Abstract
GATA4 is expressed in the proximal 85% of small intestine where it promotes a proximal intestinal (‘jejunal’) identity while repressing a distal intestinal (‘ileal’) identity, but its molecular mechanisms are unclear. Here, we tested the hypothesis that GATA4 promotes a jejunal vs. ileal identity in mouse intestine by directly activating and repressing specific subsets of absorptive enterocyte genes by modulating the acetylation of histone H3, lysine 27 (H3K27), a mark of active chromatin, at sites of GATA4 occupancy. Global analysis of mouse jejunal epithelium showed a statistically significant association of GATA4 occupancy with GATA4-regulated genes. Occupancy was equally distributed between down- and up-regulated targets, and occupancy sites showed a dichotomy of unique motif over-representation at down- vs. up-regulated genes. H3K27ac enrichment at GATA4-binding loci that mapped to down-regulated genes (activation targets) was elevated, changed little upon conditional Gata4 deletion, and was similar to control ileum, whereas H3K27ac enrichment at GATA4-binding loci that mapped to up-regulated genes (repression targets) was depleted, increased upon conditional Gata4 deletion, and approached H3K27ac enrichment in wildtype control ileum. These data support the hypothesis that GATA4 both activates and represses intestinal genes, and show that GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of H3K27.
Keywords: GATA4, intestinal epithelium, chromatin occupancy, transcriptional repression, H3K27ac, histone modification
1. Introduction
Cellular identity in adult organisms is fundamentally determined by the unique panel of genes expressed within that cell. Regulation of a distinct set of genes requires precise spatial and temporal coordination of a multitude of general and specific transcription factors at cis-regulatory elements within DNA[1]. Recognition and binding of transcription factors to specific DNA sequences occurs within the context of chromatin, whose dynamic structural characteristics play a significant role in regulating gene expression. Cell identity is thus determined by the influences of DNA sequence, transcription factor binding, and the epigenetic chromatin state within any given cell.
The absorptive enterocyte is a highly specialized cell lineage in the small intestinal epithelium that exhibits differential identities depending on its placement along the length of small intestine. Absorptive enterocytes in jejunum express digestive enzymes, transporters, and intracellular carriers necessary for the digestion and absorption of nutrients, while absorptive enterocytes in distal ileum express a discrete set of proteins that include, among others, bile acid transporters that function in the distal re-absorption of bile acids, the first step in their necessary enterohepatic circulation. Understanding how spatial differences in absorptive enterocyte identity are determined has implications for restoring regional functions that are lost due to disease processes or intestinal resection.
GATA4, a member of an ancient family of zinc finger transcription factors that bind WGATAR motifs in DNA (W = A or T, R = A or G), is a key regulator of regional identity in absorptive enterocyte gene expression and function[2–4]. GATA4 is expressed in the proximal 85% of small intestine, but is not expressed in distal ileum, highly coincident with the demarcation in changes in intestinal gene expression and function between proximal intestine and distal ileum[2, 4, 5]. Conditional deletion of Gata4 in small intestine results in the transformation of absorptive enterocyte gene expression and function from a proximal intestinal to a distal ileal pattern[2–4]. Specifically, the subset of genes that are expressed at high levels in proximal small intestine, but not expressed in distal ileum, are down-regulated in proximal intestine. Conversely, the subset of genes not normally expressed in proximal small intestine, but highly expressed in distal ileum, are up-regulated in proximal small intestine. For example, the lactase (Lct) gene that is normally expressed in jejunum and proximal ileum, but not distal ileum, is significantly down-regulated in the proximal intestine, while the solute carrier family 10, member 2 (Slc10a2) gene that encodes the ileal-specific, apical sodium dependent bile acid transporter, is significantly up-regulated in the proximal intestine [2, 4], and bile acid absorption is induced[4]. Thus, by virtue of its restricted expression to the proximal 85% of small intestine, and its functions in both activating and repressing the expression of specific intestinal genes, GATA4 promotes a jejunal identity while repressing an ileal identity in absorptive enterocyte gene expression and function. GATA4 also functions redundantly with GATA6, which is expressed throughout the length of small intestine, including distal ileum, to regulate crypt cell proliferation and secretory cell differentiation, but due to the overlapping functions with GATA6, these processes are not altered in the proximal intestine of single Gata4 knockout mice[6]. How GATA4 confers a ‘jejunal’ identity while repressing an ‘ileal’ identity on absorptive enterocytes of the proximal small intestine is the topic of this investigation.
Chromatin structure is determined by histone proteins, which undergo a multitude of covalent modifications that influence chromatin architecture and gene expression. One such modification is acetylation of histone H3, lysine 27 (H3K27ac), a histone modification mark that is highly correlated with open chromatin and gene transcription[7–10]. In cardiac and hematopoietic systems, GATA factors have been shown to interact with CBP/p300[11–14], a transcriptional coactivator which has intrinsic histone acetyl-transferase activity and H3K27 as its substrate[8, 9]. Chromatin occupancy of GATA1 in hematopoietic cells is highly correlated with H3K27ac enrichment [15–17] but little is known about the relationship between GATA4 and H3K27ac. In the present study, we tested the hypothesis that conditional deletion of Gata4 results in the ‘ilealization’ of the H3K27ac chromatin mark. To test this hypothesis, we determined the global occupancy of GATA4 in mouse jejunal epithelium using an efficient in vivo biotinylation approach, mapped this occupancy to genes down- and up-regulated by conditional Gata4 deletion, and compared H3K27ac enrichment at these loci in wild-type control jejunum to conditional Gata4 knockout jejunum, and to wild-type control ileum. Our data implicate GATA4 as both an activator and repressor of specific subsets of target genes within the small intestine, and show that GATA4 activates a subset of genes by a process that is independent of H3K27ac modification, but represses a different subset of genes by inhibiting the acetylation of H3K27. These data implicate novel mechanisms of gene regulation by GATA factors, and contribute to our understanding of transcriptional regulatory mechanisms in the intestinal epithelium.
2. Material and methods
2.1. Mice
Wild-type mice, and previously established and confirmed Gata4flapflap [18] transgenic VillinCreERt2 [19], Gata4flbio/flbio [20] and Rosa26BirA/BirA [21] mice were used to establish the following groups of mice:
| WT Ctl: | Gata4wt/wt |
| G4ΔIE: | Gata4flap/flap, VillinCreERT2-positive |
| BirA Ctl: | Rosa26BirA/BirA |
| G4flbio: | Gata4flbio/flbio, Rosa26BirA/BirA |
Wild-type control (WT Ctl) mice express wild-type GATA4 under the control of the endogenous Gata4 gene; Gata4ΔIE mice, after treatment with tamoxifen to excise floxed (fl) Gata4 and induce expression of alkaline phosphatase (ap) from the Gata4flap allele[20], do not express Gata4 in the intestinal epithelium (IE); BirA control (BirA Ctl) mice express the bacterial biotinylation ligase enzyme BirA under the control of the Rosa26 gene; and Gata4flbio mice express a modified GATA4 containing a FLAG (fl) and biotin ligase tag (bio) on its COOH-terminus that is efficiently biotinylated by BirA in vivo[22]. DNA was extracted from tail biopsies, and genotypes were determined by semiquantitive polymerase chain reaction (PCR) using previously validated primers[19–21]. Adult (6–8 wks of age) WT Ctl and Gata4ΔIE mice were injected intraperitoneally with 1 mg tamoxifen (Sigma Chemical Co, Inc, St. Louis, MO) in sunflower seed oil (Sigma) once per day for 5 days to induce recombination of conditional alleles. At the time of tissue collection, mice were anesthetized and tissue was dissected as previously described[23]. Approval was obtained from the Institutional Animal Care and Use Committee.
2.2. RNA isolation and qRT-PCR
Mouse intestinal epithelial cells were isolated for RNA extraction by incubating freshly dissected segments (cut into 1 cm pieces) of small intestine in 15 ml conical tubes containing 30 mM ethylenediaminetetraacidic acid (EDTA) in1× Weiser solution A[24], and intestinal epithelial cells were released from the lamina propria by vigorously shaking three times for 30 sec and collected by centrifugation at 4°C[25]. RNA was isolated from the small intestinal epithelial cells using the RNeasy kit (Qiagen Sciences, Germantown, MD) according to the manufacturer’s instructions. To quantify mRNA abundances, quantitative reverse transcriptase (qRT)-PCR was conducted as described previously[23]. GAPDH mRNA abundance was measured for each sample and used to normalize the data. All data were expressed relative to the averages of the wild-type samples.
