Abstract
INTRODUCTION
Many missense variants in G protein-coupled receptors (GPCRs) involved in the neuroendocrine regulation of reproduction have been identified by phenotype driven or large scale exome sequencing. Computational functional prediction analysis is commonly performed to evaluate their impact on receptor function.
METHODS
To assess the performance and outcome of functional prediction analyses for these GPCRs, we performed statistical analysis of the prediction performance of SIFT and PolyPhen-2 for variants with documented biological function as well as variants retrieved from Ensembl. We obtained missense variants with documented biological function testing from patients with reproductive disorders from a comprehensive literature search. Missense variants from individuals with known reproductive disorders were retrieved from the Human Gene Mutation Database. Missense variants from the general population were retrieved from the Ensembl genome database.
RESULTS
The accuracies of SIFT and PolyPhen-2 were 83% and 85%, respectively. The performance of both prediction tools was superior in predicting loss-of-function variants (SIFT: 92%; Polyphen-2: 95%) than in predicting variants that did not affect function (SIFT: 54%; Polyphen-2: 57%). Concordance between SIFT and PolyPhen-2 did not improve accuracy. Surprisingly, approximately half of the variants retrieved from Ensembl were predicted as loss-of-function by SIFT (47%) and Polyphen-2 (54%).
CONCLUSION
Our findings provide new guidance for interpreting the results and limitations of computational functional prediction analyses for GPCRs and will help to determine which variants require biological function testing. In addition, our findings raise important questions regarding the link between genotype and phenotype in the general population.
Keywords: G protein-coupled receptors, Neuroendocrinology, Exome sequencing, Computational analysis, Functional prediction, Reproductive disorders, Missense variants
Introduction
Phenotype-driven exome sequencing [1] and large scale next generation DNA sequencing of the general population [2] have revealed numerous rare nonsynonymous missense variants in protein-coding DNA sequences. The identification of causal missense variants that alter human phenotypes, in particular to induce disease states, is one of the fundamental goals of human genetics, with the objective of providing crucial insights into the biology connecting genotype and phenotype and potentially facilitating the prediction of disease onset. Performing biological testing to determine the effect of a missense variant on the function of the encoded protein usually produces reliable results, but is laborious and time-consuming. In this context, the search for alternative and reliable methods for assigning effects of novel variants on protein function is of primary importance. There are various computational in silico prediction tools available for predicting the function of variants, using information derived from sequence similarity [3] and phylogenetic profiles [4]. However, the performance of available functional prediction tools varies among proteins in different functional categories [5–8]. Delineating the effects of missense variants identified in G protein-coupled receptors (GPCRs), a large family of receptors involved in signal transduction and cellular response to outside signals and the most common drug targets (the targets account for 27% of all FDA approved drugs) [9] faces this challenge. Sequence alignment of 94 GPCRs revealed many highly conserved amino acids in GPCRs [10], suggestive of potentially damaging effects if mutations occur in these highly conserved amino acids. Bioinformatics approaches have been used to predict intrinsically disordered regions of GPCRs - regions lacking a stable three-dimensional structure and playing a role in intra- and extracellular plasticity and protein-protein interactions of GPCRs - and regions predicting G protein coupling specificity [11, 12]. In the GPCR family, gonadotropin-releasing hormone receptor (GnRHR), kisspeptin receptor (KISS1R), prokineticin receptor 2 (PROKR2), and tachykinin receptor 3 (TACR3) have been found to play key roles in the central neuroendocrine regulation of reproductive function [13]. In these GPCRs, more than 300 missense variants have been found, either from patients with phenotypic reproductive disorders or from large-scale genomic sequencing of general populations. Some of the variants identified in patients with reproductive disorders have undergone in vitro biological function testing [13, 14], which has aided in the interpretation of the pathogenic relationship between genotype and phenotype. Computational prediction results from Sorting Intolerant From Tolerant (SIFT) and Polymorphism Phenotyping-2 (PolyPhen-2) are available for most of the variants, but the correlation between in silico prediction and in vitro biological function for these GPCR missense variants has not been well established.
Materials and Methods
Data and materials
The missense variants of GnRHR, KISS1R, PROKR2, and TACR3 identified in patients with known phenotypes were acquired from The Human Gene Mutation Database (HGMD) (http://www.hgmd.org). The overall missense variants in representative populations were obtained from the Ensembl genome database (http://useast.ensembl.org/index.html). The data from HGMD and Ensembl were retrieved in July 2014. The missense variants with documented biological function tests were retrieved through a comprehensive literature search in PubMed of articles published between 1997 and July 2014.
Computational Analysis
Computational functional prediction results for each variant with documented biological function testing were obtained using SIFT [8] and PolyPhen-2 [15] analyses. We defined the function predicted by SIFT and PolyPhen-2 following the programs’ parameters. For SIFT, the predicted function of a variant is characterized as tolerated or deleterious based on the SIFT score (0–1). A score ≤ 0.05 is defined as deleterious and a score > 0.05 is defined as tolerated. For PolyPhen-2, the predicted function of a variant is characterized as benign (score 0–0.5), possibly damaging (score > 0.5–0.9), or probably damaging (score > 0.9) based on a score scale of 0–1. For in vitro biological function tests, we characterized the function as normal (maximum response of a variant at least 80% of the maximum response of the corresponding wild type receptor), partial loss-of-function (maximum response of a variant between 20–80% of the maximum response of the wild type receptor), or complete loss-of-function (maximum response less than 20% of the maximum response of the wild type receptor).
