Abstract
Drosophila sechellia relies exclusively on the fruits of Morinda citrifolia, which are toxic to most insects, including its sibling species Drosophila melanogaster and Drosophila simulans. Although several odorant binding protein (Obp) genes and olfactory receptor (Or) genes have been suggested to be associated with the D. sechellia host shift, a broad view of how chemosensory genes have contributed to this shift is still lacking. We therefore studied the transcriptomes of antennae, the main organ responsible for detecting food resource and oviposition, of D. sechellia and its two sibling species. We wanted to know whether gene expression, particularly chemosensory genes, has diverged between D. sechellia and its two sibling species. Using a very stringent definition of differential gene expression, we found a higher percentage of chemosensory genes differentially expressed in the D. sechellia lineage (7.8%) than in the D. simulans lineage (5.4%); for upregulated chemosensory genes, the percentages were 8.8% in D. sechellia and 5.2% in D. simulans. Interestingly, Obp50a exhibited the highest upregulation, an approximately 100-fold increase, and Or85c—previously reported to be a larva-specific gene—showed approximately 20-fold upregulation in D. sechellia. Furthermore, Ir84a (ionotropic receptor 84a), which has been proposed to be associated with male courtship behavior, was significantly upregulated in D. sechellia. We also found expression divergence in most of the chemosensory gene families between D. sechellia and the two sibling species. Our observations suggest that the host shift of D. sechellia was associated with the enrichment of differentially expressed, particularly upregulated, chemosensory genes.
Keywords: host shift, Drosophila sechellia, chemosensory genes, RNA-seq, antennal transcriptome
Introduction
Most Drosophila species utilize a wide variety of food sources (Rio et al. 1983; Louis and David 1986), but Drosophila sechellia, which is endemic to Seychelles (Tsacas and Bachli 1981), exclusively uses the fruit of Morinda citrifolia, commonly known as Tahitian Noni fruit, as its food source and for oviposition. Interestingly, the fruit of M. citrifolia is toxic to other insects, including most of the Drosophila species (Rkha et al. 1991). How D. sechellia evolved the ability to sense its specific host plant is not well understood.
Chemosensory genes are responsible for sensing odors and thus are essential for survival (finding food sources) and reproduction (finding oviposition) of most animal species. In hexapods (insects), the chemosensory gene superfamily comprises the odorant binding protein (Obp) genes, the chemosensory protein (Csp and CheA/B) genes, and three transmembrane receptor gene families: the olfactory receptor (Or) genes, the gustatory receptor (Gr) genes, and the ionotropic receptor (Ir) genes (Vosshall et al. 1999; Clyne et al. 1999, 2000; Galindo and Smith 2001; Xu et al. 2002; Benton et al. 2009; Vieira and Rozas 2011).
OBPs transport hydrophobic odorant molecules across the aqueous lymph surrounding the olfactory sensory neurons (OSNs) on the sensilla of fly antennae and lymph of other chemosensory sensilla. To be noted, CSPs and CHEA/B shared the same protein description, that is, chemosensory proteins, in the previous studies (Xu et al. 2002; Starostina et al. 2009; Vieira and Rozas 2011). Vieira and Rozas (2011) identified four CSPs and proposed that CSPs are homologous to OBPs. CHEA/B were found in the front legs of mature male flies, and thought to interact directly with lipid-like pheromones; they therefore are thought to be involved in male-specific mating behaviors (Xu et al. 2002; Starostina et al. 2009).
ORs and GRs were responsible for “smelling” and “tasting,” respectively, whereas IRs, which were identified on the basis of their structural similarity with ionotropic glutamate receptors (iGluRs), were recently found to be responsible for salt detection in Drosophila (Zhang et al. 2013). Or genes and Ir genes are expressed mostly in OSNs on different types of sensilla in antennae, and Gr genes are expressed in other body parts including proboscis, wing margins, legs, and ovipositors. As Ir genes show sequence homology to iGluRs, they are evolutionarily unrelated to Or and Gr genes (Benton et al. 2009). Ir genes are expressed in OSNs, different from those expressing Or or Gr genes, and are generally thought to mediate responses to acids and amines.
Previous studies suggested that the host shift of D. sechellia was associated with changes of several chemosensory genes either at the DNA sequence level or at the messenger RNA expression level. Examining interspecies hybrids between D. sechellia and Drosophila melanogaster deficiency strains, Matsuo et al. (2007) suggested that Obp57d and Obp57e (Obp57d/e) are responsible for the host preference in D. sechellia. They further showed that transferring Obp57d/e of D. sechellia to the Obp57d/e knockout D. melanogaster changed their preference to higher concentrations of hexanoic acid (HA) and octanoic acid (OA), the toxins contained in the ripe fruit of M. citrifolia. They proposed that the functional alteration in D. sechellia was due to a 4-bp insertion upstream of Obp57e. Dworkin and Jones (2009) showed that the D. sechellia food specialization was related to several cases of loss-of-function in genes involved in detoxification, metabolic pathways, and olfaction. They found that D. sechellia Obp56e harbors a premature stop codon, which might play a role in D. sechellia’s preference of M. citrifolia because a knockdown of this gene in D. melanogaster reduced the avoidance of M. citrifolia. Hungate et al. (2013) identified an approximately 170-kb region related to the tolerance of OA by testing the D. sechellia introgression lines under the Drosophila simulans genetic background. This locus comprises 18 genes belonging to the Obp and Osiri (Osi) gene families.
