Abstract
Background
Two apolipoprotein L1 gene (APOL1) renal-risk variants in donors and African American (AA) recipient race are associated with worse allograft survival in deceased-donor kidney transplantation (DDKT) from AA donors. To detect other factors impacting allograft survival from deceased AA kidney donors, APOL1 renal-risk variants were genotyped in additional AA kidney donors.
Methods
APOL1 genotypes were linked to outcomes in 478 newly analyzed DDKTs in the Scientific Registry of Transplant Recipients. Multivariate analyses accounting for recipient age, sex, race, panel reactive antibody level, HLA match, cold ischemia time, donor age, and expanded-criteria donation were performed. These 478 transplantations and 675 DDKTs from a prior report were jointly analyzed.
Results
Fully-adjusted analyses limited to the new 478 DDKTs replicated shorter renal allograft survival in recipients of APOL1-two-renal-risk-variant kidneys (HR 2.00; p=0.03). Combined analysis of 1153 DDKTs from AA donors revealed donor APOL1 high-risk genotype (HR 2.05; p=3×10−4), older donor age (HR 1.18; p=0.05), and younger recipient age (HR 0.70; p=0.001) adversely impacted allograft survival. Although prolonged allograft survival was seen in many recipients of APOL1-two-renal-risk-variant kidneys, follow-up serum creatinine concentrations were higher than in recipients of zero/one APOL1-renal-risk variant kidneys. A competing risk analysis revealed that APOL1 impacted renal allograft survival, but not recipient survival. Interactions between donor age and APOL1 genotype on renal allograft survival were non-significant.
Conclusions
Shorter renal allograft survival is reproducibly observed after DDKT from APOL1-two-renal-risk-variant donors. Younger recipient age and older donor age have independent adverse effects on renal allograft survival.
Introduction
Kidneys transplanted from deceased African American (AA) donors fail more rapidly than those from deceased European American (EA) donors.(1) However, kidneys from AA deceased donors with two apolipoprotein L1 gene (APOL1)-renal-risk variants are at increased risk for early allograft failure relative to kidneys from AAs with zero or one APOL1-renal-risk variants.(2,3) Thirteen percent of AAs have two APOL1-renal-risk variants and are at high risk for nephropathy.(4,5) Presence of two APOL1-renal-risk variants in donors, but not recipients, appears to translate into heightened risk for earlier allograft failure after deceased-donor kidney transplantation (DDKT).(2,3,6)
APOL1 genotyping in deceased donors may therefore improve the prediction of transplantation outcomes relative to the AA donor ethnicity variable in the Kidney Donor Profile Index (KDPI).(7,8) KDPI treats all kidneys of potential AA deceased donors as at equivalent high risk for allograft failure. In a recent report, kidneys from AA donors with fewer than two APOL1-renal-risk variants appeared to fare similarly after DDKT as kidneys from EA donors.(3) Population-based estimates suggest that 87% of AA deceased kidney donors lack two APOL1-renal-risk variants,(4,5) i.e. their kidneys may be less likely to fail after DDKT. Appropriate assessment of the likelihood for long-term allograft survival remains critical given limitations in the availability of donor kidneys.
Effects of donor age on outcomes of DDKT from APOL1-two-renal-risk-variant donors have not been explored. Absence of proteinuria and normal estimated glomerular filtration rate (eGFR) at organ procurement in younger donors may not reflect future risk for allograft failure after transplantation.(9,10) Younger donors of APOL1 genetically high-risk kidneys may not yet demonstrate their final renal phenotype, whereas older donors may have escaped second hits necessary for initiation of nephropathy and have lower risk of allograft failure after transplantation.(11,12) The present analyses tested for replication of the previously reported adverse relationship between APOL1-renal-risk variants on outcomes in an independent sample of DDKTs from AA donors.(3) The larger sample in a combined analysis with the prior report enhanced statistical power to detect additional factors impacting renal allograft survival from AA donors, including age at donation based on APOL1 genotypes.
Methods
Samples and Outcomes
DNA from deceased AA kidney donors at Emory University School of Medicine and from Genomics of Deterioration of Kidney Allograft study (DeKAF Genomics) was sent to Wake Forest School of Medicine (WFSM) for APOL1-renal-risk-variant genotyping. DeKAF Genomics received samples from Organ Procurement Organizations (OPOs) including LifeSource (Minnesota), LifeQuest (Florida), New Jersey Organ & Tissue Sharing Network, Organ Donor Center of Hawaii, Southwest Transplant Alliance (Texas), One Legacy (California), New England Organ Bank (Massachusetts), LifeBanc (Ohio), and Louisiana Organ Procurement Agency. Samples were identified by United Network of Organ Sharing (UNOS) identification numbers. This study used data from the Scientific Registry of Transplant Recipients (SRTR). WFSM received Institutional Review Board approval for genotyping DNA samples and linking outcomes to kidney recipients based on UNOS identification numbers in SRTR.(13) The SRTR data system includes data on all donors, wait-listed candidates, and transplant recipients in the United States, as submitted by the members of the Organ Procurement and Transplantation Network (OPTN). The Health Resources and Services Administration in the United States Department of Health and Human Services provides oversight to the activities of the OPTN and SRTR contractors. The clinical and research activities reported are consistent with the Principles of the Declaration of Istanbul as outlined in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
The main analysis was performed in 478 transplantations of kidneys recovered and/or transplanted at Emory University (N=230) and DeKAF Genomics (N=248), combined with results previously published in 675 DDKTs from Wake Forest School of Medicine and University of Alabama at Birmingham, a total of 1153 DDKTs performed at 113 centers.(3) In addition, an analysis limited to the 478 new transplantations from Emory University and DeKAF Genomics sources was performed.
Genotyping
Two single nucleotide polymorphisms (SNPs) in the APOL1 G1-renal-risk allele (rs73885319; rs60910145) and an insertion/deletion for the G2-renal-risk allele (rs143830837) were genotyped using a custom assay designed at WFSM on the Sequenom platform (San Diego, California). Genotype calls were visually inspected for quality control.(3,14) Genotyping of 15 blind duplicates resulted in a concordance rate of 100% and the genotyping efficiency for the three SNPs was >99% in all 1153 DDKTs.