Statistical Analysis
In order to perform statistical analyses, we simplified the characterization to be either benign or damaging for both computational prediction tests and biological function test results. As such, benign was defined as tolerated in SIFT, benign in PolyPhen-2, and normal (> 80% of wild type) in biological function tests. Damaging was defined as deleterious in SIFT, either possibly damaging or probably damaging in PolyPhen-2, and either partial or complete loss-of-function (< 80% of wild type) in biological function tests. Concordance for the in silico analyses was defined as both computational prediction tools having the same functional prediction outcome. Statistical analyses, including true positive (TP), false positive (FP), true negative (TN), false negative (FN), sensitivity/, specificity, positive prediction value (PPV), negative prediction value (NPV), accuracy (AC), and Matthews correlation coefficient (MCC), were calculated using the following formulas:
Sensitivity:
Specificity:
Positive predictive value:
Negative predictive value:
Accuracy:
Matthews correlation coefficient:
Results
Summary of variants in GnRHR, KISS1R, PROKR2, and TACR3 with documented biological function test results and their correlation with the computational functional prediction tools
A total of 52 missense variants with documented biological function testing were identified in GnRHR, KISS1R, PROKR2, and TACR3 through a literature search in PubMed. All of the variants with biological function tests were identified in patients with reproductive disorders. Table 1 summarizes the nucleotide (position and exon) and amino acid (position and receptor domain) changes, computational function prediction results by SIFT and PolyPhen-2, and biological function test results. Nineteen of 52 variants were identified in GnRHR, 7 in KISS1R, 17 in PROKR2, and 9 in TACR3. Ten of the variants had normal biological function testing, 21 had partial loss of function and 21 had complete loss of function (Table 1). All 10 variants reported with normal biological function were either PROKR2 or TACR3 variants, except one in KISS1R.
Table 1.
Summary of missense variants with documented biological function tests retrieved from a literature search.
| Variant | cDNA change | SIFT | PolyPhen-2 | Exon | Domain | Function | REF |
|---|---|---|---|---|---|---|---|
| GnRHR | |||||||
| Asn10Lys | c.30T>A | tolerated | benign | 1 | N-term | partial loss | [30] |
| Thr32Ile | c.95C>T | deleterious | probably damaging | 1 | N-term / TM1 | complete loss | [31, 32] |
| Glu90Lys | c.268G>A | deleterious | probably damaging | 1 | TM2 | complete loss | [33, 34] |
| Thr104Ile | c.311C>T | deleterious | probably damaging | 1 | ECL1 | partial loss | [35, 36] |
| Gln106Arg | c.317A>G | deleterious | probably damaging | 1 | ECL1 | partial loss | [37] |
| Tyr108Cys | c.323A>G | deleterious | probably damaging | 1 | ECL1 | complete loss | [35, 36] |
| Ala129Asp | c.386C>A | deleterious | probably damaging | 1 | TM3 | complete loss | [38] |
| Arg139His | c.416G>A | deleterious | probably damaging | 1 | TM3 | complete loss | [30] |
| Ser168Arg | c.504T>A | tolerated | benign | 1 | TM4 | complete loss | [39] |
| Ala171Thr | c.511G>A | deleterious | possibly damaging | 1 | TM4 | complete loss | [40] |
| Cys200Tyr | c.599G>A | deleterious | possibly damaging | 2 | ECL2 | complete loss | [31, 32] |
| Ser217Arg | c.651C>A | tolerated | benign | 2 | TM5 | complete loss | [41] |
| Arg262Gln | c.785G>A | deleterious | probably damaging | 3 | ICL3 | partial loss | [37] |
| Leu266Arg | c.797T>G | deleterious | probably damaging | 3 | ICL3 | complete loss | [31, 32] |
| Cys279Tyr | c.836G>A | deleterious | probably damaging | 3 | TM6 | complete loss | [31, 32] |
| Pro282Arg | c.845C>G | deleterious | probably damaging | 3 | TM6 | complete loss | [34] |
| Tyr284Cys | c.851A>G | deleterious | probably damaging | 3 | TM6 | partial loss | [42] |
| Pro320Leu | c.959 C>T | deleterious | probably damaging | 3 | TM7 | complete loss | [43] |
| Tyr323Cys | c.968A>G | deleterious | probably damaging | 3 | TM7 | complete loss | [34] |
| KISS1R | |||||||
| Leu102Pro | c.305T>C | deleterious | possibly damaging | 2 | ECL1 | partial loss | [44] |
| Leu148Ser | c.443T>C | deleterious | probably damaging | 3 | ICL2 | complete loss | [16] |
| Cys223Arg | c.667T>C | tolerated | benign | 4 | TM5 | partial loss | [45] |
| Glu252Gln | c.754G>C | tolerated | benign | 5 | ICL3 | normal | [46] |
| Phe272Ser | c.815T>C | deleterious | probably damaging | 5 | TM6 | partial loss | [47] |
| Arg297Leu | c.890G>T | tolerated | benign | 5 | ECL3 | partial loss | [45] |
| Tyr313His | c.937T>C | deleterious | probably damaging | 5 | TM7 | complete loss | [48] |
| PROKR2 | |||||||
| Arg80Cys | c.238C>T | deleterious | probably damaging | 1 | ICL1 | partial loss | [49, 50] |
| Arg85His | c.254G>A | deleterious | probably damaging | 1 | ICL1 | normal | [50, 51] |
| Arg85Cys | c.253C>T | deleterious | probably damaging | 1 | ICL1 | normal | [50, 52] |
| Tyr113His | c.337T>C | deleterious | probably damaging | 1 | ECL1 | partial loss | [52] |
| Val115Met | c.343G>A | deleterious | probably damaging | 1 | ECL1 | partial loss | [52] |
| Arg164Gln | c.491G>A | deleterious | probably damaging | 2 | ICL2 | partial loss | [51, 52] |
| Leu173Arg | c.518T>G | deleterious | probably damaging | 2 | TM4 / ICL2 | partial loss | [51, 52] |
| Trp178Ser | c.533G>C | deleterious | probably damaging | 2 | TM4 | complete loss | [51, 53] |
| Ser188Leu | c.563C>T | deleterious | possibly damaging | 2 | TM4 | complete loss | [52] |
| Gln210Arg | c.629A>G | deleterious | probably damaging | 2 | ECL2 | partial loss | [51, 53] |