The first large-scale microarray analysis of D. sechellia antennal transcriptome found that Or22a, Or22b, and Or85b have markedly increased expression in D. sechellia (Kopp et al. 2008). However, Or22a and Or22b also differed significantly in expression level between D. simulans and D. melanogaster and Or22a showed significant expression differences between two strains of D. melanogaster as well. Microarray analysis might not be suitable for studying lowly expressed genes, such as Or genes, which are lowly expressed because one OSN expresses only 1–3 Or genes, including the universally expressed olfactory receptor co-receptor gene (Orco).
Another study suggested a 5-fold acceleration of gene loss in the D. sechellia genome by comparing genomes of five closely related Drosophila species (McBride and Arguello 2007). However, the gene-loss phenomenon may only explain the process for a species not to avoid a plant host, but is insufficient to explain the adaptation to a new host as it may require a novel receptor or changes in the expression of certain existing genes. Interestingly, no novel chemosensory genes have been discovered in the D. sechellia genome so far. Furthermore, most studies focused on the well-defined chemosensory gene families, such as Obp, Or, and Gr genes. How the other chemosensory gene families are expressed in antennae and whether they are involved in the host shift of D. sechellia are unknown.
Dekker et al. (2006) proposed that the host shift in D. sechellia was related to the large differences in the numbers of sensilla in antennae. In comparison to D. melanogaster, D. sechellia has many more ab3 sensilla at the cost of ab1 and especially ab2 sensilla (Stensmyr et al. 2003; Dekker et al. 2006). The total number of ab3 sensilla on the D. sechellia antennae is about 2.5- to 3-fold higher than that of D. melanogaster (Dekker et al. 2006). Dekker et al. (2006) also suggested that the increased sensitivity to M. citrifolia, particularly to the chemical MeHex, was achieved by increasing the expression of Or genes in ab3 antennae, including Or22a, Or22b, and Or85b.
In this study, we hypothesized that the adaptation of D. sechellia to the new host was achieved by changing expression levels of existing genes, and we expected enrichment of differentially expressed chemosensory genes in the D. sechellia lineage. We would like to know whether the adaptation involved expression divergence in all chemosensory gene families, including recently identified Ir, Csp, and Che genes. We would also like to study whether the expression levels of chemosensory genes are associated with the differences in the number of sensilla in D. sechellia.
To answer these questions, we isolated total RNA from more than 300 pairs of antennae detached from the head and we used the Next Generation RNA-seq technology to identify differentially expressed genes (DEGs) between D. sechellia and its two sibling species: D. melanogaster and D. simulans. Our data revealed many highly differentially expressed chemosensory genes in D. sechellia that had not been identified in previous studies. We discussed the implications of these observations for the host preference of D. sechellia.
Materials and Methods
Sample Collection and RNA Isolation
Drosophila simulans clade split from D. melanogaster 3 Ma (Lachaise et al. 1988), and two island endemic species, D. sechellia and D. mauritiana, independently evolved from the D. simulans-like ancestor approximately 250,000 years ago (Kliman et al. 2000; McDermott and Kliman 2008; Garrigan et al. 2012). So, D. simulans is a better candidate than D. mauritiana as D. simulans clade reference for studying the host shift of D. sechellia. Therefore, to infer the evolution of chemosensory genes in the host specialization of D. sechellia, we chose the most closely related generalist species, D. simulans, as a comparison and D. melanogaster as the outgroup generalist species.
We collected antennae from D. melanogaster (Canton-S, Bloomington Drosophila Stock Center at Indiana University: #1; collected from Canton, OH in 1920s) and D. sechellia (genome strain, UC San Diego Drosophila Stock Center: #14021-0248.25; collected from Cousin Island, Seychelles in 1980) at Academia Sinica, Taiwan and from D. simulans (genome strain, Kyorin-Fly: k-s05; originally from #14021-0251.194, collected from Wolfskill orchard, Winters, CA in 1995) and D. sechellia (Kyorin-Fly: k-s10; originally from #14021-0248.25) at Ochanomizu University, Japan. The flies in Taiwan and Japan were reared on standard cornmeal fly food under similar environmental conditions, a 12/12 h light/dark cycle at 25 °C.
About 300–700 pairs of fly antennae of each sex from each species were collected for total RNA isolation. The antennae resected each day were preserved in approximately 50–100 µl Trizol (Life Technology) and stored at −80°C before further processing. Right before the RNA isolation, we spun down antennae preserved in Trizol at the max speed (13,000 rpm) for 3 min at 4°C and transferred each sample into one MagNA Lyser Green Beads tube (Roche).
The RNA isolation protocol we developed combined steps from conventional Trizol extraction and RNeasy Micro Elute Kit (QIAGEN, Inc.) with some modifications to obtain high yield and quality of total RNA. Fly antennae were disrupted with MagNA Lyser (Roche) at 7,000 rpm for 15 s each time and repeated for 4–5 times until the tissue was almost invisible. After homogenization, we cooled down the lysate on ice for 10 s to prevent RNA from degradation. Tissue lysate was transferred to a new RNase free Eppendorf tube. We used about 400 µl Trizol to rinse the beads of each sample and combine this 400 µl Trizol with the tissue lysate for the RNA isolation. Based on the Trizol standard protocol, 200 µl of chloroform per 1 ml of tissue lysate was added to the lysate and mixed well by shaking vigorously for 15 s and set 2–3 min at room temperature. The aqueous phase was separated by centrifuging the lysate at the maximum speed (13,000 rpm) at 4°C for 15 min and carefully transferred to a new Eppendorf tube after centrifuging.