Statistical Analysis
The distribution of demographic variables for recipients and deceased kidney donors, based on donor APOL1-risk genotypes, was contrasted using Wilcoxon two-sample tests (continuous variables) and chi-square tests (binary variables).(3) The main outcome was time to renal allograft failure, determined by the interval between the date of transplantation and the date of allograft loss. In those with a functioning allograft, the final observation date was censored for death with function or at last follow-up prior to November 30, 2013. Cox proportional hazard models were subsequently fitted restricting analyses to the first six years of follow-up.(15–17) The sandwich estimator was used to obtain a robust estimation of covariance matrix associated with the parameter estimates to account for the correlation between allograft failure rate and time to failure of kidneys donated by a single individual to two recipients.(18)
Competing risk models were also fitted where we defined two causes of failure: death and kidney allograft failure. Coding was then revised such that date of final observation was censored at death for individuals who died with a functioning allograft or at most recent follow-up before November 30, 2013 for living individuals with a functioning allograft. Fine and Gray’s model tested for association between APOL1 and allograft failure or death with allograft function.(19) This model was fitted using the R package (crrSC), which uses weighted estimating equations to account for the correlation between kidneys donated by a single individual to two recipients.(20) Missing genotype and phenotype data were excluded. The variables considered in this analysis have low counts of missing data (<5%), limiting the appeal for data imputation techniques. Deceased-donor age and recipient age were categorized using the outcome-oriented approach of Contal and O’Quigley,(21) suggesting cut-points for donor age at 20, 35 and 45 years, and recipient age at 30 and 45 years. Therefore, analyses treated donor-age groups 0–20, 20–35, 35–45, and 45+ years and recipient-age groups 0–30, 30–45, and 45+ years as ordinal variables.
Results
Table 1 lists demographic characteristics of the full sample of 1153 kidney transplantations from Wake Forest (N=454), University of Alabama at Birmingham (N=221), Emory University (N=230) and DeKAF Genomics (N=248), based on the number of APOL1-renal-risk variants (Supplementary Table S1 lists demographic characteristics of recipients and deceased AA kidney donors from Emory University and DeKAF Genomics). Unique kidney transplantations were analyzed; there was no overlap between recipients at these transplant centers. Both kidneys from 529 donors were engrafted separately; one kidney was engrafted from 95 donors (624 unique donors). Of the DDKTs from these four sources, 1014 were first-transplantations and 139 were re-transplantations. Immunosuppression varied between patients and centers, but typically included antibody induction with calcineurin inhibitor and an anti-proliferative agent, with or without corticosteroids. The median (first quartile, third quartile) follow-up duration after engraftment was 36.0 months (23.5, 60.0 months) for the 981 kidneys from donors with fewer than two APOL1-renal-risk variants and 36.0 months (17.9, 60.1 months) for the 172 kidneys from donors with two APOL1-renal-risk variants. In addition to higher rates of allograft failure in recipients of kidneys from donors with two APOL1-renal-risk variants, the four-source combined results (as well as results from Emory University + DeKAF Genomics) demonstrated higher serum creatinine concentration at most recent follow-up in recipients of genetically high-risk kidneys (Table 1; Supplementary Table S1). Relative to recipients of kidneys from donors with fewer than two APOL1-renal-risk-variants, recipients of two-renal-risk-variant kidneys did not have significantly higher rates of acute rejection or delayed allograft function (Table 1).
Table 1.
Demographic data for 1153 deceased-donor kidney transplant recipients, based on APOL1 genotypes of the African American donor.
| Variable | APOL1=0, N=462 | APOL1=1, N=519 | APOL1=2, N=172 | P-value |
|---|---|---|---|---|
| Panel reactive antibodies (%) | 24.5 (33.6) 5 | 24.2 (32.9) 6 | 26.9 (35.3) 7.5 | 0.35 |
| Donor age (years) | 34.4 (16.8) 34 | 36.1 (16.6) 38 | 35.5 (15.1) 37 | 0.41 |
| Donor terminal serum creatinine concentration (mg/dl) | 1.2 (0.6) 1.1 | 1.3 (0.7) 1.2 | 1.2 (0.5) 1.2 | 0.002 |
| Recipient age (years) | 48.0 (14.9) 50 | 48.7 (15.7) 51 | 47.8 (13.8) 49 | 0.51 |
| Recipient body mass index (kg/m2) | 28.0 (5.7) 27.6 | 26.9 (5.5) 26.5 | 28.2 (5.7) 27.8 | 0.007 |
| Cold ischemia time (hours) | 20.7 (10.6) 18.9 | 20.7 (10.8) 18.6 | 20.3 (8.8) 20 | 0.78 |
| HLA mismatches (N) | 4.3 (1.5) 5 | 4.3 (1.3) 5 | 4.2 (1.5) 4 | 0.56 |
| Allograft survival (months) | 42.2 (29.7) 36 | 43.1 (27.7) 36 | 43.4 (35.0) 36 | 0.52 |
| Recipient last follow-up serum creatinine (mg/dl) | 1.6 (0.9) 1.3 | 1.6 (0.8) 1.4 | 1.9 (1.5) 1.5 | 0.05 |
| Recipient last follow-up MDRD eGFR (ml/min/1.73m2) | 63.8 (36.3) 59.6 | 62.6 (50.7) 56 | 57.6 (41.3) 51.5 | 0.06 |
| Recipient gender male (N, %) | 272 (58.9%) | 311 (59.9%) | 108 (62.8%) | 0.67 |
| Donor gender male (N, %) | 275 (59.5%) | 312 (60.1%) | 97 (56.4%) | 0.69 |
