Abstract
The leucine-rich repeat kinase (LRRK)-2 protein contains nonoverlapping GTPase and kinase domains, and mutation in either domain can cause Parkinson disease. GTPase proteins are critical upstream modulators of many effector protein kinases. In LRRK2, this paradigm may be reversed, as the kinase domain phosphorylates its own GTPase domain. In this study, we found that the ameba LRRK2 ortholog ROCO4 phosphorylates the GTPase domain [termed Ras-of-complex (ROC) domain in this family] of human LRRK2 on the same residues as the human LRRK2 kinase. Phosphorylation of ROC enhances its rate of GTP hydrolysis [from kcat (catalytic constant) 0.007 to 0.016 min−1], without affecting GTP or GDP dissociation kinetics [koff = 0.093 and 0.148 min−1 for GTP and GDP, respectively). Phosphorylation also promotes the formation of ROC dimers, although GTPase activity appears to be equivalent between purified dimers and monomers. Modeling experiments show that phosphorylation induces conformational changes at the critical p-loop structure. Finally, ROC appears to be one of many GTPases phosphorylated in p-loop residues, as revealed by alignment of LRRK2 autophosphorylation sites with GTPases annotated in the phosphoproteome database. These results provide an example of a novel mechanism for kinase-mediated control of GTPase activity.—Liu, Z., Mobley, J. A., DeLucas, L. J., Kahn, R. A., West, A. B. LRRK2 autophosphorylation enhances its GTPase activity.
Keywords: G-protein, phosphorylation, GTP-hydrolysis, ROC, Parkinson disease
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common known genetic cause of Parkinson disease (PD) (1, 2). Pathogenic mutations are located within each of the 2 catalytic cores of the LRRK2 protein: the GTPase and kinase domains. LRRK2 protein consists of 5 defined regions or domains. From N to C terminus, these are a leucine-rich region, the Ras-of-complex (ROC) GTPase domain, a conserved linker that is found C-terminal to the ROC domain, the serine/threonine protein kinase domain (3), and a WD40 domain (Fig. 1A). LRRK2 is known to homodimerize, although it is not clear which domains are directly involved in dimerization, at least in part because multiple domains in LRRK2 stably interact with one another (4–8). The arrangement of domains is very highly conserved throughout eukaryotic evolution. For example, a close ortholog of LRRK2, ROCO4, can be readily identified in the single-cell organism Dictyostelium (9). The pathogenic mutations R1441C/G/H in the ROC domain decrease GTPase activity (10–12) and increase autophosphorylation in some assays (13, 14). Pathogenic LRRK2 mutations in the kinase domain, including G2019S and I2020T, likewise increase autophosphorylation activity (10, 14–16). The observation that mutations in the ROC and kinase domains have similar consequences suggests that they act in tandem.
Figure 1.
Generation and characterization of phosphorylated ROC (pROC). A) Domain arrangement of full-length human LRRK2 and the ameba LRRK2 ortholog ROCO4. B) Recombinant (human) His6-ROC was subjected to phosphorylation in the presence of γ-[32P]ATP by GST-[Δ970] LRRK2G2019S (bottom) or GST-ROCO4 (top). Phosphorylation of His6-ROC was followed at 30 min intervals in immunoblots using an antibody directed toward Thr(P)1503 (13). Total His6-ROC is shown by Coomassie blue staining of SDS gels below each immunoblot. C) Quantification of the phosphorylated His6-ROC bands shown in (B) by scintillation counting. D) GST-ROCO4 (5 μg protein/lane) and His6-ROC (10 μg protein per/lane) were analyzed by Coomassie blue staining 3 h after incubation in a kinase reaction and after GST-ROCO4 depletion from the kinase reaction by glutathione beads (His6-pROC lane). E) Phosphorylated residues (Table 1) are aligned with consensus GTP-binding motifs. Shown are all phosphothreonine residues detected in ROC as compared to autophosphorylation sites in full-length human LRRK2 protein. Previously annotated autophosphorylated residues in the ROC domain (+human LRRK2) are indicated and were derived from other studies [Table 2; (13, 21, 29, 49)]. F) Stoichiometry of His6-ROC or His6-ROC Thr-Ala mutants was determined in kinase reactions that included γ-[32P]ATP. Overall, ROCO4 catalyzed the transfer of ∼1 phosphate per every 2 ROC proteins in the kinase reaction in the 3 h reactions shown. All data are averaged from 3 independent experiments or are representative of 3 independent experiments. Data are means ± sem; significances by 1-way ANOVA with Tukey’s post hoc test. **P < 0.01, ***P < 0.001.
In some GTPases, activation of the protein by binding GTP promotes direct binding and allosteric activation of downstream effector kinases. For example, the GTPase RhoA regulates the Rho kinases (ROCK) in the regulation of the cytoskeleton (17), and the Ras proteins can activate Raf protein kinases in affecting a variety of mitogen-activated signaling cascades (18). In these examples, the GTPase and kinase domains are found on distinct gene products. LRRK2 has both GTPase and kinase activity, and LRRK2 GTPase activity may regulate LRRK2 kinase activity. In experimental settings, guanine nucleotide binding and hydrolysis by the ROC domain have been described, at least indirectly, to modulate LRRK2 kinase activity (10, 19). Such experiments are compromised by the unusual instability of LRRK2, which requires guanine nucleotide binding to stabilize both the ROC domain and kinase activity (20).
It was discovered recently that the LRRK2 kinase domain phosphorylates its own ROC domain and that this effect can occur in both cis and trans (4, 6, 13, 21). The proportion of LRRK2 that is phosphorylated in the ROC domain appears to be very low in cells and tissues (13, 14). Therefore, discovering the functional consequences of autophosphorylated residues in the LRRK2 GTPase domain has been difficult. Using novel biochemical assays and approaches, we were able to perform a series of biochemical experiments to help understand the impact of LRRK2 autophosphorylation on ROC domain function.
MATERIALS AND METHODS
Protein purification
The N-terminal GST-tagged kinase domain of ameba LRRK2 (GST-ROCO4, corresponding to residues 1–287 of PDB accession 4F0F_A) and the N-terminal His6-tagged human LRRK2 ROC domain (His6-ROC, corresponding to residues 1333–1516 of NP_940980.3) were created through custom gene synthesis (Genscript, Piscataway, NJ, USA), expressed in Escherichia coli, and purified (9, 22). GST-[Δ970] LRRK2 and GST-[Δ970] LRRK2G2019S recombinant proteins were purified from Sf9 insect cells after infection with a baculovirus (Thermo Fisher Scientific; catalog no. PV4881). Purities of all proteins were assessed by visual inspection of Coomassie blue–stained gels; purities for the recombinant protein exceeded 90% based on ImageJ [National Institutes of Health (NIH), Bethesda, MD, USA] analysis of band intensities. Concentrations of GST-ROCO4, GST-[Δ970] LRRK2, and GST-[Δ970] LRRK2G2019S were determined by absorbance at 280 nm with a Nanodrop ND-1000 (Thermo Fisher Scientific). Concentrations of His6-ROC and His6-pROC, which had been copurified with nucleotides, were determined with a Pierce BCA Protein Assay Kit and a Synergy 2 plate reader (Biotek, Winooski, VT, USA).