2.3. Nuclear extracts, biotin-streptavidin pull-down assays and Western blot
Intestinal epithelial cells were isolated from the middle 8 cm of the mouse small intestine (jejunum) as described above, and nuclear extracts were prepared as reported previously[26]. For biotin-streptavidin pull-down of GATA4, nuclear extracts were pre-cleared using protein A/G magnetic beads (Life Technologies), and incubated with streptavidin magnetic beads (M280, Life Technologies) overnight at 4°C. Beads were washed 3×5 min in 1 ml buffer B (0.1% NP-40, 5 mM Tris-HCl, 100 mM KCl and 10% glycerol) at 4°C and boiled in 30 µl 2× sample buffer for 10 min. Western blotting was performed as previously described[5] by transferring nuclear extracts to nitrocellulose membranes (Invitrogen) that were previously blocked with 5% nonfat dried milk in PBS. Membranes were then incubated with anti-mouse GATA4 (1:5000; SC-25310; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-mouse β-actin (1:5000; A5441; Sigma) overnight at 4°C, washed, incubated with horseradish peroxidase secondary antibodies and developed with SuperSignal West Femto ECL chemiluminescence solution (Thermo Fisher Scientific, Inc., Waltham, MA).
2.4. Biotin-streptavidin chromatin pull-down assays chromatin immunoprecipitation assays, and deep sequencing analysis
GATA4 chromatin occupancy was defined using biotin-streptavidin chromatin pull-down (BioChIP) assays[27], and H3K27ac enrichment was determined using chromatin immunoprecipitation (ChIP) assays[25]. In both assays, epithelial cells were isolated from the middle 8 cm (jejunum) and/or the distal 6 cm just proximal to the ileocecal junction. Epithelial fractions were cross-linked using 1% formaldehyde in phosphate-buffered saline (PBS) for 15 min at 4°C, then for 28 min at room temperature, followed by sonication in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1) using a Branson Sonifier (Branson Ultrasonic Corporation, Danbury CT) fitted with a microprobe until most DNA fragments were between 500–1000bp in length, as determined by agarose gel electrophoresis. Cell lysates were diluted 5-fold in binding buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1). For BioChIP analysis, cell lysates were incubated with streptavidin magnetic beads (Life Technologies) for 1 h at 4°C and further processed as described previously[27]. Briefly, streptavidin beads were washed 2× in 2% SDS, 1× in high salt buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 1 mM disodium EDTA, 0.1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100), 1× in LiCl buffer (10 mM Tris·Cl, pH 8.1, 250 mM LiCl, 1 mM disodium EDTA, 0.5% (v/v) Nonidet P-40 (NP-40), 0.5% (w/v) sodium deoxycholate), and 2× in TE buffer. For ChIP assays, cell lysates were incubated with anti-H3K27ac (ab4729, Abcam, Cambridge, MA) coupled to protein A/G magnetic beads (Life Technologies) for 16 h at 4°C, and washed 6 times in PBS. For both BioChIP and ChIP assays, DNA was recovered and cross-links were reversed in ChIP elution buffer (50 mM Tris·Cl, pH 8.1, 10 mM disodium EDTA, 1% (w/v) SDS) and then treated with RNase A. Enrichment at distinct loci was determined by quantitative PCR using specified primers (Supplemental Fig. S1).
For BioChIP-seq and ChIP-seq, intestinal epithelial cells from three different mice were pooled and epithelial fractions were sonicated until most fragments were between 200–400bp in length. BioChIP and ChIP was conducted as described above, and DNA libraries on pull-down material were prepared using the NEB Next kit according to the manufacturer’s instructions (New England Biolabs, Inc., Ipswich, MA). Libraries were tested for enrichment of expected fragments and amplified, and the DNA was sequenced, using the manufacturer’s protocols (Illumina, Inc., San Diego, CA) and 50 bp single-end reads. Sequences were mapped to reference genome Mus musculus build 9 (mm9) using ELAND tools, allowing 0 to 2 mismatches (Illumina), and binding peaks were identified by model-based analysis of ChIP-seq (MACS), an efficient peak calling tool that removes redundant reads, accurately adjusts peak summits based on tag length and sequencing reads, calculates peak enrichment, and estimates the empirical false discovery rate (FDR)[28]. A biological replicate confirmed the reliability of both GATA4 BioChIP-seq and H3K27ac ChIP-seq assays (Supplemental Fig. S2).
2.5. Data analysis
Analysis of BioChIP-seq and ChIP-seq data was done using tools within the Cistrome pipeline (http://cistrome.dfci.harvard.edu/ap/)[29] and Microsoft Excel. Microarray data was processed using dChip (http://www.hsph.harvard.edu/cli/complab/dchip/) with a fold enrichment cut-off of ± 1.1. Genomic Region Enrichment Annotation Tool (GREAT) was used as an advanced analytical tool to capture distal binding events[30]. The Cis-Regulatory Element Annotation System (CEAS) is a meta-gene analysis tool that was used to estimate enrichment levels across specific genomic regions (e.g., promoters, introns, etc) with respect to the whole genome[29]. Gene ontology (GO) analysis, which groups gene products by their associated biological processes, cellular components, and molecular functions, was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID)[31, 32]. GO groups were clustered using the functional annotation clustering tool within DAVID and enrichment scores were calculated for each cluster as an average of Expression Analysis Systematic Explorer (EASE) scores of GO groups (modified Fisher exact tests that reflect overlap of genes within GO groups) [33]. Data was visualized using the Integrated Genome Viewer 2.3 (Broad Institute, http://www.broadinstitute.org/igv/)[34]. The SeqPos motif tool finder, an algorithm available within Cistrome[29], was used to calculate motif enrichment within a standard 500bp distance of the summit of GATA4 peaks. H3K27ac signal profiling was conducted using the SitePro aggregation plot tool within Cistrome, at a standard profiling resolution of 50 bp. The number of GATA4 peaks analyzed by different programs varied slightly (<0.1%) due to algorithmic differences within each program.
3. Results
3.1. GATA4 chromatin occupancy in mouse small intestinal epithelium
GATA4 specifies regional identity in the mammalian small intestine, but knowledge of its direct target genes and mechanism of action in this organ have been deficient, in large part due to lack of ChIP-quality GATA4 antibodies. To overcome this limitation, GATA4 chromatin occupancy in mouse small intestinal epithelium was defined using an in vivo GATA4 biotinylation tagging, chromatin pull-down (BioChIP) approach[20, 27]. In this model, G4flbio mice express under the control of the endogenous GATA4 gene a modified GATA4 containing a FLAG peptide and biotinylation tagging site on its COOH- terminus (GATA4flbio). GATA4flbio is biotinylated by BirA, expressed under the control of the Rosa26 gene.
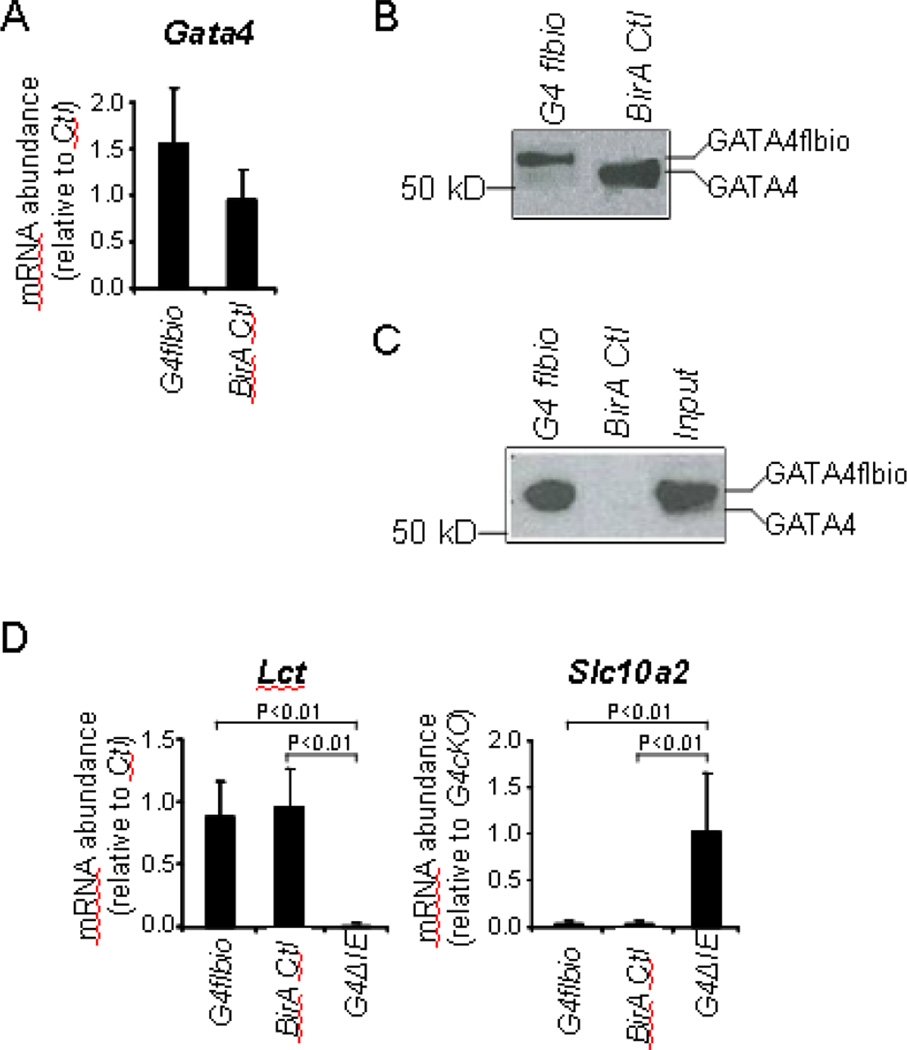
In small intestine, Gata4flbio mRNA abundance in G4flbio mice was not significantly different from Gata4 mRNA abundance in BirA Ctl mice (Fig. 1A). Using a GATA4 antibody that recognizes sequence within the COOH-terminal domain, the signal in Western blot analysis for GATA4flbio protein in G4flbio mice, though somewhat less intense, showed the expected slower mobility as compared to GATA4 in BirA Ctl mice (Fig. 1B). Western analysis of precipitates pulled down using streptavidin beads showed efficient pull-down of GATA4flbio (Fig. 1C), demonstrating that the GATA4flbio is biotinylated in vivo in the small intestinal epithelium. Importantly, Lct and Slc10a2, targets of GATA4 in jejunum that are down-regulated and up-regulated, respectively, in conditional Gata4 knockout mice (G4ΔIE)[2, 4], were expressed normally in G4flbio mice (Fig. 1D), indicating that GATA4flbio is functional in the intestine and GATA4-mediated jejunal-ileal patterning is preserved.
