Abstract
Genetic analysis of development and function of the gonadotrope cell lineage within mouse anterior pituitary has been greatly facilitated by at least three currently available Cre strains in which Cre was either knocked into the Gnrhr locus or expressed as a transgene from Cga and Lhb promoters. However, in each case there are some limitations including CRE expression in thyrotropes within pituitary or ectopic expression outside of pituitary, for example in some populations of neurons or gonads. Hence, these Cre strains often pose problems with regard to undesirable deletion of alleles in non-gonadotrope cells, fertility and germline transmission of mutant alleles. Here, we describe generation and characterization of a new Fshb-iCre deleter strain using 4.7 kb of ovine Fshb promoter regulatory sequences driving iCre expression exclusively in the gonadotrope lineage within anterior pituitary. Fshb-iCre mice develop normally, display no ectopic CRE expression in gonads and are fertile. When crossed onto a loxP recombination-mediated red to green color switch reporter mouse genetic background, in vivo CRE recombinase activity is detectable in gonadotropes at more than 95 % efficiency and the GFP-tagged gonadotropes readily purified by fluorescence activated cell sorting. We demonstrate the applicability of this Fshb-iCre deleter strain in a mouse model in which Dicer is efficiently and selectively deleted in gonadotropes. We further show that loss of DICER-dependent miRNAs in gonadotropes leads to profound suppression of gonadotropins resulting in male and female infertility. Thus, Fshb-iCre mice serve as a new genetic tool to efficiently manipulate gonadotrope-specific gene expression in vivo.
Keywords: Pituitary, Gonadotrope, Cre, Dicer, Testis, Ovary
1. Introduction
The anterior pituitary consists of a heterogeneous population of several key trophic hormone-producing cells. These include POMC/ACTH-producing corticotropes, GH-producing somatotropes, PRL-producing lactotropes, TSH-producing thyrotropes and LH/FSH producing gonadotropes (Asa and Ezzat, 2004, Bousfield, Jia and Ward, 2006, Childs, 1997, Zhu and Rosenfeld, 2004, Keegan and Camper, 2003). Gonadotropes are the least abundant cell type and represented by only 4 – 5 % of all cells within the mouse anterior pituitary (Wu, Su, Safwat et al., 2004). During embryonic mouse pituitary development, a combinatorial action of transcription factors including PROP-1, PIT-1, GATA-2 and SOX-2 directs a set of common progenitor cells into SF-1+ and FOXL2+ gonadotrope- and SF-1− and FOXL2− thyrotrope lineages (Zhu and Rosenfeld, 2004, Drouin, 2006, Mollard, Hodson, Lafont et al., 2012, Scully and Rosenfeld, 2002, Watkins-Chow and Camper, 1998, Zhu, Gleiberman and Rosenfeld, 2007). Once established, both these cell types express Cga that encodes the common glycoprotein hormone subunit (α-GSU). The α-GSU non-covalently combines with the hormone-specific LHβ or FSHβ subunits resulting in production of the corresponding functional LH and FSH heterodimers in gonadotropes (Bousfield et al., 2006, Seeburg, Mason, Stewart et al., 1987, Seminara and Crowley, 2002, Seminara, Hayes and Crowley, 1998). The common α - subunit also combines with TSHβ in thyrotropes resulting in the formation of TSH heterodimer that regulates thyroid hormone production (Bousfield et al., 2006). Gonadotropes respond to the hypothalamus-derived gonadotropin-releasing hormone (GnRH), which binds to G-protein coupled GnRH receptors on these cells (Bousfield et al., 2006). In the mouse, Gnrhr expression is initiated around E13.5 during pituitary development (McGillivray, Bailey, Ramezani et al., 2005, Xie, Cherrington, Meadows et al., 2013, Xie, Hoffmann, Meadows et al., 2015).
Temporal analysis of anterior pituitary hormone/hormone-subunit encoding mRNAs by in-situ hybridization (ISH) revealed that each of the mRNAs is sequentially expressed initially in a spatially restricted “zone” within the developing gland (Japon, Rubinstein and Low, 1994). Cga mRNA is the first marker that is expressed beginning at embryonic day (E) 11.5 followed by Lhb at E16.5 and Fshb at E17.5 in a small number of cells (Japon et al., 1994). Qualitative assessment of the ISH data indicated that it is only after birth, expression levels of gonadotropin subunit mRNAs reach those observed in adult mouse pituitary (Japon et al., 1994). Although analyses of several mouse mutants identified defects in temporal expression of gonadotropin subunit genes during anterior pituitary development (Brinkmeier, Davis, Carninci et al., 2009, Douglas and Camper, 2000), their corresponding protein expression and localization during normal mouse pituitary development have not been systematically mapped. Large-scale expression studies including transcriptome, micro-transcriptome and proteome analyses using embryonic mouse pituitaries at different stages or cell lines (Brinkmeier et al., 2009, Douglas and Camper, 2000, Ye, Xi, Qi et al., 2013, Yuen, Ruf, Chu et al., 2009, Zhang, Cai, Wei et al., 2013, Asa and Ezzat, 2005, Wu, Taylor, Lane et al., 2000) have identified several novel candidate genes/proteins. The functional relevance of many of these candidates to gonadotrope physiology remains unknown. It is desirable to develop loss of function mutations at these candidate loci by inactivating them specifically in the gonadotrope lineage by Cre-lox technology and analyzing the in vivo functional consequences.
The temporally regulated gonadotrope “hallmark” gene expression during pituitary gland development has led to the generation of useful Cre deleter strains of mice. However, these models have limitations including Cre expression not exclusively restricted to gonadotrope lineage within pituitary (Perez-Millan, Zeidler, Saunders et al., 2013), or leaky expression of Cre primarily in gonads resulting in poor fertility (Charles, Mortensen, Potok et al., 2008, Wang, Graham, Hastings et al., 2015), or Cre expression from a knock-in allele with undesirable extrapituitary expression in Gnrhr-expressing neurons and testis (Wen, Schwarz, Niculescu et al., 2008). An inducible Cre -expressing line specific to gonadotrope lineage has also been described (Naik, Pittman, Wolfe et al., 2006), but its routine application to deletion of specific alleles in gonadotropes is lacking. Here, we describe generation and characterization of an Fshb-iCre deleter mouse strain and its utility by inactivating Dicer specifically in gonadotropes. We further compare our new deleter line of mice with two other existing gonadotrope-targeted Cre lines (Perez-Millan et al., 2013, Charles et al., 2008) and in the context of our recently characterized mice in which Dicer was inactivated in gonadotropes using one of the existing Cre deleter strains (Wang et al., 2015). Our data indicate that Fshb-iCre mice are efficient Cre deleters and can be routinely used to delete desired floxed alleles specifically in the pituitary gonadotrope lineage.
2. Materials and methods
2.1. Generation of Fshb-iCre transgenic mice
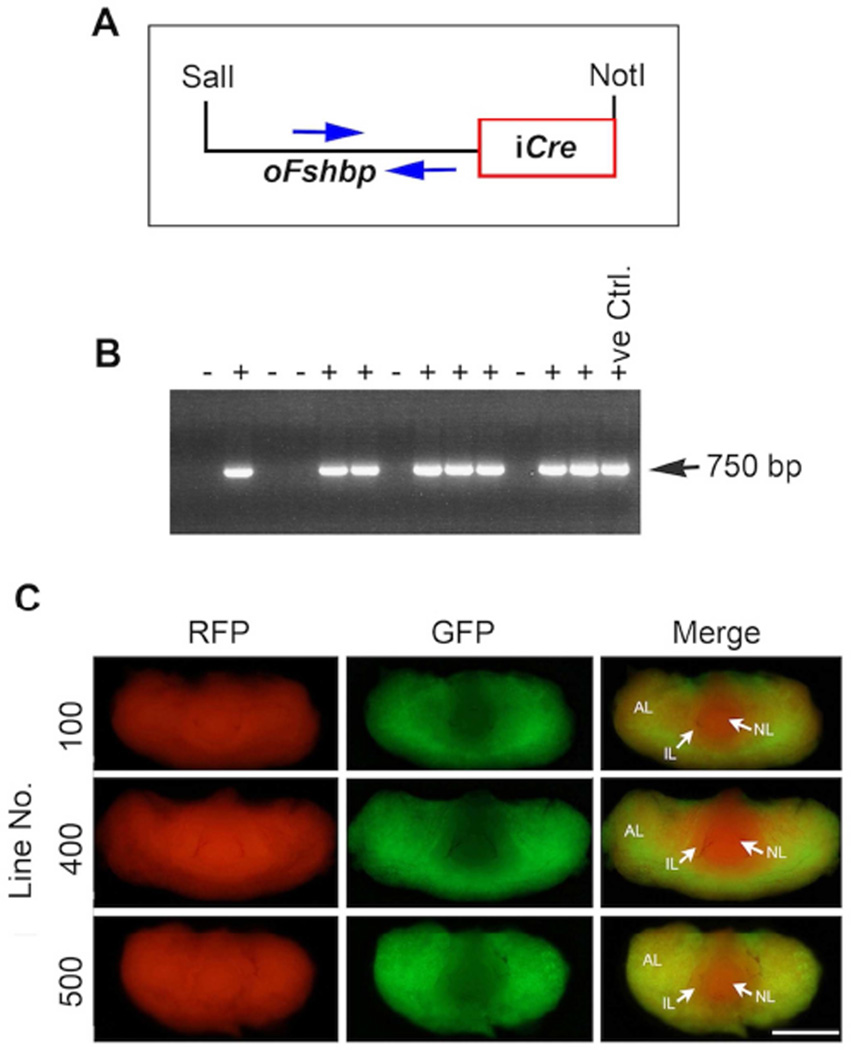
To achieve gonadotrope-specific expression of Cre, we first PCR amplified the codon-improved Cre (iCre) cDNA from the pDIRE (Addgene plasmid # 26745) plasmid by engineering BamHI restriction sites on both ends. The amplified ~ 2.0 kb of iCre fragment was first purified, later confirmed that no mutations were introduced by bi-directional DNA sequencing, digested with BamHI and cloned into the same site located downstream of the 4.7 kb of ovine Fshb regulatory sequences contained in a poFshbp plasmid. The resulting plasmid designated oFshb-iCre was purified on Midi Prep columns (Qiagen), from which the 6.7 kb transgene linear fragment was released by SalI/NotI restriction enzyme digestion. The transgene fragment was further purified on Millipore Montage DNA columns, and microinjected into one-cell FVB mouse embryos as described (Kumar, Larson, Wang et al., 2009). Tail DNA genomic PCR employing specific primers within the oFshb promoter that amplify a ~ 750 bp fragment was used to identify transgenic founder pups. Transgene-carrying (hereafter referred to as Fshb-Cre) founder mice were bred to wild-type littermates to establish independent transgenic lines and confirm stable transmission of the transgene up to F2 generation and further used in all experiments.
2.2. Other mouse strains
Dicerf/f mice were provided by Dr. Brian Harfe (Harfe, McManus, Mansfield et al., 2005). These mice were intercrossed with Rosa26mT/mGtg/tg transgenic line (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J ; purchased from Jackson Labs and designated as, mT/mG+/tg or tg/tg or mT/mG mice). These mice harbor loxP sites on either side of a membrane-targeted tdTomato (mT) reporter cassette and ubiquitously express strong red fluorescence. When intercrossed with Cre recombinase-expressing mice, the resulting offspring have the mT cassette deleted in the Cre expressing tissue/cell (s), allowing expression of the membrane-targeted EGFP (mG) located immediately downstream. These dual color reporter mice designated mT/mG were initially used to first generate Dicerf/+ mT/mG+/tg mice. These mice were subsequently intercrossed with Fshb-Cre transgene-carrying mice to generate Dicer+/f, mT/mG+/tg Cre+ mice. Finally, Dicer+/f mT/mG+/tg Cre+ male and female mice were intercrossed to generate the desired Dicerf/f mT/mG+/tg ortg/tg Cre+ (designated as Dicer cKO) mice. Dicer cKO mice were also generated using bLhb-Cre driver line and characterized as described before (Wang et al., 2015). Dr. Sally Camper, University of Michigan Medical Center, Ann Arbor, MA (USA) generously provided Cga-Cre+ and Lhb-Cre+ transgenic mouse strains. From these, we also generated the corresponding Cga-Cre+ mT/mG+/tg and Lhb-Cre+ mT/mG+/tg mice. For convenience, the red florescence due to tdTomato reporter has been designated as RFP throughout the text. Mice lacking Fshb or Lhb or both were generated and genotyped as described (Wang, Larson, Jablonka-Shariff et al., 2014). Mice were maintained on a 12h light: 12h dark light cycle with food and water provided ad libitum. The University of Kansas Medical Center IACUC approved all of the experimental animal procedures that were in compliance with the NIH guidelines.