Kinase assays
ATP (200 μM) was combined with His6-ROC, GST-ROCO4, GST-[Δ970] LRRK2, or GST-[Δ970] LRRK2G2019S in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, and 5% glycerol (buffer A). Proteins were incubated, with or without 200 μM ATP (supplemented with γ-[32P]ATP, as indicated), in buffer A for 1 h at 30°C. His6-ROC samples were concentrated to 3 μM and desalted on Zeba Spin Desalting Columns with a 7 kDa molecular mass cut-off (Thermo Fisher Scientific). Laemmli sample loading buffer was added to each reaction before analysis by SDS-PAGE. After incubation at 30°C for 5 h, no additional phosphorylation was observed. For some experiments involving γ-[32P]ATP, Coomassie blue–stained protein bands were excised, and radioactivity was measured by scintillation counting. To generate equivalently handled nonphosphorylated protein, for all preparations of GST-ROCO4 phosphorylated His6-ROC (His6-pROC) and pGST-[Δ970] LRRK2G2019S, we incubated the proteins in the same kinase reactions but without the inclusion of GST-ROCO4 kinase and ATP, respectively. All kinase assay results were repeated in at least 3 independent experiments, and consistent results were obtained.
GTPase assays
The GTP hydrolysis (GTPase) rate was determined as the amount of free phosphate formed with time and was measured with EnzChek Phosphate Assay Kit (Thermo Fisher Scientific) in the presence of 20 μM His6-ROC or His6-pROC; 2 μM GST-[Δ970] LRRK2G2019S; or pGST-[Δ970] LRRK2G2019S, in buffer A. The absorbance at 360 nm was measured every 5 min for 3 h on a microplate reader (Synergy 2; BioTek). The amount of inorganic phosphate was determined by extrapolating A360 data based on the standard curve generated using 2–150 μM KH2PO4. GTPase activities were then expressed as nanomoles phosphate formed per minute per micromole protein. The amount of inorganic phosphate (Pi) generated in the absence of any added protein was measured as the GTP self-hydrolysis background, and the amount generated in the absence of added GTP was measured as background. These signals were subtracted from each data point. All GTPase assay results were repeated in at least 3 independent experiments, and consistent results were obtained.
Size-exclusion chromatography
Proteins were loaded onto a Superdex 200 10/300 column on an Akta fast protein liquid chromatography instrument (GE Healthcare Life Sciences, Pittsburgh, PA, USA). For separation of monomeric and dimeric His6-ROC, a Superdex 75 10/300 column was used. For quantification of the dimeric His6-ROC and His6-pROC, 1 mg of each protein was loaded onto the Superdex 75 10/300 column in 3 independent runs. The peaks were analyzed with UNICORN software, version 4.12 (GE Life Sciences).Sites of phosphorylation within the His6-ROC and His6-pROC domains were discovered by analyzing phosphopeptides with liquid chromatography tandem mass spectrometry (LC/MS-MS) with an Orbitrap Velos Pro (Thermo Fisher Scientific) hybrid ion-trap mass spectrometer (13). Besides peptides identified as having phosphothreonine modifications, several lower confidence phosphoserine modifications were identified. However, each of these was excluded from further analyses because it was also identified in the His6-ROC from the (−) kinase control reactions, which should not have any phosphorylations, or was a low-confidence match.
GTP and GDP dissociation rate analyses
His6-ROC or His6-pROC were bound to Ni-NTA agarose (Thermo Fisher Scientific) before incubation with 10 μCi · ml−1 α-[32P]GTP (PerkinElmer, Waltham, MA, USA) or [3H]GDP (American Radio Labeled Chemicals, St. Louis, MO, USA) for 2 h at 22°C in buffer A. Unbound nucleotides were then removed through washing in buffer A 5 times on ice in the presence of 20 μM GTP in the wash buffer. The beads were then resuspended, divided into aliquots, and placed into tubes in a thermal vortex mixer set to 22°C at 1300 rpm. At different time points, triplicates of His6-ROC, His6-pROC, and control beads were collected. The beads were pelleted by centrifugation and the supernatants removed. The radioactivity remaining on the beads was determined by scintillation counting. The values on the control beads were subtracted from those of His6-ROC and His6-pROC to obtain the specific binding values. All measurements were repeated in at least 3 independent experiments, and consistent results were obtained.
Biophysical methods
Differential scanning calorimetry was performed by loading 1 mg · ml−1 protein samples to a VP-capillary machine (MicroCal; GE Healthcare Life Sciences) with scans from 30 to 75°C. Data collected were analyzed with Origin7 software (OriginLab, Northampton, MA, USA). Circular dichroism (CD) samples were loaded onto a photospectrometer cell with a 0.2 mm path length and scanned with the CD DSM-20 system (Olis, Bogart, GA, USA), with a wavelength from 190 to 260 nm.
In silico structure modeling
The pROC structure and variations were created in PyMOL based on PDB ID 2zej and 4wnr. The surface electrostatic model was made with APBS tools (www.poissonboltzmann.org). The phosphorylated p-loop of RAN was modeled based on the GTP-bound crystal structure [Protein Data Bank (PDB; http://www.wwpdb.org/) ID: 4C0Q] (23). The phosphorylated p-loop of RhoA is modeled on the GTPγS-bound crystal structure (PDB ID: 1A2B) (24), and the phosphorylated p-loop of G(α)q is modeled on the GDP-AlF4-bound crystal structure (PDB ID: 4QJ5) (25).
Bioinformatics analysis of phosphorylated GTPase and statistics
The phosphorylation database PhosphoSitePlus (www.phosphosite.org; Cell Signaling Technology, Inc., Danvers, MA, USA) was searched for the term “GTPase,” which returned 198 individual proteins. Each entry corresponding to a human protein was individually evaluated for phosphosites in the conserved p-loop motif, identified by the sequence GXXXXGK (26, 27), within the sequence alignments in PhosphoSitePlus.
Kinetic and statistical analyses were performed with Prism 5.0 (GraphPad, San Diego, CA, USA). Graphs were generated in Prism 5.0 and arranged in Illustrator 9.0 (Adobe, San Diego, CA, USA).
RESULTS
Phosphorylation of the human LRRK2 ROC domain by the ameba LRRK2 ortholog ROCO4
To obtain sufficient quantities of the isolated and phosphorylated ROC domain from human LRRK2 for functional biochemical analyses, we purified the ROC domain as an N-terminally His6-tagged protein from bacteria (22). When this ROC domain preparation was incubated with GST-[Δ970] LRRK2G2019S (Fig. 1B), we observed efficient phosphorylation of the isolated His6-ROC domain, consistent with previous results (6). This reaction was slow, but saturated after 5 h, and supplementation of additional ATP during the reaction did not result in additional phosphorylation. However, the ROC domain stably binds to several domains in human LRRK2 protein (4, 6, 28), making isolation of the phosphorylated ROC domain problematic. The kinase domain from an ortholog of LRRK2 in ameba (ROCO4) was recently crystallized and proposed as a biochemical and structural tool that would be useful in studies of human LRRK2 (9). With similar protein domain structure (Fig. 1A) and an overall sequence homology of ∼30% identity between the ROCO4 and human LRRK2 kinase domains, the 2 proteins are expected to fold similarly and retain basic enzymatic activities. We found that GST-ROCO4 phosphorylated the human ROC domain as efficiently as did human LRRK2 (Fig. 1B, C), but did not stably bind to His6-ROC. Thus, His6-pROC can be readily purified from GST-ROCO4 (Fig. 1D) for use in subsequent analyses.