| Recipient ethnicity African American (N, %) | 235 (50.9%) | 290 (55.9%) | 111 (64.5%) | 0.008 |
| Standard-criteria donor (N, %) | 385 (83.3%) | 417 (80.3%) | 148 (86.0%) | 0.19 |
| Non-heart-beating donor (N, %) | 21 (4.6%) | 16 (3.1%) | 5 (2.9%) | 0.40 |
| Allograft failure within 6 months (N, %) | 17 (3.7%) | 18 (3.5%) | 14 (8.1%) | 0.02 |
| Allograft failure within 1 year (N, %) | 22 (4.8%) | 31 (6.0%) | 15 (8.7%) | 0.17 |
| Allograft failure, total (N, %) | 61 (13.2%) | 93 (17.9%) | 41 (23.8%) | 0.005 |
| Recipient death (N, %) | 72 (15.6%) | 99 (19.1%) | 30 (17.4%) | 0.36 |
| Recipient return to dialysis (N, %) | 61 (13.2%) | 95 (18.3%) | 37 (21.5%) | 0.02 |
| Death with functioning allograft (N, %) | 48 (10.4%) | 51 (9.8%) | 16 (9.3%) | 0.91 |
| Recipient acute rejection (N, %) | 74 (16.0%) | 106 (20.4%) | 36 (20.9%) | 0.15 |
| Recipient first-week dialysis (N, %) | 100 (21.6%) | 120 (23.1%) | 47 (27.3%) | 0.32 |
| Recipient induction immunosuppression (N, %) | 411 (90.5%) | 432 (84.5%) | 143 (85.6%) | 0.02 |
Data presented as mean (SD) median, unless otherwise specified
Multivariate association analyses between allograft failure and APOL1 genotypes (recessive model) in the 1153 DDKTs are presented in Table 2. For all transplanted kidneys from deceased AA donors, a multivariate analysis adjusting for recipient age, sex, race, center and the race by center interaction term (center is defined as site of kidney procurement and/or transplantation and where DNA was collected), HLA match, cold ischemia time (CIT), panel reactive antibodies (PRA), donor age, and donor type (the full model) revealed significant effects on time to allograft failure for APOL1-two-renal-risk-variant-donor kidneys (HR 2.05; p=3×10−4), older donor age (HR 1.18, p=0.05), and younger recipient age (HR 0.70, p=0.001). We observed that 66.1%, 59.1%, and 53.0% of AA deceased donor kidney DNA samples provided from UAB, Emory, and Wake Forest, respectively, were engrafted in AA recipients, compared to 44.3% in DeKAF (Minnesota) donors, p= 2.3×10−5. This observation motivated the inclusion of the recipient race by center interaction term in the model. This interaction term had a p-value of 0.06, suggesting a marginal interaction effect between these two variables on allograft survival (Table 2). No evidence of an interaction effect was observed between induction immunosuppression and transplant center, or between induction immunosuppression and APOL1 status of the deceased kidney donor on allograft survival (interaction p-values 0.98 and 0.40, respectively). A similar multivariate survival analysis limited to the 478 new DDKTs linked to Emory University and DeKAF Genomics revealed that kidneys from deceased donors with two APOL1-renal-risk alleles failed more rapidly than did those from donors with fewer than two APOL1-nephropathy alleles (HR 2.00; p=0.03), with independent adverse effects of recipient AA race, younger recipient age, and receipt of an expanded-criteria donor (ECD) kidney (Table 2).
Table 2.
Multivariate association results for time to renal allograft failure, based on APOL1 genotype (recessive model) in the full model (excluding recipient diabetes mellitus, BMI, dialysis vintage and induction immunosuppression)
| Dataset | Variable | Hazard Ratio |
95% Confidence Interval |
P-value |
|---|---|---|---|---|
| Full dataset: Emory University, DeKAF Genomics, Wake Forest + University of Alabama at Birmingham (N=1153) 881 in analysis 178 graft failures |
APOL1 (recessive model) | 2.05 | (1.39,3.02) | 0.0003 |
| Increasing donor age | 1.18 | (1.00,1.40) | 0.05 | |
| Maximum panel reactive antibodies | 1.00 | (1.00,1.01) | 0.45 | |
| Increasing recipient age | 0.70 | (0.56,0.87) | 0.001 | |
| Number of HLA mismatches | 0.94 | (0.85,1.04) | 0.25 | |
| Cold ischemia time | 1.00 | (0.99,1.02) | 0.58 | |
| Standard-criteria donor (Yes) | 0.70 | (0.43,1.12) | 0.13 | |
| Recipient gender (Female) | 0.97 | (0.70,1.35) | 0.87 | |
| Source/Recipient ethnicity (ref WFU/Other) | Overall interaction p-value 0.06 | |||
| WFU/AA | 1.36 | (0.86,2.15) | 0.18 | |
| DeKAF Genomics/AA | 1.12 | (1.23,6.96) | 0.01 | |
| DeKAF Genomics/Other | 0.28 | (0.13,0.60) | 0.001 | |
| Emory/AA | 0.80 | (0.40,2.18) | 0.89 | |
| Emory/Other | 0.62 | (0.31,1.25) | 0.18 | |
| UAB/AA | 0.10 | (0.49,11.32) | 0.28 | |
| UAB/Other | 0.17 | (0.04,0.75) | 0.02 | |
| Emory University + DeKAF Genomics (N=478) 360 in analysis 77 graft failures |
APOL1 (recessive model) | 2.00 | (1.06,3.74) | 0.03 |
| Increasing donor age | 1.24 | (0.96,1.60) | 0.10 | |
| Maximum panel reactive antibodies | 1.00 | (0.99,1.01) | 0.91 | |
| Increasing recipient age | 0.50 | (0.35,0.71) | <0.0001 | |
| Number of HLA mismatches | 0.89 | (0.76,1.05) | 0.17 | |
| Cold ischemia time | 1.00 | (0.97,1.04) | 0.79 | |
| Standard-criteria donor (Yes) | 0.49 | (0.25,0.97) | 0.04 | |
| Recipient gender (Female) | 1.13 | (0.68,1.86) | 0.64 | |
| Recipient ethnicity (African American) | 2.31 | (1.33,4.01) | 0.003 | |
| Source of subjects (ref: DeKAF Genomics) | 1.11 | (0.68,1.83) | 0.67 | |
| Emory University (N=230) 133 in analysis 36 graft failures |
APOL1 (recessive model) | 2.20 | (0.95,5.10) | 0.07 |
| Increasing donor age | 1.20 | (0.85,1.69) | 0.30 | |
| Maximum panel reactive antibodies | 0.99 | (0.98,1.01) | 0.31 | |