We added 1 volume of 5 µl Carrier RNA (QIAGEN, Inc.) and 1 ml of 70% EtOH to the aqueous phase and mixed well by inverting the tubes carefully. To obtain greater amount of RNA, we kept the samples at −80°C for overnight to enhance RNA precipitation for greater yield. RNA isolation followed manufacturer’s protocol of the RNeasy Micro Elute Kit with increased volumes (700 µl) of RPE buffer and 80% ethanol to wash RNA. Genomic DNA was removed by applying on-column DNase I treatment at room temperature for 15 min.
Paired-End mRNA-seq
For paired-end mRNA-seq library preparation, we used the TruSeq v2 kits from Illumina. Input of 4 µg of total RNA was used for mRNA enrichment by oligo-dT beads followed by cation-catalyzed fragmentation for 7.5 min at 94 °C. The mRNA fragments were then converted into double-stranded cDNA by random priming followed by end repair and A-tailing. The fragments were then ligated to the barcoded paired-end adaptors and subjected to ten cycles of polymerase chain reaction (PCR) amplification and purified by Ampure XP beads (Beckman Agencourt). The absolute concentrations of the libraries were determined by Qubit fluorometer (Invitrogen) and profiled by BioAnalyzer 2100 with High Sensitivity DNA Kit (Agilent). The six barcoded cDNA libraries were pooled together at equal molar ratio after quantitative PCR normalization (KAPA Library Quantification Kits) and loaded into three lanes of flow cell, and paired-end 2*100 nt multiplexed sequencing was conducted on Illumina HiSeq2000, yielding an average of 0.5 lanes of sequencing reads in total. The raw sequencing data reported in this work have been deposited in the NCBI GEO with accession numbers GSE67587, GSE67861 and GSE67862 for D. sechellia (Tuson, #14021-0248.25), D. sechellia (k-s10; #14021-0248.25) and D. simulans (k-s05; #14021-0251.194), respectively.
The raw reads data of D. melanogaster from Shiao et al. (2013) were included for analysis. Reference genomes version r5.48 (D. melanogaster), r1.3 (D. simulans), and r1.3 (D. sechellia) were used. For RNA-seq analysis, the 100-bp paired-end sequencing reads were mapped to the reference genome using TopHat 2.0.6 (Trapnell et al. 2009), with allowance of two mismatches in read mapping to the reference genome. The expression levels of genes were measured in Fragments Per Kilobase of exon per Million fragments mapped (FPKMs) by Cufflinks with bias correction (“–multi-read-correct” and “–frag-bias-correct”).
Generating Orthologous Gene Set and Identifying DEGs
Gene annotation tables of the three species were retrieved from FlyBase (version 2012_06) with individual versions 5.48, 1.3 and 1.3 for D. melanogaster, D. simulans and D. sechellia, respectively. The orthologous gene set (OGS) was manually curated by excluding genes duplicated in any of the species and only single copy genes in all three species were selected.
The potential bias from sequencing between different samples was normalized by upper quartile implemented in NOISeq (Tarazona et al. 2012). NOISeq requires technical replicates to perform the calculation. To identify DEGs, we used sequencing data from three lanes as technical repeats. As Dmel_TW had RNA-seq data from only a single sequencing lane for each sex, we generated technical replicates of Dmel_TW artificially in the software for comparison, using one function of NOISeq, NOISeq-sim.
Quantitative Reverse Transcription Polymerase Chain Reaction
We used NanoString quantitative reverse transcription polymerase chain reaction (qRT-PCR) technology to verify the gene expression levels estimated with RNA-seq. Hundred nanograms of total RNA from the same pool of total RNA for RNA-seq was used to determine the expression levels of 43 candidate genes simultaneously and three technical replicates of each gene were performed. Targeting sequences are shown in supplementary table S1, Supplementary Material online. Five genes, Actin 42A (Act42A), Cyclophilin-33 (cyp33), mitochondrial ribosomal protein L20 (mRpL20), Ribosomal protein L32 (RpL32) and Succinate dehydrogenase A (SdhA), that were found to have relatively consistent normalized FPKM values across all the samples (no significant differential expression was detected) and were used as commercialized products by some companies (such as Qiagen) were selected as endogenous genes for qRT-PCR experiments. Lush (Obp76a) was used as a positive control.
Identification of Gene Sequences of Or22a and Or22b
Or22a and Or22b were not included in the OGS table because of the presence of duplicate gene IDs. In the gene annotation files we used for analyses (version FB2012_06), Or22a has two distinct gene IDs in D. simulans (FBgn0068650 and FBgn0194492) and in D. sechellia (FBgn0171736 and FBgn0259897), whereas Or22b has two FlyBase IDs in D. simulans (FBgn0068649 and FBgn0194493). This resulted in the removal of Or22a and Or22b from the OGS in our data set. In the latest version of the gene annotation file (FB2015_02), FBgn0194492, FBgn0171736 and FBgn0194493 were assigned to other genes. We therefore manually curated the data set, and assigned FBgn0068650 to Or22a and FBgn0068649 to Or22b in D. simulans, and FBgn0259897 to Or22a in D. sechellia. Furthermore, we aligned DNA sequences of these two IDs to confirm that Or22a and Or22b of the three species form a monophyletic clade in the phylogenetic analysis (data not shown).
Results
Expression Profiles of Orthologous Genes in the Three Species
To obtain total RNA fully representing the transcriptomes of fly antennae, we detached antennae from heads under a microscope within one hour of eclosion. Antennal RNA of the three species (∼300–700 pairs of antennae from each species) was collected independently in two laboratories. First, antennae of D. sechellia and D. melanogaster for both sexes were collected in Academia Sinica, Taiwan. Second, antennae of D. sechellia and D. simulans for both sexes were collected at Ochanomizu University, Japan.
