Abstract
Recent studies have identified family with sequence similarity member 20C (FAM20C) as a kinase that phosphorylates the Ser in Ser-X-Glu/phospho-Ser (pSer) motifs in the small-integrin–binding ligand N-linked glycoproteins (SIBLINGs). There is no in vivo evidence that validates this finding, and it is unclear whether FAM20C is the only kinase for SIBLINGs. We extracted bone noncollagenous proteins (NCPs) from Fam20C-knockout (KO) mice and analyzed the phosphorylation levels. The total NCPs were separated into osteopontin-, bone sialoprotein-, and dentin matrix protein-1–enriched fractions by anion-exchange chromatography and analyzed by SDS-PAGE, native PAGE, and Western immunoblot analysis. The NCP phosphorylation level in the KO mice was lower than that in the wild-type (WT). On the native gel, the SIBLINGs from KO mice showed a lower migration rate (Mr) than those from the WT. Calf intestine phosphatase treatment shifted SIBLINGs from the WT mice to the level adjacent to the KO, but failed to shift the latter, suggesting a phosphorylation loss of SIBLINGs in the KO mice. Mass spectrometry identified less pSers in the SIBLINGs from the KO mice [including the region of the acidic Ser- and aspartate-rich motif (ASARM) peptides]. In an intriguing finding, several pSers in the Ser-X-Glu motifs in the KO mice maintained their phosphorylation, whereas several others in non–Ser-X-Glu motifs did not. Phospho-Tyrs and phospho-Thrs in the SIBLINGs did not appear to be associated with FAM20C. Our results indicate that FAM20C is the primary, but not the only, kinase for the SIBLINGs.—Yang, X., Yan, W., Tian, Y., Ma, P., Opperman, L. A., Wang, X. Family with sequence similarity member 20C is the primary but not the only kinase for the small-integrin–binding ligand N-linked glycoproteins in bone.
Keywords: phosphorylation, biomineralization, mass spectrometry, hypophosphatemic rickets, ASARM
The organic phase of the extracellular matrix (ECM) in the bone is mainly composed of type I collagen and noncollagenous proteins (NCPs) synthesized and secreted by osteoblasts. A prominent category of NCPs in bone ECM is termed the small-integrin–binding ligand N-linked glycoprotein (SIBLING) family, which includes osteopontin (OPN), bone sialoprotein (BSP), and dentin matrix protein (DMP)-1. These polyanionic proteins play pivotal roles in osteogenesis and biomineralization, and their biologic functions are closely related to posttranslational modifications, such as phosphorylation (1–7).
OPN is commonly recognized as an effective inhibitor of hydroxyapatite formation and growth (8). Rat bone OPN contains 12 phospho-Sers (pSers) and 1 phospho-Thr, and has considerable heterogeneity in degree of phosphorylation (9). In contrast, bovine milk OPN has 27 pSers and 1 phospho-Thr and appears to be maximally phosphorylated (10). The extent of phosphorylation of OPN, as well as of the specific sites, may play an important role in its physiologic function. The activity of OPN in regulating biomineralization is lost after the removal of the phosphates (2, 11). BSP in bone has been reported to have 11 phosphates. The role of BSP as a potent nucleator of hydroxyapatite crystals is highly dependent on its phosphorylation state (3, 12,13). Studies of OPN and BSP in their phosphorylated and dephosphorylated states indicate the importance of the phosphate groups on these proteins during osteoclast attachment (14), and the phosphorylation-dependent inhibitory action of OPN on the calcification of vascular smooth muscle cells (15). DMP1 is proteolytically processed into 37 kDa N-terminal and 57 kDa C-terminal fragments. The C-terminal fragment is much more highly phosphorylated than the N-terminal fragment (1). Phosphate analysis indicates that the 37 kDa fragments have 12 phosphates per mole, whereas the 57 kDa fragments have 41 (6). Supramolecular assembly studies have shown that the phosphorylation of DMP1 significantly affects the size and direction of apatite crystal growth (7, 16). In addition, a mineral-inhibiting functional domain identified in bone NCPs—the acidic Ser- and aspartate-rich motif (ASARM)—is believed to bind to hydroxyapatite and inhibit mineralization on a dose-dependent basis (17–19) and to modulate bone mineralization through phosphaturic effects by binding to and inhibiting a protein encoded by phosphate-regulating gene with homologies to endopeptidase on the X chromosome (PHEX) (19, 20).
The hunt for the kinase that phosphorylates SIBLINGs has continued for several decades. Family with sequence similarity member 20C (FAM20C), was found to be an evolutionarily conserved molecule expressed in various tissues, including bone and dentin (21–23). Loss-of-function mutations in the human FAM20C gene caused Raine syndrome, an osteosclerotic bone dysplasia (24), whereas Fam20C-knockout (KO) mice developed severe hypophosphatemic rickets due to renal phosphate wasting that was likely attributable to an increase in circulating fibroblast growth factor (FGF)-23 (25). Recent in vitro analyses identified FAM20C as a Golgi-enriched kinase that phosphorylates Ser in the Ser-X-Glu/pSer motifs in SIBLINGs and many other proteins on the secretory pathway (26, 27), implying an association of the assumed phosphorylation failure in SIBLINGs with the severe bone defects in the FAM20C-inactivated subjects. However, the lack of in vivo evidence for the loss of phosphorylation in SIBLINGs, the fewer Ser-X-Glu motifs than the actual number of phosphates in the SIBLINGs (1), and the presence of phospho-Thrs in the SIBLINGs that do not conform to a substrate for FAM20C kinase (3, 28) led us to argue that FAM20C may not be the only kinase phosphorylating the SIBLINGs. This evidence requires further investigation to gain more insight into the consequences of Fam20C inactivation.
To address these questions, we extracted the NCPs from the bone matrix of Fam20C-KO mice and purified the SIBLING proteins to examine their phosphorylation status by using multipronged biochemical approaches.
MATERIALS AND METHODS
Kinase assay of recombinant FAM20C
The recombinant FAM20C protein was produced by using baculovirus (25). For the kinase assay, 3 µCi [γ–32P]ATP (PerkinElmer, Houston, TX, USA) was added to the 25 µl reactions containing 2 µg bacterium-made OPN or dephosphorylated casein (SignalChem, Richmond, BC, Canada), with or without 0.3 µg FAM20C, respectively. The reaction products were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining, the destained gel was dried, and the incorporated radioactivity was visualized by autoradiography.