| Increasing recipient age | 0.61 | (0.38,0.97) | 0.04 | |
| Number of HLA mismatches | 0.86 | (0.70,1.06) | 0.17 | |
| Cold ischemia time | 1.01 | (0.97,1.06) | 0.57 | |
| Standard-criteria donor (Yes) | 1.06 | (0.38,2.96) | 0.91 | |
| Recipient gender (Female) | 1.02 | (0.50,2.08) | 0.95 | |
| Recipient ethnicity (African American) | 1.23 | (0.57,2.62) | 0.60 | |
| DeKAF Genomics (N=248) 227 in analysis 41 graft failures |
APOL1 (recessive model) | 2.29 | (0.87,6.03) | 0.09 |
| Increasing donor age | 1.17 | (0.79,1.71) | 0.43 | |
| Maximum panel reactive antibodies | 1.00 | (0.99,1.01) | 0.56 | |
| Increasing recipient age | 0.41 | (0.24,0.70) | 0.001 | |
| Number of HLA mismatches | 0.95 | (0.74,1.21) | 0.67 | |
| Cold ischemia time | 1.00 | (0.94,1.05) | 0.90 | |
| Standard-criteria donor (Yes) | 0.27 | (0.10,0.73) | 0.001 | |
| Recipient gender (Female) | 1.19 | (0.59,2.40) | 0.63 | |
| Recipient ethnicity (African American) | 3.85 | (1.72,8.63) | 0.001 | |
AA – African Americans; Other – non-African Americans; WFU – Wake Forest University; UAB – University of Alabama at Birmingham;
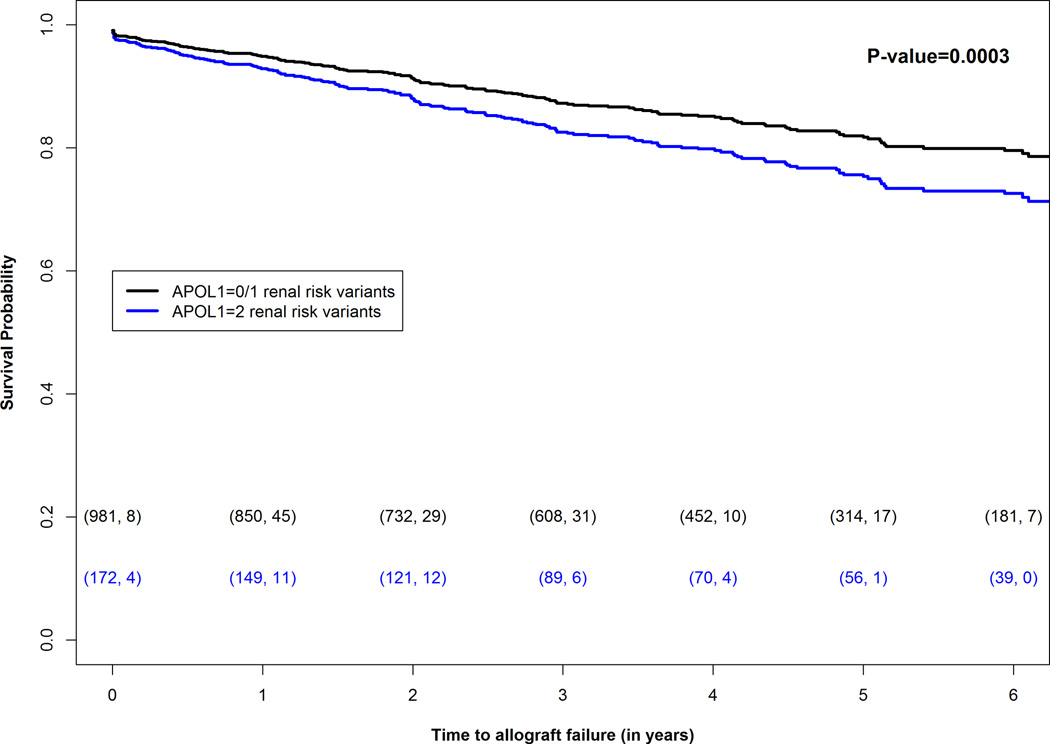
Further adjustment for recipient diabetes mellitus, dialysis vintage, induction immunosuppression, and body mass index (BMI) was also considered. These four variables had a larger percentage of missing data, which should reduce statistical power to assess effects of donor APOL1 genotypes in the model. Therefore, this analysis was performed to evaluate the magnitude of the change in estimated effect sizes between the full model and this further-adjusted model. The analysis was repeated including these four variables in the model, which led to the exclusion of 40 allograft failures: 138 included in the further-adjusted analysis including the four variables compared with 178 allograft failures in the initial full model that excluded these covariates. In the further-adjusted model (Table 3), CIT (HR 1.02, p=0.01), ECD donor kidneys (HR 1.66, p=0.04), acute rejection (HR 5.39, p=5.7×10−22), and delayed allograft function (HR 2.03, p=5.4×10−5) were significantly associated with allograft survival, in addition to APOL1-two-renal-risk-variant-donor kidneys (HR 1.65, p=0.02). The overall interaction p-value for transplant center by recipient race/ethnicity also significantly impacted time to renal allograft failure (p=0.02). Table 3 shows the HR and confidence intervals estimated from the full model and the further-adjusted model in a dataset that excludes observations where any of the additional covariates (recipient diabetes, BMI, dialysis vintage or induction immunosuppression) was missing. The association between allograft survival and donor APOL1 genotype changed appreciably for recipient age and CIT. A Kaplan-Meier plot showed significantly shorter allograft survival in recipients of APOL1-two-renal-risk-variant kidneys, relative to that from donors with fewer than two renal-risk variants, in the first five years of follow-up (Figure 1).
Table 3.
Multivariate association results for time to renal allograft failure, based on APOL1 genotype (recessive model), with data set reduced to common sample size
| Further-Adjusted Model (Full Model + 4 additional covariates)* |
Full Model | |||||
|---|---|---|---|---|---|---|
| Variable | Hazard Ratio | 95% Confidence Interval |
P-value | Hazard Ratio |
95% Confidence Interval |
P-value |
| APOL1 (recessive model) | 1.65 | (1.08,2.52) | 0.02 | 1.65 | (1.14,2.38) | 0.007 |
| Increasing donor age | 1.12 | (0.93,1.34) | 0.22 | 1.17 | (0.99,1.38) | 0.07 |
| Maximum panel reactive antibodies | 1.00 | (1.00,1.01) | 0.20 | 1.00 | (1.00,1.01) | 0.47 |
| Increasing recipient age | 0.78 | (0.60,1.02) | 0.07 | 0.71 | (0.57,0.88) | 0.002 |
| Number of HLA mismatches | 0.98 | (0.87,1.10) | 0.74 | 0.98 | (0.88,1.09) | 0.66 |
| Cold ischemia time | 1.02 | (1.00,1.04) | 0.01 | 1.01 | (0.99,1.02) | 0.26 |
| Expanded-criteria donor (Yes) | 1.66 | (1.03,2.67) | 0.04 | 1.62 | (1.04,2.52) | 0.03 |
| Recipient gender (Female) | 1.14 | (0.81,1.60) | 0.46 | 0.99 | (0.72,1.36) | 0.96 |
| Recipient diabetic kidney disease (Yes) | 1.45 | (0.94,2.24) | 0.09 | - | - | - |
| Recipient body mass index | 1.01 | (0.98,1.04) | 0.50 | - | - | - |
| Acute rejection (Yes) | 5.39 | (3.82,7.59) | 5.7×10−22 | - | - | - |
| Delayed allograft function (Yes) | 2.03 | (1.44,2.86) | 5.4×−10−5 | - | - | - |
| Induction immunosuppression (Yes) | 0.78 | (0.50,1.22) | 0.28 | - | - | - |
| Source/Recipient ethnicity (reference WFU/Other) | Overall interaction p-value 0.02 | Overall interaction p-value 0.04 | ||||
| WFU/AA | 1.00 | (0.61,1.64) | 1.00 | 1.40 | (0.89,2.20) | 0.14 |
| DeKAF Genomics/AA | 1.11 | (0.71,1.73) | 0.65 | 1.60 | (1.25,2.04) | 0.0002 |
| DeKAF Genomics/Other | 0.32 | (0.14,0.72) | 0.01 | 0.37 | (0.17,0.80) | 0.01 |
| Emory/AA | 0.70 | (0.46,1.08) | 0.11 | 1.04 | (0.83,1.30) | 0.74 |
| Emory/Other | 0.64 | (0.29,1.39) | 0.26 | 0.73 | (0.36,1.47) | 0.38 |
| UAB/AA | 0.91 | (0.41,2.03) | 0.83 | 0.77 | (0.48,1.25) | 0.29 |
| UAB/Other | 0.24 | (0.06,1.01) | 0.05 | 0.24 | (0.06,1.00) | 0.05 |
Additional covariates: recipient diabetic kidney disease, body mass index, dialysis vintage and induction immunosuppression;
AA – African American; Other – non-African Americans
Figure 1.