The transcriptomes of D. melanogaster (∼110 million paired-end reads) collected in Taiwan were published earlier in Shiao et al. (2013). To obtain similar sequencing amounts as in Shiao et al. (2013), we barcoded the six samples and pooled them to perform sequencing using three lanes on Illumina HiSeq2000. We obtained about 88–116 million paired-end sequencing reads for each (supplementary table S2, Supplementary Material online).
To compare the expression levels between species, we first generated an OGS without duplicate genes or gene loss in any of the three species, and obtained 10,034 annotated genes in OGS, including 51 Obp genes, 48 Or genes, 45 Gr genes, 34 Ir genes, 4 Csp genes, and 20 CheA/B genes (supplementary tables S3 and S4, Supplementary Material online). Eight chemosensory genes were excluded from OGS, including 5 Or genes (Or22a, Or22b, Or46a, Or65c, and Or67c), 1 Gr gene (Gr59b), and 2 Ir genes (Ir40a and Ir93a); most of these Or and Gr genes were proposed to be gene losses in D. sechellia (McBride and Arguello 2007). There were two copies of Ir40a and Ir93a in D. sechellia and D. simulans but only one copy in D. melanogaster based on gene annotation. We therefore removed these two Ir genes because of the uncertainty of orthology.
The expression levels of genes in OGS were then calculated in terms of FPKM. We used the three lanes as technical replicates, thus obtaining three FPKM values for each gene. The FPKM values of each gene from any pair of sequencing lanes were highly correlated (Pearson correlation r > 0.99), indicating no technical bias between different sequencing lanes (data not shown).
As observed in Shiao et al. (2013), the chemosensory genes that were expressed most abundantly in the antennae of D. melanogaster included the Obp gene family and a universal Or gene, Orco (also named Or83b). In total, 13 Obp genes, 2 Or genes, 1 Ir gene, and 1 Gr gene were consistently expressed at moderately high levels (FPKM > 50) in all eight samples, including D. melanogaster (supplementary table S4, Supplementary Material online).
There were ten genes consistently expressed at FPKM > 1,000 in all three species: a10, PebIII, Obp19a, Obp19d, Obp28a, Obp56d, Obp69a, Obp83a, Obp83b, and lush. The most highly expressed genes in antennae were all ligand binding protein genes. The gene lush is known to respond to 11-cis-vaccenyl acetate and acts as a sex and aggregation pheromone in D. melanogaster (Kim et al. 1998; Xu et al. 2005; Ha and Smith 2006). Obp19a and Obp19d belong to an Obp gene cluster that includes Obp19a, Obp19b, Obp19c and Obp19d, spanning an approximately 50-kb region on the X chromosome, according to the annotation of the D. melanogaster genome. The absence of Obp19b in our OGS was due to the missing annotation of D. simulans in the gene annotation file; it was expressed at moderate levels (FPKMs between 40.75 and 70.75) in D. melanogaster and D. sechellia. Obp19c was expressed at much lower levels in all three species, compared with Obp19a and Obp19d. A previous study found that in Obp19d, one nucleotide variation in the 5′-untranslated region and a synonymous substitution in the fourth exon were associated with variation in life span (Arya et al. 2010). Obp28a was proposed to be responsible for bitter taste in D. melanogaster, as suggested by the effect of Obp28a knockdown in D. melanogaster that resulted in an increased intake of bitter tartans, particularly quinine (Swarup et al. 2014). Obp69a, Obp83a, and Obp83b were proposed to be involved in “hearing” courtship song of males in female D. melanogaster (Immonen and Ritchie 2012).
Among the Or genes, Orco was expressed at the highest level in all samples, at FPKM > 1,000 in both D. melanogaster and D. simulans and >500 in D. sechellia. The high expression level of Orco is expected because it is universally expressed in OSN cells (Stengl and Funk 2013), including in the trichoid and basiconic sensilla of antennae (Larsson et al. 2004). Other highly expressed Or genes include Or42b, Or43b, Or47b, Or59b, and Or92a.
For the other chemosensory gene families, only Gr63a and Ir76b were expressed at moderate levels in all species. These two genes have been found to detect carbon dioxide and low salt, respectively, in D. melanogaster (Jones et al. 2007; Zhang et al. 2013).
DEGs in D. sechellia Antennae
We looked for genes involved in dietary shift, assuming that these genes should show differential expression between D. sechellia and the other two species, but not between male and female flies within a species because there is no sex difference in sensing the fruits of M. citrifolia. Under this hypothesis, genes related to the host shift in D. sechellia should show differential expression patterns in all of the following four comparisons: 1) Male D. melanogaster and male D. sechellia, 2) female D. melanogaster and female D. sechellia, 3) male D. simulans and male D. sechellia, and 4) female D. simulans and female D. sechellia.
The probability of significantly differential expression suggested by the authors of NOISeq software is q ≥ 0.8. A gene with q = 0.8 means it is four times more likely differentially expressed than nondifferentially expressed. Using this criterion, we obtained approximately 5,500 nonredundant single copy DEGs either between D. sechellia and D. melanogaster or between D. sechellia and D. simulans (supplementary tables S5–S8, Supplementary Material online). Among these genes, 1,532 genes are differentially expressed in D. sechellia compared with both sibling species. They contain 41 chemosensory genes (out of 177 chemosensory genes in OGS), including 14 Obp genes, 19 Or genes (including Orco), 3 Gr genes, and 5 Ir genes (supplementary table S9, Supplementary Material online).