| Arg248Gln | c.743G>A | tolerated | benign | 2 | ICL3 | normal | [52] |
| Arg268Cys | c.802C>T | deleterious | probably damaging | 2 | ICL3 | partial loss | [51, 53] |
| Pro290Ser | c.868C>T | deleterious | probably damaging | 2 | TM6 | complete loss | [51, 53] |
| Met323Ile | c.969G>A | tolerated | benign | 2 | TM7 | normal | [51, 53] |
| Val331Met | c.991G>A | tolerated | benign | 2 | C-term | normal | [51, 52] |
| Arg357Trp | c.1069C>T | deleterious | benign | 2 | C-term | normal | [52] |
| Gly371Arg | c.1111G>A | tolerated | benign | 2 | C-term | normal | [54] |
| TACR3 | |||||||
| Gly18Asp | c.53G>A | tolerated | benign | 1 | N-term | normal | [14, 55] |
| Gly93Asp | c.278G>A | tolerated | benign | 1 | TM1 | partial loss | [56] |
| His148Leu | c.443A>T | deleterious | probably damaging | 1 | ECL1 | partial loss | [57] |
| Ile249Val | c.745A>G | tolerated | benign | 3 | TM5 | normal | [14, 55] |
| Tyr256His | c.766T>C | deleterious | probably damaging | 3 | TM5 | complete loss | [14, 55] |
| Tyr267Asn | c.799T>A | deleterious | probably damaging | 3 | TM5 | complete loss | [58] |
| Arg295Ser | c.885A>C | deleterious | probably damaging | 3 | ICL3 | partial loss | [14, 55] |
| Tyr315Cys | c.944A>G | deleterious | probably damaging | 4 | TM6 | partial loss | [14, 55] |
| Pro353Ser | c.1057C>T | deleterious | probably damaging | 4 | TM6 | partial loss | [56] |
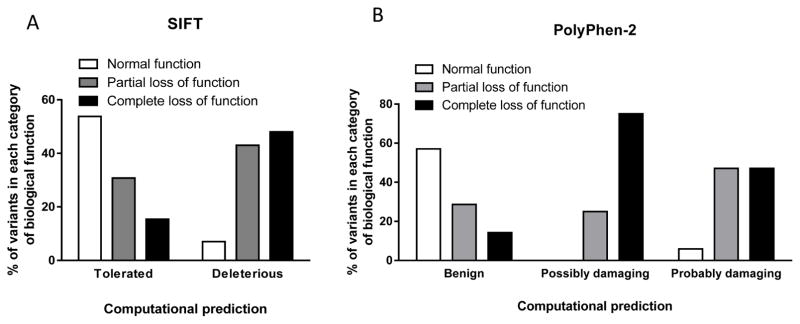
To evaluate the performance of the individual computational prediction tools, we analyzed the outcome and compared the findings with the results of biological function tests. Figure 1 summarizes the outcome of SIFT and PolyPhen-2 computational prediction testing and the comparison with biological function test results. Among variants that were predicted by SIFT to be tolerated, 53% had normal results on biological function testing (Figure 1A), while 57% of variants predicted by PolyPhen-2 to be benign had normal results on biological function testing (Figure 1B). When SIFT predicted the variants to be deleterious, 93% of them had impaired function, based on documented in vitro biological testing (Figure 1A). Similarly, among variants predicted by PolyPhen-2 to be possibly or probably damaging, 100% and 94% of the variants had impaired function based on in vitro biological test results (partial or complete loss of function), respectively (Figure 1B). Overall, both tools performed better in predicting loss-of-function variants. In contrast, the rate for correctly predicting a variant to have normal function was only slightly above 50% for both prediction programs.
Figure 1. Comparison of computational predictions of the function of GPCR variants by SIFT and PolyPhen-2 with documented biological function test findings.
(A) SIFT and (B) PolyPhen-2 were used to predict the function of 52 variants in GnRHR, KISS1R, PROKR2, and TACR3, for which the results of biological function tests are available. SIFT categorized variants as tolerated or deleterious; PolyPhen-2 categorized variants as benign, possibly damaging, or probably damaging. These predictions were compared with the results of in vitro biological function testing for each variant. Biological function was categorized as normal, partial loss of function, or complete loss of function. There were 10 variants with normal function, 21 with partial loss of function, and 21 with complete loss of function. The percentage of mutations predicted by each program in each category was then calculated.
Concordance between the prediction tools
Since there is no single perfect tool that can guarantee correct computational prediction of biological function of a GPCR variant, many scientists tend to use more than one prediction tool with the expectation that concordance between or among the prediction tools increases the accuracy of the computational predictions.
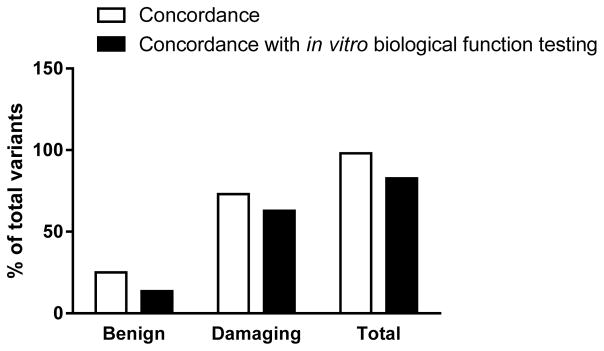
To evaluate whether the use of more than one prediction tool improved the computational prediction tool performance, we analyzed the concordance between the prediction tools assessed and matched the concordance rate with the results of biological function testing of the variants. Figure 2 shows the results of the concordance analysis. The concordance rates were calculated as the number of the variants predicted by SIFT and PolyPhen-2 to have the same functional outcome, divided by the total 52 variants with verified in vitro biological function. The overall concordance rate was 98%, but decreased to 83% after matching the computational predictions for concordance with biological function test results (Figure 2). Polyphen-2 and SIFT concordantly predicted 25% of 52 variants as benign. This decreased to 13.5% for concordance among the two computational programs and the documented biological function test results (Figure 2). Both tools concordantly predicted 73% of the total 52 variants as damaging. This rate again decreased, to 62.8%, upon matching the concordance with biological function test results (Figure 2).
Figure 2. Concordance rates between predictions by SIFT and PolyPhen-2 and between the in silico prediction programs and in vitro biological function testing.
The concordance between SIFT and PolyPhen-2 in predicting variants to be either benign or damaging was calculated and represented as the percentage of the total number of variants. The variants with concordant computational predictions were then compared with their in vitro biological function test results. The concordance among the two programs and the biological function test results was then calculated as the percentage of the total 52 variants with known biological function.