2.3. Timed matings and collection of embryos
Adult female mice at 42d of age were set up for mating with proven fertile males (2 females per male per cage) and the vaginal plugs were monitored daily in the morning. The day of the plug was considered embryonic day (E) 0.5. Mouse embryos at desired time points were recovered into cold PBS (pH7.4) from pregnant mothers under isoflurane anesthesia, and tail snips were collected for genotyping.
2.4. Fluorescence imaging of tissues
Pituitaries from mouse embryos or adult mice and gonads from adult mice were collected into and washed with cold PBS (pH 7.4). Gross images of pituitaries in PBS were visualized with green and red fluorescence emission filters using an epifluorescence microscope (Zeiss), digitally captured and merged.
2.5. Purification of gonadotropes by fluorescence activated cell sorting
Pituitaries from adult mice were pooled, sliced into pieces using a sterile blade, and collagenase-dispersed at 37C as described (Wang et al., 2015). The final cell suspension was filtered through 100µm nylon cap containing tubes (Becton Dickinson) to remove cell clumps. An aliquot was checked for red/green fluorescence on an epifluorescence microscope (Zeiss) and the cells were subjected to FACS analysis using an AriaIIU flow sorter (Becton Dickinson). A UV laser set at 488nm was used to excite GFP protein. The GFP+ and GFP− pools of cells were separately collected into RNAse-free tubes on ice, an aliquot checked for red/green fluorescence, the cell suspensions centrifuged at 3000 × g for 5 minutes at 4C and the cell pellet was used for total RNA isolation. Alternatively, FACS-sorted GFP+ and RFP+ cells were formalin-fixed at RT for 5’ and processed for immunofluorescence using antibodies against anterior pituitary cellspecific hormones as described below. When appropriate, bright field images were recorded. The percentages of GFP+-hormone+ and RFP+-hormone+ cells were calculated in each case.
2.6. RNA and miRNA isolation and real time quantitative PCR assays
Total RNA from FACS-sorted cells was isolated using either RNA mini-columns (Qiagen), reverse transcribed with random hexamer primers using Super Script III reverse transcriptase. MicroRNA was isolated using miRNeasy mini kits (Qiagen), single stranded miRNAs were reverse transcribed with specific forward primers, the second strand synthesized with universal reverse primer using microScript RT kit according to the manufacturer’s instructions and as described (Wang et al., 2015). Both SYBR green and Taqman real time qPCR assays were performed using custom-made primers or pre-inventoried primer/probe combos (Life Technologies and IDT). For mRNA quantification, Ppil1 and for miRNA quantification, 5s rRNA expression was used as internal control using our previously published methods (Wang et al., 2015). PCR assays were run on triplicate cDNA samples from at least 3 mice per genotype.
2.7. Western blot analysis
Total proteins from individual adult mice were extracted using radioimmunoassay protein extraction buffer system with protease/phosphatase inhibitors (Santa Cruz Biotechnology), denatured, separated on 12 % polyacrylamide gels and transferred to PVDF membranes (Bio- Rad) as described (Wang et al., 2015, Wang, Su, Sanai et al., 2012). Western blot analysis was performed by published methods (Wang et al., 2015, Wang et al., 2012) using primary antibodies specific to FSHβ (1:2,000; goat polyclonal antibody against human FSHβ), LHβ (1:2,000; goat polyclonal antibody against human LHβ ; Santa Cruz Biotechnology), β - tubulin (1: 2,000; rabbit polyclonal antibody, Santa Cruz Biotechnology) and appropriate secondary antibodies conjugated with HRP (Santa Cruz Biotechnology). The antigen-antibody complexes were detected with an enhanced chemiluminescence kit (GE Healthcare) and the signals visualized on Biomax films (ISC Biosciences). The signals were quantified by densitometry and the data were represented as ratios with respect to expression of β -tubulin. Recombinant hFSH was a generous gift from Dr. Irving Boime, Washington University School of Medicine. Purified bovine LH standard was purchased from Dr. A.F. Parlow, National Hormone and Peptide Program, UCLA Medical Center. Western bot analyses were performed 3 independent times using a total of 3 mice per genotype.
2.8. Gross and Histology analyses
Testes, epididymis and female reproductive tracts were harvested from adult mice at or above 42d of age, dissected free of adhering fat tissue, wet weights were recorded, rinsed in cold PBS and imaged by digital scanning. For histological analysis, gonads were fixed in Bouin’s reagent overnight at 4C and rinsed in cold 70 % ethanol several times, paraffin embedded, 6 µm sections were cut, processed in graded series of alcohol and stained with periodic acid Schiff’s reagent/hematoxylin as described (Wang et al., 2015, Wang et al., 2014). Multiple sections from at least 5 mice were analyzed and digitally photographed using a microscope (Zeiss). Vaginal opening and estrus cycles were monitored in female mice as described (Behringer, Gertsenstein, Kristina et al., 2014).
2.9. Immunofluorescence assays
Pituitaries or testes were collected from adult mice under anesthesia, fixed overnight in buffered formalin (pH 7.4) at 4C and processed as described above. Approximately 6 µm sections were cut onto glass slides, rinsed in phosphate buffered saline pH 7.4 (PBS), and blocked in ready-to-use normal goat or rabbit serum (Invitrogen) at RT for 1h and incubated overnight with pairs of primary antibodies at 4C. Immunofluorescence was performed exactly as described (Wang et al., 2015) using the following primary antibodies: Mouse monoclonal antibody against CRE (Covance, MMS-106P), mouse monoclonal anti-hFSH 4B (a gift from Dr. Irving Boime, at 1:500 dilution), rabbit polyclonal anti-hCG serum that recognizes mouse LH (a gift from Dr. Irving Boime, at 1:500 dilution), rabbit antiserum against SOX9 (1:1000, a gift from Dr. Ken Morohashi), rat monoclonal antibody against GCNA1 (no dilution, a gift from Dr. George Enders), mouse monoclonal antibody against PLZF (1:200, Calbiochem OP128L), rabbit antibody against phospho- Histone H3 (1:200, Santa Cruz, SC8656) and guinea pig antiserum against SP10 (1:200, a gift from Dr. Prabhakara Reddi). Dr. A.F. Parlow provided primary antibodies against pituitary hormones from the National Hormone and Pituitary Program. All the Alexa fluorochrome-conjugated secondary antibodies (Life Technologies) were used at a final dilution of 1:200. Sections were finally mounted in Antifade reagent (Life Technologies). The nuclei were stained red with ethidium homodimer (Life Technologies). Multiple sections from at least 3 adult mice were visualized with an epifluorescence microscope (Zeiss) and digitally photographed.
2.10. Serum hormone Assays
Serum was collected from adult mice (n=6 per genotype) as described (Wang et al., 2014). Serum FSH and LH were measured by a rat multiplex gonadotropic hormone assay (Millipore) according to the manufacturer’s instructions and using a Luminex model 200 automated analyzer. Serum steroid hormones (E, P and T) were measured using ELISA kits purchased from Calbiotech and IBL-America according to the manufacturers’ instructions (Wang et al., 2015, Wang et al., 2014).
2.11. Fertility tests
Pairs of adult mice at 42d of age were caged (one male with one female; a total of 5 pairs) over a period of 4–6 months. The number of litters and litter sizes (number of pups per litter) were used to evaluate the breeding performance.
2.12. Quantification of sperm
Cauda epididymis fragments obtained from adult male mice were minced into small fragments in 1 ml of sterile M2 medium (Sigma), incubated at 37C for 15’ to release the sperm. An aliquot of the medium containing sperm was diluted at 1:10 in PBS and counted with a hemocytometer as described (Wang et al., 2015, Wang et al., 2014).
2.13. Statistical analysis
Data are represented as Mean ± SEM. Statistical analysis was performed by PRISM program using Student’s T-Test. When appropriate, one-way ANOVA followed by Turkey’s pothoc test was used. A P value < 0.05 was considered statistically significant.
3. Results
3.1. Temporal expression of gonadotropin subunits during embryonic and postnatal mouse pituitary development
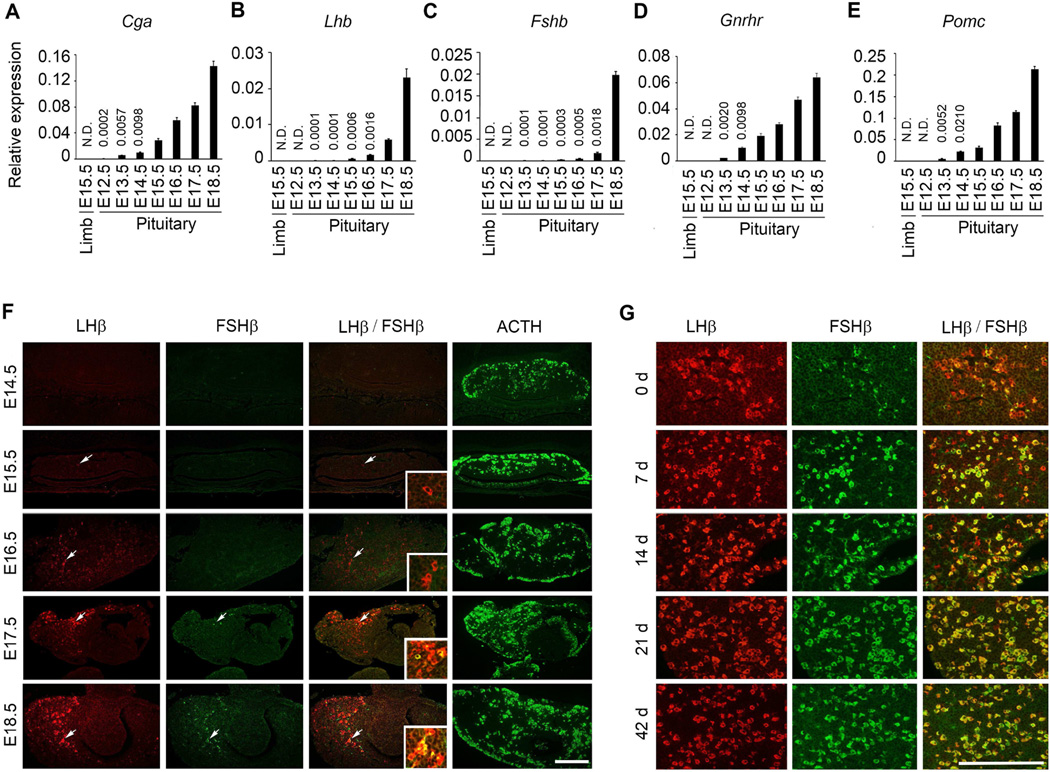
Expression of anterior pituitary hormone or hormone subunit-encoding mRNAs during mouse embryogenesis was previously analyzed qualitatively (Japon et al., 1994). To quantitatively evaluate their expression, we first performed Taqman real time PCR assays on isolated pituitaries from individual mouse embryos (collected from pregnant females) at different gestational ages. We used expression of Pomc mRNA as a positive control and RNA isolated from limb as a negative control for PCR assays since gonadotropin subunits are not expressed in this tissue. A physically defined pituitary was recognizable with the aid of a microscope and routinely isolated from E12.5 but not < E12.5 embryos. Expression of Cga was barely detectable at E12.5, but steadily increased thereafter and by E18.5 the expression increased by more than 700 times (Fig. 1A). Expression of Lhb mRNA was initiated at E13.5 and stayed minimal until E15.5. Between E16.5 and E18.5, Lhb mRNA expression was significantly increased by 40 times (Fig. 1B). Although expression of Fshb mRNA was also initiated at E13.5, the levels remained low and barely detectable until E.16.5 (Fig. 1C). By E18.5, it was also significantly increased by 40 times (Fig. 1C). Expression of Gnrhr, a gonadotrope-specific marker was detectable beginning at E13.5. Thereafter, it steadily increased and reached significantly higher levels by E18.5 (Fig. 1D). Expression of gonadotropin subunit-encoding mRNAs was undetectable in the limb at E15.5 (Fig. 1A–1E) or other embryonic time points tested (data not shown) and confirmed the specificity of the corresponding Taqman real time qPCR assays. Expression of Pomc mRNA beginning at E13.5 and thereafter was readily detectable (Fig. 1E) and further confirmed that we correctly dissected pituitaries at each embryonic stage.