As Orco is a universally expressed gene, the significantly lower expression level of Orco in D. sechellia might have resulted from the normalization process. We therefore conducted qRT-PCR by the NanoString technology to verify the differential expression of Orco between species (see “Materials and Methods”). We did not find a significant fold change in the expression levels of Orco in any of the pairwise comparisons (supplementary table S10, Supplementary Material online): −0.46 (in log2 scale) for D. sechellia versus D. melanogaster males, −0.76 for D. sechellia versus D. melanogaster females, 0.07 for D. sechellia versus D. simulans males, and −0.17 for D. sechellia versus D. simulans females. This observation suggested that the criterion of q ≥ 0.8 might not be high enough to define the truly DEGs in D. sechellia. In addition to Orco gene, we conducted qRT-PCR for some randomly selected chemosensory genes with q values between 0.8 and 0.95 or lower than 0.8. The log2-ratios of fold changes did not show consistent expression levels among species and most of the log2-ratios were not significant (supplementary table S10, Supplementary Material online). Furthermore, if no technical replicates were available, the authors of NOISeq recommended to use a higher threshold such as q = 0.9. For these reasons, we decided to use q = 0.95 in our study.
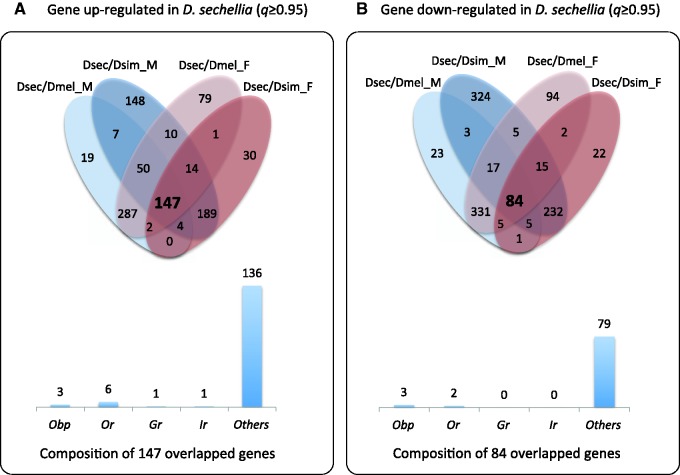
As mentioned above, we chose q ≥ 0.95 to define DEGs. In total, we identified 986 and 1,156 nonredundant single copy genes up- and downregulated, respectively, in D. sechellia antennae from the four independent comparisons: 1) 985 DEGs between males of D. melanogaster and D. sechellia, including 516 up- and 469 downregulated genes; 2) 1,142 DEGs between females of D. melanogaster and D. sechellia, including 591 up- and 551 downregulated; 3) 1,254 DEGs between males of D. simulans and D. sechellia, including 570 and 684 genes up- and downregulated; and 4) 751 DEGs between females of D. simulans and D. sechellia, including 384 up- and 367 downregulated. There were 147 genes upregulated and 84 genes downregulated in D. sechellia compared with D. melanogaster and D. simulans, including 13 and 5 chemosensory genes, respectively (fig. 1, table 1, supplementary tables S11 and S12, Supplementary Material online).
Fig. 1.—
Venn diagrams showing genes expressed differentially in D. sechellia (Dsec) in comparing with D. melanogaster (Dmel) or D. simulans (Dsim). A total of 147 genes were significantly upregulated (A) and a total of 84 genes were significantly downregulated (B) in D. sechellia. Males and females are presented in blue and red, respectively. The 147 upregulated genes include 3 Obp genes, 6 Or genes, 1 Gr gene, 1 Ir gene, 2 CheA/B genes and 134 other genes in the transcriptome (A). The 84 downregulated genes include 3 Obp genes, 2 Or genes, and 79 other genes in the transcriptome (B).
Table 1.
Expression Levels (FPKMs) and Log2-Ratios of Chemosensory Genes Differentially Expressed in Drosophila sechellia (Dsec) Compared with Drosophila melanogaster (Dmel) or Drosophila simulans (Dsim)
| Male |
Female |
Male |
Female |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Dsec | Dmel | Log2-Ratio | Dsec | Dmel | Log2-Ratio | Dsec | Dsim | Log2-Ratio | Dsec | Dsim | Log2-Ratio |
| Upregulated | ||||||||||||
| Obp19a | 8,457.39 | 3,030.22 | 1.48 | 14,437.39 | 4,482.95 | 1.69 | 6,512.19 | 3,393.15 | 0.94 | 9,217.77 | 4,944.62 | 0.90 |
| Obp50a | 45.16 | 0.35 | 7.02 | 47.24 | 0.75 | 5.98 | 23.85 | 1.79 | 3.74 | 22.26 | 1.65 | 3.75 |
| Obp56d | 18,266.52 | 2,260.36 | 3.01 | 16,892.33 | 2,426.42 | 2.80 | 13,876.18 | 4,499.63 | 1.62 | 10,827.04 | 4,812.76 | 1.17 |
| CheA75aa | 61.82 | 20.36 | 1.60 | 59.02 | 22.74 | 1.38 | 29.27 | 0.60 | 5.61 | 17.26 | 0.00 | 18.39 |
| CheA87a | 226.03 | 6.97 | 5.02 | 279.17 | 7.25 | 5.27 | 149.12 | 16.98 | 3.13 | 153.23 | 18.24 | 3.07 |
| Or23a | 44.99 | 4.42 | 3.35 | 44.62 | 4.16 | 3.42 | 40.34 | 14.91 | 1.44 | 34.92 | 13.72 | 1.35 |
