Abstract
Congenital malformations are the major cause of infant mortality in the US and Europe. Due to rapid advances in human genomics, we can now efficiently identify sequence variants that may cause disease in these patients. However, establishing disease causality remains a challenge. Additionally, in the case of congenital heart disease, many of the identified candidate genes are either novel to embryonic development or have no known function. Therefore, there is a pressing need to develop inexpensive and efficient technologies to screen these candidate genes for disease phenocopy in model systems and to perform functional studies to uncover their role in development. For this purpose, we sought to test F0 CRISPR based gene editing as a loss of function strategy for disease phenocopy in the frog model organism, Xenopus tropicalis. We demonstrate that the CRISPR/ Cas9 system can efficiently modify both alleles in the F0 generation within a few hours post fertilization, recapitulating even early disease phenotypes that are highly similar to knockdowns from morpholino oligos (MOs) in nearly all cases tested. We find that injecting Cas9 protein is dramatically more efficacious and less toxic than cas9 mRNA. We conclude that CRISPR based F0 gene modification in X. tropicalis is efficient and cost effective and readily recapitulates disease and MO phenotypes.
Keywords: Xenopus tropicalis, CRISPR, Cas9 protein, tyrosinase, dnah9, pax8, ccdc40, beta-catenin, foxj1, morpholino, fragment analysis
Introduction
Congenital malformations are a major cause of morbidity and mortality in infants and children. Of the congenital malformations, congenital heart disease (CHD) is the most common (Dolk et al., 2011; Pediatric Cardiac Genomics et al., 2013; van der Linde et al., 2011). Despite the enormous impact CHD has on child health, our understanding of the causes of CHD remains rudimentary. Recently, a number of studies have employed human genomics to identify over 350 candidate genes that may cause CHD (Fakhro et al., 2011; Glessner et al., 2014; Greenway et al., 2009; Hitz et al., 2012; Priest et al., 2012; Warburton et al., 2014; Zaidi et al., 2013; Zhao et al., 2013). However, due to high locus heterogeneity and a lack of second alleles, the disease relevance of the vast majority of these gene variants is uncertain. In addition, many of the genes identified are either novel to cardiac development, novel to embryonic development, or have no known function (Fakhro et al., 2011; Glessner et al., 2014; Greenway et al., 2009; Zaidi et al., 2013). Functionally testing these CHD candidate genes for a developmental role would improve our understanding of the genes that pattern the heart and the early embryo. Subsequent studies could then explore their developmental mechanisms. Therefore, given the large number of CHD candidate genes, there is a pressing need for a rapid and inexpensive loss of function strategy to test candidate disease genes for phenocopy in model systems.
Xenopus is an ideal model for studying human embryonic development. Compared to mammalian systems, Xenopus is rapid and cost effective, which is essential in order to evaluate the large number of genes emerging from human genomics studies of birth defect patients. In addition, Xenopus have a three-chambered heart with a highly trabeculated myocardium and a partially septated atria. The evolutionary conservation between Xenopus and humans offers additional opportunities to model human disease compared to other high-throughput aquatic vertebrates.
Traditionally, knockdown in Xenopus is achieved using translation or splice-site blocking morpholino oligos (MOs). MOs can target maternal and zygotic transcripts in order to achieve substantial or partial knockdown in F0 embryos (i.e. embryos that are injected with the MO). However, there are some disadvantages. MOs cost ~$400 per oligo and can become prohibitively expensive when screening hundreds of candidate disease genes as is the case now for CHD. Also in zebrafish, the specificity of MOs has recently been challenged (Kok et al., 2015). Unfortunately, it can be difficult to assess the off-target effects of MOs, and with start site MOs, efficacy of knockdown cannot be examined without an effective antibody. Therefore, correlating phenotype with knockdown can be challenging.
Recently, a number of genome editing tools have dramatically transformed our ability to generate mutations in the genome. Of these, clustered regularly inter-spaced short palindromic repeats (CRISPRs) are truly revolutionizing gene-editing technology in almost every model organism including Xenopus (Bassett and Liu, 2014; Blitz et al., 2013; Friedland et al., 2013; Guo et al., 2014; Hwang et al., 2013; Li et al., 2013; Nakayama et al., 2013; Wang et al., 2014). Due to its high efficacy and simplicity, CRISPR based gene editing appears amenable for large-scale genome screening. In the CRISPR/ Cas9 system, a site-specific small guide RNA (sgRNA) targets the genomic sequence adjacent to a PAM sequence, “NGG”, and recruits the Cas9 endonuclease to create a double stranded break. This is then repaired by non-homologous end joining, leading to insertions and deletions that can cause a frame shift, often leading to a null allele. Commonly in zebrafish and Xenopus, cas9 mRNA is co-injected with the target sgRNA to induce gene modifications using CRISPRs. However, recently three papers in zebrafish suggest that injecting Cas9 protein is faster and yields more mutagenesis than injecting cas9 mRNA (Gagnon et al., 2014; Kotani et al., 2015; Sung et al., 2014). However, this remains unexplored in Xenopus.
In this study, we sought to determine if CRISPR based gene editing could recapitulate disease phenotypes in F0 Xenopus embryos. Previous work in Xenopus has shown that F0 gene modifications occur in at least eleven different genes; however, combined, these studies examined embryonic phenotypes for only three loci in F0 animals (tyrosinase, six3, ptf1a/p48) (Blitz et al., 2013; Guo et al., 2014; Nakayama et al., 2013). In each case, phenotypes were detected at stage 40 or later in development roughly three days after fertilization. However, the patterning events that lead to many disease and developmental phenotypes, including cardiac development, occur much earlier. In addition, the development of Xenopus is rapid, and X. tropicalis, our diploid Xenopus model, thrives at warmer temperatures making its development more rapid than X. laevis. Therefore, we wondered if CRISPR/ Cas9 could modify the genome rapidly enough to affect early developmental events in F0 embryos and recapitulate those disease phenotypes seen in birth defects patients, including CHD.