Fig. 1.
Taqman real time qPCR assays (A–E) and dual label immunofluorescence (F and G) show temporal expression of gonadotropin subunit-encoding mRNAs and the β -subunit proteins during mouse embryo (F) and postnatal (G) pituitary development. Expression of Pomc mRNA (E) is used as a positive and limb RNA as a negative control for qPCR assays. For qPCR assays, values shown are mean ± SEM. * P < 0.01. For each time point, individual pituitaries from at least three embryos were used. N.D.= not detectable.
Expression of RNA does not always correlate with the corresponding protein expression. To test how gonadotropin β -subunit mRNA expression would correlate with their corresponding protein expression, we simultaneously analyzed localization of gonadotropin β-subunits on formalin-fixed embryonic pituitary sections by double label immunofluorescence. Expression of LHβ protein closely matched that of the corresponding mRNA and was not readily obvious in pituitary sections until E15.5 (Fig. 1E). Even at E15.5, only a rare immunostained LHβ+ cell was apparent in the entire embryonic pituitary. Within 24h, i.e. by E16.5, LHβ expression was significantly upregulated and continued to markedly increase by 18.5. Expression of FSHβ was delayed and became more obvious by E18.5 although at E17.5, very few cells expressed both LHβ and FSHβ (Fig. 1F). In contrast, Fshb mRNA expression was detectable as early as E15.5 (Fig. 1C). Several gonadotrope cells expressed only LHβ at E17.5 (Fig. 1F). These cells persisted although the number of gonadotropes co-expressing LHβ and FSHβ beginning at E18.5 and continued until postnatal day 14. By postnatal days 21 and 42, co-localization of LHβ and FSHβ was observed in nearly all gonadotropes (Fig. 1F and 1G). Visualization of ACTH immunoreactivity validated the immunofluorescence experiments and confirmed that we correctly dissected and immunolabeled the embryonic pituitaries (Fig. 1F). Together, these data indicate a strong correlation between expression of Lhb mRNA and LHβ protein, and there was discordance between Fshb mRNA and the corresponding FSHβ protein expression during embryonic mouse pituitary development.
3.2. Generation of an Fshb-Cre deleter mouse strain
To produce transgenic mice with gonadotrope - targeted expression of Cre, we performed two independent sets of microinjections of Fshb-iCre transgene DNA (Fig. 2A) into fertilized one-cell mouse embryos. From a total of 31 pups produced, we identified seven founders (four males and three females) by genomic PCR assays. We used primers that specifically amplified a 750bp fragment within the ovine Fshb promoter region thus distinguishing transgene-carrying mice from wild-type littermate control mice (Fig. 2B). Two of the seven founders did not transmit the transgene and were not further analyzed. Of the remaining five, both male and female founders successfully transmitted the transgene to F2 generation. From these, we established 3 independent lines designated Line # 100, Line # 400 and Line # 500. To check how CRE expression varied among the three lines, we separately introduced the transgene onto mT/mG genetic background and harvested pituitaries from the resulting mice at 42d of age. These mice express red fluorescence (from tdTomato = mT) prior to, and green fluorescence (enhanced green fluorescent protein = mG) following, CRE-mediated recombination and thus allow monitoring the in vivo Cre recombinase activity in specific cell types. As shown in Fig. 2C, CRE activity in pituitaries visualized by GFP fluorescence was indistinguishable among the three lines. In pituitaries of all the lines, CRE activity was restricted only to a sub-population of cells (GFP+) within the anterior lobe (AL in Fig. 2C) and both neural (NL in Fig. 2C) and intermediate (IL in Fig. 2C) lobes remained RFP+ (representing the tdTomato) and showed only red fluorescence. Thus, Fshb-iCre transgene is appropriately targeted to and expressed in mouse anterior pituitary. Both male and female Fshb-Cre deleter mice did not display any overt phenotypes and were fertile. Based on the better breeding characteristics i.e. litter size and number of litters (data not shown), we chose to perform all the subsequent analyses with either line 100 or line 400 mice.
Fig. 2.
Generation of Fshb-Cre deleter mice is achieved by pronuclear microinjection of the transgene DNA (A). The transgene is stably transmitted to F2 progeny as demonstrated by tail DNA genomic PCR (B) using specific primers indicated by blue arrows (A) that amplify a 750bp fragment. + = Transgene positive; − = WT. CRE recombinase activity monitored in vivo by GFP fluorescence in whole pituitary is restricted to only the anterior lobe from three independent lines of transgenic mice on a dual color reporter mouse mT/mG genetic background (C). White bar = 1 mm. AL= anterior lobe; IL= intermediate lobe; NL= neural lobe.
3.3. Cre expression during embryonic and postnatal pituitary development in Cga-Cre, Lhb-Cre and Fshb-Cre deleter strains
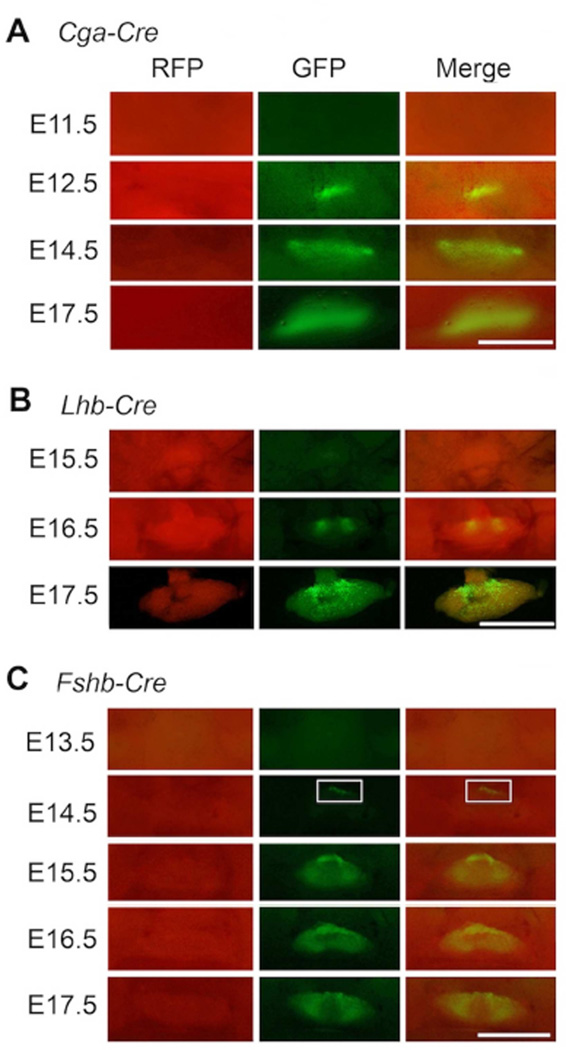
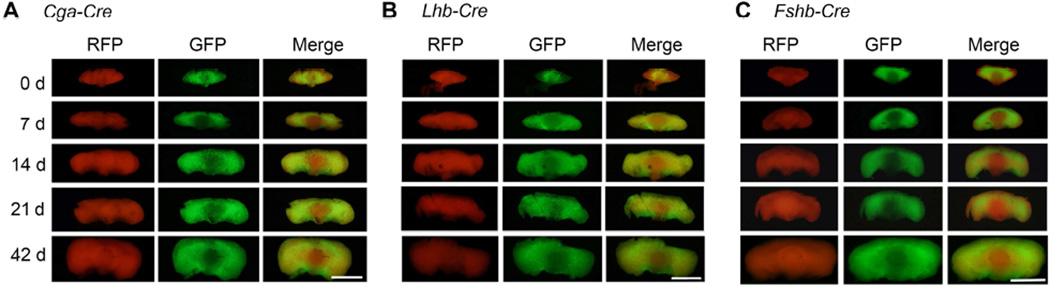
Temporally regulated activation of gonadotropin subunit promoters permits in vivo monitoring of Cre reporter transgenes expressed downstream of these regulatory elements (Perez-Millan et al., 2013, Charles et al., 2008, Brinkmeier, Gordon, Dowding et al., 1998). To compare the developmental onset of activation of each of the gonadotropin subunit promoters, we intercrossed Cga-Cre, Lhb-Cre and Fshb-Cre strains separately onto mT/mG background and visualized GFP fluorescence as an in vivo assay for CRE activity in pituitaries of the resulting mice. Expression of a Cga-Cre mouse BAC transgene, consistent with the activation of the endogenous Cga promoter (Fig. 1A), was evident by E12.5 with GFP fluorescence noted in very few cells (Fig. 3A). Expression of bLhb-Cre transgene was initiated at E 16.5 in a cluster of cells and it became more widespread by E17.5 as visualized by GFP fluorescence (Fig. 3B). This activation of bLhb promoter closely matched that of endogenous Lhb mRNA expression as described above (Fig. 1B). Finally, expression of Fshb transgene was initiated and GFP fluorescence was barely detectable in few cells at E14.5 (Fig. 3C). A greater number of GFP+ cells were visible by E16.5 (Fig. 3C) and GFP fluorescence was clearly more widespread by E17.5 (Fig. 3C), consistent with the endogenous Fshb mRNA around this embryonic time point (Fig. 1C). Despite these differences in CRE expression resulting from the activation of different gonadotropin subunit promoters at different times during embryonic pituitary development, CRE activity qualitatively assessed by visualization of GFP fluorescence was nearly identical in pituitaries of the three Cre deleter mice at birth (day 0) and postnatal ages 7d, 14d, 21d and 42d (Fig. 4A–C). Together, these data indicate that the three Cre deleter strains differ in expression of CRE mostly during embryonic but not postnatal pituitary development.
Fig. 3.
Comparison of CRE recombinase activity assayed in vivo by fluorescence visualization of tdTomato (indicated as RFP) to GFP conversion during embryonic pituitary development in three different gonadotrope-specific Cre-deleter mouse strains on an mT/mG genetic background (A–C). A representative example of pituitary harvested at each embryonic time point from each strain is shown. White box (in C) shows a group of GFP+ gonadotropes in pituitary of an Fshb-Cre+ mouse embryo at E14.5. White bar = 2mm.
Fig. 4.
Comparison of CRE recombinase activity assayed in vivo by fluorescence visualization of RFP to GFP conversion in postnatal pituitaries in three different gonadotrope-specific Credeleter mouse strains on an mT/mG genetic background shows identical pattern (A–C). White bar = 2mm.