Animals
Fam20C-KO mice mediated by Sox2-Cre were generated by a published process (25). The animal procedures were performed in accordance with the U.S. National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of Texas A&M University Baylor College of Dentistry. Genotyping was performed as has been described (25).
Extraction of bone NCPs and measurement of attached phosphates
The NCPs were extracted from the long bones of 4-wk-old mice (8 WT vs. 12 KO) with 4 M guanidine-hydrochloride/0.5 M EDTA containing protease and phosphatase inhibitors, followed by buffer exchange into 6 M urea (29). After they were dialyzed against water, the NCP samples (0.06 mg of each) were hydrolyzed in 0.2 M NaOH, and the concentration of inorganic phosphate (Pi) liberated from the phosphoproteins was determined (6, 30–32). The Pi concentration was expressed as the number of Pi per milligram total protein. Student’s t test was used to determine the difference between the WT and KO groups. Differences reaching P < 0.05 were statistically significant.
Separation of NCPs by anion-exchange chromatography and Western immunoblot analyses
Subsequent to the protein concentration determination by the bicinchoninic acid assay (Thermo Scientific, Rockford, IL, USA), the same amounts of NCPs from the WT and KO mice were loaded on a fast protein liquid chromatograph (GE Healthcare, Pittsburgh, PA, USA) connected to a Q-Sepharose anion-exchange column (GE Healthcare). The NCPs were separated into 120 fractions of 0.5 ml with a gradient of 0.1–0.8 M NaCl in 6 M urea. The fractions were examined by SDS-PAGE followed by staining with Stains-All (Sigma-Aldrich, St. Louis MO, USA) (11, 33).
The OPN-, BSP-, and DMP1-enriched fractions in 6 M urea were loaded onto PD-10 desalting columns (GE Healthcare) to exchange the urea into 10 mM PBS. A 30 µl aliquot of each SIBLING fraction was treated with calf intestine phosphatase (CIP) (Sigma-Aldrich) and loaded onto 12% SDS-PAGE or native PAGE (without SDS) gel. Equal amounts of untreated samples were loaded onto the same gels for comparison. OPN, BSP, and DMP1 were visualized by Western immunoblot with an anti-OPN monoclonal antibody (Santa Cruz Biotechnology, Dallas, TX, USA) at 0.05 µg IgG/ml, an anti-BSP monoclonal antibody (10D9.2) at 1.7 µg IgG/ml, and an anti–DMP1-C polyclonal antibody (857) at 0.2 µg IgG/ml, as has been described (34).
Immunoprecipitation and mass spectrometry
Fifty microliters each of OPN-, BSP-, and DMP1-enriched fractions was incubated with 1.5 µg of anti-OPN monoclonal antibody (Santa Cruz Biotechnology), 1.5 µg anti-BSP monoclonal antibody (10D9.2), and 2.0 µg anti–DMP1-C monoclonal antibody (8G10.3), respectively. The antigen–antibody complexes were precipitated by incubation with protein G-agarose beads (Santa Cruz Biotechnology), in accordance with the manufacturer’s instructions. After 3 washes with PBS, the beads were resuspended in loading buffer; boiled to disassociate the complex of protein, antibody, and beads; and repelleted by centrifuge. The supernatants were loaded on 10% SDS-PAGE to separate the protein from the antibody, followed by Stains-All staining.
The protein bands of the OPN, BSP, and DMP1-C fragments were cut off from the SDS-PAGE gels for subsequent in-gel tryptic digestion and mass spectrometry (MS) analysis with a DecaXP ion-trap mass spectrometer (Thermo Fisher, Waltham, MA, USA) at the Protein Chemistry Lab (Texas A&M University, College Station, TX, USA) or using the 6540 UHD Accurate-Mass quadrupole time-of-flight system (Agilent Technologies, Santa Clara, CA, USA). All MS samples were analyzed with Mascot (version 2.4.1; Matrix Science, London, United Kingdom) and X! Tandem [version 2010.12.01.1 (Cyclone); The Global Proteome Machine (GPM), thegpm.org]. Mascot was set up to search the SwissProt_2012_11 database, assuming the digestion enzyme semi-Trypsin. X! Tandem was set up to search a subset of the database, assuming the digestion enzyme trypsin. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 20 ppm. Peptide identifications were accepted if they could be established at greater than 95.0% probability. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. The posttranslational modifications (PTMs) were assigned among peptide-to-spectrum matches (PSMs) with q ≤ 0.01. Each site listed was the best location with the highest PTM score (highest quality match) in at least 1 PSM considered by the modification location scoring (ModLS) algorithm.
RESULTS
Recombinant FAM20C phosphorylates OPN and casein in vitro
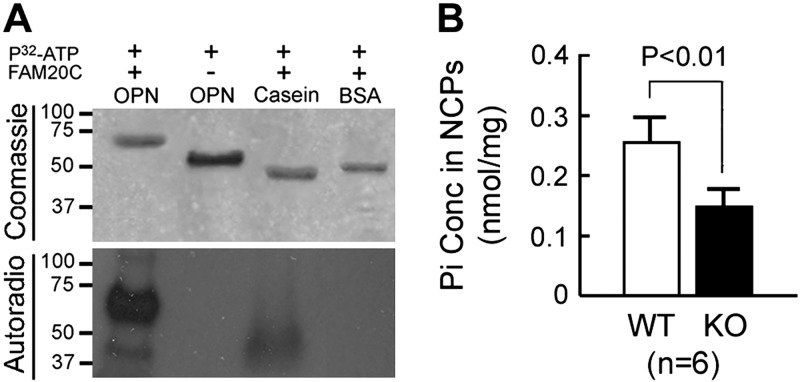
The OPN and casein incorporated [32P] from [32P]ATP in the presence of FAM20C kinase. The phosphorylated OPN showed a higher molecular weight than the nonphosphorylated OPN (Fig. 1A).
Figure 1.
