s binding site on the large ribosomal subunit partially overlaps with the binding site of the translocon with the ribosome. Considering the mutually exclusive binding sites, it is clear that a large structural rearrangement and/or displacement of both SRP and FtsY is required for RNC-translocon engagement. A major insight into a possible mechanism for the SRP-mediated engagement of RNC-translocon complexes came from the crystal structure [10] of SRP in complex with the FtsY NG domain in the presence of GMPPCP and the nonionic detergent C12E8 acting as a signal peptide mimic [54]. This structure, and a second one trapped with GDP:AlF4 [55], revealed the SRP (Ffh) and FtsY GTPase complex bound at the 5’,3’-end of the SRP RNA hairpin, hereafter referred to as the distal end [10] (Figure 2A).
Figure 2. SRP-FtsY GTPase complex relocalization to the SRP RNA distal end.
A. Crystal structure of SRP in complex with FtsY trapped with GMPPCP. Full-length 4.5S SRP RNA in tan, with the tetraloop and distal end indicated. Ffh in violet with M and NG domains indicated, and FtsY(NG) in green. PDB: 2XXA. The figure was prepared using PyMOL. B. E. coli 4.5S SRP RNA secondary structure. The tetraloop and distal end GTPase complex binding site are indicated in tan boxes.
The importance of this distal binding site of the SRP-FtsY GTPase complex was confirmed biochemically [10,11,55]. By mutagenesis and truncation analysis, it was shown that the distal end of the SRP RNA has residues critical for the SRP RNA-mediated stimulation of GTPase activation and GTP hydrolysis by the SRP-FtsY GTPase complex after initial assembly at the tetraloop end of the SRP RNA. The distal end of the SRP RNA contains two docking sites for the SRP-FtsY GTPase complex: a primary site containing residues G14, U15, G96, and U98, and an auxiliary site at C87, which together ensure complex recruitment and activation (Figure 2B) [11].
By modeling the SRP-FtsY crystal structure on the ribosome, it became apparent that this conformation of the GTPase complex exposes ribosomal proteins L23 and L29, which are the main site of translocon-RNC binding. Thus, a potential mechanism for the exchange of an RNC from SRP to a translocon emerged: by relocalizing the GTPase complex from the proximal, tetraloop binding site to this distal site upon SRP-FtsY GTPase complex assembly, the translocon binding site on the ribosome is vacated, thus facilitating RNC-translocon binding.
Work with mammalian SRP hints at a similar mechanism for exposing the translocon-binding site on the ribosome. Protein crosslinking studies of SRP-RNC interactions in the presence and absence of the SRP receptor [56], showed that in the absence of the SRP receptor, SRP54 is bound to the ribosome near the exit site, but is rearranged upon receptor binding such that it is no longer in the vicinity of the exit site. Thus, the exit site may be vacated for translocon-binding by mammalian SRP in a mode similar to the model for E. coli SRP, though detailed biochemical and structural studies are needed to confirm this due to mammalian SRP’s extra structural features compared with E. coli SRP.