Bands corresponding to His6-ROC (∼25 kDa; Fig. 1D) were excised and subjected to MS analysis to identify phosphorylated peptides and residues after trypsin or Lys-C protease digestion. Spectral counts of each peptide with respect to total peptide (phosphorylated peptide plus nonphosphorylated peptide) and phosphorylated peptide are shown in the 3 rightmost columns of Table 1. The (−) kinase column indicates the analysis of His6-ROC protein in control reactions that were not treated with GST-ROCO4. Amino acid positions are indicated with respect to full-length human LRRK2 protein (NCBI NP_940980.3), and the dissociation methods of collision-induced dissociation (CID) and electron-transfer dissociation (ETD) ion trap tandem MS are indicated. In this analysis, the ROC phosphopeptides from GST-ROCO4-phosphorylated ROC, and autophosphorylated GST-[Δ970] LRRK2G2019S, and full-length LRRK2 proteins were nearly identical, irrespective of the source of the kinase (Fig. 1E; Tables 1 and 2). The only difference we could detect with ROCO4-phosphorylated ROC was at residue 1452, where we had already annotated a phosphorylated threonine in GST-[Δ970] LRRK2G2019S.
TABLE 1.
Phosphorylated peptides identified by mass spectrometry in His6-ROC treated with ROCO4 kinase
| Spectral Counts |
|||||||
|---|---|---|---|---|---|---|---|
| Peptide sequence | Corresponding aa no. | XCorr score | Dissociation | Enzyme | Total | P (+) kinase | P (−) kinase |
| (K)LMIVGN pT GSGK(T) | T1343 | 3.4 | CID | Lys-C | 426 | 20 | 1 |
| (K)LMIVGNTGSGK pT TLLQQLMK(T) | T1348 | 7.7 | ETD | Trypsin | 331 | 20 | 0 |
| (K)LMIVGNTGSGKT pT LLQQLMK(T) | T1349 | 3.8 | CID | Trypsin | 331 | 11 | 0 |
| (K)TTLLQQLMK pT K(K) | T1357 | 3.5 | ETD | Trypsin | 53 | 35 | 0 |
| (K)KSDLGMQSA pT VGIDVK(D) | T1368 | 3.9 | CID | Trypsin | 1343 | 103 | 0 |
| (K)DLVLNVWDFAGREEFYS pT HPHFMTQR(A) | T1404 | 4.7 | ETD | Trypsin | 179 | 15 | 0 |
| (R)EEFYSTHPHFM pT QR(A) | T1410 | 3.4 | CID | Trypsin | 179 | 11 | 0 |
| (R)DYHFVNA pT EESDALAK(L) | T1491 | 5.0 | CID | Trypsin | 560 | 8 | 0 |
| (K)LRK pT IINESLNFK(I) | T1503 | 4.8 | ETD | Trypsin | 378 | 43 | 0 |
Phosphorylated residues are indicated by italicized font, and the corresponding amino acid position for the phosphorylated residue is with respect to NP_940980.3. XCorr, cross-correlation.
TABLE 2.
LRRK2 ROC autophosphorylation sites identified in vitro
Estimation of phosphorylation stoichiometry in His6-pROC by using spectral counts from total peptides (phospho and nonphospho) compared with spectral counts from phosphopeptides demonstrated that none of the 9 threonine sites is particularly abundant, despite the kinase reaction proceeding to saturation. Thr(P)1357 was the most heavily phosphorylated site in His6-pROC, consistent with a study that also found this site to be the most abundant phosphorylated residue in the ROC domain, by using full-length autophosphorylated LRRK2 enzyme (29).
To more accurately quantify overall ROC phosphorylation stoichiometries, γ-[32P]ATP was incorporated into the reactions. Despite the presence of multiple phosphoacceptor sites, an overall total of only 0.5 mole phosphate per mole His6-ROC domain was measured (Fig. 1F). We have reported this same overall autophosphorylation stoichiometry in GST-[Δ970] LRRK2G2019S (13). Therefore GST-ROCO4 is nearly as efficient in phosphorylating His6-ROC as GST-[Δ970] LRRK2G2019S is in autophosphorylation. We identified several threonine-to-alanine mutants (T1357A and T1348A-T1343A double mutant) that did not affect our ability to purify soluble His6-ROC protein. In these mutant His6-ROC proteins, overall ROCO4-directed phosphate incorporation was reduced by ∼30 and ∼20% (Fig. 1F), respectively. These data are consistent with our MS results and suggest that there is no single preferred phosphorylated residue in ROC that dominates; rather, this modification is distributed among the 9 sites identified. Thus, the kinase domains from ameba ROCO4 and human GST-[Δ970] LRRK2G2019S support phosphorylation of the isolated ROC domain of human LRRK2, which has similar kinetics and stoichiometries.
Enhanced GTPase activity in pROC and autophosphorylated GST-[Δ970] LRRK2G2019S
We measured GTPase activities and calculated steady-state kinetic parameters to generate a comparison of His6-ROC and His6-pROC. Initial velocities were plotted as a function of GTP concentration (Fig. 2A). We determined that His6-ROC has kcat (catalytic constant) = 0.007 ± 0.0004 min−1 and Km (Michaelis-Menton constant) = 154 ± 39 μM, comparable to previously published results for ROC (30). In contrast, His6-pROC had ∼2-fold increased GTPase activity with respect to His6-ROC, with a kcat of 0.016 ± 0.002 min−1 and a higher Km of 319 ± 48 μM. We also determined the maximum rates of GTPase activity. In the presence of 1 mM GTP, His6-ROC hydrolyzed GTP at 6.4 ± 0.1 nmol · min−1 per micromole protein, whereas His6-pROC hydrolyzed GTP at 12.3 ± 0.1 nmol · min−1 per micromole protein. Thus, both the turnover number (kcat) and velocity of His6-pROC were enhanced ∼2-fold, compared to His6-ROC (Fig. 2A, B; Table 3).
Figure 2.
ROC phosphorylation enhances its GTPase activity. A) Comparison of kinetics of GTP hydrolysis by ROC and pROC. Equal concentrations (20 μM) of His6-ROC or His6-pROC were assayed for GTP hydrolysis by using the various concentrations of GTP indicated. Data were fit to nonlinear regression kcat curves, and the rate constants were determined. B) Time course for GTP-hydrolysis by His6-ROC or His6-pROC (each at 20 μM) in the presence of saturating (1 mM) GTP. GTP off rates of His6-ROC and His6-pROC proteins (each at 0.3 μM) were measured using α-[32P]GTP (C) or [3H]GDP (D) and found to be indistinguishable. E) Time course for GTP hydrolysis by GST-[Δ970] LRRK2G2019S or pGST-[Δ970] LRRK2G2019S (each at 2 μM) in the presence of saturating (1 mM) GTP. All rate constants were calculated from the average of 3 independent experiments. Data are means ± sem; significances by unpaired Student’s t test.
TABLE 3.
Kinetic values for selected GTPase proteins
| GTPase | kcat | koff-GTP | koff-GDP | Reference |
|---|---|---|---|---|
| ROC | 0.007 | 0.093 | 0.148 | This study |
| 0.02 | 30 | |||
| pROC | 0.016 | 0.092 | 0.138 | This study |
| CDC42 | 1380 | 0.015 | 0.017 | 31 |
| Rac1 | 50.5 | 0.031 | 0.015 | 31 |
| RhoA | 95.9 | 0.01 | 0.029 | 31 |
| Ras | 0.0072 | 0.0026 | 0.0038 | 42–44 |
| Dynamin | 2.6 | 126 | 5580 | 45, 46 |
| Drp1 | 6.5 | 47 | ||
| Arl2 | 0.43 | 1.61 | 48 | |
| Arl3 | 2.1 | 0.01 | 48 |
Data are expressed per minute.