In vitro kinase assay and in vivo phosphate measurement of bone NCPs. A) Kinase assay of recombinant FAM20C. In these experiments, we incubated bacterium-made OPN, dephosphorylated casein, and BSA with [γ–32P]ATP, along with recombinant FAM20C. Top: Coomassie blue staining showed a higher molecular mass of the phosphorylated OPN (+FAM20C lane) than the nonphosphorylated OPN (bacterium-made OPN without the addition of FAM20C) on 12% SDS-PAGE. Casein and BSA served as positive and negative controls for the incorporation of [32P]. Bottom: the Coomassie-stained gel was subjected to an autoradiography assay. The OPN and casein incorporated [32P] in the presence of FAM20C, whereas the OPN without FAM20C did not incorporate [32P]. B) Measurement of Pi attached to the bone NCPs. The phosphate concentration was expressed as the number of phosphates per milligram of total protein. The NCPs extracted from the bone matrix of 4-wk-old Fam20C-KO mice (filled bar) had significantly less Pi liberated by NaOH hydrolysis compared with those from their WT littermates (open bar). Error bars, se.
The NCPs from the bone of Fam20C-KO mice had fewer phosphates than normal
The NCPs extracted from the Fam20C-KO mice showed significantly fewer phosphates liberated by NaOH hydrolysis than in the WT. However, the phosphoproteins in the NCPs from the KO mice retained a considerable amount of phosphorylation, despite the significant Pi loss (Fig. 1B).
The OPN, BSP, and DMP1 from Fam20C-KO mice showed major phosphorylation loss
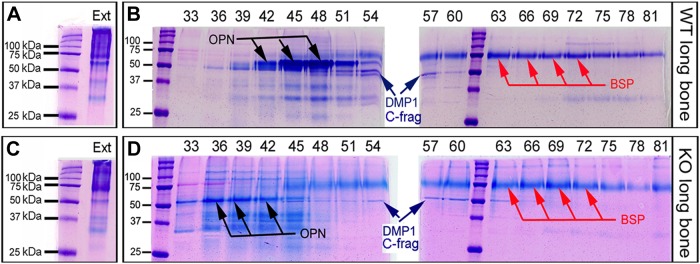
Anion-exchange chromatography separated the NCP extracts into 120 fractions, each containing 0.5 ml of sample in 6 M urea. OPN was eluted primarily into fractions 42–48 in the samples from the WT mice; in the samples from the KO mice, the OPN was mainly in fractions 36–42 (Fig. 2). Similarly, an earlier elution of BSP was found in the NCPs from KO mice than in those from the WT. These findings suggest that the OPN and BSP from KO mice have less negative charge than those from the WT mice and that the assumed reduction in negativity of OPN and BSP from the KO mice is attributable to a lower level of phosphorylation in these 2 proteins.
Figure 2.
Separation of bone NCPs by anion-exchange chromatography. A) Stains-All staining showed a smear pattern of bone NCP total extracts (Ext) from the WT mice. B) Isolation of SIBLING-enriched fractions from the NCPs of WT mice with a Q-Sepharose anion-exchange column. Staining showed that OPN (black arrows) was mainly eluted in fractions 42–48, whereas BSP (red arrows) was primarily in fractions 57–81. The C-terminal fragments of DMP1 (blue arrows) were coeluted with OPN and BSP in fractions 51–57. C) Staining showed the smear pattern of bone NCP total extracts (Ext) from the KO mice. D) Isolation of SIBLING-enriched fractions from the NCPs of KO mice. Staining showed that OPN (black arrows) was mainly in fractions 36–42, whereas BSP (red arrows) was in fractions 48–81. The DMP1-C fragments (blue arrows) were coeluted with OPN and BSP in fractions 48–60.
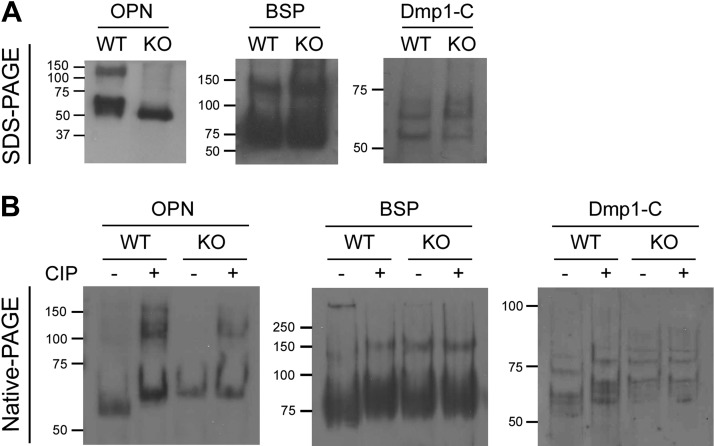
Western immunoblot analysis indicated that the OPN from KO mice had a slightly higher migration rate (Mr) than that from the WT mice (Fig. 3A), whereas BSP and DMP1-C did not show an appreciable difference in Mr between the KO and WT mice. In contrast, OPN from the KO mice on native PAGE (without SDS) displayed a lower Mr than did WT. The CIP treatment delayed the Mr of OPN from the WT mice to a position adjacent to that in the KO mice, but had no apparent impact on the OPN from the KO mice (Fig. 3B). Similar results were observed in the Western immunoblot analyses of the BSP and DMP1-C fragments between the CIP-treated and nontreated samples from the WT and KO mice. These results suggest a significant loss of phosphorylation in SIBLINGs from Fam20C-KO mice.
Figure 3.
Western immunoblot analyses of the SIBLING-enriched fractions. A) Western immunoblot analysis after SDS-PAGE. The OPN from WT mice was heterogenous and showed a higher molecular mass than that from the KO mice, whereas BSP and DMP1-C did not show a difference in Mr between the WT and KO mice. B) Western immunoblot analysis after native PAGE. OPN from the KO mice showed a slower Mr (less negative charge) than that from the WT. Dephosphorylation by CIP delayed the Mr of OPN from the WT mice to the level of that in the KO mice. CIP treatment had no effect on OPN from the KO mice. Similar results were appreciable in the Western immunoblot analyses on BSP and DMP1-C fragments treated with or without CIP.