Fig. 1.
GATA4flbio is expressed at near endogenous levels, efficiently biotinylated, and functional in jejunal epithelium of G4flbio mice. (A) Gata4 mRNA abundance, determined by qRT-PCR using mouse Gata4 cDNA primers (mean ± SD, n = 4 in each group), and (B) Western blot analysis using a GATA4 antibody shows that Gata4 is expressed at near endogenous levels in mouse epithelial cells of G4flbio mice as compared to BirA Ctl mice. (C) Streptavidin-biotin pull-down assays showing efficient pull-down of GATA4flbio from nuclear extracts of jejunal epithelium fromG4flbio mice. GATA4flbio was detected by Western analysis using a GATA4 antibody. Input is the extracts isolated from G4flbio mice that have not undergone biotin-streptavidin pull-down, and represents 10% of that used in the pull-down assay. (D) Messenger RNA abundance of the Gata4 target genes, Lct and Slc10a2, in G4flbio mice reveals no difference in gene expression from BirA Ctl mice, demonstrating that GATA4flbio is functional. As a control for impaired GATA4 function, G4ΔIE mice demonstrate the expected down-regulation and up-regulation of Lct and Slc10a2, respectively. Data are shown as mean ± SD, n = 4 in each group.
We next determined the effectiveness of utilizing the GATA4 BioChIP model to define GATA4 chromatin occupancy in the small intestine. Because existing GATA4 antibodies are not effective for immunoprecipitation (IP) of GATA4 bound to chromatin making it impossible to compare GATA4 occupancy using ChIP with GATA4flbio occupancy using BioChIP, we validated our GATA4 BioChIP by predicting occupancy at selected GATA4 targets genes and comparing binding detected by qPCR with called peaks defined by the global BioChIP-seq assay. We also assessed the most highly represented motif in the BioChIP-seq analysis.
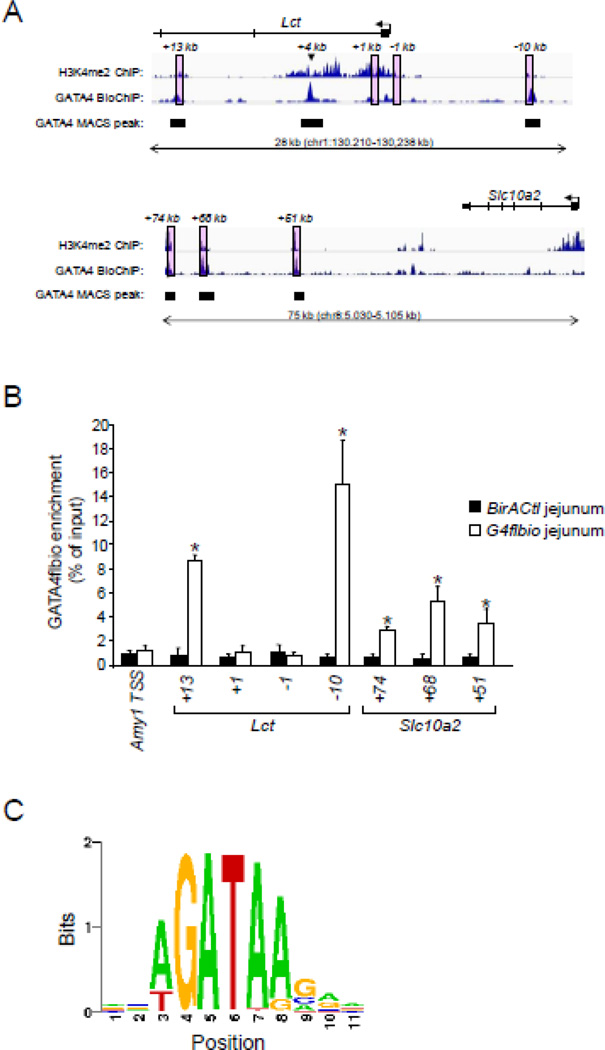
We did not know a priori where GATA4 bound chromatin in mouse small intestinal epithelium, but we reasoned that GATA4 would likely occupy sites that mapped to putative GATA4 target genes, such as Lct and Slc10a2. We further reasoned that such sites were likely to be positioned at genomic loci that were in an ‘open’ configuration and also contain at least one WGATAR consensus binding motif. Since di-methylation of histone H3, lysine 4 (H3K4me2) is indicative of open chromatin[7, 35], and global data on H3K4me2 enrichment in chromatin from mouse small intestinal epithelium are publicly available[25], we scanned the Lct and Slc10a2 genomic loci for WGATAR motifs localized within H3K4me2-enriched domains. We chose four such domains in Lct, and three in Slc10a2 (Fig. 2A, pink boxes) for further analysis.
Fig. 2.
GATA4 occupies chromatin loci in H3K4me2-enriched regions that map to the GATA4 target genes Lct and Slc10a2. (A) Sequencing tag density (revealed as Integrated Genome Browser (IGV) traces) of H3K4me2 and GATA4flbio enrichment in the lactase (Lct) and solute carrier family 10, member 2 (Slc10a2) genes. H3K4me2 enrichment was obtained from publicly available data[25], and GATA4flbio occupancy was determined by BioChIP-seq analysis as described in Methods. The statistically significant called MACS peaks are shown as filled boxes. The pink boxes indicate H3K4me2-enriched loci that contain at least one WGATAR motif. Locations are shown relative to the transcription start site (TSS) (+1 bp) above the pink boxes. One called GATA4flbio peak in Lct not within a pink box is indicated with an arrowhead (+4 kb). (B) Enrichment of GATA4flbio in H3K4me2-enriched loci containing WGATAR motifs by qPCR. BioChIP with qPCR quantification was conducted on jejunal epithelium of BirA Ctl and G4flbio mice as described in Methods. Sites analyzed are those in the pink boxes in (A). The TSS of Amy1, a gene that is not expressed in intestinal epithelium, was used as a negative control. All data are expressed relative to the mean of the BirA Ctl of the Amy1 TSS. Data are represented as mean ± SD from four independent experiments each from four individual mice (mice were also independent of those used for BioChip-seq in (A)). *P<0.05, as compared to BirA Ctl. (C) Sequencing logo plot showing that (A/T)GATA(A/G) is the most highly represented motif within GATA4-occupancy sites.
To determine GATA4 occupancy at these genomic loci, we performed BioChIP assays on chromatin isolated from jejunal epithelium of adult G4flbio mice. As controls, we included samples that contained the biotinylating enzyme BirA but lacked the epitope-tagged GATA4 (BirA Ctl). We also included ‘input’ chromatin which did not undergo pull-down. Using a qPCR detection approach on pulled-down, reverse cross-linked chromatin, GATA4 was found to be significantly enriched at two of the four sites mapped to Lct, and all three sites mapped to Slc10a2 (Fig. 2B).
On separate extracts from different mice, we subsequently conducted a deep-sequencing analysis (BioChIP-seq) using the model-based analysis of ChIP-seq (MACS) peak-calling algorithm[36], a false discovery rate of 0.05, and a P-value cutoffs of 10−5 to identify regions (‘peaks’) in which the experimental BioChIP sample was enriched for tags compared with input sample. These parameters led to the identification of 15,081 peaks, with a peak sequence length of 1450 bp, as determined by the MACS peak finding algorithm[28]. In contrast, the BirA control sample did not yield enough DNA to perform deep-sequencing. We found that the two sites enriched for GATA4 via the qPCR approach that mapped to Lct (+13 kb & −10 kb), and all three sites that mapped to Slc10a2, demonstrated a significant enrichment in sequencing tags via BioChIP-seq, while the two non-enriched sites that mapped to Lct (+1 kb & −1 kb) were not statistically called peaks in the global BioChIP-seq analysis (Fig. 2A, filled boxes), precisely matching our qPCR enrichment data (Fig. 2B). We noted an additional called peak in Lct (+4 kb) that was not analyzed by qPCR; this site was surrounded by H3K4me2 enrichment (Fig. 2A, arrowhead) and also contained a WGATAR motif.