In this study, we use the CRISPR/ Cas9 system to test six human disease genes (galnt11, dnah9, ccdc40, foxj1, pax8, and beta-catenin) with diverse phenotypes and compare them to established MO or mutant phenotypes. Given the importance of ciliopathies in human disease including CHD, we chose four genes that affect cilia in different ways (galnt11, dnah9, ccdc40, and foxj1) and consequently have different mechanisms to affect cardiac development (Brueckner, 2007; Li et al., 2015; Sutherland and Ware, 2009). In each case, these genes affect development by acting at stages 16-19 (within 20 hours of development). To examine organogenesis in an organ system that has yet to be tested using CRISPR, we examined kidney development with pax8, which also plays a critical role as early as stage 18 or 15 hours of development. Finally, to test the limitations of the technology, we examined beta-catenin, which has a known maternal role that can be effectively targeted with a translation blocking MO to create a ventralized phenotype (Heasman et al., 1994; Heasman et al., 2000; Wylie et al., 1996). We demonstrate that F0 CRISPR analysis is a powerful and dramatically less expensive loss of function screening tool than MOs. It readily recapitulates disease phenotypes that are nearly identical to MOs in most cases tested. Cas9 protein creates gene modifications more rapidly and effectively than cas9 mRNA with dramatically less toxicity enabling the detection of early embryonic phenotypes. We conclude that F0 CRISPR is an efficient screening tool for testing congenital malformation candidate genes in Xenopus.
Results
Fragment Analysis (FA) is a rapid and efficient way to detect gene modification
During a screen for gene function via genome modification using CRISPR/Cas9, there are two possible outcomes: 1) We detect a phenotype in which case we want to confirm that gene modification of the targeted site correlates with the phenotype; 2) We fail to detect a phenotype in which case, we want to confirm that there is evidence for gene modifications at the target site and that those gene modifications are such that they would be deleterious (i.e. not 3 base pair deletions). If not, we can target the loci with another sgRNA and if there is still no phenotype, we can conclude the screen of that gene. To begin our studies, we first sought to adapt a simple assay to test the efficacy of CRISPR mediated genome modification in Xenopus. Previous work has employed PCR amplification of the modified locus and either 1) direct sequencing of the PCR amplicons or 2) cloning of the PCR products followed by sequencing. The first method is limited by mosaicism, which scrambles the sequence traces making it difficult to determine the size and type of mutation (insertion/deletion) (Blitz et al., 2013; Nakayama et al., 2013). The second method is more time consuming due to multiple cloning steps and samples a relatively small number of PCR products. For screening purposes, the most critical information is the size of the indel, since early frameshift mutations are most likely to cause a loss of function allele. Additionally, an inexpensive and rapid method to genotype embryos would be powerful to correlate novel phenotypes with expected deleterious genotypes. Fragment analysis (FA), where PCR products are run on a standard capillary gel in order to determine product size at single base pair resolution, can provide this information cheaply and efficiently (Schuelke, 2000; Yang et al., 2015).
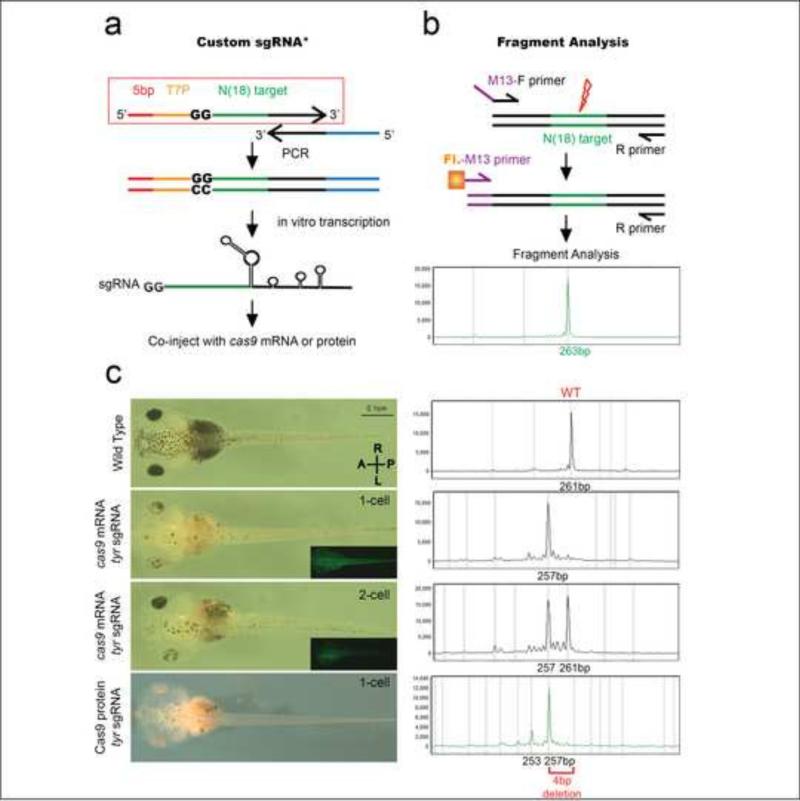
We began our studies with the tyrosinase (tyr) locus and used FA to test CRISPR efficiency. We employed the same sgRNA and PCR amplicon in previous studies (Blitz et al., 2013; Nakayama et al., 2013), a 261 base-pair (bp) product that is easily resolved using FA (Fig. 1(c)). When we inject embryos at the one cell stage with cas9 mRNA + tyr sgRNA, and then assay their genotype by FA, we observe a characteristic 4bp deletion peak. In Xenopus, if only 1 cell at the 2-cell stage is injected, we can select embryos targeting only one side of the embryo. In this case, FA reveals both the Wild Type (WT) and the 4bp deletion alleles. Interestingly, if we substitute Cas9 protein for mRNA, we find additional peaks corresponding to an 8bp deletion, as well as some insertions. These additional peaks were very rarely observed when the embryos were injected with cas9 mRNA (Supp. Fig. 1).