Because the rate at which GDP dissociates after GTP hydrolysis can be rate-limiting for turnover of a GTPase [e.g., Ras or Arf1 (31, 32)], we also assessed the impact of the phosphorylation of His6-ROC on this property. We measured the GDP off rate by using [3H]GDP as a ligand. The GDP off rates were determined to be koff-GDP (dissociation rate constant) = 0.148 ± 0.018 and 0.138 ± 0.015 min−1 for His6-ROC and His6-pROC, respectively (Fig. 2C; Table 3), and thus were not significantly different. We also measured the GTP off rates by using α-[32P]GTP. The koff-GTP was 0.093 ± 0.06 min−1 for ROC and 0.092 ± 0.01 min−1 for pROC, and no significant difference was observed (Fig. 2D; Table 3). Together, these data reveal that His6-ROC and His6-pROC display very similar guanine nucleotide–binding properties, making it likely that the ∼2-fold difference in GTPase activities resulted from a change in the rate of catalysis. It is also worth considering that stoichiometry of phosphorylation is only ∼0.5 for the His6-pROC domain and is considerably less than that at any one site, so that efficient phosphorylation of a key residue in a localized pool of LRRK2 would be expected to result in a much larger fold increase in the rate of GTP hydrolysis.
Given our results with the ROCO4-phosphorylated ROC domain, we next sought to replicate the result in the 200 kDa GST-[Δ970] LRRK2G2019S protein fragment that can incorporate ROC phosphorylation through autophosphorylation. The nonphosphorylated GST-[Δ970] LRRK2G2019S hydrolyzed GTP at 19.5 ± 0.8 nmol · min−1 per micromole protein in the presence of 1 mM GTP, comparable to the GTP hydrolysis velocity of His6-ROC (Fig. 2E). The phosphorylated GST-[Δ970] LRRK2G2019S showed significantly increased GTP hydrolysis activity of 39.6 ± 0.4 nmol · min−1 per micromole protein, a ∼2-fold increase consistent with increases observed with His6-pROC.
Mutation analysis of the GTP-hydrolysis pocket
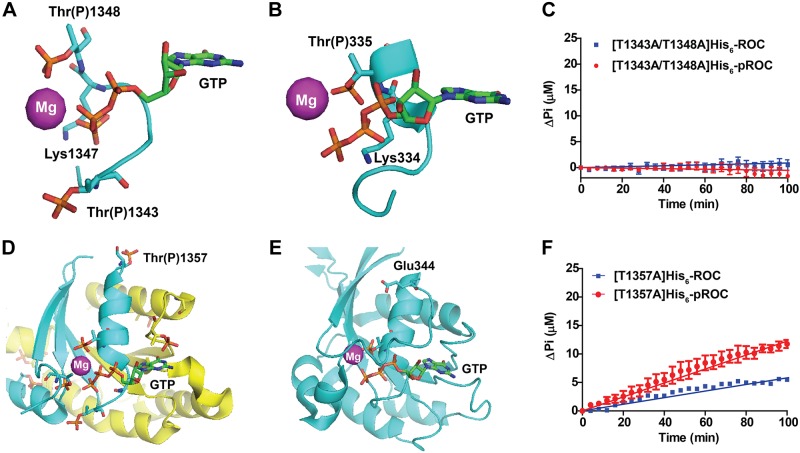
Mass spectrometry analysis of His6-pROC showed 3 phosphorylation sites that are in proximity to the Mg2+-binding pocket: Thr(P)1368, Thr(P)1343, and Thr(P)1348. The introduction of phosphates at these residues, either 1 at a time or in combination, would be expected to cause conformational changes in the guanine nucleotide-binding pocket. To understand what these changes might be, we used the crystal structure of the ROC GDP-bound dimer to model them. Although it was suggested that this structure could be critically confounded by a domain-swapped crystal artifact (33), the phosphate-binding pocket in the ROC domain crystal structure that is subject to phosphorylation is contributed by only 1 chain of the dimer, and structural alignment of this region against Ras demonstrates strong structural similarity for this part of the protein (22). Addition of phosphates to either Thr1348 or Thr1343 p-loop residues resulted in a steric clash with the bound Mg2+. Because enhanced, not diminished, GTPase activity is associated with His6-pROC, additional conformational changes could occur to accommodate the addition of phosphate groups to the p-loop. Molecular sculpting of the pROC model showed that with a shift of the p-loop away from the pocket, Mg2+ fits adjacent to the phosphates into a novel salt bridge interaction network (Fig. 3A). Thr(P)1348 plausibly interacts with Mg2+, whereas Thr(P)1343 may promote Mg2+ action with the catalytic lysine residue.
Figure 3.
Requirement for ROC p-loop phospho-residues for GTPase activity. A) Structural model based on human LRRK2 ROC domain crystal structure (PDB ID: 2zej) shows possible involvement of the 1343 and 1348 phosphorylation substrate residues in recruiting Mg2+ to the GTP-binding pocket, with respect to the catalytic lysine. B) Structural model based on M. barkeri ROCO2 (PDB ID 4wnr) with the Thr1348 equivalent residue Thr335. Phosphorylation of Thr335 may change the interaction in the Mg2+-binding pocket. C) GTPase activity of p-loop site mutants T1343A and T1348A in His6-ROC and His6-pROC. No hydrolysis activity was detected. D) The Thr1357 phosphoresidue appeared to be the preferential (i.e., most abundant) phosphorylated residue in His6-pROC and is distant from the GTP-binding pocket. Phosphorylation of this residue may induce a conformational change on adjacent α-helices (blue and yellow) that alter metal or GTP binding. E) Similar to the human LRRK2 ROC domain structure, Thr1357 equivalent residue Glu344 in M. barkeri ROCO2 is distant from the GTP-binding pocket. F) GTPase activity of the most abundant phospho-residue Thr1357 mutated to alanine (T1357A) in His6-ROC and His6-pROC protein. T1357A and pT1357A showed different GTP hydrolysis velocities of hydrolyzed GTP at 3.1 ± 0.05 and 6.4 ± 0.15 nmol · min−1 per micromole protein, respectively. P < 0.001, unpaired Student's t test, calculated from 3 independent experiments. All GTPase reactions were conducted in the presence of 1 mM GTP at each time point. All traces are calculated from 3 independent experiments. Data are means ± sem. Atoms on key residues are color coded (magnesium: magenta; phosphorus: orange; oxygen: red; nitrogen: blue; carbon on protein: cyan; carbon on GTP: green).
Recently, a crystal structure for an LRRK2 homolog in Methanosarcina barkeri, known as ROCO2 (PDB ID: 4wnr), was identified that shows a p-loop conformation similar to that of the GDP-bound human LRRK2 ROC dimer (34). The residue equivalent to human LRRK2 Thr1348 on ROCO2 (Thr335) interacts directly with Mg2+ (Fig. 3B). Phosphorylation of Thr335 in ROCO2 would also result in a steric clash with the bound Mg2+, and similar to ROC, molecular sculpting suggests the presence of a novel salt bridge that recruits metal. To test the importance of these p-loop residues for GTPase activity, we generated T1343A-T1348A double mutant His6-ROC protein. The T1343A-T1348A mutant ROC is highly soluble but enzymatically dead (Fig. 3C). These experiments demonstrate that Thr1343 and Thr1348 are critical for catalysis, and there can be no substitutions for them.
Thr1357 is the most abundant phosphorylated ROC residue in LRRK2, yet the residue is distant from the catalytic pocket (Fig. 3D). Likewise, the equivalent residue Glu344 on ROCO2 is distant from the catalytic pocket (Fig. 3E). To investigate the potential role of this residue in catalysis, we purified [T1357A]His6-ROC and [T1357A]His6-pROC and measured their GTPase activities, with and without phosphorylation (Fig. 3F). We again observed a ∼2-fold increase in GTPase activity after incubation of [T1357A]His6-ROC with GST-ROCO4 kinase (3.1 ± 0.05 and 6.4 ± 0.15 nmol GTP hydrolyzed per minute per micromole protein). Because the overall velocities of the [T1357A]His6-ROC preparations were half those of His6-ROC, there were likely structural constraints outside the guanine nucleotide–binding pocket that exerted changes that were reflected in diminished catalytic activity.