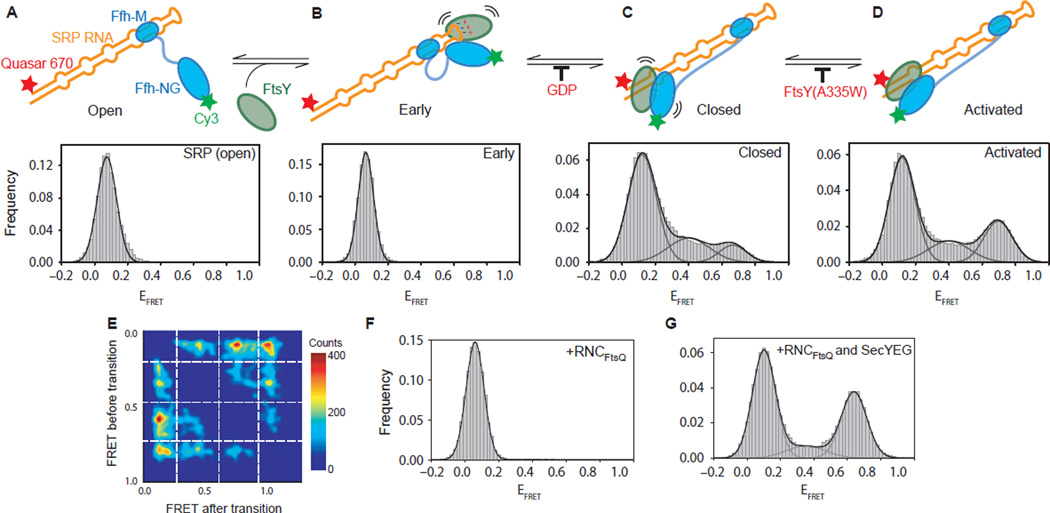
To test more directly the model proposed from the SRP-FtsY crystal structure, single-molecule FRET experiments were performed with fluorescently labeled 4.5S RNA and Ffh-FtsY GTPase complex (labeled either on FtsY-NG or Ffh-NG) to monitor the conformational dynamics of individual E. coli SRP molecules during the membrane targeting cycle [9]. In these experiments, SRP alone was observed to stably bind a low, ~0.1 FRET efficiency (EFRET) state that was assigned to the proximal, tetraloop SRP RNA binding site (Figure 3A). The same observation was made in the presence of FtsY (Figure 3B), however in the presence of FtsY and GMPPNP, the SRP-FtsY GTPase complex showed dynamic transitions between the proximal conformational state and one with high, ~0.8 EFRET (Figure 3C & D). The high EFRET state was assigned to the distal binding site, 100 Å away from the tetraloop (Figure 2A & 2B). The GTPase complex rapidly sampled multiple binding sites on the path between its distal and proximal SRP RNA binding sites as evidenced by numerous intermediate states in the single-molecule FRET (smFRET) transition density plot (Figure 3E) [9]. This was also suggested by the absence of strong electron density at either the distal or proximal binding sites in the cryo-EM reconstruction of the RNC-SRP-FtsY GTP-bound closed state complex [12]. Addition of an RNC carrying a bona fide SRP substrate, RNCFtsQ, further altered this conformational equilibrium by abolishing the GTPase movements on the RNA and stabilizing the complex in just the low EFRET, proximal state (Figure 3F). This was specific to correct SRP substrates, as an RNC with a nascent chain lacking a signal sequence, RNCluciferase, had no effect. RNCs delay GTPase activation [57,58], but this pausing was reversed in the presence of detergent-solubilized SecYEG (Figure 3G). SecYEG restored the distal, high EFRET state and restored efficient GTP hydrolysis. Thus, RNCs negatively regulate the GTPase complex’s movement to the distal state, preventing premature GTP hydrolysis, but the addition of SecYEG drives relocalization to the distal site and reactivates GTP hydrolysis, demonstrating that SRP’s conformational dynamics are regulated and critical to ensuring the timing of GTP hydrolysis and fidelity of protein targeting.
Figure 3. Conformational rearrangements within the SRP-FtsY GTPase complex drive its movement to the RNA distal site.
A-D. smFRET histograms of (A) free SRP in the open state and of the Ffh-FtsY complex in the (B) early, (C) closed, and (D) activated states. (E) Transition density plot depicting the range of EFRET values sampled by the Ffh-FtsY (GMPPNP) GTPase complex movements. The plot depicts idealized EFRET before a transition versus EFRET after the transition as 2-D population histograms. At least two distinct intermediate states, M1 and M2, are sampled by the Ffh-FtsY GTPase complex along the path between the proximal and distal sites of the SRP RNA. (F) SRP-FtsY complex bound to RNCFtsQ, and (G) SRP-FtsY complex bound to RNCFtsQ and SecYEG. Figure modified from (Shen et al. 2012 Nature).