Figure 1.
Strategy for custom sgRNA design, Fragment Analysis, and study of tyrosinase CRISPR phenotype and genotype. (a) Strategy for custom sgRNA design (PCR method) with the red box outlining the forward primer for any target gene of interest. *This strategy was modified from that described by Nakayama et al. by addition of 5bp anchor sequence and use of Ambion's MegaShortscript kit, which greatly increases RNA yield (See Methods). The forward primer consists of 5bp anchor sequence (red), T7 promoter (yellow), 18bp target sequence (green), and 20bp complimentary overhang with reverse primer (black). The stock reverse primer consists of 20bp complementary overhang (black) and the remaining sgRNA sequence (blue). With one PCR step we generate the full sgRNA template. We then use in vitro transcription to generate the sgRNA which is co-injected with cas9 mRNA or protein. (b) Strategy for fragment analysis. Forward primer with a 5’-m13 overhang sequence is used with a R-primer to PCR amplify the target locus. A 2nd PCR step with a florescent m13 Primer and the same R-primer are used to generate a fluorescently tagged PCR amplicon, which is run on a capillary electrophoresis gel and analyzed. (c) Stage 42 embryos and their FA genotype (dorsal view, anterior to the left). FA genotype – X-axis size of amplicon in basepairs (bp), Y-axis relative intensity of PCR signal to loading control. WT tadpole and the corresponding 261bp PCR amplicon; Tadpole injected at 1-cell stage with cas9 mRNA+tyr sgRNA and corresponding 257bp FA peak (4 bp deletion). GFP co-expression is observed in whole embryo; Tadpole injected at 1-of-2-cell stage with cas9 mRNA+tyr sgRNA on left side (GFP co-expression only on left) and both deletion and WT peaks detected with FA; Tadpole injected with Cas9 protein and tyr sgRNA at 1-cell stage with 257bp deletion peak on FA.
When we clone and sequence these PCR products, we find two, three, four, and eight bp deletions in the target region. The majority of the clones (58%) show 4 or 8bp deletions and the proportion of these colonies roughly correlate with the height of the peaks on FA. Although sometimes an array of peaks are observed, each sgRNA seems to have a characteristic deletion peak which is shared in all injected embryos. For example, all embryos injected with our tyrosinase sgRNA have a characteristic 4bp deletion peak, while most of those injected with galnt11 sgRNA have a 10bp deletion peak, dnah9 sgRNA embryos have a 11bp deletion peak, ccdc40 sgRNA embryos have a 16bp deletion peak, and pax8 sgRNA embryos have a 17bp deletion peak (Supp. Fig. 3(a)-(e)). Furthermore, the same deletion peaks are detected in the DNA of adult frogs previously injected with Cas9 and sgRNA. Therefore, these deletions are stable and detectable over time, and FA can serve as a rapid and inexpensive F0 genotyping assay.
Cas9 protein is less toxic and more efficacious than cas9 mRNA
In all Xenopus reports to date, cas9 mRNA is co-injected with the target sgRNA to induce gene modifications using CRISPRs (Blitz et al., 2013; Guo et al., 2014; Nakayama et al., 2014; Nakayama et al., 2013; Wang et al., 2015). In Xenopus, we must inject 2-4ng/embryo of cas9 mRNA to generate an F0 phenotype at the tyrosinase locus (albinism). However, cas9 mRNA at a dose of 2ng/embryo is fairly toxic leading to >50% death in embryos (Supp. Fig. 2). In addition, a substantial percentage of surviving embryos are malformed which is not expected if mutations were restricted to the tyr locus. A similar degree of toxicity has been reported in a previous study (Guo et al 2015). This toxicity is dose dependent where more embryos survive at lower doses. However, injecting lower doses of cas9 mRNA dramatically reduces the incidence of F0 phenotypes. If we instead inject Cas9 protein directly into the embryo, we observe no toxic effects above uninjected controls with close to 90% of embryos surviving. Furthermore, the degree of albinism is much more pronounced with Cas9 protein co-injection compared to cas9 mRNA (Fig. 1(c), Supp. Fig. 2).
Cas9 gene modifications occur early in development
Next, in order to recover F0 phenotypes, especially phenotypes that occur early in development, the timing of the CRISPR/Cas9 mediated gene modifications is important. Once cas9 mRNA is injected, it needs to be translated, the protein folded, and the Cas9/ sgRNA complex assembled before genome editing can occur. We would expect this process to occur more rapidly if Cas9 protein was injected, especially if the Cas9 protein was incubated with the sgRNA prior to injection, although the kinetics of this process is not clear. Two studies in zebrafish suggest that injecting Cas9 protein induces mutations faster than cas9 mRNA (Kotani et al., 2015; Sung et al., 2014). Since Xenopus development is rapid, slow kinetics could preclude detecting early phenotypes in F0 animals. Clearly, phenotypes at later stages can be detected such as albinism at stage 40, but whether F0 phenotypes can be detected in Xenopus at earlier stages such as stage 20 is not known.