Statistical analyses of the performance of the computational prediction tools compared to biological function test outcomes
We analyzed sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and Matthews correlation coefficient (MCC) to further evaluate the performance of the prediction programs. Of note, damaging (possibly or probably) or deleterious was defined as positive (loss of function) while benign or tolerated was defined as negative (normal function). Table 2 summarizes the results of these statistical analyses. The concordant prediction group is defined as the group in which both in silico tools concordantly predict the variants to have the same function. Among 42 variants with documented impaired biological function based on in vitro testing, each prediction program alone, as well as the combined prediction from both tools, predicted 36 variants to be damaging. Among 10 variants with normal biological function test results, PolyPhen-2 predicted 8 to be benign while SIFT predicted 7 to be benign; a concordant prediction from both computational tools also correctly predicted 7 to be benign. One variant with normal biological function tests had discordance in functional prediction by the two in silico prediction tools.
Table 2.
Statistical analysis of missense variants with documented biological function testing.
| SIFT | PolyPhen-2 | Concordant Prediction | |
|---|---|---|---|
| True positive (N) | 36 | 36 | 36 |
| True negative (N) | 7 | 8 | 7 |
| False positive (N) | 3 | 2 | 2 |
| False negative (N) | 6 | 6 | 6 |
| Total (N) | 52 | 52 | 51 |
| Sensitivity (%) | 85.7 | 85.7 | 85.7 |
| Specificity (%) | 70.0 | 80.0 | 77.8 |
| Positive predictive value (%) | 92.3 | 94.7 | 94.7 |
| Negative predictive value (%) | 53.8 | 57.1 | 53.8 |
| Accuracy (%) | 82.7 | 84.6 | 84.3 |
| MCC | 0.5 | 0.6 | 0.6 |
N: Number of variants. MCC: Matthews correlation coefficient
The sensitivity (the ability to identify variants with loss of function) was 86% in all groups. The specificity (the ability to identify variants with normal function) was 70%, 80% and 78% for SIFT, PolyPhen-2, and the concordant prediction group, respectively. The positive predictive values (confidence that the prediction of a variant to be damaging in biologic testing was correct) were 92%, 95% and 95% for the SIFT, PolyPhen-2, and the concordant prediction groups, respectively. The negative predictive values (confidence that a prediction of a variant to be benign was correct) were 54%, 57%, and 54%, respectively. The prediction accuracies were 83%, 85% and 84% for SIFT, PolyPhen-2 and the concordant prediction groups, respectively. The Matthews correlation coefficient (MCC) was 0.5, 0.6 and 0.6, respectively.
Proportion and distribution of missense variants with documented or unknown phenotype and biological function, and functional prediction results
We retrieved nonsynonymous missense variants in GNRHR, KISS1R, PROKR2, and TACR3 from Ensembl, one of the largest databases for human variants. After excluding duplicates and the variants with known pathogenic clinical significance, there were a total of 322 missense variants derived from the Ensembl database. Among the variants, 16% (52), 12% (40), 30% (98), and 41% (132) were identified in GnRHR, KISS1R, PROKR2 and TACR3 respectively (Table 3). In comparison, 95 variants identified by phenotype-driven sequencing were retrieved from the Human Gene Mutation Database (HGMD) (Table 3). Among phenotype-related variants retrieved from HGMD, 36% (34), 13% (12) 38% (36), and 14% (13) were identified in GnRHR, KISS1R, PROKR2 and TACR3, respectively. Among 52 variants identified by phenotype-driven sequencing with documented in vitro biological function test results, 37% (19), 13% (7) 33% (17), and 17% (9) were identified in GnRHR, KISS1R, PROKR2 and TACR3, respectively (Table 3).
Table 3.
Summary of all missense variants retrieved from Ensembl, HGMD, and literature search.
| Unknown Phenotype (from Ensembl) | Known Phenotype (from HGMD) | Documented biological function | |
|---|---|---|---|
| GnRHR | 52 | 34 | 19 |
| KISS1R | 40 | 12 | 7 |
| PROKR2 | 98 | 36 | 17 |
| TACR3 | 132 | 13 | 9 |
| Total | 322 | 95 | 52 |
HGMD: Human gene mutation database
We took advantage of the availability of SIFT and PolyPhen-2 prediction results in Ensembl and analyzed the computational functional predictions of the nonsynonymous missense variants retrieved from Ensembl. Interestingly, SIFT and PolyPhen-2 predicted 53% (171/322) and 46% (146/316) of the variants to be benign, respectively, with the other 47% (151/322) and 54% (170/316) as deleterious or damaging (Table 4). PolyPhen-2 prediction results for 6 variants were not available. We compared the concordance between the two prediction programs. Among 169 variants that were predicted to be benign (tolerated) by SIFT, 72% (122/169) were concordantly predicted to be benign by PolyPhen-2. Among 150 variants that SIFT predicted to be damaging (deleterious), 145 (97%) of them were predicted to be damaging by Polyphen-2 as well. In turn, when we calculated the concordance based on the PolyPhen-2 predictions, 86% (126/146) and 82% (155/170) were concordantly predicted by SIFT to be benign (tolerated) and damaging (deleterious), respectively (Supplementary Table S1).
Table 4.