MS identified both phosphorylated and nonphosphorylated residues in the SIBLINGs from Fam20C-KO mice
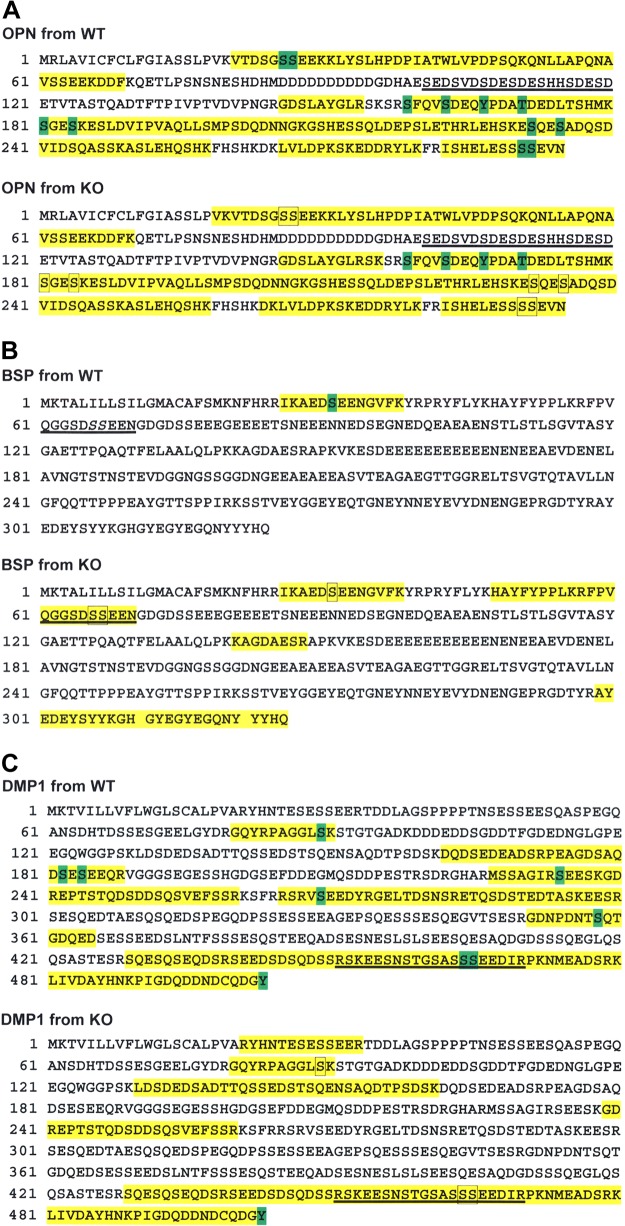
The MS coverage for the tryptic peptides of OPN from the WT mice was 63%. Eight Sers located in Ser-X-Glu motifs were phosphorylated. Two Sers that were not located in a Ser-X-Glu motif, a Tyr and a Thr, were also phosphorylated (Fig. 4A). The OPN from the KO mice had a 66% MS coverage. Among the 8 Sers in Ser-X-Glu motifs that were phosphorylated in the OPN of WT mice, 7 did not have attached phosphates in the OPN from KO mice; 1 Ser (pSer163 located in a Ser-X-Glu motif) was phosphorylated. pSer235, which is in a Ser-Ala-Asp motif, was phosphorylated in OPN from WT mice, but not in the OPN from KO mice (Fig. 4A; Supplemental Fig. S1). A phospho-Tyr (pTyr167), a phospho-Thr (pThr171), and a pSer (pSer159) were phosphorylated in the OPN from both the WT and KO mice.
Figure 4.
MS analysis of SIBLING proteins in the WT and Fam20C-KO mice. A) MS of OPN. Yellow: MS coverage; green: phosphorylated residues; framed amino acids: residues that lost phosphorylation in the KO mice; underlined sequences: ASARM motif. Eight Sers in the Ser-X-Glu motifs—Ser26, Ser27, Ser163, Ser181, Ser184, Ser232, Ser290, and Ser291—were phosphorylated in WT mice. Two Sers in non–Ser-X-Glu motifs, Ser159 and Ser235, as well as a Tyr, Tyr167, and a Thr, Thr171, were phosphorylated in both WT and KO mice. Seven of the 8 Sers in Ser-X-Glu motifs that were phosphorylated in the WT mice—Ser26, Ser27, Ser181, Ser184, Ser232, Ser290, and Ser291—were not phosphorylated in the KO mice. Ser163 retained phosphorylation in the KO mice, although it is located in a Ser-X-Glu motif, whereas Ser235, a Ser located in an Ser-Ala-Asp motif, lost its phosphorylation in the KO mice. B) MS of BSP. Indicators are the same as described in (A). Underlined italic S: the phosphorylated Ser in ASARM reported in another study (35). Ser31 was phosphorylated in the BSP from WT mice, but was not phosphorylated in the KO mice. The Sers in the ASARM motif of BSP were not phosphorylated in the KO mice. C) Indicators are as described in (A). In the DMP1 from the WT mice, 6 Sers located in the Ser-X-Glu/pSer motifs—Ser182, Ser184, Ser234, Ser269, Ser464, and Ser465—were phosphorylated; Ser89 and Ser358, which are located in Ser-Lys-Ser and Ser-Gln-Thr motifs, were phosphorylated; a phospho-Tyr, Tyr503, was identified at the very end of the DMP1 C terminus. In the DMP1 from the KO mice, the MS coverage did not overlap well with that from the WT mice. Ser89, which was phosphorylated in the WT mice, was not phosphorylated in the KO mice. Ser464 and Ser465, 2 Sers in the ASARM motif, lost their phosphorylation in the KO mice. Tyr503 was phosphorylated in the KO mice.
The MS coverage for BSP from the WT mice was only 4%, wherein a pSer (pSer31) was identified in a Ser-X-Glu motif. The MS coverage of BSP from the KO mice was 21%; the residue Ser31 in BSP from the KO mice was not phosphorylated (Fig. 4B; Supplemental Fig. S2).