Adjusted Kaplan-Meier survival plots (full model) in 1153 deceased-donor kidney transplantations from African American donors based on donor APOL1 genotypes. Plots compare survival of kidneys from donors with two renal-risk variants versus that for kidneys from donors with fewer than two renal-risk variants. The numbers within the parentheses below the curves reflect (number of transplantations at the start of each year, number of allograft failures within that year).
Results of the competing risk analyses are displayed in Table 4. Donor APOL1 genotypes did not significantly impact risk for death with a functioning allograft, nor did any other factor in this analysis except transplant center. In contrast, donor APOL1 genotype, acute rejection, induction immunosuppression and delayed allograft function had significant effects on allograft failure in transplantations from deceased AA donors.
Table 4.
Competing risk model for association between time to allograft failure and death in the full sample
| Event | Label | Hazard Ratio | 95% CI | P-value |
|---|---|---|---|---|
| Allograft failure | APOL1 (recessive model) | 1.61 | (1.05,2.47) | 0.028 |
| Panel reactive antibodies | 1.00 | (1.00,1.01) | 0.24 | |
| Increasing donor age | 1.13 | (0.96,1.32) | 0.14 | |
| Increasing recipient age | 0.88 | (0.75,1.04) | 0.13 | |
| Number of HLA mismatches | 0.97 | (0.88,1.07) | 0.53 | |
| Cold ischemia time | 1.01 | (1.00,1.03) | 0.09 | |
| Expanded-criteria donor (Yes) | 1.39 | (0.84,2.30) | 0.21 | |
| Recipient gender (Female) | 1.03 | (0.74,1.43) | 0.88 | |
| Acute rejection (Yes) | 4.27 | (3.00,6.09) | <0.0001 | |
| Delayed allograft function (Yes) | 1.92 | (1.37,2.68) | 0.0002 | |
| Induction immunosuppression (Yes) | 0.68 | (0.46,1.00) | 0.049 | |
| Recipient diabetic kidney disease (Yes) | 1.36 | (0.91,2.03) | 0.13 | |
| Recipient WFU/AA* | 1.10 | (0.71,1.71) | 0.68 | |
| Recipient Emory University/Other* | 0.63 | (0.31,1.31) | 0.22 | |
| Recipient UAB/Other* | 0.22 | (0.05,0.97) | 0.046 | |
| Recipient DeKAF Genomics/Other* | 2.65 | (1.08,6.51) | 0.034 | |
| Recipient Emory University/AA* | 0.51 | (0.30,0.88) | 0.016 | |
| Recipient UAB/AA* | 0.97 | (0.60,1.56) | 0.88 | |
| Recipient DeKAF Genomics/AA* | 1.07 | (0.01,107.23) | 0.98 | |
| Death with functioning allograft | APOL1 (recessive model) | 1.07 | (0.61,1.89) | 0.80 |
| Panel reactive antibodies | 1.00 | (0.99,1.00) | 0.38 | |
| Increasing donor age | 1.22 | (0.97,1.53) | 0.09 | |
| Increasing recipient age | 1.15 | (0.93,1.41) | 0.21 | |
| Number of HLA mismatches | 1.06 | (0.88,1.27) | 0.55 | |
| Cold ischemia time | 0.99 | (0.97,1.01) | 0.30 | |
| Expanded-criteria donor (Yes) | 0.73 | (0.37,1.46) | 0.37 | |
| Recipient gender (Female) | 0.96 | (0.61,1.50) | 0.85 | |
| Acute rejection (Yes) | 0.58 | (0.32,1.07) | 0.08 | |
| Delayed allograft function (Yes) | 0.94 | (0.57,1.55) | 0.81 | |
| Induction immunosuppression (Yes) | 1.75 | (0.86,3.58) | 0.12 | |
| Recipient diabetic kidney disease (Yes) | 1.21 | (0.78,1.87) | 0.41 | |
| Recipient WFU/AA* | 0.68 | (0.34,1.36) | 0.27 | |
| Recipient Emory University/Other* | 1.61 | (0.79,3.30) | 0.19 | |
| Recipient UAB/Other* | 0.42 | (0.12,1.42) | 0.16 | |
| Recipient DeKAF Genomics/Other* | 1.09 | (0.35,3.38) | 0.88 | |
| Recipient Emory University/AA* | 0.55 | (0.37,0.82) | 3.3×10−3 | |
| Recipient UAB/AA* | 1.94 | (1.35,2.76) | 2.9×10−4 | |
| Recipient DeKAF Genomics/AA* | 0.90 | (0.15,5.57) | 0.91 |
Reference group is WFU/Other (Wake Forest University non-African American Recipients)
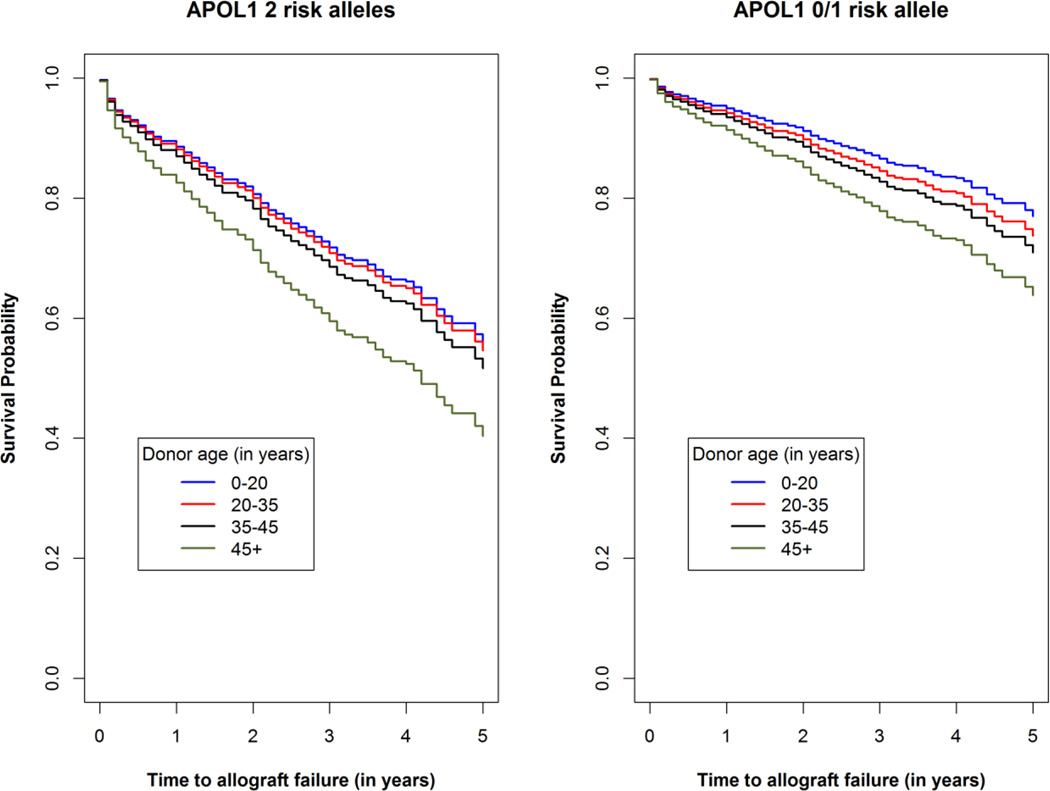
Analyses contrasting the impact of the age at kidney donation based on the number of APOL1-renal-risk variants in donors were performed in the full sample of 1153 transplantations. Graphs displaying the survival probability over time in donors of age <20, 20–35, 35–44, and >45 years are displayed in Figure 2, separately for kidneys from donors with fewer than two APOL1-renal-risk variants versus kidneys from donors with two APOL1-renal-risk variants. Allograft survival is shown for the first six years of follow-up. The interaction test between APOL1 renal-risk variants and donor age was not statistically significant (p=0.51); suggesting that allograft survival did not differ significantly based on donor age within the samples receiving two- or zero/one-APOL1-renal-risk-variant kidneys.
Figure 2.
Renal allograft survival based on donor age, by number of APOL1 renal-risk alleles. Adjusted Kaplan-Meier survival curves (full model) for kidneys from deceased African American donors with two APOL1-renal-risk alleles (Panel a) and zero or one APOL1-renal-risk alleles (Panel b). The APOL1-donor-age interaction p-value was 0.51, revealing that the effect of donor age is not modified by APOL1.
Supplementary Table S2 displays predictive abilities of the fully-adjusted model, including and excluding effect of donor APOL1 genotype (recessive model) on outcomes. Although effects of APOL1 were not marked, we were unable to account for competing effects of other risk factors (e.g., BK virus infection, donor specific antibodies, etc.) that might have clarified the effects of donor genotype.