In silico prediction of the variants derived from the Ensembl database.
| SIFT | Polyphen-2 | |||
|---|---|---|---|---|
|
| ||||
| Tolerated | Deleterious | Benign | Damaging | |
| GnRHR | 22 | 30 | 25 | 31 |
| KISS1R | 23 | 17 | 12 | 18 |
| PROKR2 | 48 | 50 | 53 | 51 |
| TACR3 | 78 | 54 | 56 | 70 |
| Total | 171 | 151 | 146 | 170 |
| Percentage * | 53.1 | 46.9 | 46.2 | 53.8 |
Percentage of total receptors predicted by SIFT or Polyphen-2
Discussion
With the advances in genetic sequencing methodologies, the list of new nonsynonymous missense variants continues to increase rapidly. Accurate determination of the effect of each missense variant on protein function is a fundamental step to provide the connection between genotype and phenotype. Biological function testing is an effective way to determine the pathogenicity of a newly identified missense variant, which can provide revolutionary advances in our knowledge of human physiology and pathophysiology. For example, after a novel Leu148Ser variant of KISS1R was identified in a patient with hypogonadotropic hypogonadism (HH), our laboratory performed biological function testing and confirmed that the variant caused loss of function of KISS1R [16]. This finding revealed the key role of KISS1R and its ligand, kisspeptin, in the regulation of reproductive function and advanced our understanding of reproductive control upstream of GnRH. Measuring changes in second messengers involved in GPCR-mediated signal transduction is an effective way to assess the impact of variants on receptor function. GnRHR, KISS1R, PROKR2, and TACR3 are Gq-coupled receptors; activation of these receptors is reflected by changes in inositol phosphate production (IP), intracellular calcium concentrations ([Ca2+]i), and ERK phosphorylation. The assays for measurement of these second messengers have been well established [17–19]. However, biological function testing can be costly, labor-intensive, and time consuming. As shown in this study, only 50% of the missense variants identified to date from phenotype-driven sequencing have had biological function tests (Table 3). As an alternative, computational functional prediction tools have been used widely to predict the impact of missense variants on protein function, in an attempt to identify disease-causing variants. However, the accuracy of each prediction tool, while impressive, is not yet ideal. In a recent comprehensive analysis, the accuracies of nine widely used prediction methods were in the range of 60%–82% [5].
In this study, we analyzed the performance of two frequently used prediction tools, SIFT and PolyPhen-2, in predicting the functional effects of missense variants of GPCRs involved in the central neuroendocrine regulation of reproduction. Another important rationale for our selection of these two prediction programs for this study is that the results of computational analyses of GPCR variants by SIFT and Polyphen-2 are available in the Ensembl database. Analysis of the performance of these two programs will aid investigators in the proper interpretation of the results of computational analysis of variants retrieved from Ensembl. There are additional computational prediction programs available, such as Panther (http://www.pantherdb.org/tools/csnpScoreForm.jsp), and Mutation Taster (http://www.mutationtaster.org). Computational prediction programs principally use several attributes related to protein structure, evolutionary conservation, phylogeny, biophysical characteristics of the substitution, secondary structural information, and chain flexibility. The programs share some similarities but have some distinct features [5]. SIFT makes inferences from sequence similarity using mathematical operations [8], while PolyPhen2 employs a combination of sequence- and structure-based attributes for the description of an amino acid substitution, and the effect of a mutation is predicted by a naive Bayesian classifier [15, 20].
Although the mechanisms of predictions differ between these two programs, both performed well in predicting loss-of-function variants (Figure 1). In contrast, however, the capability for precisely identifying normal function variants was inferior for both SIFT and PolyPhen-2, and as such, more than 40% of variants with demonstrated impaired receptor function by biologic testing were falsely predicted to be benign by each method (Figure 1). This is an important consideration, because investigators may stop pursuing biological function tests if in silico programs predict a novel variant to be benign. This statement is supported by our finding that only a small proportion (13/52 in SIFT, 14/52 in PolyPhen-2) of variants with documented biological function testing were predicted to be benign (Table 1), whereas half of all missense variants from Ensembl were predicted to be benign by both programs (Table 4). Alternatively, this difference in the proportion of variants predicted to be benign in the two datasets may be because the first set were identified in patients with reproductive phenotypes, and were therefore more likely to be damaging, whereas variants from large scale sequencing were found in general populations with unknown phenotypes – since HH and central precocious puberty are rare, variants in these populations are unlikely to come from individuals with an underlying reproductive phenotype, so the variants in this population should be more likely to be benign. This tendency for investigators to stop pursuing biological function tests if in silico programs predict a novel variant to be benign may also explain why the benign variants with documented biological function reported were primarily in PROKR2 and TACR3, but not in GnRHR – since most of the studies of GnRHR were done earlier than the more recent studies of PROKR2 and TACR3, at a time when it was less accepted to report benign variants. In addition to loss-of-function mutations reported in these GPCRs in patients with GnRH deficiency, a gain-of-function mutation, R386P KISS1R, has been reported in association with central precocious puberty [21]. However, neither SIFT nor Polyphen-2 is designed to predict variants with gain of function.
To increase the accuracy of computational functional prediction analysis, a common approach is to apply more than one prediction tool. Interestingly, in this study, when the concordance between SIFT and PolyPhen-2 predictions was matched with biological function testing outcomes, the performance of the concordant predictions was not superior to the performance of a single program (Table 2). Evidently, both programs can concordantly predict the incorrect function in a variant, since concordance rates were always higher for the two in silico prediction tests than the rates when concordance of the prediction tests were further matched with biological function testing outcomes, although these differences did not reach statistical significance (Figure 2). It is even more difficult to interpret the function of a variant if the program predictions are discordant.
In our study, sensitivity represents the ability to identify loss-of-function variants. Both in silico prediction tools had the same sensitivity, using biologic function testing as the gold standard (Table 2). Although even biological testing may not be perfect, there is no other better method to be used as the gold standard. In contrast, our statistical analysis showed that the ability to identify variants with normal function by in silico prediction (specificity and negative predictive value) was inferior to the ability to identify loss-of-function variants (sensitivity and positive predictive value) (Table 2). Given the fact that more than 40% of variants predicted to be benign were actually deleterious/damaging in biological function testing (Figure 1 and Table 2), if a novel GPCR variant is predicted to be benign by in silico prediction tools, a biological function test may nonetheless still be indicated, particularly if there is a strong phenotype correlation. On the other hand, performance of biological function testing may not be necessary if a variant is predicted to be damaging or deleterious, as more than 90% of variants predicted to be damaging were true loss-of-function variants in biologic function testing assays (Figure 1 and table 2). The interpretation of concordant prediction analysis is complex. After excluding one variant with discordant predictions, the combined prediction analysis by SIFT and PolyPhen-2 had comparable performance outcomes when compared with the single program predictions (Table 2). The accuracies range from 83% to 86% for SIFT and Polyphen-2 respectively which, interestingly, were better than an earlier comprehensive analysis [5].