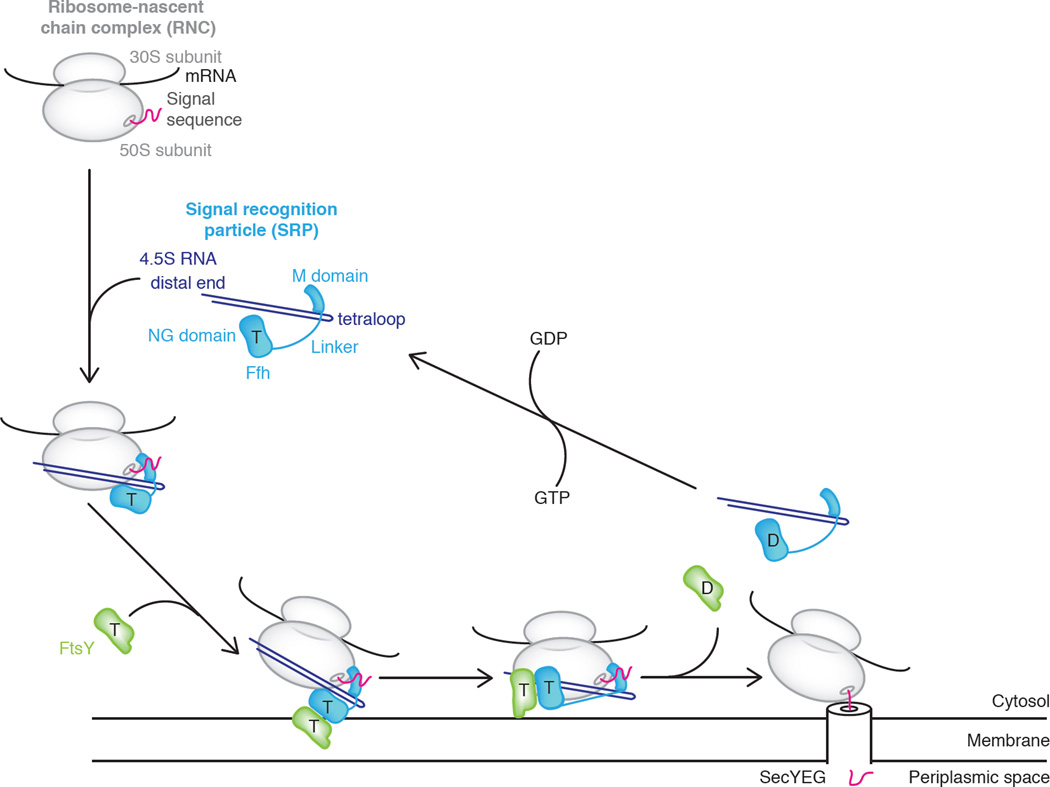
The molecular gymnastics of the SRP-FtsY GTPase complex – its reversible relocalization from the proximal binding site on the SRP RNA to the distal binding site through a trial and error search process – are likely a critical step in ensuring productive exchange of the targeting and translocation machineries at the ribosomal exit tunnel binding site (Figure 4). It remains to be seen, however, precisely how the SRP-mediated RNC-translocon engagement is executed after the GTPase complex is stabilized at the SRP RNA distal site, as well as when and how the signal sequence is transferred from SRP’s M domain to the translocon, and the timing of SRP’s and the SRP receptor’s dissociation from the RNC relative to translocon engagement. There is support for the existence of a transient quaternary RNC-SRP-SR-translocon complex [9,59] and thus support for a model involving concerted exchange of binding partners, however structural information on this complex, which would truly inform this mechanism, is lacking.
Figure 4. Mechanistic model of co-translational protein targeting.
E. coli SRP is composed of 4.5S RNA (violet) and Ffh (blue). SRP binds a ribosome-nascent chain complex (RNC) (gray) and exposed signal sequence (magenta) in its GTP-bound state (“T”). Ffh associates with FtsY (green), driving membrane localization. SecYEG promotes a conformational rearrangement of the Ffh-FtsY GTPase complex from the proximal to the distal site on 4.5S RNA, allowing transfer of the RNC onto SecYEG. GTP hydrolysis triggers SRP-FtsY disassembly and recycling (GDP = “D”).
Old dog, new tricks
In the cell, there is one SRP molecule for every 100 ribosomes [60]. For many years, the field has tried to reconcile this large imbalance in concentrations and explain how SRP is able to rapidly sample all these ribosomes and effectively identify its cargo in a crowded cellular environment. The simple comparison of in vivo concentrations may be oversimplifying the picture, however. It remains to be seen whether SRP targets all membrane-targeted ribosomes, or only the “pioneering” ribosome within a polysome translating any given mRNA. Additionally, it is debated whether ribosomes are recycled to the cytosol after translation termination [61], or remain bound to the membrane for subsequent translation initiation [62], which would further alter the number of ribosomes that SRP needs to scrutinize.