Discussion
A recent report indicated that variation in the APOL1 nephropathy susceptibility gene in deceased AA kidney donors and AA recipient race independently reduced allograft survival.(3) APOL1 genotypes account for much of the increased risk for non-diabetic nephropathy in individuals with recent African ancestry, relative to those with European ancestry.(22) The present study replicates and extends the observation in DDKT by assessing the outcomes of 478 additional DDKTs from AA donors and performing a combined analysis in 1153 DDKTs. Results strongly support the initial observations that recipients of DDKTs from individuals with recent African ancestry possessing two APOL1-renal-risk variants have shorter allograft survival (effects of two renal risk variants [recessive model] were of stronger significance than possession of a single risk variant).(2,3) Effects of APOL1 were independent of other traditional risk factors known to adversely impact renal allograft survival. Novel findings include that donor age did not significantly impact allograft survival based on the number of APOL1-renal-risk variants and recipients of APOL1-two-renal-risk-variant kidneys had higher serum creatinine concentrations at latest follow-up. Finally, this well-powered analysis in 1153 DDKTs from AA donors detected multiple additional factors that independently impacted allograft survival in the APOL1 era that were not apparent in an initial analysis of 675 DDKTs.(3)
The multivariable analysis in the full sample of 1153 transplantations demonstrated that the risk for allograft failure was significantly increased for APOL1-two-renal-risk-variant donor kidneys, older donors, and younger recipients. Significant effects of the center where DNA was procured by recipient race/ethnicity were also observed, potentially related to different proportions of African American kidney transplant recipients and different rates of acceptance of marginal organs for transplantation between centers. Sensitization based on the maximal PRA titer, CIT, degree of HLA match, recipient gender, diabetic kidney disease, BMI, induction immunosuppression, and dialysis vintage did not exert significant effects on overall allograft survival. However, the competing risk model limited to “allograft failure” revealed that, in addition to the effects of donor APOL1 genotype, acute rejection, induction immunosuppression, and delayed allograft function also had significant effects on allograft failure in transplantations from deceased AA donors. These effects were masked by the censored outcome of “death with allograft function”. The effect of recipient AA race at each transplant center is likely not related to APOL1,(6) but could involve socioeconomic factors and/or differences in immune response.(23,24) The shorter renal allograft survival in younger transplant recipients was somewhat surprising, but could reflect better medication compliance, less vigorous immune response (particularly in the elderly), and higher rates of death with allograft function (censored outcomes) among older recipients. Similar to the prior report,(3) most APOL1-two-renal-risk-variant kidneys did not fail early after engraftment; 72.6% functioned beyond five years and 56.9% for more than 10 years. As in native-kidney disease, allograft failure in recipients of organs from donors with two APOL1-renal-risk variants may result from additional modulating factors or second hits.(11,12) Identifying modifiable genetic and environmental factors is critical to determining the mechanisms that may lead to premature allograft failure in recipients of APOL1-two-renal-risk-variant kidneys.(25,26)
An important consideration in transplanting kidneys from AA deceased donors with two APOL1-renal-risk variants is donor age. This opinion is based on the “normal for now” observation in live-donor kidney transplantation, where lack of albuminuria and normal eGFR in young donors (child to parent, young sibling to sibling) fails to consider final renal phenotypes.(9) Genetically susceptible younger donors may appear to lack kidney disease when evaluated for nephrectomy, yet develop nephropathy after follow-up. A striking example was reported in a non-diabetic Afro-Caribbean man.(10) He received a live-donor kidney transplant from his identical twin brother at 21 years of age. The donor lacked nephropathy at evaluation, then developed nephrotic syndrome with focal segmental glomerulosclerosis (FSGS) seven years post-nephrectomy. The transplanted kidney also failed due to FSGS after five years. Both siblings possessed two APOL1-renal-risk-variants. Not all genetically high-risk individuals develop APOL1-associated nephropathy. We initially postulated that APOL1-two-risk-variant kidneys from older phenotypically normal donors may have escaped modifying factors initiating nephropathy and might function for longer periods after DDKT. Herein, we found that transplantations from younger deceased donors had better allograft survival, relative to older donors. Donor age did not impact allograft survival within APOL1 genotype risk groups.