The MCC [22] is an important statistical parameter, as it is unaltered by differing proportions of benign and damaging variants, while PPV and NPV may be affected by the prevalence in varied populations [23]. Because of its insensitivity to variation in sample size, the MCC gives a more balanced assessment of performance than the other performance measures [24]. In light of the reference range (−1 to 1), a coefficient of +1 represents a perfect prediction, 0 equals random prediction and −1 resemble complete discrepancy between prediction and observation. Compared with an earlier study [5], the MCC in this study (Table 2) indicates a fair performance of SIFT, PolyPhen-2, and concordant predictions, without any differences among the different analyses.
Ensembl is one of several well-known genome browsers for the retrieval of genomic information. We chose Ensembl to retrieve missense variants of GPCRs involved in the neuroendocrine regulation of reproductive function because it also provides information on the functional prediction results by SIFT and PolyPhen-2. Analysis of the computational prediction results revealed a surprising finding that half of the variants were predicted to be deleterious/damaging by SIFT and PolyPhen-2 (Table 4). Interpretation of the results of these prediction analyses in this setting is even more difficult, since the majority of the missense variants were found in large-scale genomic sequencing of general, unphenotyped populations rather than from phenotype-driven sequencing (Table 3). In view of the fact that both SIFT and PolyPhen-2 accurately predict more than 90% of variants with documented impaired function as deleterious/damaging (Figure 1 and Table 2), it is likely that most variants retrieved from Ensembl and predicted by SIFT and PolyPhen-2 to be deleterious/damaging, do have impaired function. Linking these genotype findings to phenotype is difficult since the majority variants from Ensembl were not identified by phenotype-driven sequencing. In the general population, the prevalence of HH is extremely low, estimated to be about 0.01–0.025% [25]. Individuals harboring loss-of-function variants may have a subclinical phenotype or no phenotype at all, in light of diverse pathogenetic mechanisms and inheritance patterns (autosomal recessive [26], autosomal dominant [14], haploinsufficiency, digenic inheritance [27, 28] and oligogenic inheritance [29]) of GPCR loss-of-function variants. We speculate that if a variant co-segregates with an HH phenotype in a family, one might expect that it is more likely to be deleterious, but we did not specifically test this hypothesis in this study.
In conclusion, we have found that the performance of the computational prediction programs, SIFT and PolyPhen-2, is effective in predicting loss of function for variants of GPCRs involved in the neuroendocrine regulation of reproduction. In contrast, both programs were less effective for predicting benign variants - more than 40% of variants predicted to be benign showed loss of function in biologic function tests. Based on these findings, we recommend performing biological function testing even for variants predicted to be benign, especially if there is a close phenotype correlation. A surprising finding that a significant number of the variants identified in large-scale sequencing were predicted to be loss-of-function variants creates an immense challenge for the interpretation of their clinical significance.
Supplementary Material
Acknowledgments
This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (NIH), through cooperative agreement U54 HD28138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and by R01 HD19938 (to U.B.K.), and by NICHD/NIH K08 HD070957 (to L.M.).
Footnotes
The authors have nothing to disclose.
References
- 1.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 2.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 3.Zhang MQ. Promoter analysis of co-regulated genes in the yeast genome. Comput Chem. 1999;23:233–250. doi: 10.1016/s0097-8485(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 4.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 6.Chan AO. Performance of in silico analysis in predicting the effect of non-synonymous variants in inherited steroid metabolic diseases. Steroids. 2013;78:726–730. doi: 10.1016/j.steroids.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Nakken S, Alseth I, Rognes T. Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes. Neuroscience. 2007;145:1273–1279. doi: 10.1016/j.neuroscience.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews Drug discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 10.Smith KA, Komuniecki RW, Ghedin E, Spiro D, Gray J. Genes encoding putative biogenic amine receptors in the parasitic nematode brugia malayi. Invert Neurosci. 2007;7:227–244. doi: 10.1007/s10158-007-0058-y. [DOI] [PubMed] [Google Scholar]
- 11.Moller S, Vilo J, Croning MD. Prediction of the coupling specificity of g protein coupled receptors to their g proteins. Bioinformatics. 2001;17 (Suppl 1):S174–181. doi: 10.1093/bioinformatics/17.suppl_1.s174. [DOI] [PubMed] [Google Scholar]