Phosphorylation enhances dimerization of GDP-bound ROC
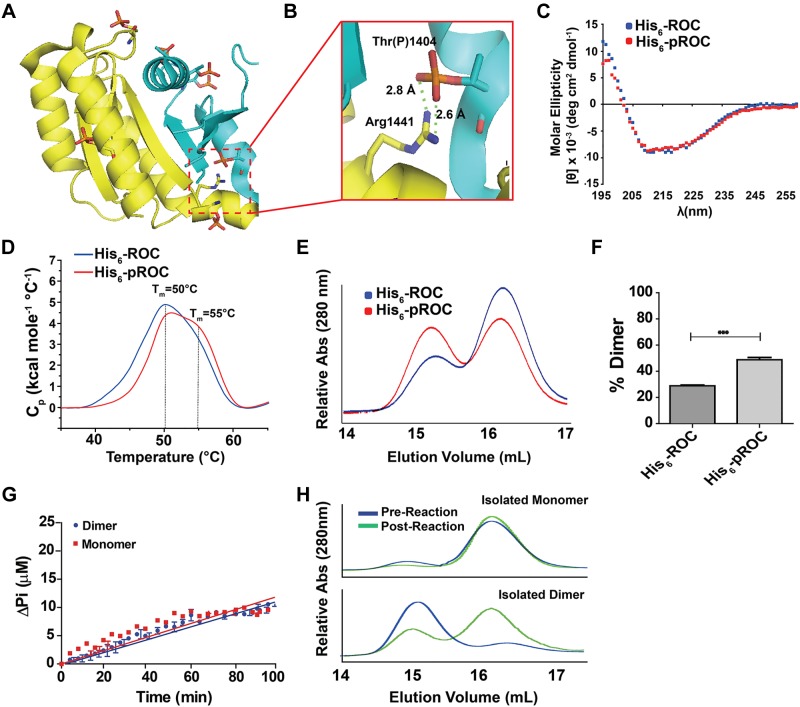
Dimerization of LRRK2 or the isolated ROC domain may be a primary mechanism for the regulation of ROC GTPase activity. Deng et al. (22) proposed that dimerization is necessary for GTP hydrolysis, based in part on the dimer observed in the ROC crystal structure. Because we observed enhanced GTPase activity with His6-pROC, we investigated whether phosphorylation of ROC influences dimerization. Modeling the phosphorylated residues identified in Fig. 1E on the ROC dimer crystal structure showed that none of the phosphorylation sites interrupted the interaction between the 2 monomers (Fig. 4A). Instead, Thr(P)1404 may have formed a salt bridge with Arg1441 on the paired monomer that enhanced dimerization (Fig. 4B). In addition, phosphorylation at other sites may have evoked conformational changes that influenced dimerization.
Figure 4.
Phosphorylation of ROC enhances dimer formation. A) Overlay of all phosphorylation occurring in His6-pROC based on ROC crystal structure (PDB ID 2zej). Yellow and cyan represent chain A and B in the dimer, respectively. Atoms on key residues are color coded (phosphorus: orange, oxygen: red, nitrogen: blue, carbon: yellow/cyan). B) Enlargement of the dimerization interface, with the calculated distances indicated. The Thr(P)1404 is predicted to form a salt bridge with the Arg1441 residue. C) CD profiles. No secondary structure differences were detected between His6-ROC and His6-pROC. D) Differential scanning calorimetry profiles. Both His6-ROC and His6-pROC displayed peaks at ∼50°C, whereas the His6-pROC also had a broad shoulder peak with predicted Tm ≅ 55°C. C, D) Traces are representative of at least 3 independent experiments. E) Representative elution profiles of His6-ROC and His6-pROC, immediately after a 3 h kinase reaction (Fig. 1B). E) GTPase assay with purified monomer and dimer His6-ROC. Near equivalent velocities of hydrolyzed GTP at 5.8 ± 0.1 and 6.1 ± 0.2 nmol · min−1 per micromole protein, respectively, were noted. F) Comparative analysis of the proportion of dimer to monomer. Maximum relative peak heights were calculated from 3 independent runs and are displayed as column graphs with bars representing sem. ***P < 0.00; unpaired t test. G) Elution profiles of purified monomer (top) and dimer (bottom) for His6-ROC before (blue) and after (green) the 2 h GTPase assay. Profiles shown are representative of 3 independent runs.
Because Thr1357 and other residues distant from the GTP-binding pocket (e.g., Thr1404) can alter GTPase activity, we hypothesized that ROC phosphorylation causes an overall conformational change that affects GTPase activity. CD did not detect any differences between His6-ROC and His6-pROC (Fig. 4C). Differential scanning calorimetry revealed that both preparations displayed melting temperature (Tm) of ∼50°C, but the His6-pROC data included a broadened shoulder and second peak at ∼55°C (Fig. 4D). Gel filtration analysis showed that the percentage of dimers in His6-pROC (48.7 ± 1.9%) increased ∼1.7-fold compared to His6-ROC (28.8 ± 0.7%; Fig. 4E, F). These results are consistent with an enhanced proportion of a more thermostable dimer present in the phosphorylated preparation of the ROC domain.
To test the hypothesis that the increased proportion of dimer in His6-pROC underlies the increased catalytic activity (Fig. 2), we used gel filtration to purify preparations of ROC monomers and dimers and compared GTP hydrolysis activity. In the presence of 1 mM GTP, monomeric His6-ROC and dimeric His6-ROC preparations showed near equivalent GTPase activities of hydrolyzed GTP at 5.8 ± 0.1 and 6.1 ± 0.2 nmol · min−1 per micromole protein, respectively, and these were not significantly different (P > 0.1; Fig. 4G). It is noteworthy that, although our monomer preparation appeared reasonably stable and remained monomeric throughout the GTPase reaction, a proportion of dimer dissociated into monomer during the GTPase reaction (Fig. 4H). Although we cannot rule out subtle effects of the dimer on GTPase activity, the data suggest that there are no striking differences in GTPase activity between dimer or monomer conformations.
In silico predictions of phosphorylation modifications of GTP hydrolysis pockets
Because of recent advances in MS, several well-curated phosphoproteome databases are now publically available. We searched these for phosphorylation sites on GTPases and identified 42 sties with p-loop phosphorylation annotations comparable to the Thr(P)1343 and Thr(P)1348 modifications found in LRRK2 (Table 4). Some of these proteins are well-described members of the Rab, Rho, Arf, Ras-like, dynamin-like, and heterotrimeric G-protein family. Overlay of the phosphorylation modifications in well-annotated crystal structures all resulted in significant steric or electrostatic clashes that would be predicted to destroy the binding of metal and nucleotide and consequently kill the catalytic activity of the GTPase (Fig. 5). However, by shifting the p-loop conformation, the phosphate groups aligned into a network where they could coordinate Mg2+ through salt bridges or adjust the position of bound GTP through electrostatic repulsion. Further experimental data would be needed to validate these models and test the possible functional effects of phosphorylation on these GTPase proteins.
TABLE 4.