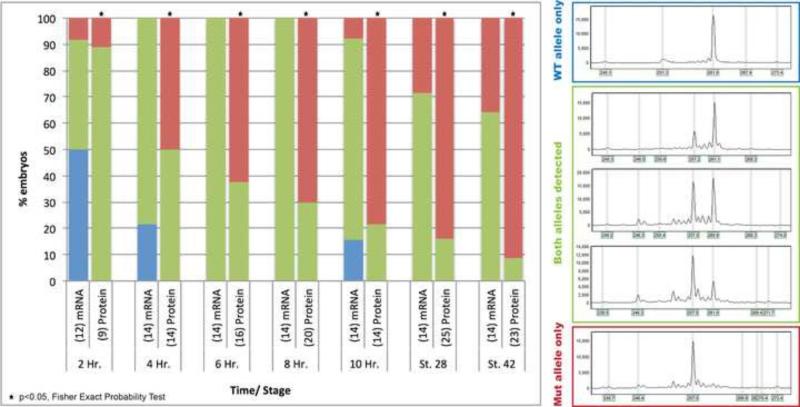
Therefore, we sought to determine the timing of deletions after injecting Cas9+sgRNA. To test this, we performed a time course experiment by injecting cas9 mRNA or protein with the tyrosinase sgRNA at the one cell stage, and collected embryos every 2 hours post injection. We used FA to assay for the presence of deletions. Importantly, even as early as two hours post injection, deletions were detected in 50% of embryos injected with cas9 mRNA and 100% of embryos injected with Cas9 protein. With time, a greater percentage of embryos had deletions, and we note that at stage 9 (5 hours post fertilization), when the maternal to zygotic transition (MZT) occurs, embryos co-injected with Cas9 protein/sgRNA had more evidence of biallelic modifications than embryos co-injected with cas9 mRNA. At every time point, a greater percentage of embryos had deletions with Cas9 protein compared to cas9 mRNA (p<0.05 at all time points) (Fig. 2). These results also correlate with the greater degree of albinism seen in Cas9 protein vs. mRNA-injected embryos. Taken together, we conclude that Cas9 protein is less toxic and more efficacious than cas9 mRNA and can potentially target genes that are expressed at the onset of MZT. For all subsequent experiments we injected Cas9 protein + sgRNA which we abbreviate as CRISPRp.
Figure 2.
Time course of CRISPR/Cas9 deletions. All embryos injected with tyr sgRNA and either injected with cas9 mRNA or Cas9 protein. X-axis = time/ stage of embryos post-injection (number of embryos in parenthesis). Y-axis = % embryos with corresponding FA genotype (blue = WT allele only, green = both alleles detected, red = mutant allele only). Statistical analysis performed by Fisher exact probability test, * = p<0.05.
CRISPRp injected embryos phenocopy established mutant or MO knockdown phenotypes in the F0 generation
We next wanted to test if F0 CRISPR mediated gene deletion could recapitulate known mutant or MO knockdown phenotypes. We sought to examine disease-causing genes that create a diverse array of disease phenotypes including, ciliopathies that can cause CHD and heterotaxy (foxj1, ccdc40, dnah9, galnt11) and an organ system that has not yet been targeted in Xenopus, kidneys (pax8). In each case, these genes affect development well before stage 20 providing a test of how early CRISPRp can affect development as well as a diversity of phenotypes.
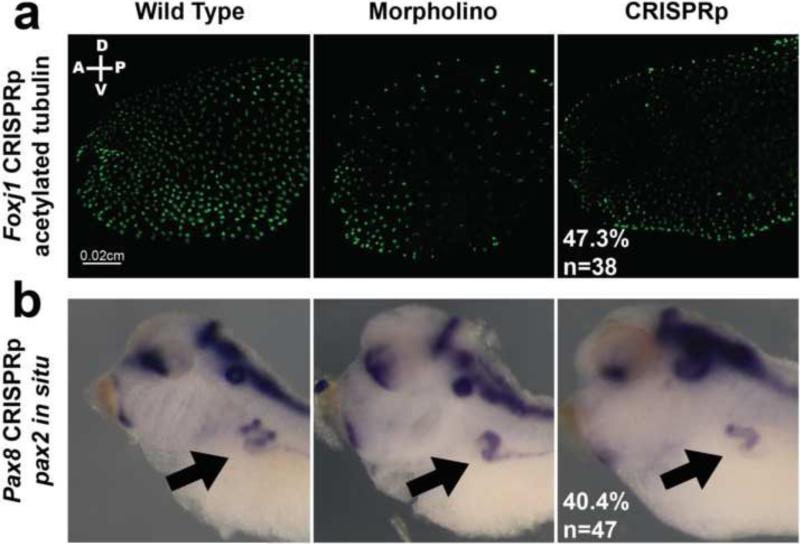
First we looked at a range of ciliopathy genes. We chose this set of genes for the diversity of ciliopathies; in some cases, loss of function leads to less ciliated cells (ccdc40, foxj1), more ciliated cells (galnt11), or normal cilia morphology but reduced motility (dnah9). Foxj1 MO knockdown leads to decreased epidermal cilia (Stubbs et al., 2008) and 68% of our foxj1 CRISPRp injected embryos (n=38) had grossly reduced epidermal ciliated cells (Fig. 3, Table 1). The endogenous FoxJ1 protein levels, as assayed by western blot were decreased in both MO and CRISPR injected embryos (Supp. Fig. 3(f)). Similarly, ccdc40 mutations cause primary ciliary dyskinesia that leads to decreased and abnormal cilia in patients and in Xenopus (Becker-Heck et al., 2011; del Viso et al., 2012). Injecting ccdc40 CRISPRp readily recapitulated this decreased and abnormal cilia phenotype in 48% of injected embryos (n=40), and they had a corresponding 16bp deletion peak (Supp. Fig. 3(c) and (g), Table 1).
Figure 3.
CRISPR injected embryos phenocopy established mutant or MO knockdown phenotypes in the F0 generation. Left panel = WT embryo, Middle panel = MO injected embryo, Right panel = CRISPRp injected embryo. (a) Foxj1 CRISPRp – Immunohistochemistry of stage 28 embryos with acetylated tubulin to mark surface cilia. (b) Pax8 CRISPRp – in situ hybridization of stage 38 embryos with pax2 probe to stain kidney tubule. Black arrow points to pronephros.
Table 1.