There is also a growing body of work that points to SRP-independent membrane targeting pathways [61,63] and new roles for SRP beyond its well-studied canonical membrane targeting pathway, thus expanding what we know about protein targeting and SRP’s cellular functions. For example, it has recently been shown that σ32, the central transcription factor driving the heat shock response, requires membrane-, not cytosolic-, localization for proper signaling function and that membrane delivery is accomplished via a direct interaction with SRP and its receptor despite σ32’s lack of a canonical signal sequence [64]. This finding points to an important new regulatory role for SRP in protein-folding homeostasis, and raises the possibility that the cell may also use the co-translational targeting machinery to functionally juxtapose other components without engaging the translocation machinery.
Conclusions
Despite many years of study, SRP continues to surprise us with its varied and sophisticated mechanisms for recognizing and targeting proteins destined for the membrane. Recent studies on the structure and dynamics of the SRP-FtsY GTPase complex, as well as detailed analysis of the SRP RNA features that regulate these conformational rearrangements, indicate another critical role for the SRP RNA in regulating SRP-FtsY GTPase complex movements and catalyzing GTP hydrolysis in addition to its other roles in the Ffh-FtsY GTPase cycle [25]. Dynamic assays have also revealed SRP’s interactions with actively translating ribosomes – a critical aspect of its mechanism that has long been overlooked – and shown that its recruitment to RNCs is dependent on the emergence of a signal sequence from the ribosome’s exit tunnel. It will be exciting to see what additional mechanistic features SRP has hidden from view as more tools become available to dissect this molecular machine.
Acknowledgements
This work was supported by the Howard Hughes Medical Institute and National Institutes of Health grant GM032384 (to M.M.E. and P.W.). The authors thank Thomas R. Noriega and Ciara M. Gallagher for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of special interest (*) or outstanding interest (**)
- 1.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 2.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 3.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 6.Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 7.Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- 8.Samuelsson T, Zwieb C. The Signal Recognition Particle Database (SRPDB) Nucleic Acids Res. 1999;27:169–170. doi: 10.1093/nar/27.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen K, Arslan S, Akopian D, Ha T, Shan SO. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature. 2012;492:271–275. doi: 10.1038/nature11726. A single-molecule FRET-based study of the Ffh-FtsY GTPase complex’s conformational dynamics and their regulation by RNCs and SecYEG.
- 10. Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. SRP-FtsY (GMPPCP) complex showing re-localization of the GTPase complex from the proximal, tetraloop end of 4.5S RNA to distal end.
- 11. Shen K, Wang Y, Hwang Fu YH, Zhang Q, Feigon J, Shan SO. Molecular mechanism of GTPase activation at the signal recognition particle (SRP) RNA distal end. J Biol Chem. 2013;288:36385–36397. doi: 10.1074/jbc.M113.513614. Systematic analysis of the SRP RNA residues that are essential to Ffh-FtsY GTPase complex activation and GTP hydrolysis.
- 12.von Loeffelholz O, Jiang Q, Ariosa A, Karuppasamy M, Huard K, Berger I, Shan SO, Schaffitzel C. Ribosome-SRP-FtsY cotranslational targeting complex in the closed state. Proc Natl Acad Sci U S A. 2015;112:3943–3948. doi: 10.1073/pnas.1424453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci U S A. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc Natl Acad Sci U S A. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buskiewicz I, Kubarenko A, Peske F, Rodnina MV, Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 22.Valent QA, Scotti PA, High S, de Gier JW, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 24.Connolly T, Gilmore R. The signal recognition particle receptor mediates the GTPdependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- 25. Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. A comprehensive recent overview of SRP and co-translational protein targeting.