GTPases annotated in phosphoproteome databases
| Family | Protein | p-Loop |
|---|---|---|
| ROC | LRRK2 | GN T GSGK T T |
| INA | GIMAP2 | GKTGTGK S A |
| GIAMP5 | GK T GCGKSA | |
| Ras-like | ATPBD1C | GPAG S GKST |
| DIRAS2 | GAGGVGK S S | |
| Arf | SAR1B | GQT T GKGSI |
| Rho | RhoA | GDGACGK T C |
| RhoC | GDGACGK T C | |
| Rab | Ran | GDGG T GK T T |
| Rab15 | GD S GVGKTC | |
| Rab17 | GSG S VGKSS | |
| Rab1A | GD S GVGKSC | |
| Rab1B | GD S GVGKSC | |
| Rab2 | GD T GVGKSC | |
| Rab22A | GD T GVGKSS | |
| Rab24 | GKE Y VGKTS | |
| Rab2B | GD T GVGKSC | |
| Rab33A | GDSNVGK T C | |
| Rab34 | GDL S VGKTC | |
| Rab35 | GDSGVGK S S | |
| Rab38 | GDLGVGK T S | |
| Rab3A | GNSSVGK T S | |
| Rab4 | GNAG T GKSC | |
| Rab40AL | GDRDVGK S E | |
| Rab41 | GEQSVGK T S | |
| Rab5A | GE S AVGKSS | |
| Rab7 | GD S GVGKTS | |
| Rab8A | GD S GVGKTC | |
| Rab8B | GD S GVGKTC | |
| RabL2A | GDSAVGK S K | |
| RabL2B | GDSAVGK S K | |
| RabL3 | GD S GVGKSS | |
| Dynamin-like | Opa1 | GDQ S AGK T S |
| Drp1 | GTQSSGKS S | |
| Heterotrimeric G protein | G α (s) | GAGE S GKST |
| G α 12 | GAGE S GKST | |
| G α i1 | GAGE S GKST | |
| G α i2 | GAGE S GKST | |
| G α t1 | GAGE S GKST | |
| G α t2 | GAGE S GKST | |
| G α t3 | GAGE S GKST | |
| G α (o) | GAGE S GKST | |
| G α (olf) | GAGE S GKST | |
| G α (q) | GTGESGK S T | |
| G α (z) | GT S NSGKST |
Phosphorylation sites in p-loop residues are shown in italicized font.
Figure 5.
In silico models of phosphorylated p-loop residues in other GTPases. Phosphorylated residues imposed on crystal structures of RAN (A), RhoA (B), and G(α)q (C). Steric (black arrows) and electrostatic (red arrows) clashes are indicated. Molecular sculpting of the structures to prevent clashes shows possible salt-bridge networks that may stabilize and promote Mg2+ binding. Atoms on key residues are color coded (magnesium: magenta; phosphorus: orange; oxygen: red; nitrogen: blue; carbon on protein: cyan; carbon on GTP/GDP: green; sulfur: yellow; aluminum: gray; and fluorine: pale cyan).
DISCUSSION
Our results center on 4 novel observations. First, the evolutionarily distant ROCO4 protein kinase phosphorylated the human LRRK2 ROC domain with substrate specificity that almost precisely matched that of LRRK2 autophosphorylation of its ROC domain. Second, phosphorylation of the ROC domain stimulated GTP hydrolysis without affecting nucleotide affinity. To our knowledge, this is the first report of p-loop phosphorylation that regulates GTP catalytic activity of any GTPase. Third, phosphorylation of the ROC domain promoted protein dimerization, although the ROC monomer possessed catalytic activity comparable to that of the ROC dimer. Fourth, phosphorylations analogous to those found in ROC were found in a diverse group of GTPase proteins, suggesting a more general means of kinase-mediated control of GTPase proteins.
LRRK2-mediated autophosphorylation of the ROC GTPase domain has been observed by using several different experimental approaches. First, several phosphopeptides were identified directly within a kinase-active LRRK2 fragment, GST-[Δ970] LRRK2, and the same phosphopeptides were identified in GST-ROC domain incubated with GST-[Δ970] LRRK2 (6). A second study (21) purified full-length GST-LRRK2 directly from mammalian cells, incubated the recombinant protein in a kinase assay, and subsequently identified the same phosphorylated residues in the ROC domain as the first study. Likewise, we independently identified these sites in both GST-[Δ970] LRRK2 and GST-[Δ970] G2019S-LRRK2, which harbors the pathogenic G2019S PD mutation (13).
Although ROC phospho residues are identified in multiple in vitro studies (Tables 1 and 2), the detection of phosphorylation modifications in vivo has been challenging. First, the stoichiometry of phosphorylation on each phospho residue is low, even in vitro, thereby challenging most phospho-specific antibodies that still have the potential to interact with non-phospho peptides. Second, ROC phosphorylation may be subject to rapid dephosphorylation in cellular lysates by abundant and active phosphatases, especially during tissue processing and LRRK2 purification (13). Third, kinase-active LRRK2 phosphorylated in the ROC domain may reside in cellular compartments that are challenging to isolate from cells and tissues, and the process may require harsh detergents that hinder the ability to affinity purify or enrich autophosphorylated LRRK2 protein for analysis. However, a recent study demonstrated that the Thr1357 residue is the most abundantly phosphorylated site in the ROC domain, and antibodies directed to Thr(P)1357 could detect this modification directly from cellular lysates (29). The authors of that study also found that a brief incubation of the cellular lysates with ATP dramatically up-regulates ROC phosphorylation. Thr(P)1357 is also the only ROC phosphorylation identified by global phosphoproteomic analysis, as annotated in the PhosphoSitePlus database. Our own attempts to detect other sites such as Thr(P)1503 and Thr(P)1343 directly from cellular lysates were not successful (13), but these residues show a much lower stoichiometry of phosphorylation. Antibodies directed to Thr(P)1357 may be used in future studies to help illuminate the stoichiometry of ROC phosphorylation in particular tissues and cells and the conditions necessary to measure LRRK2 kinase activation.
LRRK2 can be purified as a homodimer from mammalian cells and tissue, and ROC phosphorylation activity is associated with dimerization (5). Although kinase-active and kinase-dead LRRK2 can each form homodimers, kinase-active LRRK2 does not phosphorylate kinase-dead LRRK2 proteins (4). However, when the isolated His6-ROC domain from human LRRK2 is used in kinase reactions with human GST-[Δ970] LRRK2 or GST-ROCO4, 9 of 10 threonines are phosphorylated in trans. We speculate that the ROC domain is normally conformationally constrained by other domains in LRRK2, to prevent other LRRK2 proteins from phosphorylating ROC residues. In contrast, when the ROC domain is presented on its own to the LRRK2 kinase or is conformationally altered, the threonine sites are subject to phosphorylation. As expected, the ROC domain stably interacts with other domains within LRRK2 (6), and we were therefore unable to isolate phosphorylated His6-ROC protein from the full-length LRRK2 protein for use in biochemical assays. Whereas the isolated kinase domain of human LRRK2 is inactive, the ROCO4 kinase domain is highly active (9, 35). Because we found that the ROCO4 kinase phosphorylates His6-ROC at the same residues as the full-length human protein, and because ROCO4 does not stably interact with His6-ROC, we were able to purify His6-pROC for the biochemical analyses performed in the current study.
The ROC domain shares the highest sequence similarity to members of the Ras GTPase superfamily (36). However, kinetic values of nucleotide exchange and GTP hydrolysis activity are highly variable in the Ras superfamily, as well as for other GTPases (Table 3). ROC and pROC have kcat values similar to Ras GTPase, but their nucleotide off rates are closer to members of the ARF family (e.g., Arl2, Table 3). As the nucleotide off rate of ROC is high, there appears to be no clear requirement for a guanine nucleotide exchange factor among Ras superfamily GTPases to speed the release of tightly bound GDP. In contrast, the intrinsic GTPase activity associated with ROC is low (kcat = 0.007 min−1 for His6-ROC and 0.016 min−1 for His6-pROC). Such a slow rate of intrinsic GTP hydrolysis is consistent with the existence of other factors capable of speeding hydrolysis, comparable to the GTPase-activating proteins that are downstream of many Ras superfamily GTPases that can act as effectors and terminators in signaling (37, 38). We therefore hypothesize that the primary regulatory proteins of LRRK2 enzymatic activity are GTPase-activating proteins that have yet to be identified.