Summary of genes and CRISPR phenotypes
| Gene/Mutation | OMIM Disease Phenotype | Morpholino/ Mutant line Phenotype | CRISPR Phenotype |
|---|---|---|---|
| FoxJ1 | Allergic rhinitis, Autoimmune disease | 90% Decreased epidermal cilia(Stubbs et al., 2008) | 68% Decreased epidermal cilia |
| Ccdc40 | Primary ciliary dyskinesia | Decreased and abnormal epidermal cilia(Becker-Heck et al., 2011; del Viso et al., 2012) | 48% Decreased and abnormal epidermal cilia |
| Dnah9 | Primary ciliary dyskinesia | 30% laterality defect(Vick et al., 2009) | 28% A+L Heart Loops |
| Galnt11 | Heterotaxy | 25% A+L Heart Loops | 23% A+L Heart Loops |
| Increased Epidermal Cilia(Boskovski et al., 2013) | 50% Increased Epidermal Cilia | ||
| Pax8 | Kidney agenesis, Hypothyroidism | 59% Malformed pronephros(del Viso et al., 2012) | 40% Malformed pronephros |
| Tyrosinase | Oculocutaneous albinism | Albinism(Blitz et al., 2013; Guo et al., 2014; Nakayama et al., 2013) | ~ 100% Albinism |
| Beta-Catenin | Cancer, Intellectual disability | Ventralization of embryo(Heasman et al., 2000; Khokha et al., 2002) | No ventralization |
Next, we looked at two genes, dnah9 and galnt11, that both affect left-right patterning of the developing embryo and cardiac development (Boskovski et al., 2013; Vick et al., 2009). Dnah9 is an important component of the ciliary axoneme that affects cilia motility, but not ciliogenesis, and causes laterality defects when knocked down (Vick et al., 2009). 28% of dnah9 CRISPRp injected embryos (n=32) had heart-looping defects (A or L loops) and all of these embryos contained a characteristic 11bp deletion peak (Table 1, Supp. Fig. 3(b)). Furthermore, as expected, the injected embryos had normal epidermal cilia staining (Supp. Fig. 3(g)). Galnt11, on the other hand, is a Notch regulator identified from a patient with heterotaxy, and its loss leads to a left-right patterning defect as well as an increase in multiciliated epidermal cell density (Boskovski et al., 2013; Fakhro et al., 2011). 23% of our galnt11 CRISPRp injected embryos (n=35) had A and L heart looping defects, which is comparable to the 25% heart looping defects observed with MO knockdown (Boskovski et al., 2013). 50% of these injected embryos also successfully recapitulated the increase in epidermal ciliated cell density and all injected embryos contained a characteristic 10bp deletion peak when analyzed by FA (Table 1, Supp. Fig. 3(a) and (g)).
Next, to examine kidney development, we tested pax8, a transcription factor critical for the specification of the pronephros and proper kidney development. MO knockdown as well as the previously published pax8 mutant, Ruby (del Viso et al., 2012), leads to abnormal pronephros development as assayed by pax2 in situ hybridization. 40% of pax8 CRISPRp injected embryos (n=47) showed abnormal pronephros development, and we detected a 17bp deletion peak in these injected embryos by FA (Fig. 3, Supp. Fig. 3(e), Table 1).
Finally, we designed a CRISPR to beta-catenin, a maternally expressed gene, which, when knocked down using a translation blocking MO, leads to a severe ventralization of the embryos (Heasman et al., 2000; Khokha et al., 2002; Khokha et al., 2005). However, in CRISPRp injected embryos we do not observe gross ventralization of embryos, even though these embryos show an array of deletions on FA (Supp. Fig. 3(d)).
Discussion
Here, we show that the CRISPR/ Cas9 system can identify F0 phenotypes for human disease genes and has a number of advantages. First, FA is an efficient strategy to detect deletions and insertions in most genes. Second, Cas9 protein is less toxic and more efficient in creating gene modifications than cas9 mRNA. Additionally, with Cas9 protein, there is a high degree of biallelic gene modification by the MZT, allowing for the study of early zygotic gene function in F0 embryos. Third, CRISPRp injected embryos recapitulate a diverse array of mutant or MO knockdown phenotypes in the F0 generation, and these embryos can be analyzed using standard molecular techniques like in situ hybridization, immunohistochemistry, and western blotting, similar to MO injected animals.
Furthermore, unlike MOs, it is simple to assay the efficacy of CRISPR mediated gene disruption by PCR. In this paper, we employ FA to distinguish wildtype alleles from deletions or insertions at a single base-pair resolution. This method is also inexpensive – at Yale, it costs $0.50 to run one lane of FA and with four flurophores commercially available, one can run up to four samples per lane reducing the cost to $0.13 per sample. FA can also be done in a 96-well plate format allowing for quick analysis of multiple embryos and targets. However, our PCR conditions are not amenable to quantitation so peaks seen on FA only correlate with the percentage of clones or cells that contain that deletion or insertion. In using FA, we were surprised to detect characteristic single deletion peaks in most embryos injected with a particular sgRNA. Cloning these PCR products indicates that while many deletions are of the same size and occur in a similar region of the target sequence, only some of them are identical. This suggests that there are independent double strand break and repairs in multiple cells during early cell division, after which those cells give rise to a clonal population of cells all with the same deletion. From our time-course, we conclude that deletions are detectable as early as 2 hours post injection and they are detected in a greater percentage of embryos when injected with Cas9 protein compared to mRNA. This builds confidence that studying early developmental events might be tractable using this technology, especially if Cas9 protein is used.
Finally, we show that the CRISPR/ Cas9 system can be used to phenocopy known mutant and MO knockdown phenotypes in a variety of genes affecting heart development, ciliogenesis or kidney development. These phenotypes are characteristic since foxj1 and ccdc40 CRISPRp decrease the number of epidermal cilia, dnah9 CRISPRp affects heart looping, but does not affect the number of epidermal cilia, while galnt11 CRISPRp causes heart looping defects and increases the number of epidermal ciliated cells. Although mosaic, we can successfully generate biallelic deletions in the F0 generation, and these embryos can be studied using standard molecular techniques like in situ hybridization, immunohistochemistry, and western blotting.