These results suggest that improvements can be made in the organ allocation process for DDKT based on APOL1 genotyping.(8) APOL1 G1 and G2 renal-risk variants provide the necessary information and ancestry-informative markers need not be genotyped or included in the decision-making process.(3) Kidneys donated by deceased AAs with two APOL1-renal-risk variants are associated with shorter allograft survival and higher serum creatinine concentrations after transplantation.(2,3) Incorporating the impact of APOL1, rather than self-reported race, would more appropriately inform physicians and potential recipients about projected outcomes of transplanted kidneys.
A shortage of deceased-donor kidneys for transplantation in the AA community remains, as well in as other populations. AA recipients of renal allografts are more likely to receive kidneys from AA deceased donors due to racial distributions of both HLA-DR alleles and blood types.(27,28) Transplantation of APOL1-two-renal-risk-variant allografts could exacerbate race-based disparities in transplant outcomes. Identification of kidneys at risk for earlier failure should result in more appropriate organ allocation with the potential to narrow racial disparities. Improved risk stratification of organs should increase confidence in the clinical assessment of organ quality and improve utilization of organs with and without two APOL1-renal-risk variants. A relative risk for graft failure ≥1.6 was used to define expanded-criteria donors; subsequently leading to a new classification of deceased donor risk. We note that the hazard ratio for the effect of APOL1-two-renal-risk-variant-donor kidneys exceeds 2.0 in the present report. In the future, rapid assessment of APOL1 G1 and G2 genotypes at time of organ recovery may become advisable.(8)
Limitations in this report include the possible shortcoming that transplant outcomes were captured using the large SRTR database which may not be complete or fully accurate. However, SRTR captures allograft loss well because it links to the United States Renal Data System, including initiation of renal replacement therapy even when information is not provided by transplant centers, with dates of re-transplantation and death. Recipient BMI, cause of nephropathy, induction immunosuppression, and dialysis vintage were missing in up to 10% of transplantations. Therefore, multivariate analyses were performed with and without these covariates. Confounding variables could include different immunosuppressive regimens across centers and changing patterns of immunosuppression over time. These factors could not be accounted for, as engraftments were performed at 113 different centers. SRTR is unable to capture medication changes over time. Finally, analyzing recipient and kidney donor APOL1 genotypes, and their interaction, would have been informative. We lack recipient data and are unaware of existing datasets containing APOL1 genotypes in large numbers of kidney donors and recipients. As such, we believe that a prospective analysis assessing effects of donor and recipient APOL1 genotypes (and their interaction) and other critical variables lacking in the SRTR should be performed.
Kidneys from deceased AA donors with two APOL1-renal-risk variants are reproducibly associated with an increased risk for early failure after transplantation. However, many such genetically high-risk kidneys functioned for prolonged periods. It is possible that replacing AA donor race with APOL1 genotype may improve the ability of the KDPI to predict transplantation outcomes, although genotyping costs need to be considered. In the future, including APOL1 genotyping in donors of self-reported AA ancestry could be incorporated into decisions regarding organ allocation. In addition to APOL1 genotypes, recipient age, donor age, and AA recipient race have significant effects on allograft survival for kidneys from deceased AA donors. We suggest that approaches to rapidly perform APOL1 genotyping in deceased AA kidney donors be developed and a national trial performed to prospectively test their impact on transplantation outcomes.(8,29)
Supplementary Material
Acknowledgments
This work was supported, in part, by NIH RO1 DK070941 (BIF), NIH RO1 DK084149 (BIF), NIH RO1 MD009055 (JD, BIF), and NIH/NIAD Genomics of Transplantation 5U19-AI070119 (AKI). The data reported here have been supplied by the Minneapolis Medical Research Foundation (MMRF) as the contractor for the Scientific Registry of Transplant Recipients (SRTR). The interpretation and reporting of these data are the responsibilities of the authors and in no way should be considered as an official policy of or interpretation by the SRTR or the United States Government.
Footnotes
The authors declare no conflicts of interest.
Contributor Information
Barry I. Freedman, Email: bfreedma@wakehealth.edu.
Stephen O. Pastan, Email: Stephen.Pastan@emoryhealthcare.org.
Ajay K. Israni, Email: isran001@umn.edu.
David Schladt, Email: DSchladt@cdrg.org.
Bruce A. Julian, Email: bjulian@uab.edu.
Michael D. Gautreaux, Email: mgautrea@wakehealth.edu.
Vera Hauptfeld, Email: vhauptfeld@uabmc.edu.
Robert A. Bray, Email: rbray@emory.edu.
Howard M. Gebel, Email: hgebel@emory.edu.
Allan D. Kirk, Email: allan.kirk@duke.edu.
Robert S. Gaston, Email: rgaston@uab.edu.
Jeffrey Rogers, Email: jerogers@wakehealth.edu.
Alan C. Farney, Email: afarney@wakehealth.edu.
Giuseppe Orlando, Email: gorlando@wakehealth.edu.
Robert J. Stratta, Email: rstratta@wakehealth.edu.