We observed a ∼2-fold enhancement of GTPase activity by His6-pROC compared to its unphosphorylated form. We also observed a ∼2-fold enhancement of GTPase activity in pGST-[Δ970] LRRK2G2019S compared to its unphosphorylated form. We attribute these results to differences in the rate of catalysis (i.e., cleavage of the β–γ phosphate bond), as no differences were observed in the rates of dissociation of GDP. If the increased GTPase activity results from phosphorylation of a key residue in the ROC domain, and if we assume that phosphate, at 0.5 mole per mole ROC, is spread equally among the 9 residues modified, then complete phosphorylation of such a residue might be expected to increase GTPase rates by 36-fold. This magnitude of change is well within the limits of what one would predict for biologically important regulation. Unfortunately, substitutions of amino acids were not tolerated for most threonine residues in the ROC domain including p-loop residues. However, our discovery that the evolutionarily distant ROCO4 kinase directs ROC phosphorylation in the same manner as human LRRK2 kinase gives further credence to ROC phosphorylation as a biologically important feature of LRRK2.
There has been a debate within the field as to whether the LRRK2 ROC domain can exist as a dimer and whether its quaternary state plays a direct role in regulating its GTPase activity (22, 33, 39). Although it has been reported that ROC exists only as a monomer, our results suggest that both ROC and pROC are in equilibrium between dimer and monomer. Of interest, a larger percentage of pROC (at the end of our kinase reactions) was found to gel filter at a size consistent with a homodimer, compared to ROC incubated under the same conditions, but without the kinase. Because we observed that pROC has both an increased propensity toward forming dimers and enhanced GTPase activity, we speculated that dimerization regulates ROC protein GTPase activity. In agreement with our hypothesis, Deng, et al. (22) reported that the pathogenic R1441G mutation, which has a decreased propensity to form a dimer, shows decreased GTPase activity. However, in our GTPase assays, both the monomeric and dimeric ROC preparations had equivalent GTPase capability. These results suggest that dimerization of ROC/pROC does not directly regulate GTP hydrolysis activity. Instead, ROC conformation may be important for ROC interaction with other proteins or in the context of the full LRRK2 protein.
In an effort to begin to assess the prevalence of phosphorylation as a regulatory factor in GTP-binding pockets, we searched phosphoproteomics databases for GTPases and then for phosphopeptides in the conserved GTP-binding motifs. We identified 42 other GTPases found to be phosphorylated in p-loop residues in phosphoproteomics databases. Twenty-four of them are members of the Rab family, and 11 are members of the heterotrimeric family of GTPases. Phosphorylation of p-loop residues may be a more common means of regulation than is currently appreciated. Our search was not exhaustive, and not every GTPase is currently represented in proteomic databases. However, several of the GTPases we identified have associated high-resolution structures available, and in every case, the p-loop phosphorylations are predicted to clash sterically with the nucleotide or magnesium in the binding pocket. In silico modeling can predict the types of conformational changes that might occur, but unfortunately cannot predict the overall effect on GTPase activity. Among the 42 GTPases identified with phosphorylated p-loops, the dynamin-like family proteins OPA1 and Drp1 have been postulated to be LRRK2 kinase substrates (40). Rab7 was reported to be a binding partner with LRRK2 (28, 41). We hypothesize that LRRK2, along with many other protein kinases, serves as a regulator of GTPase activity in important signal transduction cascades.
Acknowledgments
The authors thank C. Deivanayagam and D. Chattopadhyay [Center for Structural Biology, Department of Optometry, The University of Alabama at Birmingham (UAB)] for useful discussions and guidance in the experiments. This work was supported by U.S. National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke Grant R01 NS064934 (to A.B.W.); NIH National Cancer Institute Grant P30 CA13148 (to L.J.D.); and by the UAB Comprehensive Cancer Center Mass Spectrometry and Proteomics Shared Facility. The authors declare no conflicts of interest.
Glossary
- CD
circular dichroism
- CID
collision-induced dissociation
- ETD
electron-transfer dissociation
- kcat
catalytic constant
- koff
dissociation rate constant
- Km
Michelis-Menten constant
- LRRK
leucine-rich repeat kinase
- MS
mass spectrometry
- PD
Parkinson disease
- Pi
inorganic phosphate
- pROC
phosphorylated ROC
- ROC
Ras-of-complex
- Ser(P)
phosphorylated serine
- Thr(P)
phosphorylated threonine
REFERENCES
- 1.Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44, 595–600 [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 [DOI] [PubMed] [Google Scholar]
- 3.Cookson M. R. (2010) The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat. Rev. Neurosci. 11, 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J. M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., Thomas K. J., Baekelandt V., Cookson M. R. (2008) The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 283, 16906–16914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen S., Webber P. J., West A. B. (2009) Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 284, 36346–36356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greggio E., Taymans J. M., Zhen E. Y., Ryder J., Vancraenenbroeck R., Beilina A., Sun P., Deng J., Jaffe H., Baekelandt V., Merchant K., Cookson M. R. (2009) The Parkinson’s disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochem. Biophys. Res. Commun. 389, 449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Civiero L., Vancraenenbroeck R., Belluzzi E., Beilina A., Lobbestael E., Reyniers L., Gao F., Micetic I., De Maeyer M., Bubacco L., Baekelandt V., Cookson M. R., Greggio E., Taymans J. M. (2012) Biochemical characterization of highly purified leucine-rich repeat kinases 1 and 2 demonstrates formation of homodimers. PLoS One 7, e43472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger Z., Smith K. A., Lavoie M. J. (2010) Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry 49, 5511–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilsbach B. K., Ho F. Y., Vetter I. R., van Haastert P. J., Wittinghofer A., Kortholt A. (2012) Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations. Proc. Natl. Acad. Sci. USA 109, 10322–10327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West A. B., Moore D. J., Choi C., Andrabi S. A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K. L., Dawson V. L., Dawson T. M. (2007) Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 16, 223–232 [DOI] [PubMed] [Google Scholar]
- 11.Li X., Tan Y. C., Poulose S., Olanow C. W., Huang X. Y., Yue Z. (2007) Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J. Neurochem. 103, 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis P. A., Greggio E., Beilina A., Jain S., Baker A., Cookson M. R. (2007) The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357, 668–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webber P. J., Smith A. D., Sen S., Renfrow M. B., Mobley J. A., West A. B. (2011) Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J. Mol. Biol. 412, 94–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C. E., Dominguez S. L., Solanoy H. O., Drummond J., Zhang X., Ding X., Cai F., Song Q., Li X., Yue Z., van der Brug M. P., Burdick D. J., Gunzner-Toste J., Chen H., Liu X., Estrada A. A., Sweeney Z. K., Scearce-Levie K., Moffat J. G., Kirkpatrick D. S., Zhu H. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med. 4, 164ra161 [DOI] [PubMed] [Google Scholar]