- 26.Raine A, Ullers R, Pavlov M, Luirink J, Wikberg JE, Ehrenberg M. Targeting and insertion of heterologous membrane proteins in E. coli. Biochimie. 2003;85:659–668. doi: 10.1016/s0300-9084(03)00130-5. [DOI] [PubMed] [Google Scholar]
- 27.Gloge F, Becker AH, Kramer G, Bukau B. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol. 2014;24:24–33. doi: 10.1016/j.sbi.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Fedyukina DV, Cavagnero S. Protein folding at the exit tunnel. Annu Rev Biophys. 2011;40:337–359. doi: 10.1146/annurev-biophys-042910-155338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ariosa A, Lee JH, Wang S, Saraogi I, Shan SO. Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1422594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–1775. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 32.Holtkamp W, Lee S, Bornemann T, Senyushkina T, Rodnina MV, Wintermeyer W. Dynamic switch of the signal recognition particle from scanning to targeting. Nat Struct Mol Biol. 2012;19:1332–1337. doi: 10.1038/nsmb.2421. [DOI] [PubMed] [Google Scholar]
- 33.Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 34. Noriega TR, Chen J, Walter P, Puglisi JD. Real-time observation of signal recognition particle binding to actively translating ribosomes. Elife. 2014;3 doi: 10.7554/eLife.04418. This study harnessed the power of single-molecule FRET to study individual SRP molecules’ interactions with translating ribosomes, capturing both the spatial and temporal dimensions of SRP-RNC binding.
- 35.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millman JS, Andrews DW. A site-specific, membrane-dependent cleavage event defines the membrane binding domain of FtsY. J Biol Chem. 1999;274:33227–33234. doi: 10.1074/jbc.274.47.33227. [DOI] [PubMed] [Google Scholar]
- 37.Luirink J, ten Hagen-Jongman CM, van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam VQ, Akopian D, Rome M, Henningsen D, Shan SO. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J Cell Biol. 2010;190:623–635. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Leeuw E, te Kaat K, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelini S, Boy D, Schiltz E, Koch HG. Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J Cell Biol. 2006;174:715–724. doi: 10.1083/jcb.200606093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 2005;6:476–481. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch HG. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic. 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- 43.Tajima S, Lauffer L, Rath VL, Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz T, Blobel G. Structural basis for the function of the beta subunit of the eukaryotic signal recognition particle receptor. Cell. 2003;112:793–803. doi: 10.1016/s0092-8674(03)00161-2. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conti BJ, Devaraneni PK, Yang Z, David LL, Skach WR. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell. 2015;58:269–283. doi: 10.1016/j.molcel.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voorhees RM, Hegde RS. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife. 2015;4 doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 49.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 50. Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. High-resolution cryo-EM structure of mammalian RNC-translocon complex, exemplifying the ability to achieve high-resolution structural information from a relatively small number of heterogeneous particles.
- 51. Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. SecYEG was reconstituted in a lipid environment in nanodiscs, and the structure of the RNC-translocon was solved with cryo-EM and molecular dynamics simulations, revealing ribosome-lipid interactions.
- 52.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, 3rd, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 54.Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voigts-Hoffmann F, Schmitz N, Shen K, Shan SO, Ataide SF, Ban N. The structural basis of FtsY recruitment and GTPase activation by SRP RNA. Mol Cell. 2013;52:643–654. doi: 10.1016/j.molcel.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Rashid R, Wang K, Shan SO. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science. 2010;328:757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akopian D, Dalal K, Shen K, Duong F, Shan SO. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J Cell Biol. 2013;200:397–405. doi: 10.1083/jcb.201208045. Kinetic evidence for a transient RNC-SRP-FtsY-SecYEG quaternary complex.
- 60.Jensen CG, Pedersen S. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: the stability of Ffh protein is dependent on the concentration of 4.5S RNA. J Bacteriol. 1994;176:7148–7154. doi: 10.1128/jb.176.23.7148-7154.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. Comprehensive analysis of mRNAs undergoing protein synthesis at the ER membrane, revealing SRP-dependent and -independent proteins.
- 62.Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16:221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aviram N, Schuldiner M. Embracing the void--how much do we really know about targeting and translocation to the endoplasmic reticulum? Curr Opin Cell Biol. 2014;29:8–17. doi: 10.1016/j.ceb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Lim B, Miyazaki R, Neher S, Siegele DA, Ito K, Walter P, Akiyama Y, Yura T, Gross CA. Heat shock transcription factor sigma32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol. 2013;11:e1001735. doi: 10.1371/journal.pbio.1001735. [DOI] [PMC free article] [PubMed] [Google Scholar]