3.4. Gonadotrope-specific Cre expression in pituitaries of Fshb-Cre deleter mice
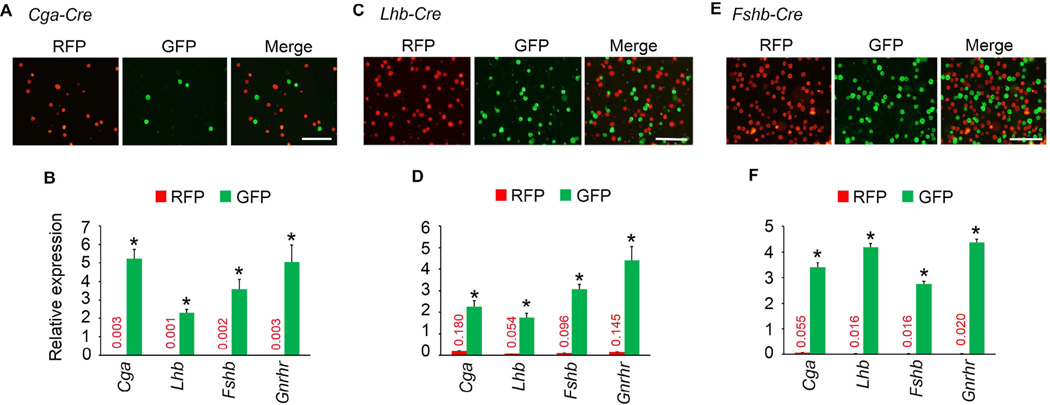
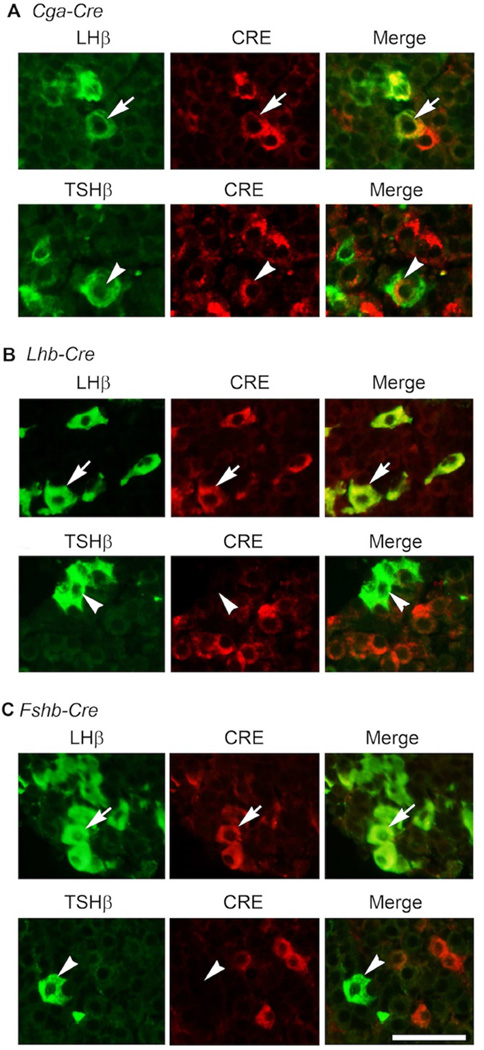
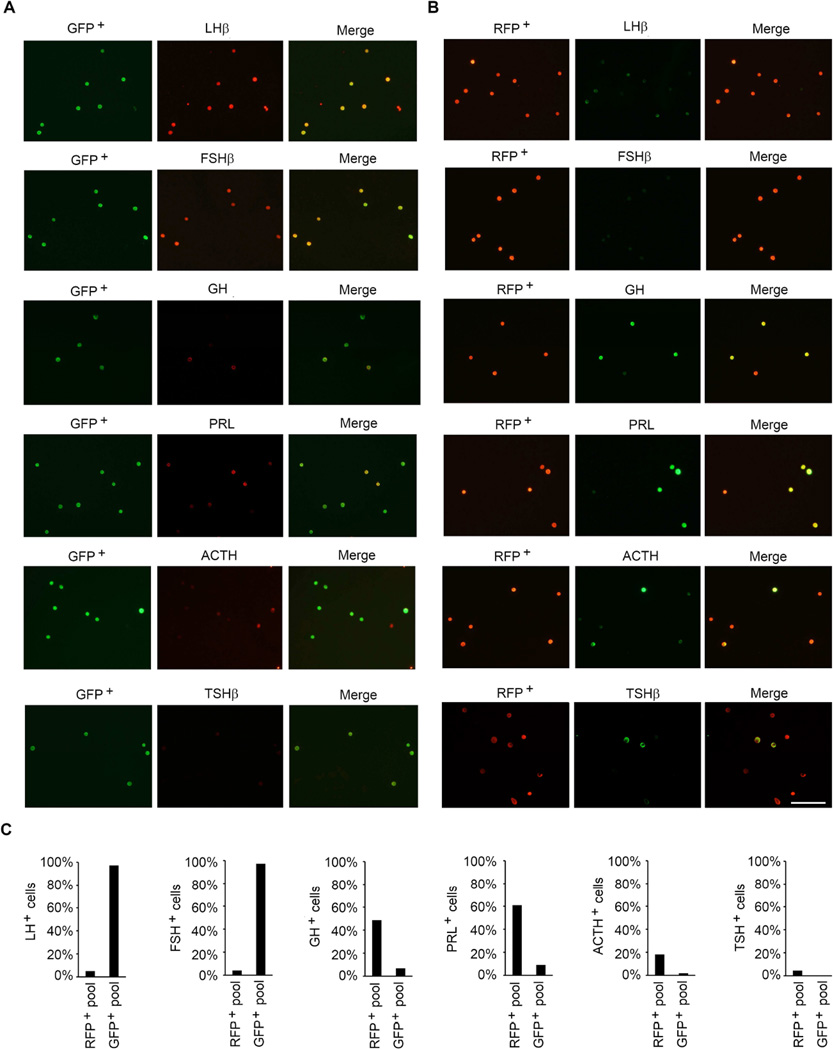
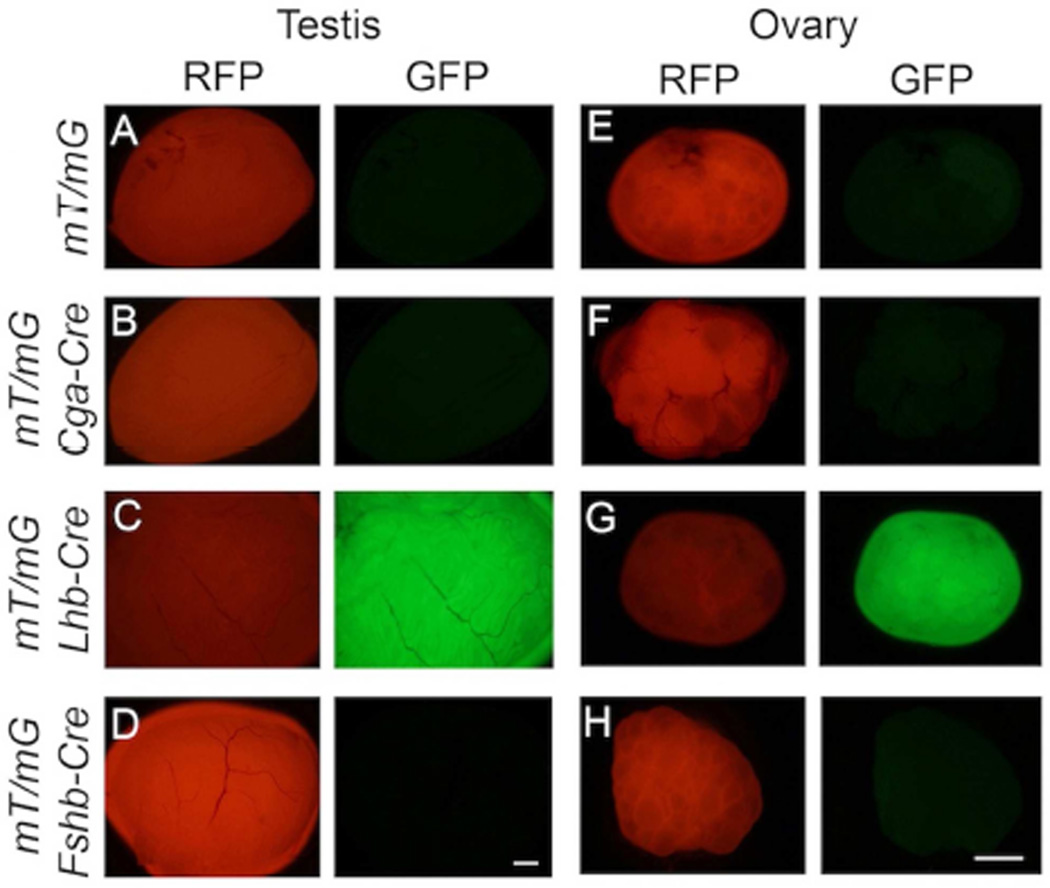
We next tested the cell-specific expression of CRE in pituitaries of the three Cre deleter mice. If CRE expression were activated in the gonadotrope lineage in pituitaries of Fshb-Cre mice on an mT/mG background, then gonadotropes should be GFP+ and the CRE− RFP+ population of cells should represent non-gonadotrope cells and should not express mRNAs encoding gonadotropin subunits. To evaluate this, first we confirmed that GFP+ and RFP+ cells exist as two different populations in the pre-sort mixture of collagenase-dissociated pituitary cells. Next, we sorted these mixtures of enzyme- dissociated GFP+ and RFP+ cells by FACS. When viewed under an epifluorescence microscope, these two cell populations appeared distinct (Fig. 5A, C and E). Next, we performed Taqman real time qPCR assays to quantify expression of gonadotropin subunit mRNAs in these purified cell fractions. As shown in Fig. 5B, D and F, Cga, Lhb, Fshb and Gnrhr mRNAs were highly expressed GFP+ cells and thus these represented purified gonadotropes. As predicted, expression of Cga, Lhb and Fshb mRNAs was very low or undetectable in RFP+ non-gonadotrope cells (Fig. 5B, D and F). Furthermore, double label immunofluorescence analysis using antibodies against CRE and LHβ or TSHβ on multiple pituitary sections (250 cells from at least 20 sections from 3 adult mice were scored) further confirmed that whereas CRE was localized to both gonadotropes and thyrotropes in Cga-Cre mice (Fig. 6A), it was localized to only gonadotropes in Lhb-Cre (Fig. 6B) and Fshb-Cre (Fig. 6C) mice. To further quantitatively determine expression of CRE activity in gonadotropes and non-gonadotrope cells, in two independent experiments, we obtained GFP+ and RFP+ pools of cells by flow sorting of pituitary cells from Fshb-Cre mice (Supplementary Fig. S1 and Fig. S2), performed immunofluorescence using antibodies specific for each of the anterior pituitary cell types and calculated percentages of each of the hormone+ cell types (Fig. 7A–C). These data indicate that nearly 97–100 % GFP+ cells represent gonadotropes (Fig. 7A and C) whereas RFP+ pool represented non-gonadotrope cells including GH+, PRL+, ACTH+ and TSH+ cells (Fig. 7B and C). Gonadotropes were on an average < 3 % in the RFP+ non-gonadotrope pool of cells separated by flow sorting. CRE activity in ~ 85 – 90 % of gonadotropes and thyrotropes in case of Cga-Cre strain (Perez-Millan et al., 2013), and in nearly similar percentage of gonadotropes in case of Lhb-Cre strain of mice was reported previously (Charles et al., 2008). We next tested ectopic expression of CRE activity in gonads from adult mice. Gonads from control mt/mG mice served as negative controls and did not show any GFP in the absence of any CRE activity (Fig. 8A and E). CRE activity was detected in gonads from only Lhb-Cre (Fig. 8C and G) but not Cga- Cre (Fig. 8B and F) or Fshb-Cre (Fig. 8D and H) mice on an mT/mG background consistent with tight regulation of the ovine Fshb promoter in vivo as reported previously (Wu et al., 2004, Huang, Sebastian, Strahl et al., 2001, Pernasetti, Spady, Hall et al., 2003). Collectively, these data indicate that CRE activity in Fshb-Cre deleter mice, unlike in Cga-Cre mice, is restricted to only gonadotropes in pituitary, and unlike in Lhb-Cre mice, not ectopically present in gonads.
Fig. 5.
FACS-sorted GFP+ (green) and RFP+ (red) pituitary cells are visualized by fluorescence microscopy in three different gonadotrope-specific Cre deleter strains on an mT/mG genetic background (A, C and E). Cells shown are visualized post-sorting. Merged images indicate no overlap in each case. White bar = 100 µm. Real time qPCR assays (B, D and F) indicate that gonadotropin subunit- and Gnrhr - encoding mRNAs are expressed at significantly higher levels in GFP+ gonadotrope cells compared to RFP+ non-gonadotrope cells (* P < 0.01 by T-Test; each sample assayed in triplicate).
Fig. 6.
Dual immunofluorescence on pituitary sections shows that in case of Cga-Cre deleter strain, CRE is also expressed in thyrotropes, immunolabeled with a TSHβ antibody (white arrowheads in lower panel in A). In contrast, CRE expression is restricted to only gonadotropes visualized by an LHβ antibody in pituitaries of Lhb-Cre (B) and Fshb-Cre (C) deleter strains. White Bar = 50 µm.
Fig. 7.
Flow-sorted GFP+ (A) and RFP+ (B) pituitary cells obtained from Fshb-Cre adult mice on mT/mG genetic background were immunolabeled with various antibodies specific for each of the anterior pituitary cell types. The percentage of hormone+ cells is calculated and shown in each case (C). Approximately 97 -100 % of GFP+ cells represent gonadotropes indicating specific CRE recombinase activity in these cells. Non-gonadotrope cells (GH+, PRL+, ACTH+ and TSH+) are enriched in RFP+ pool of cells. White Bar = 1000 µm.
Fig. 8.