- 12.Tovo-Rodrigues L, Roux A, Hutz MH, Rohde LA, Woods AS. Functional characterization of g-protein-coupled receptors: A bioinformatics approach. Neuroscience. 2014;277:764–779. doi: 10.1016/j.neuroscience.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noel SD, Kaiser UB. G protein-coupled receptors involved in gnrh regulation: Molecular insights from human disease. Mol Cell Endocrinol. 2011;346:91–101. doi: 10.1016/j.mce.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noel SD, Abreu AP, Xu S, Muyide T, Gianetti E, Tusset C, Carroll J, Latronico AC, Seminara SB, Carroll RS, Kaiser UB. Tacr3 mutations disrupt NK3R function through distinct mechanisms in gnrh-deficient patients. FASEB J. 2014;28:1924–1937. doi: 10.1096/fj.13-240630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The gpr54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 17.Ahow M, Min L, Pampillo M, Nash C, Wen J, Soltis K, Carroll RS, Glidewell-Kenney CA, Mellon PL, Bhattacharya M, Tobet SA, Kaiser UB, Babwah AV. Kiss1r signals independently of galphaq/11 and triggers lh secretion via the beta-arrestin pathway in the male mouse. Endocrinology. 2014;155:4433–4446. doi: 10.1210/en.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babwah AV, Pampillo M, Min L, Kaiser UB, Bhattacharya M. Single-cell analyses reveal that kiss1r-expressing cells undergo sustained kisspeptin-induced signaling that is dependent upon an influx of extracellular ca2+ Endocrinology. 2012;153:5875–5887. doi: 10.1210/en.2012-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min L, Soltis K, Reis AC, Xu S, Kuohung W, Jain M, Carroll RS, Kaiser UB. Dynamic kisspeptin receptor trafficking modulates kisspeptin-mediated calcium signaling. Mol Endocrinol. 2014;28:16–27. doi: 10.1210/me.2013-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The pfam protein families database. Nucleic acids research. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A gpr54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews BW. Comparison of the predicted and observed secondary structure of t4 phage lysozyme. Biochimica et biophysica acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 23.Altman DG, Bland JM. Diagnostic tests 2: Predictive values. Bmj. 1994;309:102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 25.Seminara SB, Hayes FJ, Crowley WF., Jr Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and kallmann’s syndrome): Pathophysiological and genetic considerations. Endocr Rev. 1998;19:521–539. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 26.Sarfati J, Guiochon-Mantel A, Rondard P, Arnulf I, Garcia-Pinero A, Wolczynski S, Brailly-Tabard S, Bidet M, Ramos-Arroyo M, Mathieu M, Lienhardt-Roussie A, Morgan G, Turki Z, Bremont C, Lespinasse J, Du Boullay H, Chabbert-Buffet N, Jacquemont S, Reach G, De Talence N, Tonella P, Conrad B, Despert F, Delobel B, Brue T, Bouvattier C, Cabrol S, Pugeat M, Murat A, Bouchard P, Hardelin JP, Dode C, Young J. A comparative phenotypic study of kallmann syndrome patients carrying monoallelic and biallelic mutations in the prokineticin 2 or prokineticin receptor 2 genes. J Clin Endocrinol Metab. 2010;95:659–669. doi: 10.1210/jc.2009-0843. [DOI] [PubMed] [Google Scholar]
- 27.Quaynor SD, Kim HG, Cappello EM, Williams T, Chorich LP, Bick DP, Sherins RJ, Layman LC. The prevalence of digenic mutations in patients with normosmic hypogonadotropic hypogonadism and kallmann syndrome. Fertility and sterility. 2011;96:1424–1430. e1426. doi: 10.1016/j.fertnstert.2011.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. The Journal of clinical investigation. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silveira LF, Trarbach EB, Latronico AC. Genetics basis for gnrh-dependent pubertal disorders in humans. Molecular and cellular endocrinology. 2010;324:30–38. doi: 10.1016/j.mce.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Costa EM, Bedecarrats GY, Mendonca BB, Arnhold IJ, Kaiser UB, Latronico AC. Two novel mutations in the gonadotropin-releasing hormone receptor gene in brazilian patients with hypogonadotropic hypogonadism and normal olfaction. J Clin Endocrinol Metab. 2001;86:2680–2686. doi: 10.1210/jcem.86.6.7551. [DOI] [PubMed] [Google Scholar]
- 31.Beranova M, Oliveira LM, Bedecarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, Pitteloud N, Kaiser UB, Crowley WF, Jr, Seminara SB. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2001;86:1580–1588. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 32.Bedecarrats GY, Linher KD, Janovick JA, Beranova M, Kada F, Seminara SB, Michael Conn P, Kaiser UB. Four naturally occurring mutations in the human gnrh receptor affect ligand binding and receptor function. Mol Cell Endocrinol. 2003;205:51–64. doi: 10.1016/s0303-7207(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 33.Soderlund D, Canto P, de la Chesnaye E, Ulloa-Aguirre A, Mendez JP. A novel homozygous mutation in the second transmembrane domain of the gonadotrophin releasing hormone receptor gene. Clin Endocrinol (Oxf) 2001;54:493–498. doi: 10.1046/j.1365-2265.2001.01211.x. [DOI] [PubMed] [Google Scholar]
- 34.Tello JA, Newton CL, Bouligand J, Guiochon-Mantel A, Millar RP, Young J. Congenital hypogonadotropic hypogonadism due to gnrh receptor mutations in three brothers reveal sites affecting conformation and coupling. PLoS One. 2012;7:e38456. doi: 10.1371/journal.pone.0038456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antelli A, Baldazzi L, Balsamo A, Pirazzoli P, Nicoletti A, Gennari M, Cicognani A. Two novel gnrhr gene mutations in two siblings with hypogonadotropic hypogonadism. Eur J Endocrinol. 2006;155:201–205. doi: 10.1530/eje.1.02198. [DOI] [PubMed] [Google Scholar]
- 36.Maya-Nunez G, Janovick JA, Aguilar-Rojas A, Jardon-Valadez E, Leanos-Miranda A, Zarinan T, Ulloa-Aguirre A, Conn PM. Biochemical mechanism of pathogenesis of human gonadotropin-releasing hormone receptor mutants thr104ile and tyr108cys associated with familial hypogonadotropic hypogonadism. Mol Cell Endocrinol. 2011;337:16–23. doi: 10.1016/j.mce.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 38.Caron P, Chauvin S, Christin-Maitre S, Bennet A, Lahlou N, Counis R, Bouchard P, Kottler ML. Resistance of hypogonadic patients with mutated gnrh receptor genes to pulsatile gnrh administration. J Clin Endocrinol Metab. 1999;84:990–996. doi: 10.1210/jcem.84.3.5518. [DOI] [PubMed] [Google Scholar]
- 39.Pralong FP, Gomez F, Castillo E, Cotecchia S, Abuin L, Aubert ML, Portmann L, Gaillard RC. Complete hypogonadotropic hypogonadism associated with a novel inactivating mutation of the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab. 1999;84:3811–3816. doi: 10.1210/jcem.84.10.6042. [DOI] [PubMed] [Google Scholar]