The MS coverage for DMP1 from the WT mice was 40%. Six pSers were identified in the Ser-X-Glu/pSer motifs, and 2 others were detected in Ser-Lys-Ser and Ser-Gln-Thr. A phospho-Tyr was detected at the very end of the C terminus in both the WT and the KO mice (Fig. 4C; Supplemental Fig. S3). The MS coverage for DMP1 from the KO mice was 30%, wherein most of the amino acid sequences did not overlap with the coverage for WT mice. Two pSers located in Ser-X-Glu motifs within the ASARM motif of DMP1 from WT mice were nonphosphorylated in the KO mice.
DISCUSSION
The kinase assay clearly showed that recombinant FAM20C can phosphorylate OPN and casein in vitro. For in vivo analyses, the NCPs extracted from the KO mice showed significantly fewer phosphates attached to the proteins than did the NCPs from the WT mice (Fig. 1). These results may have suggested that inactivation of Fam20C leads to a significant loss of phosphorylation in the SIBLINGs, which compose the major group of NCPs. However, a considerable amount of phosphates were retained in the NCPs from KO mice, indicating that, in addition to FAM20C, other kinases are involved in phosphorylating the SIBLINGs.
We used anion-exchange chromatography to separate the SIBLING-enriched fractions from the NCP total extracts. Notably, OPN and BSP were eluted from the KO mice earlier than from the WT mice (Fig. 2), suggesting that these polyanionic proteins in the KO mice were less negatively charged than those in the WT. Because phosphate groups are a major contributor to the negative charge in SIBLINGs, these results support the suggestion that there is a loss of phosphorylation in the SIBLINGs caused by Fam20C inactivation.
Western immunoblot analyses of the SIBLING-enriched fractions provided further support for the speculation that the proteins from KO mice had fewer phosphates. On the native PAGE (without SDS), the Mr of a protein is mainly determined by the negative charges it carries. The SIBLINGs from the KO mice having a slower Mr had a less negative charge than those from the WT (Fig. 3B). More convincing evidence showed that removing the phosphate groups from SIBLINGs of the WT mice with CIP delayed their Mr to levels close to those of the KO, whereas the CIP treatment had no apparent effects on the Mr of the proteins from the KO mice (Fig. 3B), indicating that the SIBLINGs (especially the OPN) from the KO mice may have lost a significant amount of their attached phosphates. It is notable that OPN from the KO mice showed a more prominent migration shift than DMP1 and BSP, when compared with their counterparts from WT, which is persuasive evidence that it is the primary kinase of FAM20C for OPN, but appears not to support a major involvement of FAM20C in the phosphorylation of DMP1 and BSP, especially when the fact is taken into consideration that DMP1 has much more pSers (in the Ser-X-Glu motif) than OPN (6, 9, 10).
MS provided definitive evidence regarding the phosphorylation profile in some of these SIBLINGs. However, the MS coverage of the tryptic peptides from some of the SIBLING proteins is widely variable—for example, 4% for BSP derived from WT mice and 21% for BSP derived from KO mice. Given the variability in coverage, the MS data in this study represent only part of the phosphorylation profile in the SIBLING proteins. Elucidation of a more comprehensive phosphorylation profile for these proteins is warranted in future studies. Among the 3 SIBLINGs, OPN showed nearly identical MS coverage between the WT- and KO-derived proteins, whereas DMP1 and BSP showed considerably different coverage between the WT- and KO-derived proteins. The amino acid sequences of BSP present an intrinsic difficulty in generating proper-sized peptides for MS fragmentation by trypsin or alternative digestion, and the sulfation in BSP may add difficulty to retrieving the negatively charged peptides in positive-ion mode. The better MS coverage of BSP from the KO mice may be related to a lower negative charge in the peptides because of phosphorylation loss compared with that in the WT mice. For DMP1 MS, the nonoverlapped coverage between KO- and WT-derived proteins was about 17% of the total amino acid residues. Our MS data confirmed that KO-derived DMP1 had less phosphorylation than that of WT mice. In an earlier study, we reported significant down-regulation of DMP1 in the Fam20C-KO mice (25). It is unclear whether the lower levels of expression and phosphorylation of DMP1 in the KO mice were related to the different MS coverage of the WT- and KO-derived proteins. The overlapped coverage between the WT and KO mice validated a significant loss of pSers in the Ser-X-Glu motifs of SIBLINGs caused by Fam20C inactivation (Fig. 4A–C).
Notably, a pSer (pSer163) in OPN appeared to be unaffected by Fam20C inactivation, although the location is in an Ser-X-Glu motif (Fig. 4A; Supplemental Fig. S1), suggesting that this Ser is phosphorylated by another kinase. Casein kinase (CK)-2 phosphorylates Ser in the Ser-X-Glu motif in vitro and has been reported in some studies (36–38) to phosphorylate SIBLINGs. However, CK2 is mainly located in the cytoplasm and nucleus (26), where it has little chance to encounter proteins on the secretory pathway. It remains controversial if CK2 is involved in the phosphorylation of SIBLINGs. In addition, a pSer in OPN (pSer159) located in a non–Ser-X-Glu motif (Ser-Phe-Glu) retained phosphorylation in the KO mice (Fig. 4A), indicating that this pSer was not phosphorylated by FAM20C. Another study reported several pSers located in non–Ser-X-Glu motifs in BSP (pSer9 in Ser-Ile-Leu and pSer18 in Ser-Met-Lys) (35), which appear unlikely to be phosphorylated by FAM20C. Unfortunately, we were not able to evaluate their phosphorylation status in the KO mice because of the limited MS coverage.