- 15.Ray S., Bender S., Kang S., Lin R., Glicksman M. A., Liu M. (2014) The Parkinson disease-linked LRRK2 protein mutation I2020T stabilizes an active state conformation leading to increased kinase activity. J. Biol. Chem. 289, 13042–13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. USA 102, 16842–16847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop A. L., Hall A. (2000) Rho GTPases and their effector proteins. Biochem. J. 348, 241–255 [PMC free article] [PubMed] [Google Scholar]
- 18.Avruch J., Zhang X. F., Kyriakis J. M. (1994) Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem. Sci. 19, 279–283 [DOI] [PubMed] [Google Scholar]
- 19.Smith W. W., Pei Z., Jiang H., Dawson V. L., Dawson T. M., Ross C. A. (2006) Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 9, 1231–1233 [DOI] [PubMed] [Google Scholar]
- 20.Taymans J. M., Vancraenenbroeck R., Ollikainen P., Beilina A., Lobbestael E., De Maeyer M., Baekelandt V., Cookson M. R. (2011) LRRK2 kinase activity is dependent on LRRK2 GTP binding capacity but independent of LRRK2 GTP binding. PLoS One 6, e23207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloeckner C. J., Boldt K., von Zweydorf F., Helm S., Wiesent L., Sarioglu H., Ueffing M. (2010) Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. J. Proteome Res. 9, 1738–1745 [DOI] [PubMed] [Google Scholar]
- 22.Deng J., Lewis P. A., Greggio E., Sluch E., Beilina A., Cookson M. R. (2008) Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl. Acad. Sci. USA 105, 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maertens G. N., Cook N. J., Wang W., Hare S., Gupta S. S., Öztop I., Lee K., Pye V. E., Cosnefroy O., Snijders A. P., KewalRamani V. N., Fassati A., Engelman A., Cherepanov P. (2014) Structural basis for nuclear import of splicing factors by human Transportin 3. Proc.Natl. Acad. Sci. USA 111, 2728–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273, 9656–9666 [DOI] [PubMed] [Google Scholar]
- 25.Lyon A. M., Begley J. A., Manett T. D., Tesmer J. J. (2014) Molecular mechanisms of phospholipase C beta3 autoinhibition. Structure 22, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saraste M., Sibbald P. R., Wittinghofer A. (1990) The P-loop: -a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 27.Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 28.Beilina, A., Rudenko, I. N., Kaganovich, A., Civiero, L., Chau, H., Kalia, S. K., Kalia, L. V., Lobbestael, E., Chia, R., Ndukwe, K., Ding, J., Nalls, M. A.; International Parkinson's Disease Genomics Consortium; North American Brain Expression Consortium; Olszewski, M., Hauser, D. N., Kumaran, R., Lozano, A. M., Baekelandt, V., Greene, L. E., Taymans, J. M., Greggio, E., and Cookson, M. R. (2014) Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc.Natl. Acad. Sci. USA 111, 2626–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamikawaji S., Ito G., Sano T., Iwatsubo T. (2013) Differential effects of familial parkinson mutations in LRRK2 revealed by a systematic analysis of autophosphorylation. Biochemistry 52, 6052–6062 [DOI] [PubMed] [Google Scholar]
- 30.Liao J., Wu C. X., Burlak C., Zhang S., Sahm H., Wang M., Zhang Z. Y., Vogel K. W., Federici M., Riddle S. M., Nichols R. J., Liu D., Cookson M. R., Stone T. A., Hoang Q. Q. (2014) Parkinson disease-associated mutation R1441H in LRRK2 prolongs the “active state” of its GTPase domain. Proc. Natl. Acad. Sci. USA 111, 4055–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B., Zhang Y., Wang Z., Zheng Y. (2000) The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J. Biol. Chem. 275, 25299–25307 [DOI] [PubMed] [Google Scholar]
- 32.Burstein E. S., Macara I. G. (1992) Interactions of the ras-like protein p25rab3A with Mg2+ and guanine nucleotides. Biochem. J. 282, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotthardt K., Weyand M., Kortholt A., Van Haastert P. J., Wittinghofer A. (2008) Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 27, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terheyden S., Ho F. Y., Gilsbach B. K., Wittinghofer A., Kortholt A. (2015) Revisiting the Roco G-protein cycle. Biochem. J. 465, 139–147 [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., Galemmo R. A. Jr., Fraser K. B., Moehle M. S., Sen S., Volpicelli-Daley L. A., DeLucas L. J., Ross L. J., Valiyaveettil J., Moukha-Chafiq O., Pathak A. K., Ananthan S., Kezar H., White E. L., Gupta V., Maddry J. A., Suto M. J., West A. B. (2014) Unique functional and structural properties of the LRRK2 protein ATP-binding pocket. J. Biol. Chem. 289, 32937–32951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anand V. S., Braithwaite S. P. (2009) LRRK2 in Parkinson’s disease: biochemical functions. FEBS J. 276, 6428–6435 [DOI] [PubMed] [Google Scholar]
- 37.East M. P., Kahn R. A. (2011) Models for the functions of Arf GAPs. Semin. Cell Dev. Biol. 22, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahn R. A. (2011) GAPs: Terminator versus effector functions and the role(s) of ArfGAP1 in vesicle biogenesis. Cell. Logist. 1, 49–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. (2009) It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 40.Stafa K., Tsika E., Moser R., Musso A., Glauser L., Jones A., Biskup S., Xiong Y., Bandopadhyay R., Dawson V. L., Dawson T. M., Moore D. J. (2014) Functional interaction of Parkinson’s disease-associated LRRK2 with members of the dynamin GTPase superfamily. Hum. Mol. Genet. 23, 2055–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez-Suaga P., Rivero-Ríos P., Fdez E., Blanca Ramírez M., Ferrer I., Aiastui A., López De Munain A., Hilfiker S. (2014) LRRK2 delays degradative receptor trafficking by impeding late endosomal budding through decreasing Rab7 activity. Hum. Mol. Genet. 23, 6779–6796 [DOI] [PubMed] [Google Scholar]
- 42.Gibbs J. B., Ellis R. W., Scolnick E. M. (1984) Autophosphorylation of v-Ha-ras p21 is modulated by amino acid residue 12. Proc. Natl. Acad. Sci. USA 81, 2674–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azarani A., Goltzman D., Orlowski J. (1996) Structurally diverse N-terminal peptides of parathyroid hormone (PTH) and PTH-related peptide (PTHRP) inhibit the Na+/H+ exchanger NHE3 isoform by binding to the PTH/PTHRP receptor type I and activating distinct signaling pathways. J. Biol. Chem. 271, 14931–14936 [DOI] [PubMed] [Google Scholar]
- 44.Hall B. E., Bar-Sagi D., Nassar N. (2002) The structural basis for the transition from Ras-GTP to Ras-GDP. Proc. Natl. Acad. Sci. USA 99, 12138–12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonard M., Song B. D., Ramachandran R., Schmid S. L. (2005) Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol. 404, 490–503 [DOI] [PubMed] [Google Scholar]
- 46.Binns D. D., Barylko B., Grichine N., Atkinson M. A., Helms M. K., Jameson D. M., Eccleston J. F., Albanesi J. P. (1999) Correlation between self-association modes and GTPase activation of dynamin. J. Protein Chem. 18, 277–290 [DOI] [PubMed] [Google Scholar]
- 47.Koirala S., Guo Q., Kalia R., Bui H. T., Eckert D. M., Frost A., Shaw J. M. (2013) Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA 110, E1342–E1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanzal-Bayer M., Linari M., Wittinghofer A. (2005) Properties of the interaction of Arf-like protein 2 with PDEdelta. J. Mol. Biol. 350, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 49.Kamikawaji S., Ito G., Iwatsubo T. (2009) Identification of the autophosphorylation sites of LRRK2. Biochemistry 48, 10963–10975 [DOI] [PubMed] [Google Scholar]