We do note that penetrance of the phenotype does not always correlate with the genotype detected by FA. On one hand, this is reflective of the penetrance of the phenotypes that we studied. For example, mouse null mutants of many heterotaxy genes show abnormal cardiac looping in only 50% of embryos (Li et al., 2015; Supp et al., 1997). Also our overall goal was to test if CRISPRp would work for a large scale screen of many birth defect genes; therefore, in some cases we detected patches of cells with an abnormal phenotype but unless a dramatic phenotype was present, we scored these as wildtype. These more subtle phenotypes can be difficult to detect when screening many genes especially when the phenotype is unknown. Importantly, even with this “high bar,” phenotypes were detected in substantial percentages over controls reassuring us that CRISPRp can be highly effective for large scale F0 loss of function screens.
While F0 CRISPR based gene editing has many advantages, challenges remain, and there are limitations to using CRISPR technology. First, it is essential to determine if a particular sgRNA favors indels that cause a frameshift. Second, different sgRNAs can have different DNA editing efficiency and further studies need to establish factors that determine sgRNA activity and efficiency. Third, the ability to design and use CRISPRs effectively depends upon the quality of the genome models for a particular organism. For example, the Xenopus genome models are not polished to the degree of human or rodent genomes and some genes are still not properly annotated. We initially designed an sgRNA to the annotated exon 1 of foxJ1 that was predicted to include the start of the protein. However, this sgRNA did not show any phenotype. Upon closer inspection of the conserved Foxj1 protein sequence, we realized that the actual protein start site is in the predicted exon 2. When we re-designed our sgRNA to this new start site, we successfully phenocopied the MO phenotype of decreased epidermal cilia. Incorrect genomic sequence can also limit the use of FA to assay for indels since it becomes difficult to design specific primers to the region of interest. Lastly, as demonstrated by our beta-catenin results, we were not able to recapitulate the early dorsal-ventral patterning phenotype that is well-established with a translation blocking beta-catenin MO (Heasman et al., 2000; Khokha et al., 2005) despite using two non-overlapping CRISPRp (data only shown for one). However, we still detect deletions using FA, suggesting that although gene editing is taking place, there is no phenotype likely due to presence of maternal transcript. This serves to highlight another limitation of CRISPRp where use of a MO might in some cases be more useful to target genes.
Furthermore, to test specificity, either rescue of the CRISPR phenotype or phenocopy with a second non-overlapping sgRNA is necessary. FA can be useful to test efficacy, but western blotting remains the gold standard to assay the degree of knockdown, if effective antibodies are available. These concerns over specificity are no different than MOs, although the specificity of MOs have come under scrutiny recently (Kok et al., 2015). Lastly, two studies also suggest that injecting nuclease proteins directly, instead of gene- or mRNA-mediated expression, potentially alleviates some off-target effects, further arguing for use of Cas9 protein over mRNA (Gaj et al., 2012; Izmiryan et al., 2011). Our results support the use of MOs in Xenopus, and CRISPR phenocopy offers an alternative avenue to test MO specificity, again especially with the use of Cas9 protein.
In conclusion, we recommend using Cas9 protein to maximize CRISPR efficiency, using FA as a rapid and inexpensive tool to assay for indels, and using CRISPRp to screen and validate the hundreds of candidate genes being discovered from congenital malformation patients in a rapid and efficient disease model such as Xenopus.
Materials and Methods
Frog Husbandry
X. tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by our local IACUC.
CRISPR sgRNA design
CRISPR sgRNA were designed from the v7.1 or v8 gene models on Xenbase and zifit website (http://zifit.partners.org/ZiFiT/). For the PCR method, target sites were either identified using “T7 promoter” option, which searches for 5’-GG-N(18) target sequence-3’, or “None” which excludes the 5’-GG. We generate the site-specific sgRNA by ordering a single oligo that costs ~$10 (compare to $400 for a single MO), with the sequence 5’-CTAGCTAATACGACTCACTATAGG-n(18) target sequence-GTTTAGAGCTAGAAATAGCAAG-3’. Colors correspond to Figure 1 (Red = 5bp anchor, Orange = T7 Promoter, Green = site-specific sequence). PCR was done using Phusion polymerase (NEB) using stock CRISPR-R primer (Supp. Table 1) to generate the complete 100nt sgRNA DNA template. PCR conditions were 94°C for 2 min, followed by 20 cycles at 94°C for 10 s, 45°C for 15 s and 72°C for 15 s, and final extension for 5 min at 72°C. sgRNA was transcribed using T7 MEGAShortscript kit (Ambion) by using 8μl of un-purified PCR product in a 20ul reaction at 37°C for 4 hours. In this study we modified the PCR strategy previously described (Nakayama et al., 2013) to increase RNA yield: 1) addition of a 5bp anchor sequence 5’ to the T7 promoter, and 2) use of Ambion's T7 MEGAshortScript kit for RNA transcription (instead of MEGAscript kit). For example, tyrosinase sgRNA yield increased from 34 ng to 2.5 μg with the addition of the 5bp extension, and to 45 μg when the MEGAshortScript was used concurrently. Generally, we get 10-50 μg of RNA per reaction. For cloning method (ccdc40, galnt11, dnah9, tyrosinase) we used the “T7 promoter” option in the zifit program to include BsaI overhangs. Oligos 1 and 2 were combined to a concentration of 25mM and further diluted 1/1000×. They were annealed by boiling at 94°C for 2 min and then decreasing the temperature by 0.1°C/s to 25°C. This was then treated with T4 PNK (NEB). The receiving vector pDR274 (Addgene ID 42250) was linearized with BsaI and treated with CIP (NEB). Oligos were ligated and cloned. sgRNA was transcribed by linearizing the plasmid with DraI and using T7 MEGAscript kit (Ambion). sgRNA target sequences can be found in Supp. Table 1. We inject sgRNA at a concentration of 400pg/ embryo except for ccdc40, which was injected at 200pg/ embryo due to toxicity.