Gonads from control mt/mG mice show only RFP+ red fluorescence (A and E) indicating conversion to GFP is CRE-dependent. Ectopic expression of CRE recombinase activity is noted in gonads in case of only Lhb-Cre strain (C and G) but not Cga-Cre (B and F) or Fshb-Cre (D and H) deleter strains. White bar = 1 mm.
3.5. Efficient deletion of Dicer by gonadotrope-targeted expression of Fshb-Cre transgene
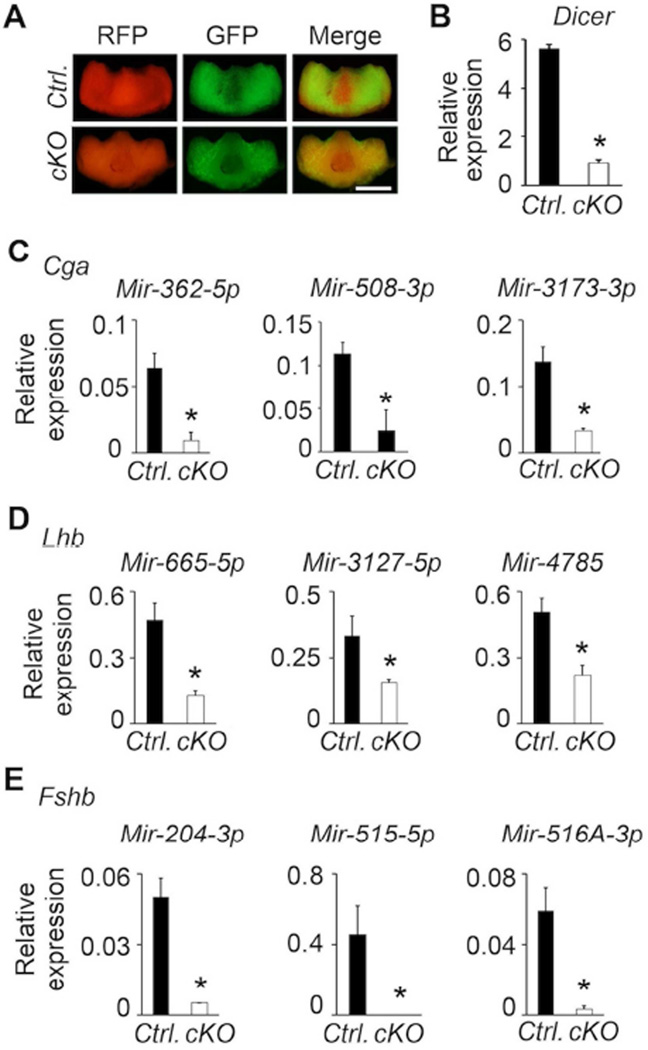
DICER is an RNAse III family member and essential for microRNA (miRNA) production (Bartel, 2004, Lewis and Steel, 2010) in gonadotropes as shown recently using Lhb-Cre deleter mice (Wang et al., 2015). To establish the feasibility of using Fshb-Cre deleter mice to conditionally inactivate a functionally important gene specifically in gonadotropes, we generated Dicerf/f Fshb-Cre+ (Dicer cKO) mice on an mT/mG+ background (Fig. 9A). We purified GFP+ gonadotropes from pituitaries of adult control and Dicer cKO mice by FACS. We quantified the expression of Dicer mRNA by a Taqman real time qPCR assay and found that it was significantly suppressed in cKO mutants compared to that in controls (Fig. 9B). Furthermore, real time qPCR assays identified that several of the miRNAs computationally predicted to bind to 3’-untranslated regions of gonadotropin subunit mRNAs (Cga, Lhb and Fshb) were significantly suppressed in GFP+ gonadotropes purified from pituitaries of Dicer cKO mice (Fig. 9C–E). Thus, Fshb-Cre deleter mice are useful to efficiently delete Dicer selectively in gonadotropes within pituitary.
Fig. 9.
Fshb-Cre deleter strain efficiently deletes Dicer alleles as shown by gross immunofluorescence visualization of pituitaries (A) and Dicer expression analysis by a real time qPCR assay (B). Several of the miRNAs predicted to bind 3’-untranslated region of gonadotropin subunit mRNAs are all significantly suppressed in pituitaries of cKO mice compared to those in control mice (C–E). Control mice (Ctrl.) belong to Fshb-Cre+ mT/mG + genotype. White bar = 1 mm. * P < 0.01 by T-Test; n= 3 mice and each sample used in triplicate.
3.6. Regulation of gonadotropins in pituitaries of Dicer cKO male mice
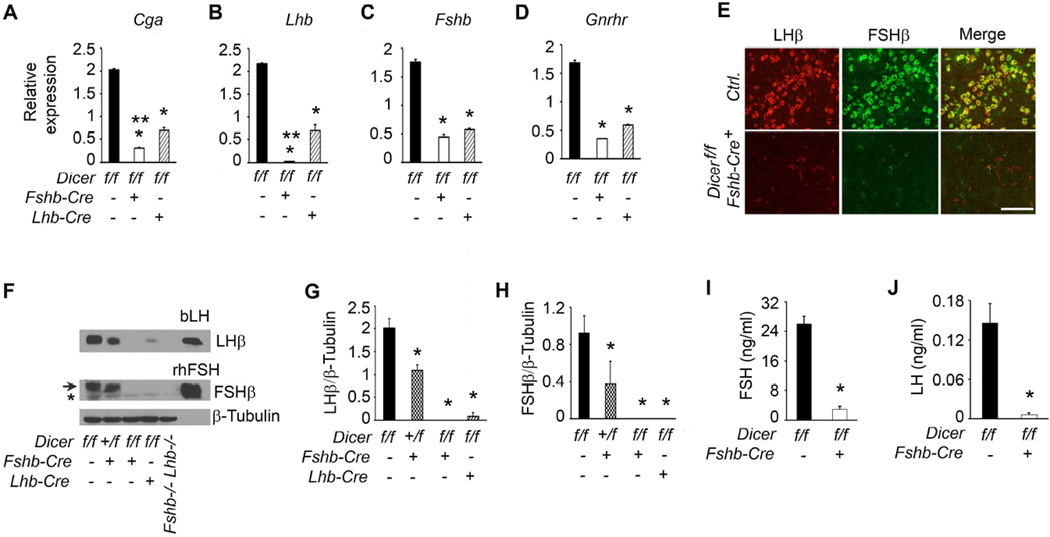
Next, we determined how CRE-mediated loss of DICER in gonadotropes affected gonadotropin subunits in pituitaries of adult male mice by Taqman real time qPCR assays. Gonadotrope-specific loss of DICER in pituitaries of mutant male mice resulted in severe suppression of Cga, Lhb and Fshb mRNAs compared to those in pituitaries of control mice (* in Fig. 10A–C). Loss of DICER also significantly suppressed Gnrhr mRNA in pituitaries of Dicer cKO mice (Fig. 10 D). The extent of Cga and Lhb mRNA suppression with Fshb-Cre deleter mice was more robust compared with that obtained using Lhb-Cre deleter mice (** Fig. 10A–D). To further evaluate how loss of DICER affected gonadotropin subunit proteins, immunofluorescence (Fig. 10E) and Western blot (Fig. 10F–H) analyses were performed on pituitaries obtained from adult control and Dicer cKO male mice. Pituitary sections from control mice (Ctrl.) showed gonadotropes intensely labeled with antibodies against both LHβ and FSHβ (Fig. 10E; upper panel). In contrast, pituitary sections from Dicer cKO mice showed gonadotropes with significantly reduced expression of LHβ and FSHβ proteins. Both LHβ and FSHβ subunits were also nearly undetectable in pituitaries of Dicer cKO mice when expression was quantified by densitometry following Western blot analysis (Fig. 10F–H). Similar to qPCR analysis, reduction of gonadotropin subunit protein expression was also more robust with Fshb-Cre deleters compared to that obtained with Lhb-Cre deleter mice (Fig. 10F–H). Loss of LHβ and FSHβ protein expression in pituitaries of double null mice lacking both Fshb and Lhb alleles and intense immunoreactivity with purified bovine LH (bLH) and recombinant human FSH (rhFSH) proteins further validated the specificity of the antibodies used for Western blot analysis (Fig. 10F). Serum ELISA analyses further confirmed that both LH and FSH were severely suppressed in Dicer cKO male mice compared to the age-matched littermate controls (Fig. 10I and J). Thus, gonadotrope-specific deletion of Dicer achieved with an Fshb-Cre deleter strain results in robust suppression of gonadotropin β - subunit encoding mRNAs and the corresponding proteins in pituitaries of Dicer cKO male mice.
Fig. 10.
Defects in gonadotropin homeostasis in Dicer cKO male mice analyzed by real time qPCR assays (A–D), dual immunofluorescence (E), Western blot analysis of pituitary proteins (F–H) and RIAs on serum samples (I and J). These data indicate that loss of DICER-dependent miRNAs results in profound suppression of gonadotropin synthesis and secretion. An asterisk below the arrow in (F) indicates a non-specific band. In A–D, G and H, * P < 0.01 compared to corresponding controls and ** P <0.01 when two Dicer mutants (Lhb-Cre and Fshb-Cre) are compared by One way ANOVA followed by Turkey’s post-hoc test. In I and J, P < 0.01 by TTest. Adult male mice at 6 weeks of age (n=4–5) are used in each experiment. Representative immunolocalization (E) and immunoblots (F) are shown. White bar in E = 100 µm.
3.7. Reproductive phenotypes of Dicer cKO male mice
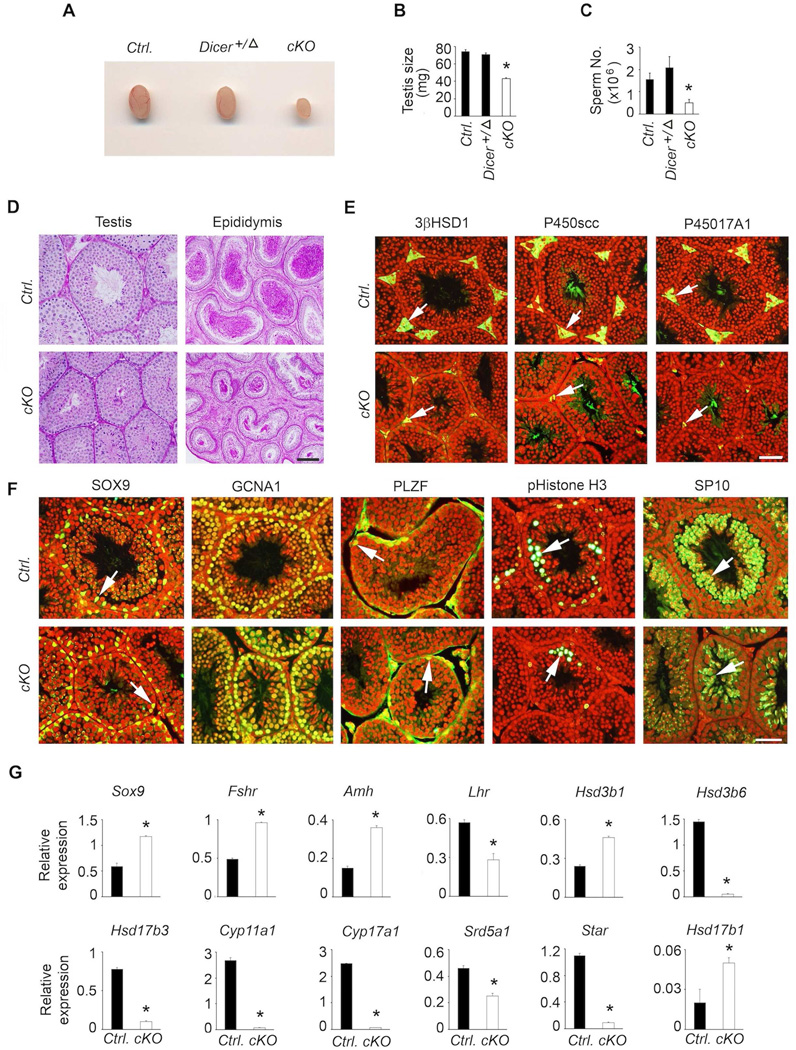
Loss or suppression of each or both of the pituitary gonadotropins leads to male fertility defects in mice (Kumar, 2007, Kumar, Wang, Lu et al., 1997, Ma, Dong, Matzuk et al., 2004, Matzuk, Kumar and Bradley, 1995). Mice with a gonadotrope-specific loss of Dicer achieved with Lhb-Cre deleter strain display aberrant and variable male reproductive phenotypes (Wang et al., 2015). However, it was not possible to determine if the infertility was a secondary consequence to suppressed gonadotropins or due to loss of DICER itself locally within the testes since Lhb-Cre was ectopically expressed in testis (Wang et al., 2015). To examine the consequences of suppressed gonadotropins alone in Dicer cKO mice achieved with Fshb-Cre deleter strain, we analyzed several aspects of male reproductive physiology. Testes were grossly smaller (Fig. 11A) and their weights significantly reduced by > 50 % (Fig. 11B) in Dicer cKO mice at 42 days of age compared to those in age-matched control and heterozygous littermates. Mating trials of Dicer cKO males (n=4) with control females (n=8) over a period of 4–6 months did not result in any litters compared to 21 litters and 8.4 ± 0.7 pups per litter from age-matched control males (n=5; P < 0.01, t-test). One of the four (25 %) Dicer cKO males sired one litter of 4 pups within the first month of the mating trial and subsequently ceased breeding. In accordance with the fertility data, epididymal sperm number was significantly reduced in Dicer cKO mutants compared to that in control males (Fig. 11C). Thus, these data indicate that gonadotrope-specific Dicer deletion results in male fertility defects as a secondary consequence to suppressed gonadotropins.