Other studies have identified several phospho-Thrs in the SIBLINGs, with phosphorylation that is not governed by FAM20C (3, 28, 39). We confirmed the existence of some of these phospho-Thrs in the Fam20C-KO mice. In addition, we identified several phospho-Tyrs in the SIBLINGs that had not been reported before. It remains to be determined whether they are catalyzed by the recently discovered secreted Tyr kinase acting in the extracellular environment (35) and their exact role in SIBLING-mediated signaling and mineralization. These results indicate that FAM20C is likely to be the primary kinase for some SIBLINGs, but not the only one.
pSer235 in OPN (located in Ser-Ala-Asp) and pSer89 in DMP1 (located in Ser-Lys-Ser) unexpectedly lost their phosphorylation in the KO mice (Figs. 4A and Supplemental Fig S1; Fig. 4C and Supplemental Fig. S3). It is unclear whether the second Ser in the Ser-Lys-Ser motif was phosphorylated; thus, the sequence still conforms to a Ser-X-pSer motif, which, however, was probably not based on the sequence context. We suspect that Ser-X-Asp/Ser, or more conservatively, Ser-Ala-Asp and Ser-Lys-Ser in these cases, may be alternative recognition motifs for FAM20C kinase, since a similar mechanism has been used by CK2 in catalyzing the phosphorylation of Sers (38). However, previous studies did not detect FAM20C-mediated phosphorylation on several synthesized peptides containing Ser-X-Asp/Ser motifs (26, 27). It remains to be determined whether these specific amino acid sequences are recognized by FAM20C kinase.
The ASARM peptides in SIBLINGs are believed to inhibit biomineralization through binding to the hydroxyapatite, and an excess of phosphorylated ASARM peptides derived from the overexpressed OPN in Hyp mice is thought to be associated with hypophosphatemic rickets (17–19). Although the MS analyses in this study did not retrieve the ASARM peptides from OPN, our MS data of BSP derived from Fam20C-KO mice compared with results reported in a previous study of BSP derived from WT mice (3), as well as our MS analyses of DMP1 derived from both WT and Fam20C-KO mice, clearly showed pSer loss in the ASARM motifs in these SIBLINGs (Fig. 4B, C). It is plausible to assume that the ASARM derived from OPN would also demonstrate deficient phosphorylation in the Fam20C-KO mice. The DMP1–PHEX–integrin complex binding on the surface of osteocytes is believed to be involved in suppressing FGF23 expression and possibly increases FGF23 protein degradation through 7B2 coactivation of proprotein convertase (PC2) (19, 40). It has been proposed that this model is involved in the elevation of FGF23 and hypophosphatemic rickets in Dmp1-KO mice and Hyp mice (19). The phosphorylation of the DMP1 ASARM motif, which is believed to be pivotal for DMP1 binding to PHEX in the formation of the DMP1–PHEX–integrin complex (19), is catalyzed by FAM20C, as supported by our MS results. In a recent study, Ramos-Molina et al. (41) demonstrated that FAM20C also plays a role in mediating PC2 proprotein activation by phosphorylating the Thr111 residue in 7B2. However, Tagliabracci et al. (42) believed that FAM20C directly phosphorylates FGF23 at Ser180 within the FGF23 Arg176-XX-Arg179/Ser180-Ala-Glu subtilisin-like PC motif, which inhibits O-glycosylation of FGF23 by polypeptide N-acetylgalactosaminyltransferase-3 (GalNAc-T3) and promotes FGF23 cleavage. Taken together, the 3 models outlined above suggest that the FGF23 elevation in Fam20C-KO mice is an accumulating effect caused by the phosphorylation failure of multiple molecules, including the ASARM peptides in DMP1, the Thr111 residue in 7B2, and the Ser180 residue in FGF23 itself. This speculation is consistent with our observation that the serum FGF23 level in Fam20C-KO mice is higher than that in the Hyp mice and the Dmp1-KO mice (25). Accordingly, the serum phosphate level in the Fam20C-KO mice is lower, and their bone defects more severe, than those in the other 2 types of mutant mice.
Although this study gave us new insight into the consequences of Fam20C inactivation, several questions remain open for further investigation. For example, more informative MS analyses are needed to clarify whether FAM20C primarily phosphorylates OPN, but has relatively minor effects on DMP1 and BSP. In addition, it is not clear whether the tissue-nonspecific alkaline phosphatase (TNAP) level in Fam20C-KO mice has changed and whether this change contributed to the phosphorylation profile of SIBLINGs, seeing that TNAP can remove phosphate groups from phosphoproteins; future study of Fam20C;TNAP double-KO mice may help resolve this argument.
Fam20C-KO mice have severe hypophosphatemia. It is not clear whether the lower phosphate level in serum contributed to the phosphorylation profile of the SIBLINGs. Future investigation of high-phosphate-diet–treated KO mice may provide a definitive answer to this question. However, several lines of evidence suggest that the phosphorylation loss in Fam20C-KO mice is FAM20C specific, but not hypophosphatemia related. First, if hypophosphatemia had contributed to the lower level of phosphorylation in the SIBLINGs, we believe that the phosphorylation loss would have been nonspecific. In other words, the phosphorylation loss would have been observed, not only in Ser-X-Glu motifs, but also in non–Ser-X-Glu motifs, such as phospho-Thrs and phospho-Tyrs. However, our results showed that most of the phosphorylation loss occurred in the Ser-X-Glu motifs, and none of the phospho-Thrs or phospho-Tyrs lost their phosphorylation, suggesting that the phosphorylation loss was site specific (or FAM20C related). Second, the phosphorylation of SIBLINGs is a process under constant real-time regulation; therefore, the SIBLINGs derived from WT mice should be heterogeneously phosphorylated. It is very common to observe a specific phosphorylation in a fragment during MS analysis, but not in another with the same amino acid sequences in normal mouse samples. If hypophosphatemia contributed to the phosphorylation loss, a heterogeneous phosphorylation in the Ser-X-Glu motifs should have been observed in SIBLINGs derived from the KO mice. However, the loss of phosphorylation was homogeneous in the KO mice, indicating that the phosphorylation deficiency arose from kinase dysfunction, not hypophosphatemia. After considering all the evidence, we believe that the phosphorylation loss in SIBLINGs derived from the Fam20C-KO mice was FAM20C specific but not hypophosphatemia related.
In conclusion, this study provided in vivo evidence that FAM20C is the primary kinase for OPN but appears less involved in the phosphorylation of BSP and DMP1. The loss of phosphorylation in these proteins was likely associated with mineralization defects in the Fam20C-KO mice. FAM20C is not the only kinase that catalyzes the phosphorylation of the SIBLINGs.
Acknowledgments
The authors thank Dr. C. Qin (Texas A&M University Baylor College of Dentistry) for critical review of the manuscript, and J. Santa Cruz for assistance with the editing of the article. This work was supported by U.S. National Institutes of Health (NIH) National Institute of Dental and Craniofacial Research Grant R03DE23873-01 (to X.W.). The authors declare no conflicts of interest.