| Or35a | 88.84 | 28.26 | 1.65 | 117.30 | 32.10 | 1.87 | 75.51 | 11.84 | 2.67 | 85.61 | 13.15 | 2.70 |
| Or56a | 98.88 | 40.70 | 1.28 | 109.11 | 48.20 | 1.18 | 111.82 | 29.27 | 1.93 | 109.05 | 33.82 | 1.69 |
| Or67ba | 44.83 | 17.05 | 1.39 | 44.11 | 20.50 | 1.11 | 36.12 | 11.17 | 1.69 | 31.19 | 11.95 | 1.38 |
| Or85b | 156.59 | 42.97 | 1.87 | 151.82 | 50.11 | 1.60 | 128.78 | 17.20 | 2.90 | 107.25 | 20.00 | 2.42 |
| Or85c | 95.97 | 4.53 | 4.41 | 161.15 | 5.80 | 4.80 | 68.71 | 4.28 | 4.00 | 103.45 | 8.54 | 3.60 |
| Gr64f | 43.67 | 18.36 | 1.25 | 57.43 | 25.85 | 1.15 | 29.77 | 3.67 | 3.02 | 35.23 | 5.85 | 2.59 |
| Ir84a | 30.10 | 5.72 | 2.40 | 22.87 | 5.74 | 1.99 | 25.92 | 2.62 | 3.30 | 19.13 | 2.75 | 2.80 |
| Downregulated | ||||||||||||
| Obp83a | 7,520.39 | 16,673.69 | −1.15 | 5,927.25 | 14,355.04 | −1.28 | 6,196.39 | 10,887.62 | −0.81 | 4,466.68 | 9,469.02 | −1.08 |
| Obp99c | 22.62 | 170.49 | −2.91 | 32.12 | 183.01 | −2.51 | 54.44 | 487.17 | −3.16 | 72.02 | 543.59 | −2.92 |
| Obp99d | 6.02 | 60.59 | −3.33 | 6.04 | 57.55 | −3.25 | 2.55 | 36.81 | −3.85 | 2.97 | 34.06 | −3.52 |
| Or9aa | 1.69 | 55.75 | −5.05 | 6.36 | 86.09 | −3.76 | 0.91 | 19.77 | −4.44 | 3.09 | 48.11 | −3.96 |
| Or42ba | 115.98 | 214.92 | −0.89 | 116.81 | 265.63 | −1.19 | 110.75 | 245.24 | −1.15 | 104.88 | 285.36 | −1.44 |
Note.—FPKMs shown in this table were upper-quartile normalized. Differential expression was defined with q ≥ 0.95 in the NOISeq test (Tarazona et al. 2012).
aThose four genes do not show consistent results between qRT-PCR and RNA-seq.
The chemosensory genes with the greatest log2-ratio are CheA75a and CheA87a. CheA75a was expressed at a very low level, almost no expression, in D. simulans, resulting in a very high log2-ratio between D. sechellia and D. simulans (5.61 and 18.39 in males and females, respectively). More interestingly, CheA87a was expressed at a high level in D. sechellia but at a low level in the two sibling species. CheA/B was proposed to be involved in male-specific pheromone sense in the front legs. Our data suggest a possible association of the CheA/B gene family with the host-plant shift of D. sechellia.
For the other chemosensory genes, the log2-ratios of FPKM values for Obp50a between D. sechellia and the two sibling species ranged from 3.74 to 7.04, indicating that the expression level of Obp50a was more than 100-fold higher in D. sechellia than in D. melanogaster (FPKM < 1) and D. simulans (FPKM = 1.79 and 1.65 in the two sexes, respectively). Obp50a belongs to a gene cluster that includes Obp50a/b/c/d/e, but only Obp50a was expressed at an intermediately high level in D. sechellia. Among the Or genes, Or85c showed the greatest differences in expression level between species: Greater than 20-fold higher in D. sechellia than in D. melanogaster and greater than 10-fold higher in D. sechellia than in D. simulans. Or85c was reported to be a larva-specific receptor in D. melanogaster (Couto et al. 2005; Kreher et al. 2005) and was proposed to be the receptor of 3-octanol (Mathew et al. 2013). More recent data indicated that it was expressed at a very low level in D. melanogaster antennae (Shiao et al. 2013). However, as there is no evidence that 3-octanol is one of the chemicals emitted by Noni fruit; the potential role of Or85c in sensing M. citrifolia by D. sechellia is unclear.
DEGs Validated by qRT-PCR
To verify DEGs, we conducted qRT-PCR using the NanoString technology. A total of 18 differentially expressed chemosensory genes were analyzed, along with 20 other genes to be discussed later in this section. The same total RNA used for RNA-seq was used for qRT-PCR. Targeting probes were designed in the consensus regions of the three species for all the genes selected. The Act42A, Cyp33, mRpL20, RpL32, and SdhA genes were selected as endogenous controls to normalize the expression levels. In addition, lush and Orco were included as positive controls. All the endogenous genes selected for qRT-PCR had similar expression levels (FPKMs) in the three species.