Fig. 11.
Analysis of male reproductive phenotypes in Dicer cKO mice indicate that grossly (A) testis size (B) and sperm number (C) are decreased compared to those in age-matched littermate controls. Histological analysis of testis by PAS/hematoxylin staining shows decreased testis tubule size and reduced sperm number in cauda epididymis (D), immunolocalization reveals hypoplasia of Leydig cells (white arrows in E) and grossly normal expression of Sertoli and germ cell markers (F), but real time qPCR assays indicate aberrant testis somatic cell gene expression (G). For each experiment, at least 4–5 adult mice at 42 days of age are used. Real time qPCR assays are done in triplicate per sample. In A–C, * P < 0.01 compared to corresponding controls by One way ANOVA followed by Turkey’s post-hoc test. For qPCR assays, * P < 0.01 by T-test. In E and F panels, merged images are shown and nuclei are stained red. Bars in D-F = 100 µm.
To further characterize male reproductive phenotypes, we performed periodic acid- Schiff’s reagent (PAS)/hematoxylin staining on formalin-fixed testis and epididymis sections. Consistent with the phenotypes described above, histological analysis of testis indicated a reduction in tubule size (Fig 10D, left panels), and that of cauda epididymis indicated significantly reduced sperm (Fig. 11D; right panels) in adult Dicer cKO mutants compared to those in age-matched controls. Interestingly, expression of Leydig cell markers including 3β HSD1, P450SCC and P45017CYPA1 was markedly reduced in the interstitium of testis sections obtained from Dicer cKO mice (Fig. 11E). While the morphology of the immunostained Leydig cells in the interstitium of control mouse testis sections indicated that there were mostly mature adult-type Leydig cells, those in the interstitium of Dicer cKO mouse testis sections mostly resembled immature and fetal-type Leydig cells, typically seen in the absence of LH in Lhb knockout mice (Ma et al., 2004). To evaluate further if testis tubule markers were affected in testis of Dicer cKO mutants, we next performed immunofluorescence analysis (Fig. 11F) using antibodies against SOX9 (specific for Sertoli cells), GCNA1 (a pan-germ cell marker), PLZF (specific for early spermatogonial progenitors), pHistone-H3 (labels mitotically dividing germ cells) and SP-10 (specific for round spermatids). We found no obvious differences in expression of various Sertoli/germ cell markers when testis sections from Dicer cKO mice were compared to those from control mice (arrows in Fig. 11F indicate similar expression pattern). Consistent with the morphological and histological data, Taqman real time qPCR assays confirmed that expression of many adult Leydig cell marker genes was severely suppressed and many marker genes in Sertoli and germ cells were aberrantly regulated in testes of Dicer cKO mutants compared to those in controls (Fig. 11G). Despite a reduction in Leydig cell marker gene expression, testosterone levels in Dicer ckO mice were only marginally suppressed compared to those in controls (Mean ± SEM, 0.26 ± 0.03 vs. Mean ± SEM, 0.2 ± .04; P > 0.05; T-test; n= 4–6). Together, these data indicate that Dicer cKO male mice display reduced testis size, severe oligospermia, defects in Leydig cell maturation resulting in impaired fertility as a secondary consequence to suppressed gonadotropins.
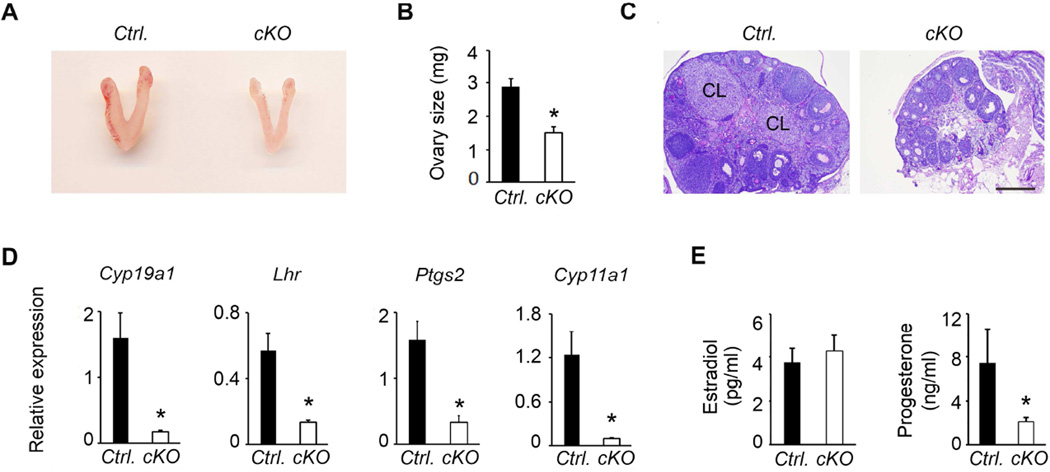
3.8. Regulation of gonadotropins in pituitaries of Dicer cKO female mice
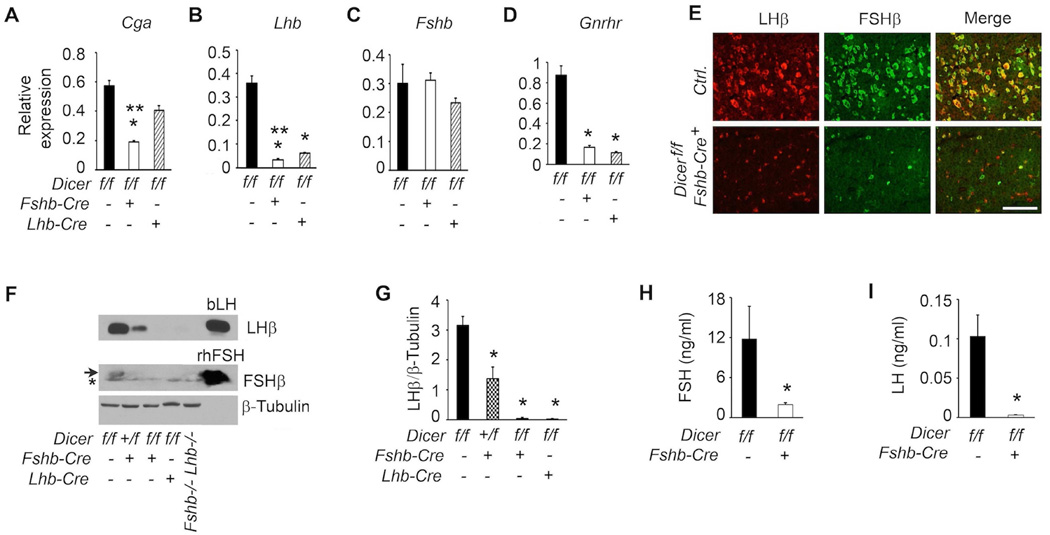
Gonadotropin regulation within pituitary is gender-specific (Kumar, Fairchild-Huntress and Low, 1992, Kumar, Schuff, Nusser et al., 2006). To test how gonadotrope-specific deletion of Dicer affected gonadotropin synthesis in pituitaries of female Dicer cKO mice, we first analyzed expression of gonadotropin subunit-encoding mRNAs by real time qPCR assays. Of the three subunit-encoding mRNAs, both those encoding α-GSU and LHβ were severely suppressed (Fig. 12A–B) whereas expression of Fshb mRNA was not affected (Fig. 12C). In addition, Gnrhr mRNA was also severely suppressed in pituitaries of Dicer cKO female mutants (Fig.12D). The suppression of Cga and Lhb mRNAs was more severe using Fshb-Cre compared to that in pituitaries of Dicer cKO female mice generated using the bLhb-Cre driver line (open versus hatched bars in Fig. 12A–B, indicated by **). Immunofluorescence analysis further confirmed that both LHβ and FSHβ protein expression was significantly suppressed in pituitary sections obtained from Dicer cKO female mice (Fig. 12E; lower panels) compared to that obtained from control (Ctrl.) mice (Fig. 12E; top panels). Finally, Western blot analysis of total proteins, consistent with the immunolabeling data, indicated that expression of LHβ was significantly suppressed in pituitaries of Dicer cKO female mice (Fig. 12F; top panel and densitometry quantification shown in Fig. 12G). In contrast to expression of LHβ, FSHβ protein was undetectable in pituitaries of Dicer cKO female mice (Fig. 11E middle panel). In accordance with the protein expression data, serum analyses confirmed that gonadotropins were severely suppressed in Dicer cKO mutant females when compared to those in age-matched littermate controls (Fig. 12H and I). Thus, gonadotrope-specific deletion of Dicer achieved with an Fshb- Cre deleter strain results in severe suppression of all aspects of gonadotropin synthesis and secretion in Dicer cKO female mice.
Fig. 12.
Defects in gonadotropin homeostasis in Dicer cKO female mice are analyzed exactly as described in Fig. 10. Real time qPCR assays (A–D), dual immunofluorescence localization (E–G) and serum RIAs (H and I) all confirm that gonadotropin synthesis and secretion are suppressed in Dicer cKO mice compared to age-matched littermate controls. An asterisk below the arrow in (F) indicates a non-specific band. In A-D, and G, * P < 0.01 compared to corresponding controls and in A, ** P < 0.01 Dicer deletion with Fshb-Cre vs. Lhb-Cre by One way ANOVA followed by Turkey’s post-hoc test. In H and I, P < 0.01 by T-Test. Adult female mice at 6 weeks of age (n=4–5) are used in each experiment. Representative immunolocalization (E) and immunoblots (F) are shown. White bar in E = 100 µm.
3.9. Reproductive phenotypes of Dicer cKO female mice
Loss (Kumar et al., 1997, Ma et al., 2004) or suppression (Matzuk et al., 1995, Matzuk and Lamb, 2008) of gonadotropins results in impaired female fertility. To determine the reproductive consequences of severe suppression of both FSH and LH, we first grossly examined the female reproductive tracts obtained from Dicer cKO adult female mice. We found hypoplastic uteri (uteri reduced by 45 % in mutants; 72.1 ± 6.6 mg in controls versus 32.4± 3.1 mg in mutants, P<0.01, n=3) and reduced size ovaries in Dicer cKO mutants compared to those in littermate control female mice (Fig. 13A and B). Ovarian histology by PAS/hematoxylin indicated follicles at all developmental stages including corpora lutea (CL in Fig. 13C; left panel) were readily apparent in ovarian sections from control mice (Ctrl.). However, ovarian sections from Dicer cKO female mice mostly contained immature and antral stage follicles and did not demonstrate any periovulatory follicles or corpora lutea (Fig. 13C, right panel) indicating arrested folliculogenesis, and failure to exhibit estrus cycles. Real time qPCR analysis further confirmed that both granulosa and theca cell marker genes (Cyp19a1, Lhr, Ptgs2 and Cyp11a1) typically regulated by gonadotropins were severely suppressed in ovaries of Dicer cKO mutant females (Fig. 13D). Serum analyses indicated that progesterone (Mean ± SEM, 7± 3 vs. 2 ± 0.4, P < 0.05 T-Test; n=6) but not estradiol (Mean ± SEM, 4 ± 0.6 vs. 4.2 ±0.8, P > 0.05 T-Test; n=6) levels were suppressed in Dicer cKO mutants compared to those in age-matched randomly cycling controls (Fig. 13 E and F). Despite the failure to demonstrate estrus cycles, Dicer ckO mutant females displayed normal puberty and vaginal opening. Consistent with ovarian histology and marker gene expression data, mating trials of Dicer cKO mutant females (5 out of 5 females) with proven fertile male mice did not result in production of any pups over a period of 4 months. Together, these data indicate that gonadotrope-specific deletion of Dicer achieved with an Fshb-Cre deleter strain results in severely suppressed gonadotropins and as a secondary consequence complete infertility in females.