Motif analysis at called peaks indicated that the most highly represented motif was the consensus binding sequence WGATAR (Fig. 2C). Using the Motif Based Interval Screener in Cistrome, we identified 14,870 WGATAR motifs at the 15,081 GATA4-occupied sites, compared with 13,605 WGATAR motifs at 15,081 randomly selected genomic sites (P<10−3). Although we are unable to directly compare GATA4flbio occupancy in Gata4flbio mice with endogenous GATA4 occupancy in wild-type mice, the detection of GATA4 occupancy at likely sites that map to putative target genes, the concordance of statistically significant binding detected by qPCR with called peaks defined independently by the global BioChIP-seq assay, and the identification of a classic GATA binding motif at called peaks, all validate the GATA4 BioChIP assay, and indicate that it is a useful approach to define global GATA4 occupancy in chromatin from mouse jejunal epithelial cells
3.2. GATA4 occupancy is enriched in intronic and intergenic regions, but densest in promoter regions
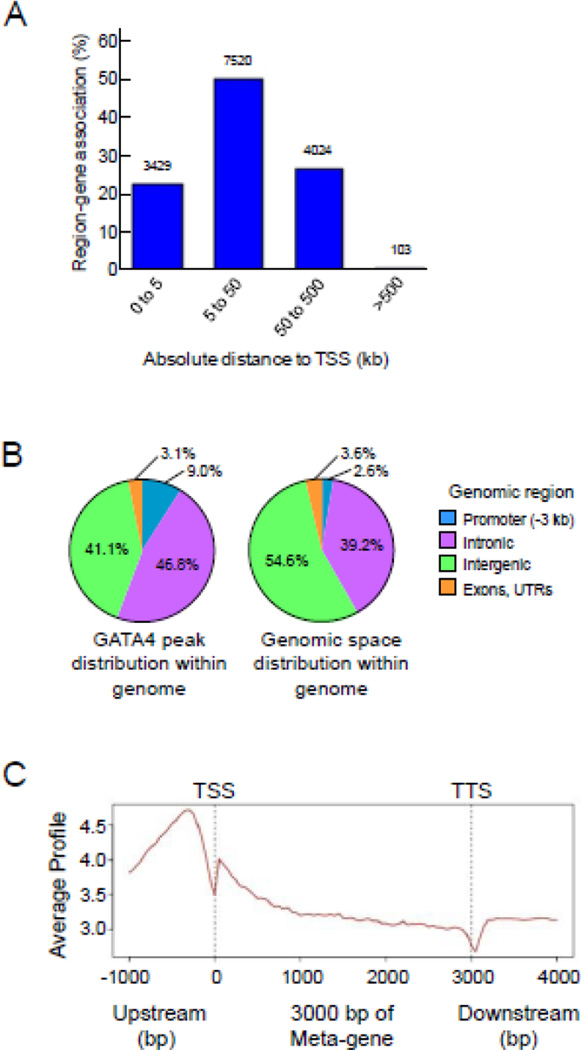
The 15,081 GATA4-occupied sites identified by BioChIP-seq were all mapped to the nearest transcription start site (TSS) of genes in the mouse genome, without limitation to distance. Accordingly, while each peak was mapped to a single gene, specific genes could have multiple peaks mapped to it. GREAT analysis showed that more than 70% of occupied sites occurred within 50 kb of a TSS, and nearly all occurred within 500 kb of a TSS (Fig. 3A). Of the 15,081 sites, 1362 (9.0%) were located on promoters (defined as the 3 kb upstream from a TSS), 7057 (46.8%) were located within introns, 6198 (41.1%) were located in the intergenic regions, and 464 (3.1%) were located in exons or untranslated regions (Fig. 3B). Using the CEAS meta-analysis tool, GATA4 occupancy was found to be densest in the region just upstream of the TSS (Fig. 3C). Noteworthy were regions of relatively lower occupancy right at the TSS and the transcription termination site (TTS). Together, these data indicate that although greater than 85% of GATA4 occupancy occurs outside of the promoter region (i.e., intronic and intergenic regions), GATA4 occupancy is densest within the promoter region nearest to the TSS.
Fig. 3.
Genome-wide distribution of GATA4 occupancy in chromatin from mouse jejunal epithelium. (A) Distribution of GATA4 occupancy relative to its nearest transcription start site (TSS), shown as absolute distance from the TSS, as determined using the Genomic Region Enrichment Annotation Tool (GREAT)[30]. (B) Distribution of GATA4 occupancy within genomic space, as determined by Cis-Regulatory Element Annotation System (CEAS) [29]. Promoter is defined as 3 kb upstream of the TSS; Intronic is introns; Intergenic is all regions that do not include the promoter (−3 kb from TSS), introns, exons, or untranslated regions (UTRs); Exons, UTRs are also shown. (C) Meta-analysis of GATA4 enrichment level across specific genomic regions was determined using the Cis-Regulatory Element Annotation System (CEAS)[29].
3.3. GATA4 occupancy is associated with GATA4-regulated genes
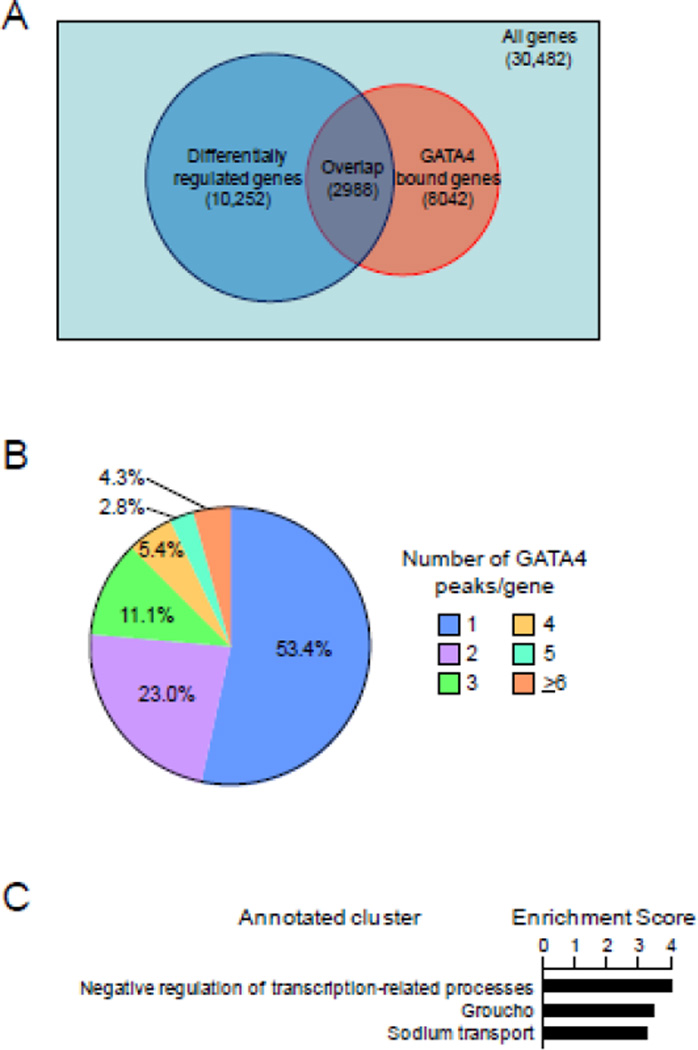
To identify the intestinal genes likely regulated directly by GATA4, we defined the overlap of GATA4-bound genes with differentially-regulated genes determined from publicly available profiling data from conditional Gata4 knockout mouse jejunum[3]. We found that the 15,081 GATA4 peaks identified from our BioChIP-seq analysis mapped to 8042 genes, an average of 1.88 peaks/gene. Of the 8042 GATA4-bound genes, 2988 (37.2%) were differentially regulated (Fig. 4A), significantly greater than that which would be expected if the distribution were random across the genome (Fisher’s exact test: P<10−16). The 2988 GATA4-bound, differentially regulated genes represent 29.1% of all differentially regulated genes; the remaining 70.9% are likely either secondary or downstream targets of GATA4, or direct targets associated with GATA4 occupancy sites that were not statistically called by our MACS peak analysis. Of the 5054 GATA4 bound genes that were not differentially regulated upon conditional Gata4 deletion, 507 (10.0%) were altered in ileum by conditional GATA6 deletion[6], suggesting that these genes might be GATA4/GATA6 redundant targets.
Fig. 4.