Sumit Mohan, Email: sm2206@columbia.edu.
Lijun Ma, Email: lima@wakehealth.edu.
Carl D. Langefeld, Email: clangefe@wakehealth.edu.
Donald W. Bowden, Email: dbowden@wakehealth.edu.
Pamela J. Hicks, Email: panderso@wakehealth.edu.
Nicholette D. Palmer, Email: nallred@wakehealth.edu.
Amudha Palanisamy, Email: apalanis@wakehealth.edu.
Amber M. Reeves-Daniel, Email: areeves@wakehealth.edu.
W. Mark Brown, Email: wmbrown@wakehealth.edu.
Jasmin Divers, Email: jdivers@wakehealth.edu.
References
- 1.U.S. Renal Data System. USRDS 2012 Annual Data Report, Vol 1: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. 2012 [Google Scholar]
- 2.Reeves-Daniel AM, Depalma JA, Bleyer AJ, et al. The APOL1 Gene and Allograft Survival after Kidney Transplantation. Am J Transplant. 2011;11:1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman BI, Julian BA, Pastan SO, et al. Apolipoprotein L1 Gene Variants in Deceased Organ Donors Are Associated With Renal Allograft Failure. Am J Transplant. 2015 doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BT, Kumar V, Williams TA, et al. The APOL1 Genotype of African American Kidney Transplant Recipients Does Not Impact 5-Year Allograft Survival. Am J Transplant. 2012;12:1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Israni AK, Salkowski N, Gustafson S, et al. New National Allocation Policy for Deceased Donor Kidneys in the United States and Possible Effect on Patient Outcomes. J Am Soc Nephrol. 2014;25:1842–1848. doi: 10.1681/ASN.2013070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman BI, Julian BA. Should kidney donors be genotyped for APOL1 risk alleles? Kidney Int. 2015;87:671–673. doi: 10.1038/ki.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steiner RW. 'Normal for now' or 'at future risk': a double standard for selecting young and older living kidney donors. Am J Transplant. 2010;10:737–741. doi: 10.1111/j.1600-6143.2010.03023.x. [DOI] [PubMed] [Google Scholar]
- 10.Kofman T, Audard V, Narjoz C, et al. APOL1 polymorphisms and development of CKD in an identical twin donor and recipient pair. Am J Kidney Dis. 2014;63:816–819. doi: 10.1053/j.ajkd.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Freedman BI, Skorecki K. Gene-Gene and Gene-Environment Interactions in Apolipoprotein L1 Gene-Associated Nephropathy. Clin J Am Soc Nephrol. 2014 doi: 10.2215/CJN.01330214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruzel-Davila E, Wasser WG, Aviram S, Skorecki K. APOL1 nephropathy: from gene to mechanisms of kidney injury. Nephrol Dial Transplant. 2015 doi: 10.1093/ndt/gfu391. [DOI] [PubMed] [Google Scholar]
- 13.Leppke S, Leighton T, Zaun D, et al. Scientific Registry of Transplant Recipients: collecting, analyzing, and reporting data on transplantation in the United States. Transplant Rev (Orlando) 2013;27:50–56. doi: 10.1016/j.trre.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Freedman BI, Langefeld CD, Turner J, et al. Association of APOL1 variants with mild kidney disease in the first-degree relatives of African American patients with non-diabetic end-stage renal disease. Kidney Int. 2012;82:805–811. doi: 10.1038/ki.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 16.Cox DR. Regression models and life tables. J Royal Stat Soc, Series B. 1972;34:187–220. [Google Scholar]
- 17.Therneau TM, Grambsch PM. Modeling Survival Data:Extending the Cox Model. New York, NY: Springer; 2000. pp. 170–174. [Google Scholar]
- 18.Lin DY, Wei LJ. The robust inference for the Cox Proportional Hazards Model. J Am Stat Assoc. 1989;84:1074–1078. [Google Scholar]
- 19.Fine J, Gray A. A proportional Hazards Model for the Subdistribution of a competing risk. J Am Stat Assoc. 1999:496–509. [Google Scholar]
- 20.Zhou B, Latouche A. Competing risks regression for Stratified and Clustered data. R package version 1.1 (2013) 2013 ttp://CRAN.R-project.org/package=crrSC. Ref Type: Electronic Citation. [Google Scholar]
- 21.Contal C, O'Quigley J. An application of changepoint methods in studying the effect of age on survival in breast cancer. Computational Statistics & Data Analysis. 1999;30:253–270. [Google Scholar]
- 22.Palmer ND, Freedman BI. APOL1 and Progression of Nondiabetic Nephropathy. J Am Soc Nephrol. 2013;24:1344–1346. doi: 10.1681/ASN.2013060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butkus DE, Meydrech EF, Raju SS. Racial differences in the survival of cadaveric renal allografts. Overriding effects of HLA matching and socioeconomic factors. N Engl J Med. 1992;327:840–845. doi: 10.1056/NEJM199209173271203. [DOI] [PubMed] [Google Scholar]
- 24.Curtis JJ. Kidney transplantation: racial or socioeconomic disparities? Am J Kidney Dis. 1999;34:756–758. doi: 10.1016/S0272-6386(99)70404-X. [DOI] [PubMed] [Google Scholar]
- 25.Divers J, Nunez M, High KP, et al. JC polyoma virus interacts with APOL1 in African Americans with nondiabetic nephropathy. Kidney Int. 2013;84:1207–1213. doi: 10.1038/ki.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Divers J, Palmer ND, Lu L, et al. Gene-gene interactions in APOL1-associated nephropathy. Nephrol Dial Transplant. 2014;29:587–594. doi: 10.1093/ndt/gft423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts JP, Wolfe RA, Bragg-Gresham JL, et al. Effect of changing the priority for HLA matching on the rates and outcomes of kidney transplantation in minority groups. N Engl J Med. 2004;350:545–551. doi: 10.1056/NEJMoa025056. [DOI] [PubMed] [Google Scholar]
- 28.Cannon RM, Brock GN, Marvin MR, Slakey DP, Buell JF. The contribution of donor quality to differential graft survival in African American and Caucasian renal transplant recipients. Am J Transplant. 2012;12:1776–1783. doi: 10.1111/j.1600-6143.2012.04091.x. [DOI] [PubMed] [Google Scholar]
- 29.Ojo A, Knoll GA. APOL1 Genotyping of African American Deceased Organ Donors: Not Just Yet. Am J Transplant. 2015 doi: 10.1111/ajt.13230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.