Fig. 13.
Gross morphology of female reproductive tract in Dicer cKO mice shows hypoplastic uteri and ovaries (A). Ovarian size is reduced in Dicer cKO mutants at 63d of age compared to age-matched controls (B). Histology of PAS/hematoxylin – stained ovaries from mutants (C) show arrested follicular development and absence of corpora lutea compared to that in ovarian sections obtained from age-matched controls, where corpora lutea are readily apparent and indicated as CL. Real time qPCR assays (D) indicate gonadotropin-responsive ovarian markers are severely suppressed in ovaries of Dicer cKO mice (* P < 0.01 cKO vs. Ctrl., n= 3 mice, each sample in triplicate). Whereas serum estradiol (E) is not affected (P > 0.05; T-test, n=6 mice), serum progesterone (E) significantly suppressed in Dicer cKO mice (* P < 0.01; T-test, n=6 mice). Bar in B = 200 µm.
4. Discussion
Temporal expression of anterior pituitary cell-specific marker genes including those encoding gonadotropin subunits during mouse embryogenesis was previously investigated by an in situ hybridization method using specific radioactive cRNA probes (Japon et al., 1994). Here, we used real time qPCR assays and quantitatively analyzed expression of gonadotropin subunit encoding mRNAs beginning at E12.5 and further refined the earlier observations. Moreover, we have systematically mapped the corresponding gonadotropin protein expression in embryonic mouse pituitary sections by dual immunofluorescence. Together, these data allowed us to design a new gonadotrope-specific Fshb-iCre deleter strain in which we used 4.7 kb of sheep Fshb promoter sequences to drive the expression of CRE recombinase specifically to the gonadotrope lineage. This promoter was previously well characterized with regard to cell-specific and hormonal regulation (Huang, Sebastian, Strahl et al., 2001) and successfully used to express several reporters including luciferase (Huang et al., 2001, Huang et al., 2001), H-2Kk (Wu et al., 2004) surface antigen, and SV40 virus large T-antigen (Pernasetti, Vasilyev, Rosenberg et al., 2001) in transgenic mice. In the present study, the in vivo CRE recombinase activity as measured by conversion of a RFP to GFP reporter was initiated as early as E 14.5 and was robust by E17.5 (Fig. 3C) consistent with the endogenous expression of FSH protein in normal mouse anterior pituitary visualized by immunofluorescence (Fig. 1F). Thus, CRE expression in our Fshb-Cre strain temporally matched the activation of the endogenous mouse Fshb promoter and hence this deleter strain should aid in recombining floxed alleles beginning at E14.5 during embryonic anterior pituitary development.
The Camper laboratory previously developed two useful CRE deleter strains namely Cga-Cre and Lhb-Cre specific for the gonadotrope lineage (Perez-Millan et al., 2013, Charles et al., 2008). CRE expression in these strains was initiated at E12.5 in the Cga-Cre line and E17.5 in the Lhb-Cre line based on visualization by either X-gal staining (in case of Cga-Cre) or immunolabeling (in case of Lhb-Cre) method, respectively. While the in vivo CRE recombinase activity in Cga-Cre strain visualized by conversion of the RFP to GFP matched as reported earlier (Perez-Millan et al., 2013), it was apparent that in case of Lhb-Cre strain, CRE recombinase activity was observed one day earlier at E16.5 (Fig. 3B). This subtle difference could be due to differences in half-life and/or turn over of different reporter mRNAs/proteins (lacZ versus GFP) used to visually monitor the in vivo CRE recombinase activity. Thus, Fshb-Cre deleter strain should prove useful when recombination of floxed alleles is desired beginning around E14.5 during anterior pituitary development.
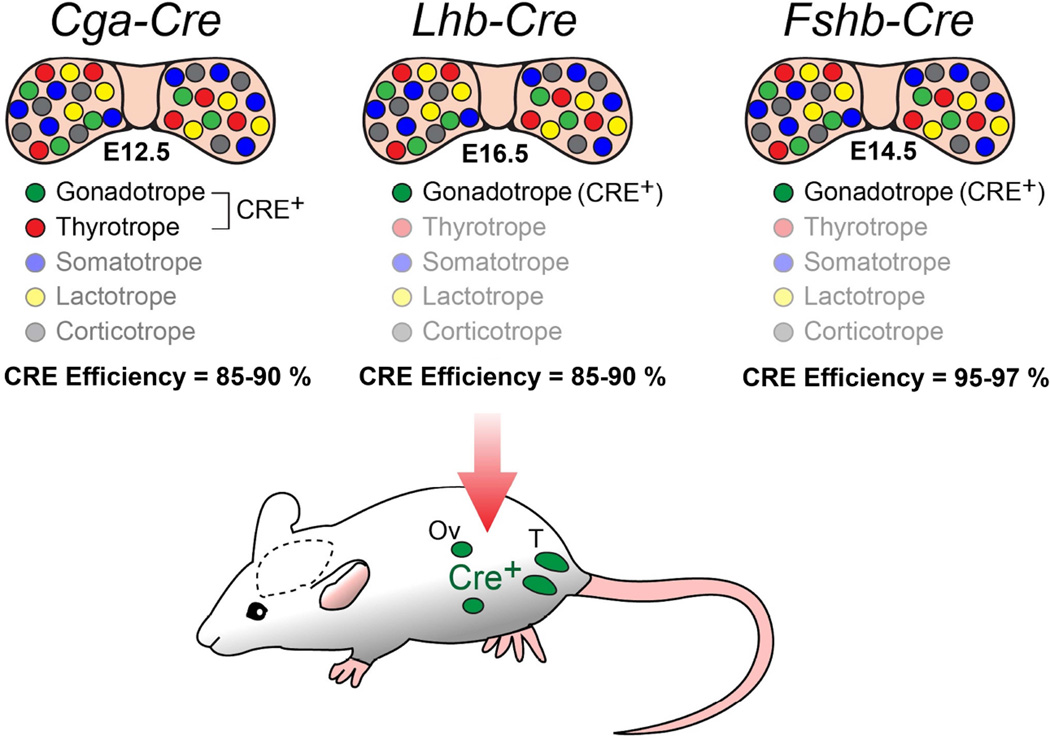
While both Cga-Cre and Lhb-Cre deleter strains have been extremely useful for achieving gonadotrope-specific deletion of several alleles of interest (Perez-Millan et al., 2013, Charles et al., 2008, Akhter, CarlLee, Syed et al., 2014, Luongo, Martin, Vella et al., 2015), both these strains have some limitations. Cga promoter is active in thyrotropes in addition to gonadotropes and Lhb promoter is ectopically active in gonads. Therefore, CRE recombination also occurs in non-gonadotrope cells (thyrotropes)/tissues (gonads) in these two delter strains. This poses the problem of undesirable lethality and infertility depending on the allele (s) deleted. Similarly, such limitations also seem apparent with the Gnrhr-CreKI mouse strain that has been used to delete alleles in the gonadotrope lineage (Wen et al., 2008, Fortin, Boehm, Deng et al., 2014). Fshb-Cre deleter strain circumvents these limitations since CRE recombination in this strain is exclusively restricted to the gonadotrope lineage and no ectopic expression was observed in gonads (Fig. 14). Together, the availability of Cga-Cre, Lhb-Cre and Fshb-Cre deleter mouse strains provides us a set of gonadotropin subunit promoter-based genetic tools to regulate CRE recombinase activity broadly from E12.5 to E16.5 during anterior pituitary gland development.
Fig. 14.
Comparative analysis of three different gonadotropin subunit promoters (Cga, Lhb and Fshb) driving Cre-expression in pituitary illustrates that Cre expression is specific to only the gonadotrope lineage with no ectopic expression in gonads in Fshb-Cre deleter strain. Different cell lineages within anterior pituitary are color-coded. CRE activation initiated during embryonic pituitary development and CRE efficiency in each strain is indicated.
Quantitative analyses indicated that a high level (> 95 %) of in vivo CRE recombinase activity was achieved specifically in gonadotropes in Fshb-Cre mice. We demonstrated the applicability of Fshb-Cre strain by specifically deleting Dicer in gonadotropes of mice. Gonadotrope-specific deletion of Dicer was chosen because we could compare these mutant mice with those we previously reported (Wang et al., 2015) in which Dicer was deleted in gonadotropes using the Lhb-Cre deleter strain. Dicer and DICER-dependent miRNAs were profoundly suppressed when GFP-tagged and FACS sorted gonadotropes were analyzed. This led to suppression of gonadotropin synthesis and secretion and resulted in various reproductive abnormalities that manifest as a secondary consequence, similarly observed in mice with Lhb-Cre - mediated Dicer deletion (Wang et al., 2015). Our present studies with Fshb-Cre driver line unequivocally confirmed that infertility in Dicer cKO male mice was secondary to the gonadotropin-specific Dicer deletion. Our previous observations with regard to Fshb mRNA regulation analyzed in Dicer cKO mutants generated using Lhb-Cre strain (Wang et al., 2015) are consistent with data obtained with Dicer cKO mice generated with Fshb-Cre deleter strain. Together, these two independent genetic models suggest the involvement of a complex array of regulatory factors including those that are DICER-independent.
Although grossly similar reproductive phenotypes manifest in both Dicer cKO mouse models, we noticed subtle differences in expression of gonadotropin subunit mRNAs and the corresponding proteins in pituitary. Both Cga and Lhb mRNAs and LHβ protein were more significantly suppressed in pituitaries of Dicer cKO mice generated with Fshb-Cre compared to those in pituitaries of mice generated with Lhb-Cre strain. Additionally, in contrast to Dicer cKO male mice generated with Lhb-Cre (Wang et al., 2015), those generated with Fshb-Cre deleter strain did not show any variable testis phenotypes. These male mutant mice also did not display any variable fertility problems because Cre was not ectopically expressed and Dicer was not locally deleted within the testis of these mice. This feature of lack of ectopic expression of CRE recombinase activity in testes of Fshb-Cre deleter strain should be particularly useful when pituitary-specific recombined alleles need to be propagated via the male germline.
In conclusion, we developed and characterized a new and efficient Cre-deleter mouse strain using ovine Fshb 5’- regulatory sequences and targeted Cre specifically to the gonadotrope lineage within anterior pituitary. These mice serve as another useful in vivo genetic tool and should complement the existing Cre-deleter strains to specifically manipulate gene expression and function in gonadotropes. The subtle differences in initiation of Cre expression in different gonadotrope-specific Cre deleter strains may prove advantageous for lineage tracing experiments and deleting genes during a narrow window of embryonic pituitary development.
Supplementary Material
Highlights.
A new Fshb-iCre deleter strain specific for gonadotrope lineage has been developed.
CRE recombinase activity is restricted to only gonadotropes within anterior pituitary.
Fshb-iCre mice are fertile and develop normally with no overt phenotypes.
Fshb-iCre mice efficiently delete Dicer selectively in gonadotropes.
Loss of DICER in gonadotropes results in defects in gonadotropin homeostasis and fertility.
Acknowledgments
We sincerely thank Dr. Sally Camper for generously providing Cga-Cre and Lhb-Cre strains of mice, Dr. Brian Harfe for Dicerf/f mice, Dr. Irving Boime for supplying us recombinant FSH standard and antisera against FSH, Dr. A.F. Parlow for pituitary hormone standards and antisera, Dr. Buck Hales, Dr. George Enders, Dr. Prabhakara Reddi and Dr. Ken Morohashi for antibody reagents, Dr. Irfan Saadi for many helpful comments on the manuscript, Mr. Ian Graham for helping with mouse genotyping and Mr. Stan Fernald for his assistance with the art work. This work was funded in part by NIH grants AG029531, HD069751, GM104936, CA166557 (to T.R.K.).