GATA4 occupancy is associated with GATA4-regulated genes. (A) Overlap of differentially regulated genes with GATA4 bound genes. A GATA4 bound gene is defined as a gene that has one or more peaks mapped to it, regardless of the peak-to-gene distance. A differentially regulated gene is defined as one that significantly changes at least 1.1-fold when Gata4 is conditionally deleted in the small intestine[3]. The distribution of GATA4 peaks was significantly different from that which would be expected if the distribution were random (P<10−16, Fisher’s exact test). (B) Distribution of peaks to differentially regulated, GATA4-bound genes, segmented by number of allocated peaks per gene reveals a subset of genes with multiple GATA4 peaks. (C) Gene ontology (GO) analysis of genes with 6 or more GATA4 peaks associated with them. The enrichment score is defined as the geometric average of Expression Analysis Systematic Explorer (EASE) scores (modified Fisher exact tests) of the individual GO groups in the cluster[57].
We next defined the relationship between peak density and gene regulation and function. Of the 2988 GATA4-bound, differentially regulated genes, over half were associated with only one GATA4 peak, and nearly one quarter were associated with two peaks; 127 (4.3%) were associated with 6 or more peaks (Fig. 4B). There was no correlation between the number of peaks associated with a specific gene, and fold change in mRNA abundance (data not shown), as observed by others[37]. However, GO analysis of only those differentially regulated genes with 6 or more GATA4 peaks showed the most enrichment for a gene set related to negative transcriptional regulation (Fig. 4C), suggesting that transcriptional repression is an essential GATA4 function [38]. Specific examples of possible downstream transcription factor targets include Gata6 (18 peaks), leucine-rich repeats and immunoglobulin-like domains 3 (Lrig3,12 peaks), BarH-like homeobox 2 (Barx2,11 peaks), Jun oncogene (Jun, 10 peaks), and intestine-specific homeobox (Isx, 9 peaks).
3.4. GATA4 occupies both activated and repressed genes
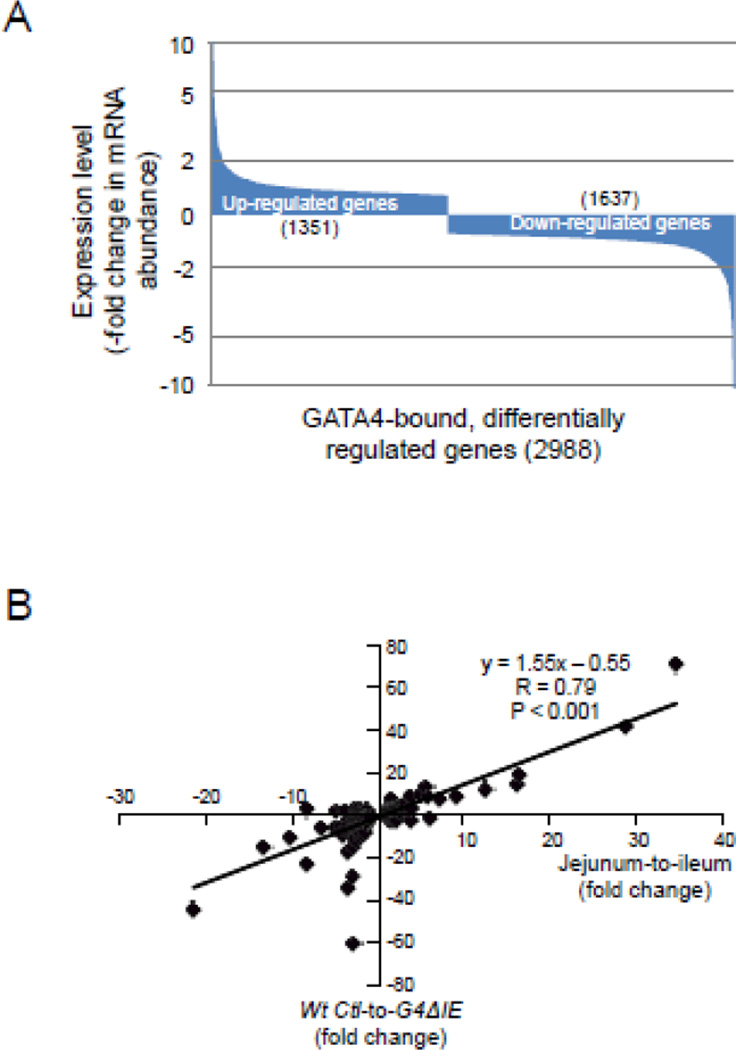
We next determined the association of GATA4 occupancy with down-regulated vs. up-regulated genes. Of the 2988 GATA4-bound, differentially regulated genes, 1351 (48%) were up-regulated, and 1637 (52%) were down-regulated upon conditional Gata4 deletion (Fig. 5A). The distribution of fold-change was similar between the two groups with 85% of up- and down-regulated genes each changing less than 2-fold, and less than 2.5% in each group changing more than 5-fold. The fold change in expression of down-regulated and up-regulated GATA4-bound genes was highly correlated with the fold change in expression between jejunum and ileum of the same set of genes (R=0.79, P<0.001) (Fig. 5B), suggesting that direct targets of GATA4 define jejuno-ileal patterning. The relatively equal distribution of GATA4 occupancy between down-regulated vs. up-regulated genes suggests that GATA4 both activates and represses genes directly in the epithelium of mouse small intestine.
Fig. 5.
GATA4 occupies both activated and repressed target genes. (A) Distribution of up-regulated and down-regulated, GATA4 bound genes. Genes are distributed from the most up-regulated to the most down-regulated. A change of 2-fold and 5-fold (up or down) is indicated by dotted lines. (B) Correlation of GATA4 regulation of direct targets with jejuno-ileal differences in gene expression. Fold changes in mRNA abundance of the 2988 GATA4-bound, differentially regulated genes were plotted against the publicly available[3] fold changes from jejunum-to-ileum of the same gene set.
Gene ontology (GO) analysis of all 2988 GATA4 bound, regulated genes revealed a marked enrichment for genes associated with cell death, signal transduction, cytoskeleton, transcription-related processes, and lipid metabolism (Supplemental Fig. S3). These functions were segregated by direction of GATA4 regulation; GATA4-bound, down-regulated genes (i.e., activation targets) demonstrated a significant enrichment for genes associated with transcription-related processes, while GATA4-bound, up-regulated genes (i.e., repression targets) showed enrichment for cell death, signal transduction, cytoskeleton, and lipid metabolism. Down-regulated genes (activation targets) also showed enrichment for genes associated with digestion/absorption processes. These data are in general agreement with that found for all GATA4-induced gene expression changes as determined by Battle et al.[3] using Ingenuity Pathway Analysis (IPA), implying that not only direct targets, but also indirect, downstream targets, regulate the same set of processes.
3.5. Specific motifs are segregated with down- and up-regulated GATA4 target genes
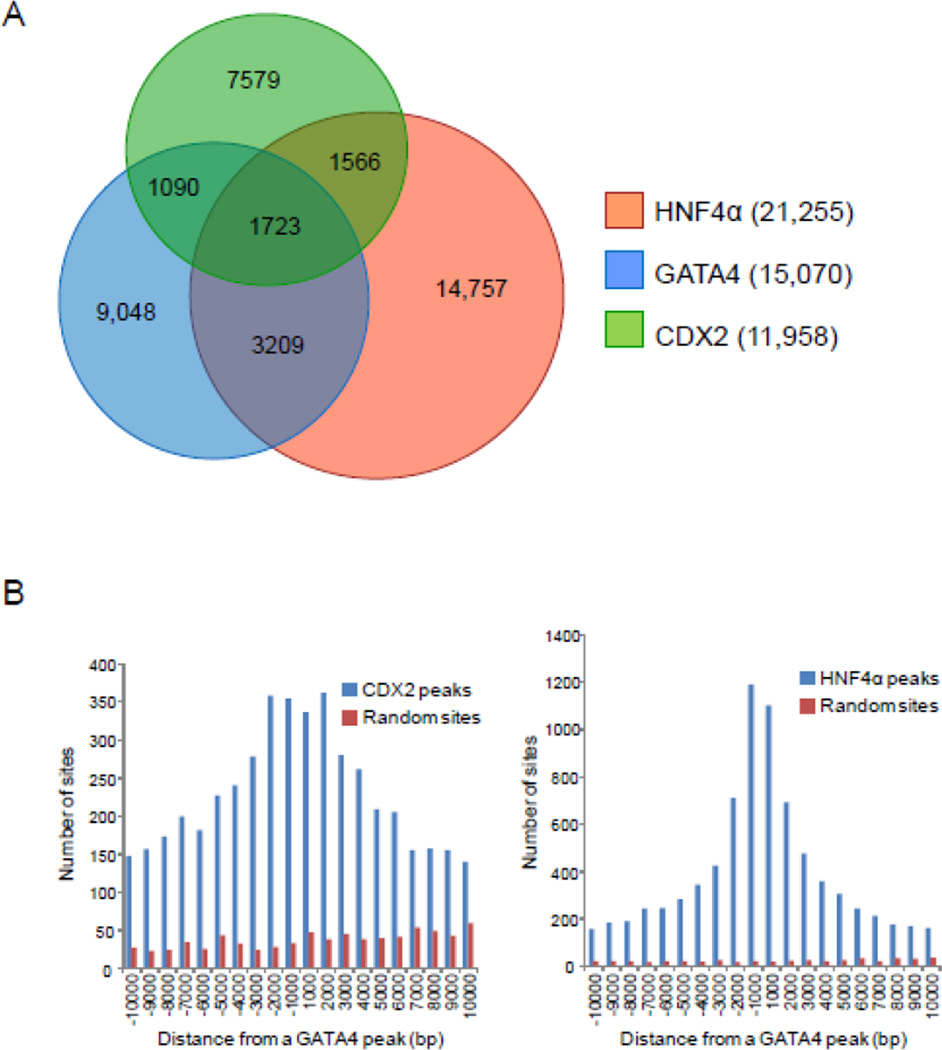
To identify possible co-regulatory transcription factors that segregate with down- and up-regulated GATA4 target genes, motif analysis was performed on the 500 bp centered on the summit of all GATA4 peaks using the SeqPos motif tool[29], a motif finding algorithm within the Cistrome pipeline. Thirty-six motifs were significantly associated with GATA4 occupancy, based on a cut-off of P<10−3(Supplemental Fig. S4). As anticipated, the most highly represented motif was WGATAR. Of the 36 motifs, 16 (44%) were common to both down-regulated and up-regulated genes suggesting possible core co-regulatory transcription factors that function in general intestinal gene expression. Notable were motifs for CDX2 and HNF4, two transcription factors known to regulate intestinal genes in vivo[39], and also occupy regions associated with GATA occupancy in Caco-2 cells[40]. Using publicly available ChIP-seq data for CDX2[25] and HNF4α[41] in mouse jejunum, together with our GATA4 BioChIP-seq data, we found a high level of overlap in occupancy among these three transcription factors (all peaks called at P<10−5, FDR<5%) (Fig. 6A). Occupancy analysis further revealed that CDX2 and HNF4α peaks were most abundant near summits of GATA4 peaks (Fig. 6B). These data suggest that GATA4 works in concert with CDX2 and HNF4α to regulate intestinal gene expression.