CRISPR Cas9 mRNA and Protein
We obtained pCasX from the Cho lab (UC Irvine). Plasmid was linearized with Asp718I and transcribed by using T7 mMessage mMachine kit (Ambion) for 2 hours at 37°C. Cas9 Protein was ordered from PNA-Bio (Item #CP01) and eluted to stock concentration of 1ng/ nl. We inject cas9 mRNA at a concentration of 2ng/ embryo, Cas9 protein at a concentration of 1.5ng/ embryo.
Fragment Analysis
Tadpoles were collected at appropriate stages and lysed with 100ul of 50mM NaOH at 95°C for 10 mins and neutralized with 20ul 1M Tris-HCl. Primers for fragment analysis were designed with a 5’-m13 tail (5’- TCCCAGTCACGACGT-) on the Forward primer. PCR was done in 2 steps. Step 1: Using 5’-m13-F primer and R-primer; PCR conditions were 94°C for 2 min, followed by 34 cycles at 94°C for 10 s, 58°C for 30 s and 72°C for 30 s, and final extension for 5 min at 72°C. Step 2: Using NED, PET, VIC, or 6-FAM-m13-F primer and original R- primer and 1/50× concentration of PCR product from Step 1; PCR conditions were 94°C for 2 min, followed by 25 cycles at 94°C for 10 s, 52°C for 30 s and 72°C for 30 s. Final extension was for 5 min at 72°C. PCR products were analyzed using fragment analysis at Yale's DNA sequencing facility. We use the GeneMarker (SoftGenetics) software to analyze the results. All primer sequences can be found in Supp. Table 1.
In vitro fertilization and microinjections
In vitro fertilization and microinjection were done as previously described and protocols available on our website (http://khokha.medicine.yale.edu). Embryos were injected at one cell stage with 0.5 or 1 ng of MO solution for Pax8, 10 ng of MO solution for Foxj1. For CRISPR, embryos were injected at one cell stage with 2ng of cas9 mRNA or 1.5ng Cas9 Protein (PNA-Bio) along with 400pg sgRNA for each gene. After injections, embryos were left in 1/9X MR + 3% Ficoll for 1 hr and then transferred to 1/9X MR supplemented with 50 μg/ml of gentamycin. Embryos were raised at 22-27°C until the appropriate stage for fixation or evaluating phenotype. Pax8 translation blocking MO was obtained from Genetools, LLC. The sequence of the MO was: 5’-ATGCTGCTGTTGGGCATCTTCCTCC-3’. FoxJ1 translation blocking MO was obtained from Genetools, LLC. The sequence of the MO was: 5’- GCAGGTCAAACATTAATAAAGCCCT-3’. MO and CRISPR were co-injected with Alexa 488 or Tetramethylrhodamine Dextran dye (Life Technologies) as a lineage tracer.
Time Course
Embryos were collected at 2-hour interval starting from the time of injection. For early stages, embryos were lysed using 5ul of 50mM NaOH at 95°C for 5 mins and neutralized with 1ul 1M Tris-HCl. Fragment analysis was performed as per protocol above. Statistics comparing cas9 mRNA vs Protein injected embryos at each time point was done using Fisher Exact probability test using a 2×3 matrix (www.vassarstats.net/fisher2x3.html).
Whole Mount in situ hybridization (WMISH)
The DNA templates were in vitro transcribed with T7 High Yield RNA Synthesis Kit (E2040S) from New England Biolabs to synthesize digoxigenin-labeled antisense probes. Whole mount in situ hybridization was performed according to the standard protocol with minor modifications (Khokha et al., 2002). We stained embryos using BM purple at room temperature or 4°C depending on each probe. If embryos needed to be genotyped after WMISH, fixation after staining was done with 4% paraformaldehyde instead of Bouin's fixative to preserve DNA. After bleaching and equilibration in 100% glycerol, embryos were examined and photographed.
Immunohistochemistry
For immunofluorescence, embryos were fixed in 4% paraformaldehyde/PBS overnight at 4°C. Embryos were labeled by immunofluorescence as described previously (Boskovski et al., 2013). All embryos were mounted in Pro-Long Gold (Invitrogen) before imaging. Imaging was performed on a Zeiss Axiovert microscope equipped with Apotome optical interference imaging to obtain optical sections. We employed the following antibodies: anti-acetylated tubulin (Sigma, no. T-6793) (Piperno and Fuller, 1985) or mouse monoclonal anti-acetylated tubulin, clone 6-11B-1 (1:1,000) (Boskovski et al., 2013). For secondary antibody we use goat anti-mouse IgG with Alexa Flour 488 conjugate (Life technologies, Item # A-11001, 1:500 dilution).
Western Blotting
Pools of 6-10 control or MO/ CRISPR injected embryos were collected and placed in 10ul/ embryo of 1× RIPA buffer. Embryos were then crushed using a pestle and spun down at 14,000 RPM for 20 minutes. Western Blots were carried out following standard protocols using an anti-foxj1 (AbCam, 108452, 1:250 dilution), or anti-GAPDH (Ambion, AM4300, 1:5000 dilution) primary antibody and an anti-mouse or anti-rabbit HRP conjugated secondary antibody (Jackson Immuno Research Laboratories, 715-035-150 or 211-032-171, 1:15000 dilution). Quantification of changes in protein level was calculated using ImageJ software from NIH.
PCR and cloning strategy
PCR was done with Phusion High-Fidelity DNA Polymerase (NEB) and conditions were as follows: 98°C for 2 min, followed by 39 cycles at 98°C for 15 s, 58°C for 30 s and 72°C for 30 s. Final extension was at 72°C for 10 min. Samples were then incubated with Taq polymerase for 10 min at 72°C to incorporate A overhangs for TOPO TA cloning (Invitrogen) and sequenced with m13R or T7P.