Glossary
- ASARM
acidic Ser- and aspartate-rich motif
- BSP
bone sialoprotein
- CIP
calf intestine phosphatase
- CK
casein kinase
- DMP
dentin matrix protein
- ECM
extracellular matrix
- FAM20C
family with sequence similarity member 20C
- FGF
fibroblast growth factor
- KO
knockout
- Mr
migration rate
- MS
mass spectrometry
- NCP
noncollagenous protein
- OPN
osteopontin
- PC
proprotein convertase
- PHEX
phosphate-regulating gene with homologies to endopeptidase on the X chromosome
- Pi
inorganic phosphate
- pSer
phospho-Ser
- PSM
peptide-to-spectrum match
- PTM
posttranslational modification
- SIBLING
small-integrin–binding ligand N-linked glycoprotein
- TNAP
tissue-nonspecific alkaline phosphatase
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Qin C., Baba O., Butler W. T. (2004) Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit. Rev. Oral Biol. Med. 15, 126–136 [DOI] [PubMed] [Google Scholar]
- 2.Gericke A., Qin C., Spevak L., Fujimoto Y., Butler W. T., Sørensen E. S., Boskey A. L. (2005) Importance of phosphorylation for osteopontin regulation of biomineralization. Calcif. Tissue Int. 77, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salih E., Flückiger R. (2004) Complete topographical distribution of both the in vivo and in vitro phosphorylation sites of bone sialoprotein and their biological implications. J. Biol. Chem. 279, 19808–19815 [DOI] [PubMed] [Google Scholar]
- 4.Richardson W. S., Munksgaard E. C., Butler W. T. (1978) Rat incisor phosphoprotein: the nature of the phosphate and quantitation of the phosphoserine. J. Biol. Chem. 253, 8042–8046 [PubMed] [Google Scholar]
- 5.Saito T., Arsenault A. L., Yamauchi M., Kuboki Y., Crenshaw M. A. (1997) Mineral induction by immobilized phosphoproteins. Bone 21, 305–311 [DOI] [PubMed] [Google Scholar]
- 6.Qin C., Brunn J. C., Cook R. G., Orkiszewski R. S., Malone J. P., Veis A., Butler W. T. (2003) Evidence for the proteolytic processing of dentin matrix protein 1: identification and characterization of processed fragments and cleavage sites. J. Biol. Chem. 278, 34700–34708 [DOI] [PubMed] [Google Scholar]
- 7.He G., Dahl T., Veis A., George A. (2003) Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein 1. Nat. Mater. 2, 552–558 [DOI] [PubMed] [Google Scholar]
- 8.Boskey A. L., Maresca M., Ullrich W., Doty S. B., Butler W. T., Prince C. W. (1993) Osteopontin-hydroxyapatite interactions in vitro: inhibition of hydroxyapatite formation and growth in a gelatin-gel. Bone Miner. 22, 147–159 [DOI] [PubMed] [Google Scholar]
- 9.Prince C. W., Oosawa T., Butler W. T., Tomana M., Bhown A. S., Bhown M., Schrohenloher R. E. (1987) Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J. Biol. Chem. 262, 2900–2907 [PubMed] [Google Scholar]
- 10.Sørensen E. S., Højrup P., Petersen T. E. (1995) Posttranslational modifications of bovine osteopontin: identification of twenty-eight phosphorylation and three O-glycosylation sites. Protein Sci. 4, 2040–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razzouk S., Brunn J. C., Qin C., Tye C. E., Goldberg H. A., Butler W. T. (2002) Osteopontin posttranslational modifications, possibly phosphorylation, are required for in vitro bone resorption but not osteoclast adhesion. Bone 30, 40–47 [DOI] [PubMed] [Google Scholar]
- 12.Curtin P., McHugh K. P., Zhou H. Y., Flückiger R., Goldhaber P., Oppenheim F. G., Salih E. (2009) Modulation of bone resorption by phosphorylation state of bone sialoprotein. Biochemistry 48, 6876–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baht G. S., O’Young J., Borovina A., Chen H., Tye C. E., Karttunen M., Lajoie G. A., Hunter G. K., Goldberg H. A. (2010) Phosphorylation of Ser136 is critical for potent bone sialoprotein-mediated nucleation of hydroxyapatite crystals. Biochem. J. 428, 385–395 [DOI] [PubMed] [Google Scholar]
- 14.Salih E., Zhou H. Y., Glimcher M. J. (1996) Phosphorylation of purified bovine bone sialoprotein and osteopontin by protein kinases. J. Biol. Chem. 271, 16897–16905 [DOI] [PubMed] [Google Scholar]
- 15.Jono S., Peinado C., Giachelli C. M. (2000) Phosphorylation of osteopontin is required for inhibition of vascular smooth muscle cell calcification. J. Biol. Chem. 275, 20197–20203 [DOI] [PubMed] [Google Scholar]
- 16.Deshpande A. S., Fang P. A., Zhang X., Jayaraman T., Sfeir C., Beniash E. (2011) Primary structure and phosphorylation of dentin matrix protein 1 (DMP1) and dentin phosphophoryn (DPP) uniquely determine their role in biomineralization. Biomacromolecules 12, 2933–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addison W. N., Masica D. L., Gray J. J., McKee M. D. (2010) Phosphorylation-dependent inhibition of mineralization by osteopontin ASARM peptides is regulated by PHEX cleavage. J. Bone Miner. Res. 25, 695–705 [DOI] [PubMed] [Google Scholar]
- 18.Barros N. M., Hoac B., Neves R. L., Addison W. N., Assis D. M., Murshed M., Carmona A. K., McKee M. D. (2013) Proteolytic processing of osteopontin by PHEX and accumulation of osteopontin fragments in Hyp mouse bone, the murine model of X-linked hypophosphatemia. J. Bone Miner. Res. 28, 688–699 [DOI] [PubMed] [Google Scholar]