Fig. 6.
GATA4 co-occupies multiple sites with CDX2 and HNF4α. (A) Overlap of GATA4, CDX2, and HNF4α binding in intestinal epithelium from WT Ctl jejunum, determined by ChIP-seq peaks called at a P-value of <10−5 and a false discovery rate (FDR) <5%. Occupancy was obtained from publicly available CDX2[25] and HNF4α[41] ChIP-seq data from mouse jejunum, and our GATA4 BioChIP-seq analysis. (B) Histogram depicting the frequency at which CDX2 (left) or HNF4α (right) ChIP-seq peaks appear within 1 kb windows of the indicated distance from the summit of a GATA4 peak. CDX2 and HNF4α (blue bars) bind near the summit of GATA4 peaks; such clustering is not evident for GATA4 and random genomic regions equal in number and length to CDX2 or HNF4α binding sites (red bars).
Of the 36 motifs associated with GATA4 peaks, 14 were specifically associated with down-regulated genes (activation targets), while only 1 (Nr1d1) was specifically associated with up-regulated genes (repression targets) (Supplemental Fig. S4), suggesting that GATA4 co-associates with distinct transcription factors to carry out its specific functions in the small intestine. The unique association of specific motifs with down- vs. up-regulated genes further supports the concept that GATA4 both activates and represses specific genes in the small intestine.
3.6. GATA4 represses genes by modulating the acetylation of H3K27
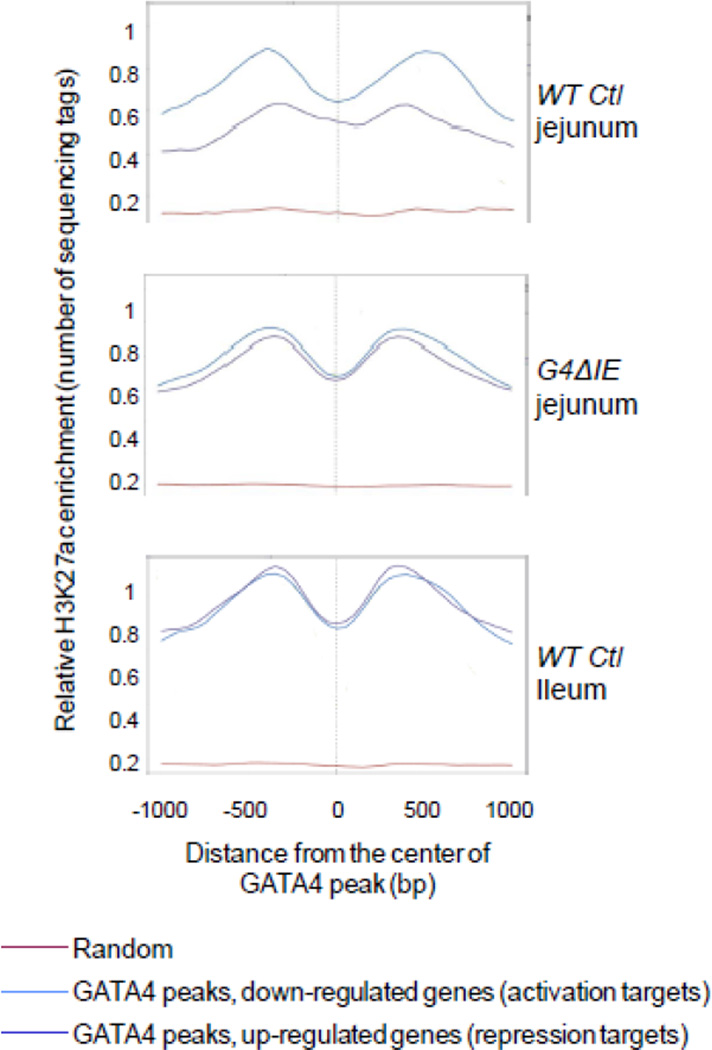
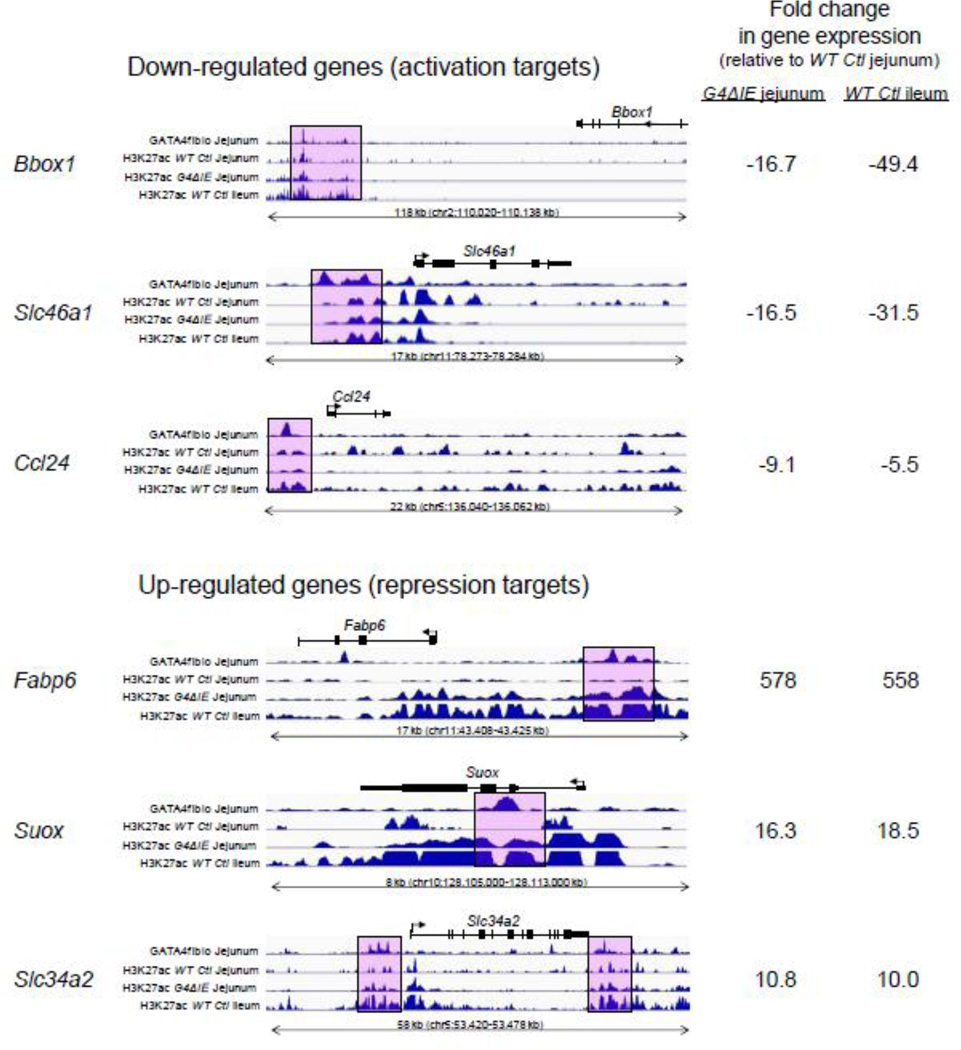
Because GATA factors have been shown to interact with CBP/p300[11–14], a transcriptional coactivator which acetylates H3K27 [8, 9], we performed ChIP-seq analysis with antibodies for H3K27ac on chromatin isolated from epithelial cells of WT Ctl jejunum, G4ΔIE jejunum, and WT Ctl ileum. We then compared H3K27ac enrichment at GATA4 peaks mapped to genes down-regulated (activation targets) and up-regulated (repression targets) by conditional Gata4 deletion using SitePro analysis[29] (Fig. 7). In all models, H3K27ac enrichment was expectedly low at random genomic loci (Fig. 7, red line). In WT Ctl jejunum (Fig. 7, top panel), H3K27ac at GATA4 peaks that mapped to activation targets (blue line) was more enriched than those that mapped to repression targets (purple line). Moreover, H3K27ac enrichment at activation targets showed a typical ‘bunny ears’ pattern consistent with an open chromatin configuration at TSSs. The ‘bunny-ears’ pattern is marked by low H3K27ac enrichment at the transcription factor binding peak due to a displaced nucleosome that is flanked by high H3K27ac enrichment on either side[40, 42]. In G4ΔIE jejunum (Fig. 7, middle panel), H3K27ac at GATA4 peaks that mapped to activation targets surprisingly remained highly enriched and in an open configuration, contrasting with the down-regulation of these targets upon conditional Gata4 deletion. H3K27ac at GATA4 peaks that mapped to repression targets became highly enriched upon conditional Gata4 deletion, and demonstrated an open configuration nearly identical to that of activation targets. In WT Ctl ileum (Fig. 7, bottom panel), H3K27ac was highly enriched and showed an open chromatin configuration for both activation and repression targets, similar to that in G4ΔIE jejunum. We visualized the raw deep-sequencing data from GATA4 BioChIP experiments from 3 representative activation and 3 representative repression targets (Fig. 8) using the publicly available Integrated Genome Viewer[34]. The fold change from profiling data is shown on the right. As shown at specific called GATA4 peaks (pink boxes) mapped to down-regulated genes (activation targets), H3K27ac enrichment changes little when Gata4 is deleted, despite a reduction in gene expression. In contrast, at specific called GATA4 peaks mapped to up-regulated genes (repression targets), H3K27ac enrichment is increased upon Gata4 deletion, consistent with activation of these genes. Taken together, these data show that conditional deletion of Gata4 in proximal small intestine results in the transformation of the H3K27ac profile at GATA4 target genes to one that approaches that in wild-type distal ileum.
Fig. 7.
Loss of GATA4 in intestinal epithelium promotes the acetylation of H3K27 at GATA4 peaks mapped to repression targets. Using SitePro, a tool embedded in the Cistrome pipeline[29], H3K27ac enrichment was determined across 2000 bp centered on sites of GATA4 occupancy (GATA4 peak) at loci mapped to genes down-regulated by conditional Gata4 deletion (activation targets, blue line), and genes up-regulated by conditional Gata4 deletion (repression targets, purple line). In order to control for variability across ChIP-seq experiments, H3K27ac enrichment was determined in a subset of random genomic loci (red line) as a negative control. H3K27ac enrichment at all TSSs in the genome was used as a positive control to normalize the data. H3K27ac enrichment patterns are shown for wild-type (WT Ctl) jejunum (top panel), conditional Gata4 knockout (G4ΔIE) jejunum (middle panel), and WT Ctl ileum (bottom panel).
Fig. 8.
Individual genes show H3K27ac enrichment patterns similar to genome-wide enrichment patterns. (A) Fold change of selected genes from profiling data of GATA4 knockout-WT and jejuno-ileal differences[3]. ChIP-seq traces at gene loci of down-regulated (B) and up-regulated (C) genes. Pink boxes indicate selected, called GATA4 peaks. Integrated Genome Browser (IGV) races for GATA4 in WT Ctl jejunum, and for H3K27ac in WT Ctl and G4ΔIE jejunum and WT Ctl ileum are shown.
4. Discussion