Abbreviations
- ACTH
Adrenocorticotrophin
- Cga
Common glycoprotein hormone subunit
- CL
Corpora lutea
- FACS
Fluorescence-activated cell sorting
- FSH
Follicle-stimulating hormone
- GFP
Green fluorescent protein
- GH
Growth hormone
- LH
Luteinizing hormone
- miRNA
Micro RNA
- mT/mG
ROSA26 mice expressing dual color reporter gene cassette
- PAS
Periodic Acid Schiff’s reagent
- PVDF
Polyvinyl difluoride
- POMC
Proopiomelanocortin
- PRL
Prolactin
- RFP
Red fluorescence representing tdTomato
- TSH
Thyroid-stimulating hormone.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors have nothing to disclose
References
- 1.Asa SL, Ezzat S. Molecular basis of pituitary development and cytogenesis. Front Horm Res. 2004;32:1–19. doi: 10.1159/000079035. [DOI] [PubMed] [Google Scholar]
- 2.Bousfield GR, Jia L, Ward DN. Gonadotropins: chemistry and biosynthesis. In: Neill JD, editor. Knobil and Neill's Physiology of Reproduction. New York: Elsevier Press; 2006. pp. 1581–1634. [Google Scholar]
- 3.Childs GV. Cytochemical studies of multifunctional gonadotropes. Microsc Res Tech. 1997;39:114–130. doi: 10.1002/(SICI)1097-0029(19971015)39:2<114::AID-JEMT3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Rosenfeld MG. Transcriptional control of precursor proliferation in the early phases of pituitary development. Curr Opin Genet Dev. 2004;14:567–574. doi: 10.1016/j.gde.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Keegan CE, Camper SA. Mouse knockout solves endocrine puzzle and promotes new pituitary lineage model. Genes Dev. 2003;17:677–682. doi: 10.1101/gad.1085903. [DOI] [PubMed] [Google Scholar]
- 6.Wu JC, Su P, Safwat NW, Sebastian J, Miller WL. Rapid, efficient isolation of murine gonadotropes and their use in revealing control of follicle-stimulating hormone by paracrine pituitary factors. Endocrinology. 2004;145:5832–5839. doi: 10.1210/en.2004-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouin J. Molecular mechanisms of pituitary differentiation and regulation: implications for hormone deficiencies and hormone resistance syndromes. Front Horm Res. 2006;35:74–87. doi: 10.1159/000094310. [DOI] [PubMed] [Google Scholar]
- 8.Mollard P, Hodson DJ, Lafont C, Rizzoti K, Drouin J. A tridimensional view of pituitary development and function. Trends Endocrinol Metab. 2012;23:261–269. doi: 10.1016/j.tem.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- 10.Watkins-Chow DE, Camper SA. How many homeobox genes does it take to make a pituitary gland? Trends Genet. 1998;14:284–290. doi: 10.1016/s0168-9525(98)01476-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Gleiberman AS, Rosenfeld MG. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol Rev. 2007;87:933–963. doi: 10.1152/physrev.00006.2006. [DOI] [PubMed] [Google Scholar]
- 12.Seeburg PH, Mason AJ, Stewart TA, Nikolics K. The mammalian GnRH gene and its pivotal role in reproduction. Recent Prog Horm Res. 1987;43:69–98. doi: 10.1016/b978-0-12-571143-2.50008-3. [DOI] [PubMed] [Google Scholar]
- 13.Seminara SB, Crowley WF., Jr Genetic approaches to unraveling reproductive disorders: examples of bedside to bench research in the genomic era. Endocr Rev. 2002;23:382–392. doi: 10.1210/edrv.23.3.0469. [DOI] [PubMed] [Google Scholar]
- 14.Seminara SB, Hayes FJ, Crowley WF., Jr Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): pathophysiological and genetic considerations. Endocr Rev. 1998;19:521–539. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 15.McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–2185. doi: 10.1210/en.2004-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie H, Cherrington BD, Meadows JD, Witham EA, Mellon PL. Msx1 homeodomain protein represses the alphaGSU and GnRH receptor genes during gonadotrope development. Mol Endocrinol. 2013;27:422–436. doi: 10.1210/me.2012-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H, Hoffmann HM, Meadows JD, Mayo SL, Trang C, Leming SS, Maruggi C, Davis SW, Larder R, Mellon PL. Homeodomain Proteins SIX3 and SIX6 Regulate Gonadotrope-specific Genes During Pituitary Development. Mol Endocrinol. 2015;29:842–855. doi: 10.1210/me.2014-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Japon MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 19.Brinkmeier ML, Davis SW, Carninci P, MacDonald JW, Kawai J, Ghosh D, Hayashizaki Y, Lyons RH, Camper SA. Discovery of transcriptional regulators and signaling pathways in the developing pituitary gland by bioinformatic and genomic approaches. Genomics. 2009;93:449–460. doi: 10.1016/j.ygeno.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas KR, Camper SA. Partial transcriptome of the developing pituitary gland. Genomics. 2000;70:335–346. doi: 10.1006/geno.2000.6400. [DOI] [PubMed] [Google Scholar]
- 21.Ye RS, Xi QY, Qi Q, Cheng X, Chen T, Li H, Kallon S, Shu G, Wang SB, Jiang QY, Zhang YL. Differentially expressed miRNAs after GnRH treatment and their potential roles in FSH regulation in porcine anterior pituitary cell. PLoS One. 2013;8:e57156. doi: 10.1371/journal.pone.0057156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuen T, Ruf F, Chu T, Sealfon SC. Microtranscriptome regulation by gonadotropin-releasing hormone. Mol Cell Endocrinol. 2009;302:12–17. doi: 10.1016/j.mce.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Cai Z, Wei S, Zhou H, Zhou H, Jiang X, Xu N. MicroRNA expression profiling of the porcine developing hypothalamus and pituitary tissue. Int J Mol Sci. 2013;14:20326–20339. doi: 10.3390/ijms141020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asa SL, Ezzat S. Genetics and proteomics of pituitary tumors. Endocrine. 2005;28:43–47. doi: 10.1385/ENDO:28:1:043. [DOI] [PubMed] [Google Scholar]
- 25.Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–975. [PubMed] [Google Scholar]
- 26.Perez-Millan MI, Zeidler MG, Saunders TL, Camper SA, Davis SW. Efficient, specific, developmentally appropriate cre-mediated recombination in anterior pituitary gonadotropes and thyrotropes. Genesis. 2013;51:785–792. doi: 10.1002/dvg.22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charles MA, Mortensen AH, Potok MA, Camper SA. Pitx2 deletion in pituitary gonadotropes is compatible with gonadal development, puberty, and fertility. Genesis. 2008;46:507–514. doi: 10.1002/dvg.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Graham I, Hastings R, Gunewardena S, Brinkmeier ML, Conn PM, Camper SA, Kumar TR. Gonadotrope-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. J Biol Chem. 2015;290:2699–2714. doi: 10.1074/jbc.M114.621565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen S, Schwarz JR, Niculescu D, Dinu C, Bauer CK, Hirdes W, Boehm U. Functional characterization of genetically labeled gonadotropes. Endocrinology. 2008;149:2701–2711. doi: 10.1210/en.2007-1502. [DOI] [PubMed] [Google Scholar]
- 30.Naik K, Pittman It, Wolfe A, Miller RS, Radovick S, Wondisford FE. A novel technique for temporally regulated cell type-specific Cre expression and recombination in the pituitary gonadotroph. J Mol Endocrinol. 2006;37:63–69. doi: 10.1677/jme.1.02053. [DOI] [PubMed] [Google Scholar]
- 31.Kumar TR, Larson M, Wang H, McDermott J, Bronshteyn I. Transgenic mouse technology: principles and methods. Methods Mol Biol. 2009;590:335–362. doi: 10.1007/978-1-60327-378-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Larson M, Jablonka-Shariff A, Pearl CA, Miller WL, Conn PM, Boime I, Kumar TR. Redirecting intracellular trafficking and the secretion pattern of FSH dramatically enhances ovarian function in mice. Proc Natl Acad Sci U. S. A. 2014;111:5735–5740. doi: 10.1073/pnas.1321404111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Su Z, Sanai N, Xue X, Lu L, Chen Y, Wu J, Zheng W, Zhuge Q, Wu ZB. microRNA expression profile and differentially-expressed genes in prolactinomas following bromocriptine treatment. Oncol Rep. 2012;27:1312–1320. doi: 10.3892/or.2012.1690. [DOI] [PubMed] [Google Scholar]
- 35.Behringer R, Gertsenstein M, Kristina VN, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. Fourth Edition. New York: COLD SPRING HARBOR LABORATORY PRESS; 2014. [Google Scholar]
- 36.Brinkmeier ML, Gordon DF, Dowding JM, Saunders TL, Kendall SK, Sarapura VD, Wood WM, Ridgway EC, Camper SA. Cell-specific expression of the mouse glycoprotein hormone alpha-subunit gene requires multiple interacting DNA elements in transgenic mice and cultured cells. Mol Endocrinol. 1998;12:622–633. doi: 10.1210/mend.12.5.0103. [DOI] [PubMed] [Google Scholar]
- 37.Huang HJ, Sebastian J, Strahl BD, Wu JC, Miller WL. The promoter for the ovine follicle-stimulating hormone-beta gene (FSHbeta) confers FSHbeta-like expression on luciferase in transgenic mice: regulatory studies in vivo and in vitro. Endocrinology. 2001;142:2260–2266. doi: 10.1210/endo.142.6.8202. [DOI] [PubMed] [Google Scholar]
- 38.Pernasetti F, Spady TJ, Hall SB, Rosenberg SB, Givens ML, Anderson S, Paulus M, Miller WL, Mellon PL. Pituitary tumorigenesis targeted by the ovine follicle-stimulating hormone beta-subunit gene regulatory region in transgenic mice. Mol Cell Endocrinol. 2003;203:169–183. doi: 10.1016/s0303-7207(02)00430-6. [DOI] [PubMed] [Google Scholar]
- 39.Kumar TR. Mouse models for gonadotropins: a 15-year saga. Mol Cell Endocrinol. 2007;260(262):249–254. doi: 10.1016/j.mce.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 41.Ma X, Dong Y, Matzuk MM, Kumar TR. Targeted disruption of luteinizing hormone beta-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility. Proc Natl Acad Sci U S A. 2004;101:17294–17299. doi: 10.1073/pnas.0404743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 43.Kumar TR, Fairchild-Huntress V, Low MJ. Gonadotrope-specific expression of the human follicle-stimulating hormone beta-subunit gene in pituitaries of transgenic mice. Mol Endocrinol. 1992;6:81–90. doi: 10.1210/mend.6.1.1738375. [DOI] [PubMed] [Google Scholar]
- 44.Kumar TR, Schuff KG, Nusser KD, Low MJ. Gonadotroph-specific expression of the human follicle stimulating hormone beta gene in transgenic mice. Mol Cell Endocrinol. 2006;247:103–115. doi: 10.1016/j.mce.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang HJ, Sebastian J, Strahl BD, Wu JC, Miller WL. Transcriptional regulation of the ovine follicle-stimulating hormone-beta gene by activin and gonadotropin-releasing hormone (GnRH): involvement of two proximal activator protein-1 sites for GnRH stimulation. Endocrinology. 2001;142:2267–2274. doi: 10.1210/endo.142.6.8203. [DOI] [PubMed] [Google Scholar]
- 47.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL. Cell-specific transcriptional regulation of follicle-stimulating hormone-beta by activin and gonadotropin-releasing hormone in the LbetaT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 48.Akhter N, CarlLee T, Syed MM, Odle AK, Cozart MA, Haney AC, Allensworth-James ML, Benes H, Childs GV. Selective deletion of leptin receptors in gonadotropes reveals activin and GnRH-binding sites as leptin targets in support of fertility. Endocrinology. 2014;155:4027–4042. doi: 10.1210/en.2014-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luongo C, Martin C, Vella K, Marsili A, Ambrosio R, Dentice M, Harney JW, Salvatore D, Zavacki AM, Larsen PR. The selective loss of the type 2 iodothyronine deiodinase in mouse thyrotrophs increases basal TSH but blunts the thyrotropin response to hypothyroidism. Endocrinology. 2015;156:745–754. doi: 10.1210/en.2014-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fortin J, Boehm U, Deng CX, Treier M, Bernard DJ. Follicle-stimulating hormone synthesis and fertility depend on SMAD4 and FOXL2. FASEB J. 2014;28:3396–3410. doi: 10.1096/fj.14-249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.