- 40.Karges B, Karges W, Mine M, Ludwig L, Kuhne R, Milgrom E, de Roux N. Mutation ala(171)thr stabilizes the gonadotropin-releasing hormone receptor in its inactive conformation, causing familial hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2003;88:1873–1879. doi: 10.1210/jc.2002-020005. [DOI] [PubMed] [Google Scholar]
- 41.de Roux N, Young J, Brailly-Tabard S, Misrahi M, Milgrom E, Schaison G. The same molecular defects of the gonadotropin-releasing hormone receptor determine a variable degree of hypogonadism in affected kindred. J Clin Endocrinol Metab. 1999;84:567–572. doi: 10.1210/jcem.84.2.5449. [DOI] [PubMed] [Google Scholar]
- 42.Layman LC, Cohen DP, Jin M, Xie J, Li Z, Reindollar RH, Bolbolan S, Bick DP, Sherins RR, Duck LW, Musgrove LC, Sellers JC, Neill JD. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet. 1998;18:14–15. doi: 10.1038/ng0198-14. [DOI] [PubMed] [Google Scholar]
- 43.Meysing AU, Kanasaki H, Bedecarrats GY, Acierno JS, Jr, Conn PM, Martin KA, Seminara SB, Hall JE, Crowley WF, Jr, Kaiser UB. Gnrhr mutations in a woman with idiopathic hypogonadotropic hypogonadism highlight the differential sensitivity of luteinizing hormone and follicle-stimulating hormone to gonadotropin-releasing hormone. J Clin Endocrinol Metab. 2004;89:3189–3198. doi: 10.1210/jc.2003-031808. [DOI] [PubMed] [Google Scholar]
- 44.Tenenbaum-Rakover Y, Commenges-Ducos M, Iovane A, Aumas C, Admoni O, de Roux N. Neuroendocrine phenotype analysis in five patients with isolated hypogonadotropic hypogonadism due to a l102p inactivating mutation of gpr54. J Clin Endocrinol Metab. 2007;92:1137–1144. doi: 10.1210/jc.2006-2147. [DOI] [PubMed] [Google Scholar]
- 45.Semple RK, Achermann JC, Ellery J, Farooqi IS, Karet FE, Stanhope RG, O’Rahilly S, Aparicio SA. Two novel missense mutations in g protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2005;90:1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- 46.Teles MG, Trarbach EB, Noel SD, Guerra-Junior G, Jorge A, Beneduzzi D, Bianco SD, Mukherjee A, Baptista MT, Costa EM, De Castro M, Mendonca BB, Kaiser UB, Latronico AC. A novel homozygous splice acceptor site mutation of kiss1r in two siblings with normosmic isolated hypogonadotropic hypogonadism. Eur J Endocrinol. 2010;163:29–34. doi: 10.1530/EJE-10-0012. [DOI] [PubMed] [Google Scholar]
- 47.Nimri R, Lebenthal Y, Lazar L, Chevrier L, Phillip M, Bar M, Hernandez-Mora E, de Roux N, Gat-Yablonski G. A novel loss-of-function mutation in gpr54/kiss1r leads to hypogonadotropic hypogonadism in a highly consanguineous family. J Clin Endocrinol Metab. 2011;96:E536–545. doi: 10.1210/jc.2010-1676. [DOI] [PubMed] [Google Scholar]
- 48.Brioude F, Bouligand J, Francou B, Fagart J, Roussel R, Viengchareun S, Combettes L, Brailly-Tabard S, Lombes M, Young J, Guiochon-Mantel A. Two families with normosmic congenital hypogonadotropic hypogonadism and biallelic mutations in kiss1r (kiss1 receptor): Clinical evaluation and molecular characterization of a novel mutation. PLoS One. 2013;8:e53896. doi: 10.1371/journal.pone.0053896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abreu AP, Trarbach EB, de Castro M, Frade Costa EM, Versiani B, Matias Baptista MT, Garmes HM, Mendonca BB, Latronico AC. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive kallmann syndrome. J Clin Endocrinol Metab. 2008;93:4113–4118. doi: 10.1210/jc.2008-0958. [DOI] [PubMed] [Google Scholar]
- 50.Abreu AP, Noel SD, Xu S, Carroll RS, Latronico AC, Kaiser UB. Evidence of the importance of the first intracellular loop of prokineticin receptor 2 in receptor function. Mol Endocrinol. 2012;26:1417–1427. doi: 10.1210/me.2012-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. Kallmann syndrome: Mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, Seminara SB, Huot C, Alos N, Speiser P, Takeshita A, Van Vliet G, Pearce S, Crowley WF, Jr, Zhou QY, Pitteloud N. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: Molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monnier C, Dode C, Fabre L, Teixeira L, Labesse G, Pin JP, Hardelin JP, Rondard P. Prokr2 missense mutations associated with kallmann syndrome impair receptor signalling activity. Hum Mol Genet. 2009;18:75–81. doi: 10.1093/hmg/ddn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCabe MJ, Gaston-Massuet C, Gregory LC, Alatzoglou KS, Tziaferi V, Sbai O, Rondard P, Masumoto KH, Nagano M, Shigeyoshi Y, Pfeifer M, Hulse T, Buchanan CR, Pitteloud N, Martinez-Barbera JP, Dattani MT. Variations in prokr2, but not prok2, are associated with hypopituitarism and septo-optic dysplasia. J Clin Endocrinol Metab. 2013;98:E547–557. doi: 10.1210/jc.2012-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gianetti E, Tusset C, Noel SD, Au MG, Dwyer AA, Hughes VA, Abreu AP, Carroll J, Trarbach E, Silveira LF, Costa EM, de Mendonca BB, de Castro M, Lofrano A, Hall JE, Bolu E, Ozata M, Quinton R, Amory JK, Stewart SE, Arlt W, Cole TR, Crowley WF, Kaiser UB, Latronico AC, Seminara SB. Tac3/tacr3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin b in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95:2857–2867. doi: 10.1210/jc.2009-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK. Tac3 and tacr3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin b in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guran T, Tolhurst G, Bereket A, Rocha N, Porter K, Turan S, Gribble FM, Kotan LD, Akcay T, Atay Z, Canan H, Serin A, O’Rahilly S, Reimann F, Semple RK, Topaloglu AK. Hypogonadotropic hypogonadism due to a novel missense mutation in the first extracellular loop of the neurokinin b receptor. J Clin Endocrinol Metab. 2009;94:3633–3639. doi: 10.1210/jc.2009-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francou B, Bouligand J, Voican A, Amazit L, Trabado S, Fagart J, Meduri G, Brailly-Tabard S, Chanson P, Lecomte P, Guiochon-Mantel A, Young J. Normosmic congenital hypogonadotropic hypogonadism due to tac3/tacr3 mutations: Characterization of neuroendocrine phenotypes and novel mutations. PLoS One. 2011;6:e25614. doi: 10.1371/journal.pone.0025614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.