In this study, we provide the first genome-wide chromatin occupancy analysis of GATA4 in mouse intestinal epithelium, and compare this to a GATA4 gene profiling dataset and H3K27ac enrichment allowing us to characterize features of GATA4 in vivo occupancy that correlate with gene context-dependent transcriptional activity and absorptive enterocytes cell identity. We used an in vivo biotinylated GATA4 chromatin pull-down model (BioChIP) allowing us to circumvent limitations of currently available antisera. This in vivo biotinylation approach has been validated in prior studies[22, 43–45] and current data suggests that it does not alter GATA4 activity in the intestine (Fig. 1). GATA4 chromatin occupancy was validated in our study by accurately predicting GATA4 occupancy at open chromatin loci containing WGATAR motifs that map to putative GATA4 targets, and subsequently confirmed by independent pull-down with PCR-based detection, and by global motif analysis that showed WGATAR as the most highly representative motif (Fig. 2). Although our dataset may fail to identify all of the bona fide GATA4 occupancy sites due to a possible bias toward high-affinity sites, determination of a large number of high-confidence sites allowed us to apply statistical methods to further understand the transcriptional activity of GATA4 in the small intestinal epithelium, and how it confer regional identity to absorptive enterocytes. Our data uncover the jejunal GATA4-specific transcriptome and implicate GATA4 as both an activator and repressor of specific subsets of intestinal genes. Our data also reveal that GATA4 maintains proximal-distal differences in H3K27 acetylation at GATA4 target genes by inhibiting H3K27 acetylation specifically at repression targets. These data contribute to our understanding of transcriptional regulatory mechanisms and cell identity in the intestinal epithelium.
We found that the binding motif that best predicts global GATA4 occupancy in the small intestine is the classic WGATAR motif, though with an emphasis for ‘A’ in the 6th position (Fig. 2C). This is identical to that found for GATA4 occupancy in cardiac cells[22] and GATA6 occupancy in Caco-2 cells[40], and similar to that found for GATA1 occupancy in hematopoietic cells, which was somewhat more extended with greater sequence preference than the classic WGATAR motif[16, 46]. A large majority of sites of GATA4 occupancy occurred outside of the promoter region (>85%), likely in distal enhancers, while the highest density of GATA4 occupancy occurred in promoter regions (Fig. 3), a pattern similar to that found for other global GATA occupancy data[16, 22, 40, 46]. These data reveal that GATA occupancy characteristics across family members and tissue and cell types are generally conserved.
Comparison of our global analysis of GATA4 occupancy with profiling data obtained from conditional Gata4 knockout mice[3] allowed us to also define the genes directly regulated by GATA4. We mapped all GATA4 peaks to the nearest gene, a well recognized method for defining transcription factor targets on a global basis[40, 47], though chromosome conformation capture [48] or other techniques are necessary to define direct regulation of specific genes. We identified a total of 2988 genes that were both bound by GATA4, and differentially regulated upon conditional Gata4 deletion, thus representing an approximation of the GATA4 intestinal epithelial transcriptome (Fig. 5A). The differentially regulated genes were highly correlated with those whose expression differs significantly between jejunum and ileum (Fig. 5B), indicating that direct targets of GATA4 define jejuno-ileal patterning.
Motifs for CDX2 were over-represented in GATA4 occupancy sites (Supplemental Fig. S4), and GATA4 and CDX2 occupancy were found to overlap significantly in small intestinal epithelium (Fig. 6). CDX2 is an intestine-specific homeodomain protein that specifies embryonic intestinal epithelium[49] and maintains adult intestinal function and identity[25, 50, 51]. CDX2 chromatin occupancy in Caco-2 cells shows a surprising lability with redistribution from sites occupied only in proliferating cells to new sites in differentiated cells, with a preponderance of co-occupancy with GATA6 in proliferating cells and HNF4α in differentiated cells[40]. In mouse intestinal epithelium, CDX2 maintains a transcription-permissive chromatin state enabling occupancy of HNF4α[41]. Our data support combinatorial interactions between CDX2 and GATA4 in intestinal epithelium (Fig. 6), though future work is necessary to determine the extent to which CDX2 and GATA4 occupancy and effects on chromatin structure show an ordered hierarchy vs. codependency.
GATA4 occupies nearly equal numbers of down-regulated and up-regulated genes (Fig. 5A) suggesting that GATA4 both activates and represses genes directly in the epithelium of mouse small intestine. This is consistent with the direct activation and repression of cardiac genes by GATA4[52] and of hematopoietic genes by GATA1[16, 46]. In cardiac cells, GATA4 has been shown to interact directly with the coactivator p300, friend of GATA (FOG) cofactors, and corepressor complexes including the nucleosome remodeling and histone deacetylase (NuRD) complex and the polycomb repressive complex 2 (PRC2)[53]. We previously showed that conditional knock-in of a GATA4 mutant that is unable to bind FOG cofactors results in a modest (10%) but significant up-regulation of Slc10a2[54], a gene shown here to be a direct repression target of GATA4 (Fig. 2). These data suggest a role for FOG cofactors in mediating GATA4 gene repression in the small intestine.