For most of these genes, the qRT-PCR data corresponded very well to the RNA-seq data except for Or9a, Or42b, Or67b, and CheA75a (supplementary table S13, Supplementary Material online). The qRT-PCR data for Or9a showed that the expression level was not significantly different between D. sechellia and the two other species because the log2-ratios ranged only from −0.13 to 0.08. On the other hand, the RNA-seq data suggested a much lower expression level for Or9a in D. sechellia. We checked the genomic location and found that a putative gene, FBgn0166364, overlapped with Or9a in the gene annotation table. We then calculated FPKMs by using a different version of annotation table (gene_orthologs_fb_2013_01.tsv.gz) that did not include FBgn166364 in the table (data not shown). The log2-ratios of FPKM values then suggested no significant differences in expression levels between species, consistent with the qRT-PCR results. Thus, some of the incongruences between RNA-seq and qRT-PCR analysis might be due to annotation artifacts (e.g., overlapping genes) and associated mapping issues. However, the reason why the qRT-PCR and the RNA-seq data for Or42b and Or67b differed is unknown.
Based on gene sequence analysis, CheA75a in D. simulans has diverged greatly from those in the other two species (∼68% divergence in peptide sequence between D. simulans and the two other species). We therefore could not find any probe-targeting consensus region in D. simulans CheA75a and expected very low or no amplification of CheA75a in D. simulans. Interestingly, we still detected expression of CheA75a in D. simulans by qRT-PCR and the expression level was almost the same as that in D. melanogaster. We performed a BLAST (Basic Local Alignment Search Tool) search using the 100-bp probe sequence of CheA75a against the D. simulans genome assembly and annotated genes in FlyBase, but no identical sequence segment longer than 20 bp was found. However, CheA75a had a sequence identify of 98% (gi190067942) and 99% (gi190008972) with two clones from a full-length cDNA in the cDNA library of D. simulans adult males in GenBank. Thus, the expression level of CheA75a in D. simulans remains unclear.
In addition to chemosensory genes, we also studied the expression levels of 18 genes with conspicuous differential expression patterns between D. sechellia and the other two species (CG16799, CG4797, and Listericin) with potentially interesting functions (supplementary table S13, Supplementary Material online), such as sense perception (nompC), olfactory learning (vsg), protein binding (Nup50), hormone catabolic process (Jheh1, Jheh2, Jheh3, and to), and insecticide metabolism (Cyp308a1 and GstE9). For most of these genes, the qRT-PCR data corresponded very well to the RNA-seq data.
For D. melanogaster, we generated artificial technical replicates, as recommended by NOISeq (see Materials and Methods). This procedure ignores the potential stochastic variation in the library preparation or sequencing reaction. Our qRT-PCR data were in agreement with the RNA-seq data.
Enrichment of Differentially Expressed Chemosensory Genes in D. sechellia
Based on our analysis, there were a total of 18 chemosensory genes among the 231 genes that showed up- or downregulation in the D. sechellia lineage (7.8%). Among the genes in OGS, there are a total of 9,844 genes including 177 chemosensory genes expressed in antennae. Note that we set the threshold of FPKM ≥ 0.05 to define that a gene is expressed in a species, following our previous papers (Shiao et al. 2012, 2013). All genes with FPKM ≥ 0.05 in at least one species were included in the analysis. In other words, a gene with FPKM < 0.05 in all three species was considered as “not expressed” in the antennae. We observed enrichment of chemosensory genes among the DEGs (18 [chemosensory DEGs]/213 [nonchemosensory DEGs] versus 159 [non-DEG chemosensory genes]/9,454 [non-DEG nonchemosensory genes], χ2 = 48.13, P < 0.00001). For comparison, we also investigated the DEGs in the D. simulans lineage (supplementary fig. S1, Supplementary Material online). We found that in D. simulans, there were 17 chemosensory genes among a total of 316 DEGs (5.4%), and compared with a total of 177 chemosensory genes among the 9,844 genes expressed in antennae, there was enrichment of chemosensory genes (17 [chemosensory DEGs]/299 [nonchemosensory DEGs] versus 160 [non-DEG chemosensory genes]/9,368 [non-DEG nonchemosensory genes], χ2 = 23.72, P < 0.00001). The analysis showed that chemosensory genes were enriched in the DEGs in the antennae of both D. sechellia and D. simulans, but there was a higher percentage of chemosensory genes differentially expressed in D. sechellia.
We further found that in D. sechellia, 13 and 5 chemosensory genes were up- or downregulated among a total of 147 upregulated and 84 downregulated genes, respectively, showing a higher percentage of upregulated chemosensory genes (8.8%) than downregulated chemosensory genes (6.0%). In contrast, the percentages of up- and downregulated chemosensory genes were quite similar in D. simulans; 10 (5.2%) and 7 (5.6%) among 191 upregulated and 125 downregulated genes, respectively. In summary, we proposed that some of these chemosensory genes have undergone Darwinian selection in the D. sechellia lineage.
Differentially Expressed Nonchemosensory Genes
Among the upregulated nonchemosensory genes, those most enriched in D. sechellia were mostly those with uncertain functions such as genes predicted to be in the subfamily of choline kinases (CG10513, CG10514, CG10553, CG32195, CG9497, and CG9498) and genes predicted to be involved in the following metabolism pathways (KEGG): Propanoate metabolism (succinyl-CoA synthetase [Sucb], CG10932, CG31075, and CG8778); butanoate and fatty acid metabolism (CG10932, CG31075, Arc42, and CG8778); tryptophan metabolism and lysine degradation (CG10932, CG31075, and CG8778) (supplementary table S14, Supplementary Material online). These genes tended to have higher fold-changes in expression between D. sechellia and its sibling species (log2-ratios ranging from 1.13 to 2.83), suggesting that these genes might be associated with new functions related to the host-plant adaptation of D. sechellia.