Supplementary Material
Research Highlights.
-
1)
F0 biallelic mutations occur rapidly enough to study early zygotic phenotypes in Xenopus
-
2)
Fragment analysis can quickly and inexpensively detect deletions or insertions
-
3)
Cas9 protein is less toxic than injecting mRNA
-
4)
Cas9 protein is far more efficacious than mRNA in gene editing
-
5)
F0 CRISPR successfully recapitulates disease phenotypes that match MO results in Xenopus
Acknowledgements
We thank Sarah Kubek and Michael Slocum for frog husbandry. DB was supported by Yale MSTP department NIH MSTP TG T32GM07205 grant. This work was supported by NIH R01HD081379 and 5R21HL120783 to MKK. MKK is a Mallinckrodt Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DB and MKK conceived and designed the experiments and wrote the manuscript. DB, CAM, DL, ML carried out the wet laboratory experiments. All the authors read and approved the final manuscript.
References
- Bassett A, Liu JL. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods. 2014;69:128–136. doi: 10.1016/j.ymeth.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, Fliegauf M, Horvath J, Reinhardt R, Nielsen KG, Marthin JK, Baktai G, Anderson KV, Geisler R, Niswander L, Omran H, Burdine RD. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nature genetics. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KW. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner M. Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation. 2007;115:2793–2795. doi: 10.1161/CIRCULATIONAHA.107.699256. [DOI] [PubMed] [Google Scholar]
- del Viso F, Bhattacharya D, Kong Y, Gilchrist MJ, Khokha MK. Exon capture and bulk segregant analysis: rapid discovery of causative mutations using high-throughput sequencing. BMC genomics. 2012;13:649. doi: 10.1186/1471-2164-13-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolk H, Loane M, Garne E, European Surveillance of Congenital Anomalies Working, G. Congenital heart defects in Europe: prevalence and perinatal mortality, 2000 to 2005. Circulation. 2011;123:841–849. doi: 10.1161/CIRCULATIONAHA.110.958405. [DOI] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nature methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J, DePalma SR, Golhar R, Sanders SJ, Yamrom B, Ronemus M, Iossifov I, Willsey AJ, State MW, Kaltman JR, White PS, Shen Y, Warburton D, Brueckner M, Seidman C, Goldmuntz E, Gelb BD, Lifton R, Seidman J, Hakonarson H, Chung WK. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circulation research. 2014;115:884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nature genetics. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Developmental biology. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D'Amours G, Lemyre E, Cullum R, Bigras JL, Thibeault M, Chetaille P, Montpetit A, Khairy P, Overduin B, Klaassen S, Hoodless P, Awadalla P, Hussin J, Idaghdour Y, Nemer M, Stewart AF, Boerkoel C, Scherer SW, Richter A, Dube MP, Andelfinger G. Rare copy number variants contribute to congenital left-sided heart disease. PLoS genetics. 2012;8:e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmiryan A, Basmaciogullari S, Henry A, Paques F, Danos O. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic acids research. 2011;39:7610–7619. doi: 10.1093/nar/gkr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC. Techniques and probes for the study of Xenopus tropicalis development. Developmental dynamics : an official publication of the American Association of Anatomists. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Developmental cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani H, Taimatsu K, Ohga R, Ota S, Kawahara A. Efficient Multiple Genome Modifications Induced by the crRNAs, tracrRNA and Cas9 Protein Complex in Zebrafish. PLoS One. 2015;10:e0128319. doi: 10.1371/journal.pone.0128319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nature biotechnology. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KW, Grainger RM. Cas9-based genome editing in Xenopus tropicalis. Methods Enzymol. 2014;546:355–375. doi: 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pediatric Cardiac Genomics, C. Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Kaski JP, Kim R, Kline J, Mercer-Rosa L, Porter G, Roberts A, Rosenberg E, Seiden H, Seidman C, Sleeper L, Tennstedt S, Kaltman J, Schramm C, Burns K, Pearson G, Rosenberg E. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circulation research. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest JR, Girirajan S, Vu TH, Olson A, Eichler EE, Portman MA. Rare copy number variants in isolated sporadic and syndromic atrioventricular septal defects. American journal of medical genetics. Part A. 2012;158A:1279–1284. doi: 10.1002/ajmg.a.35315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature biotechnology. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nature genetics. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, Lee HW, Kim JS. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. Journal of the American College of Cardiology. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Vick P, Schweickert A, Weber T, Eberhardt M, Mencl S, Shcherbakov D, Beyer T, Blum M. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Developmental biology. 2009;331:281–291. doi: 10.1016/j.ydbio.2009.05.547. [DOI] [PubMed] [Google Scholar]
- Wang F, Shi Z, Cui Y, Guo X, Shi YB, Chen Y. Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci. 2015;5:15. doi: 10.1186/s13578-015-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Ronemus M, Kline J, Jobanputra V, Williams I, Anyane-Yeboa K, Chung W, Yu L, Wong N, Awad D, Yu CY, Leotta A, Kendall J, Yamrom B, Lee YH, Wigler M, Levy D. The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease. Human genetics. 2014;133:11–27. doi: 10.1007/s00439-013-1353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Maternal beta-catenin establishes a ‘dorsal signal’ in early Xenopus embryos. Development. 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- Yang Z, Steentoft C, Hauge C, Hansen L, Thomsen AL, Niola F, Vester-Christensen MB, Frodin M, Clausen H, Wandall HH, Bennett EP. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic acids research. 2015 doi: 10.1093/nar/gkv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe'er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Niu G, Shen B, Zheng Y, Gong F, Wang X, Lee J, Mulvihill JJ, Chen X, Li S. High-resolution analysis of copy number variants in adults with simple-to-moderate congenital heart disease. American journal of medical genetics. Part A. 2013;161A:3087–3094. doi: 10.1002/ajmg.a.36177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.