Analysis of H3K27ac enrichment at GATA4 occupancy sites at activation and repression targets in control and Gata4 knockout jejunum and control ileum allowed us to interrogate the effects of GATA4 occupancy on chromatin state, and its role in maintaining a jejunal vs. ileal profile. H3K27ac can be deposited by both p300 and CREB binding protein (CBP)[8] and is associated with active enhancers in mammalian cells[55]. P300 is a transcriptional coactivator in multiple tissues, and is recruited to transcriptional enhancers in cardiac cells by GATA4[13]. We utilized H3K27ac enrichment as a measure of active chromatin state and possible p300 recruitment. We found that GATA4 loci that mapped to activation targets were highly enriched in H3K27ac. Surprisingly, however, H3K27ac enrichment at these sites was not reduced upon conditional Gata4 deletion (Figs. 7 & 8). These sites were also highly enriched in control ileum, where Gata4 is not expressed. Although many of these target genes are down-regulated, some to undetectable levels (e.g., Lct), in G4ΔIE jejunum and control ileum, their GATA4 binding loci remain in an ‘open’ chromatin state. Thus, GATA4 activates jejunal genes by a process that is independent of H3K27 acetylation, and likely independent of p300 recruitment.
GATA4 loci that mapped to repression targets were not enriched in H3K27ac, became enriched upon conditional Gata4 deletion, and approached H3K27ac enrichment in wild-type control ileum (Figs. 6 & 7). These genes were significantly up-regulated, many from undetectable levels (e.g., Slc10a2), in G4ΔIE jejunum and control ileum, consistent with the acetylation of H3K27 and establishment of an ‘open’ chromatin configuration. Thus, GATA4 represses ileal genes in the proximal small intestinal epithelium by preventing the acetylation of H3K27. Chromatin occupancy of GATA1 in hematopoietic cells is highly correlated with H3K27ac enrichment [15–17], but our data show that this is not the case for GATA4 in the intestine. While it remains possible that GATA4 could induce an open chromatin state early in intestinal development [56] and not be required for the maintenance of an open state in mature intestine, our data nonetheless show that for certain genes (i.e., repression targets), GATA4 inhibits the acetylation of H3K27 at these targets preventing their expression.
Our data provide a genome-wide analysis of GATA4 chromatin occupancy in the intestinal epithelium, enabling an examination of its transcriptional mechanisms in this tissue. The findings suggest that GATA4 directly activates and represses subsets of genes, possibly by co-association with distinct sets of transcription factors within enhancers and promoters of its targets. While GATA4 activates genes in the intestinal epithelium by a mechanism that is independent of H3K27ac modification, it represses genes by a process that inhibits the acetylation of H3K27. Future work should focus on GATA4 partners, including other transcription factors as well as specific co-activators or co-repressors, and their co-association at activation vs. repression target genes. This data set should provide a valuable resource to other investigators studying transcriptional regulation in the small intestine.
Supplementary Material
Highlights.
Intestinal GATA4 promotes a jejunal phenotype while repressing an ileal phenotype
GATA4 chromatin occupancy in intestinal epithelium maps to genes regulated by GATA4
Occupancy is equally distributed between activation and repression targets
The H3K27ac activation mark at repression targets is increased upon GATA4 deletion
GATA4 represses an ileal program of gene expression by inhibiting H3K27 acetylation
Acknowledgements
We thank L. A. T. M. Vissers, P. Cegas, and M. Battle for helpful discussions and comments on the manuscript. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK-061382 (S.D.K.) and K01-DK-088868 (M.P.V.), fellowship no. 1987 from the Crohn’s and Colitis Foundation of America (M.P.V.), the Harvard Digestive Disease Center (5P30-DK-34854), the Nutricia Research Foundation (B.E.A.), KWFKankerbestrijding (B.E.A.), Prins Bernhard Cultuurfonds (B.E.A.) in The Netherlands, and the European Society for Pediatric Research (B.E.A.) in Switzerland.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26(23):9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battle MA, Bondow BJ, Iverson MA, Adams SJ, Jandacek RJ, Tso P, Duncan SA. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135(5):1676–1686. e1671. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA, et al. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut. 2010;59(7):888–895. doi: 10.1136/gut.2009.204990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Wering HM, Bosse T, Musters A, de Jong E, de Jong N, Hogen Esch CE, Boudreau F, Swain GP, Dowling LN, Montgomery RK, et al. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physio Gastrointest Liver Physiol. 2004;287(4):G899–G909. doi: 10.1152/ajpgi.00150.2004. [DOI] [PubMed] [Google Scholar]
- 6.Beuling E, Baffour-Awuah NY, Stapleton KA, Aronson BE, Noah TK, Shroyer NF, Duncan SA, Fleet JC, Krasinski SD. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140(4):1219–1229. e1211–e1212. doi: 10.1053/j.gastro.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucl Acids Res. 2010;38(15):4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morceau F, Schnekenburger M, Dicato M, Diederich M. GATA-1: friends, brothers, and coworkers. Ann NY Acad Sci. 2004;1030:537–554. doi: 10.1196/annals.1329.064. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanazume T, Hasegawa K, Morimoto T, Kawamura T, Wada H, Matsumori A, Kawase Y, Hirai M, Kita T. Cardiac p300 is involved in myocyte growth with decompensated heart failure. Mol Cell Biol. 2003;23(10):3593–3606. doi: 10.1128/MCB.23.10.3593-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanazume T, Morimoto T, Wada H, Kawamura T, Hasegawa K. Biological role of p300 in cardiac myocytes. Mol Cell Biochem. 2003;248(1–2):115–119. doi: 10.1023/a:1024132217870. [DOI] [PubMed] [Google Scholar]
- 15.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23(4):1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36(4):682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos GL, Karkoulia E, Tsamardinos I, Porcher C, Ragoussis J, Bungert J, Strouboulis J. GATA-1 genome-wide occupancy associates with distinct epigenetic profiles in mouse fetal liver erythropoiesis. Nucl Acids Res. 2013;41(9):4938–4948. doi: 10.1093/nar/gkt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454(7200):109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 20.He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26(1):37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Driegen S, Ferreira R, van Zon A, Strouboulis J, Jaegle M, Grosveld F, Philipsen S, Meijer D. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14(4):477–482. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 22.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beuling E, Aronson BE, Tran LM, Stapleton KA, ter Horst EN, Vissers LA, Verzi MP, Krasinski SD. GATA6 is required for proliferation, migration, secretory cell maturation, and gene expression in the mature mouse colon. Mol Cell Biol. 2012;32(17):3392–3402. doi: 10.1128/MCB.00070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem. 1973;248(7):2536–2541. [PubMed] [Google Scholar]
- 25.Verzi MP, Shin H, Ho LL, Liu XS, Shivdasani RA. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol Cell Biol. 2011;31(10):2026–2039. doi: 10.1128/MCB.01250-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucl Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He A, Pu WT. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol / edited by Frederick M Ausubel [et al] 2010;Chapter 21(Unit 21):20. doi: 10.1002/0471142727.mb2120s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucl Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucl Acids Res. 2013 doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeiser S, Liebscher HV, Tiedemann H, Rubio-Aliaga I, Przemeck GK, de Angelis MH, Winkler G. Number of active transcription factor binding sites is essential for the Hes7 oscillator. Theor Biol Med Model. 2006;3:11. doi: 10.1186/1742-4682-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verzi MP, Shin H, San Roman AK, Liu XS, Shivdasani RA. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol Cell Biol. 2012 doi: 10.1128/MCB.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19(5):713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verzi MP, Shin H, San Roman AK, Liu XS, Shivdasani RA. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol Cell Biol. 2013;33(2):281–292. doi: 10.1128/MCB.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100(13):7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24(13):2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Mitra A, Dojer N, Fu S, Rowicka M, Brasier AR. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-kappaB/RelA regulatory network using ChIP-Seq. Nucl Acids Res. 2013 doi: 10.1093/nar/gkt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16(4):588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hryniuk A, Grainger S, Savory JG, Lohnes D. Cdx function is required for maintenance of intestinal identity in the adult. Dev Biol. 2012;363(2):426–437. doi: 10.1016/j.ydbio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Stringer EJ, Duluc I, Saandi T, Davidson I, Bialecka M, Sato T, Barker N, Clevers H, Pritchard CA, Winton DJ, et al. Cdx2 determines the fate of postnatal intestinal endoderm. Development. 2012;139(3):465–474. doi: 10.1242/dev.070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338(2):251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- 54.Beuling E, Bosse T, aan de Kerk DJ, Piaseckyj CM, Fujiwara Y, Katz SG, Orkin SH, Grand RJ, Krasinski SD. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008;322(1):179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.