- 19.Rowe P. S. (2012) Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway. Crit. Rev. Eukaryot. Gene Expr. 22, 61–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S., Rowe P. S., Vierthaler L., Zhou J., Quarles L. D. (2007) Phosphorylated acidic serine-aspartate-rich MEPE-associated motif peptide from matrix extracellular phosphoglycoprotein inhibits phosphate regulating gene with homologies to endopeptidases on the X-chromosome enzyme activity. J. Endocrinol. 192, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nalbant D., Youn H., Nalbant S. I., Sharma S., Cobos E., Beale E. G., Du Y., Williams S. C. (2005) FAM20: an evolutionarily conserved family of secreted proteins expressed in hematopoietic cells. BMC Genomics 6, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Hao J., Xie Y., Sun Y., Hernandez B., Yamoah A. K., Prasad M., Zhu Q., Feng J. Q., Qin C. (2010) Expression of FAM20C in the osteogenesis and odontogenesis of mouse. J. Histochem. Cytochem. 58, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Wang S., Lu Y., Gibson M. P., Liu Y., Yuan B., Feng J. Q., Qin C. (2012) FAM20C plays an essential role in the formation of murine teeth. J. Biol. Chem. 287, 35934–35942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson M. A., Hsu R., Keir L. S., Hao J., Sivapalan G., Ernst L. M., Zackai E. H., Al-Gazali L. I., Hulskamp G., Kingston H. M., Prescott T. E., Ion A., Patton M. A., Murday V., George A., Crosby A. H. (2007) Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am. J. Hum. Genet. 81, 906–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Wang S., Li C., Gao T., Liu Y., Rangiani A., Sun Y., Hao J., George A., Lu Y., Groppe J., Yuan B., Feng J. Q., Qin C. (2012) Inactivation of a novel FGF23 regulator, FAM20C, leads to hypophosphatemic rickets in mice. PLoS Genet. 8, e1002708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tagliabracci V. S., Engel J. L., Wen J., Wiley S. E., Worby C. A., Kinch L. N., Xiao J., Grishin N. V., Dixon J. E. (2012) Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa H. O., Xu A., Ogura E., Manning G., Irvine K. D. (2012) The Raine syndrome protein FAM20C is a Golgi kinase that phosphorylates bio-mineralization proteins. PLoS One 7, e42988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keykhosravani M., Doherty-Kirby A., Zhang C., Brewer D., Goldberg H. A., Hunter G. K., Lajoie G. (2005) Comprehensive identification of post-translational modifications of rat bone osteopontin by mass spectrometry. Biochemistry 44, 6990–7003 [DOI] [PubMed] [Google Scholar]
- 29.Huang B., Sun Y., Maciejewska I., Qin D., Peng T., McIntyre B., Wygant J., Butler W. T., Qin C. (2008) Distribution of SIBLING proteins in the organic and inorganic phases of rat dentin and bone. Eur. J. Oral Sci. 116, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linde A., Bhown M., Butler W. T. (1980) Noncollagenous proteins of dentin: a re-examination of proteins from rat incisor dentin utilizing techniques to avoid artifacts. J. Biol. Chem. 255, 5931–5942 [PubMed] [Google Scholar]
- 31.Van Veldhoven P. P., Mannaerts G. P. (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 161, 45–48 [DOI] [PubMed] [Google Scholar]
- 32.Qin C., Brunn J. C., Baba O., Wygant J. N., McIntyre B. W., Butler W. T. (2003) Dentin sialoprotein isoforms: detection and characterization of a high molecular weight dentin sialoprotein. Eur. J. Oral Sci. 111, 235–242 [DOI] [PubMed] [Google Scholar]
- 33.Sun Y., Prasad M., Gao T., Wang X., Zhu Q., D’Souza R., Feng J. Q., Qin C. (2010) Failure to process dentin matrix protein 1 (DMP1) into fragments leads to its loss of function in osteogenesis. J. Biol. Chem. 285, 31713–31722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C., Brunn J. C., Jones J., George A., Ramachandran A., Gorski J. P., Butler W. T. (2001) A comparative study of sialic acid-rich proteins in rat bone and dentin. Eur. J. Oral Sci. 109, 133–141 [DOI] [PubMed] [Google Scholar]
- 35.Bordoli M. R., Yum J., Breitkopf S. B., Thon J. N., Italiano J. E. Jr., Xiao J., Worby C., Wong S. K., Lin G., Edenius M., Keller T. L., Asara J. M., Dixon J. E., Yeo C. Y., Whitman M. (2014) A secreted tyrosine kinase acts in the extracellular environment. [Published correction (2014) in Cell 159, 955] Cell 158, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George A., Sabsay B., Simonian P. A., Veis A. (1993) Characterization of a novel dentin matrix acidic phosphoprotein: implications for induction of biomineralization. J. Biol. Chem. 268, 12624–12630 [PubMed] [Google Scholar]
- 37.Sfeir C., Veis A. (1995) Casein kinase localization in the endoplasmic reticulum of the ROS 17/2.8 cell line. J. Bone Miner. Res. 10, 607–615 [DOI] [PubMed] [Google Scholar]
- 38.Veis A., Sfeir C., Wu C. B. (1997) Phosphorylation of the proteins of the extracellular matrix of mineralized tissues by casein kinase-like activity. Crit. Rev. Oral Biol. Med. 8, 360–379 [DOI] [PubMed] [Google Scholar]
- 39.Christensen B., Nielsen M. S., Haselmann K. F., Petersen T. E., Sørensen E. S. (2005) Post-translationally modified residues of native human osteopontin are located in clusters: identification of 36 phosphorylation and five O-glycosylation sites and their biological implications. Biochem. J. 390, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan B., Feng J. Q., Bowman S., Liu Y., Blank R. D., Lindberg I., Drezner M. K. (2013) Hexa-D-arginine treatment increases 7B2•PC2 activity in hyp-mouse osteoblasts and rescues the HYP phenotype. J. Bone Miner. Res. 28, 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos-Molina B., Lindberg I. (2015) Phosphorylation and alternative splicing of 7B2 reduce prohormone convertase 2 activation. Mol. Endocrinol. 29, 756–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagliabracci V. S., Engel J. L., Wiley S. E., Xiao J., Gonzalez D. J., Nidumanda Appaiah H., Koller A., Nizet V., White K. E., Dixon J. E. (2014) Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. USA 111, 5520–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]