The nonchemosensory genes downregulated in D. sechellia were enriched in the GO category of toxin response and in gene families putatively involved in odorant degradation (Younus et al. 2014). These genes included glutathione S transferase E9 and D8 (GstE9 and GstD8), juvenile hormone eposide hydrolase 1, 2, and 3 (Jheh1, Jheh2, and Jheh3), and cytochrome P450 308a1 and 4d8 (Cyp308a1 and Cyp4d8) (supplementary table S15, Supplementary Material online). None of the above genes and gene families was significantly upregulated in D. sechellia, suggesting the need for further investigation of odorant degradation genes in D. sechellia antennae.
Expression Divergence of Chemosensory Genes between D. sechellia and Its Sibling Species
We generated heat maps for the Or, Ir, Obp, and Gr gene families and also the whole antennal transcriptome for each sex in each species (figs. 2–4 and supplementary figs. S2–S3, Supplementary Material online). We calculated the average expression level (without upper quartile normalization) of each gene estimated from three sequencing lanes followed by ranking within each of the above four gene families. Duplicate genes excluded in the OGS were now included for expression profile analysis. Genes with lowest expression levels (FPKM = 0 in all the samples) were ranked as 1 (colored yellow in the heat maps), whereas those with highest expression levels were colored red in the heat maps. Hierarchical agglomerative clustering was applied to cluster species as well as genes for each gene family. The significance of clustering was tested by multiscale bootstrap resampling implemented in the pvclust R package (Suzuki and Shimodaira 2006).
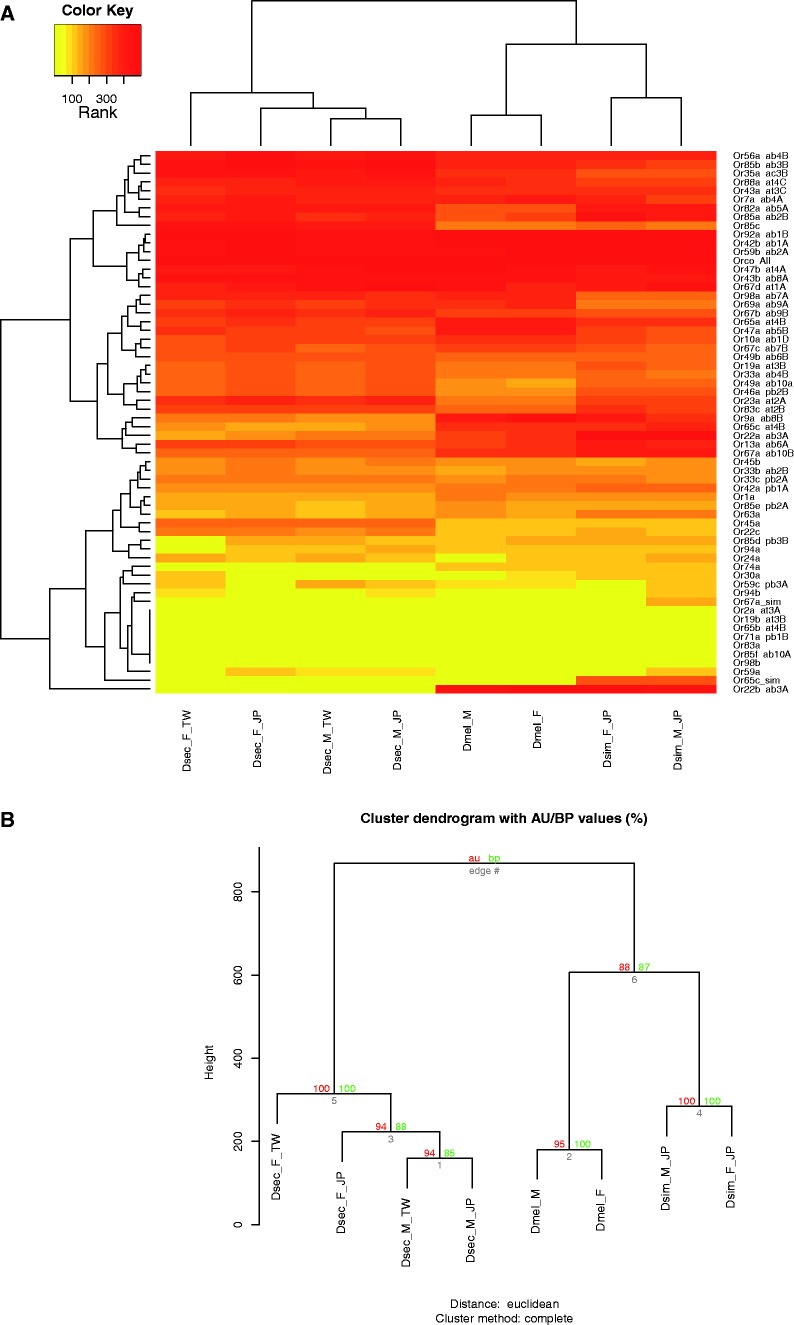
Fig. 2.—
Heat maps of expression profiles of Or genes. (A) Heat maps of ranked expression levels of Or genes. The red color indicates the highest expression level, whereas the yellow color indicates a low expression level. Clustering results of genes is shown on the left of the figure, and the gene names and expressed sensilla (Or genes) are shown on the right. Clustering of species is shown at the top of each figure and the information of each sample is shown at the bottom of the figure. Dmel, D. melanogaster; Dsim, D. simulans; Dsec, D. sechellia; M, males; F, females; TW, Taiwan; JP, Japan. (B) Statistical significance of clustering results among three species. The approximately unbiased (au) P value (%, in red) and bootstrap probability (bp) value (%, in green) are shown on each branch (in gray color).
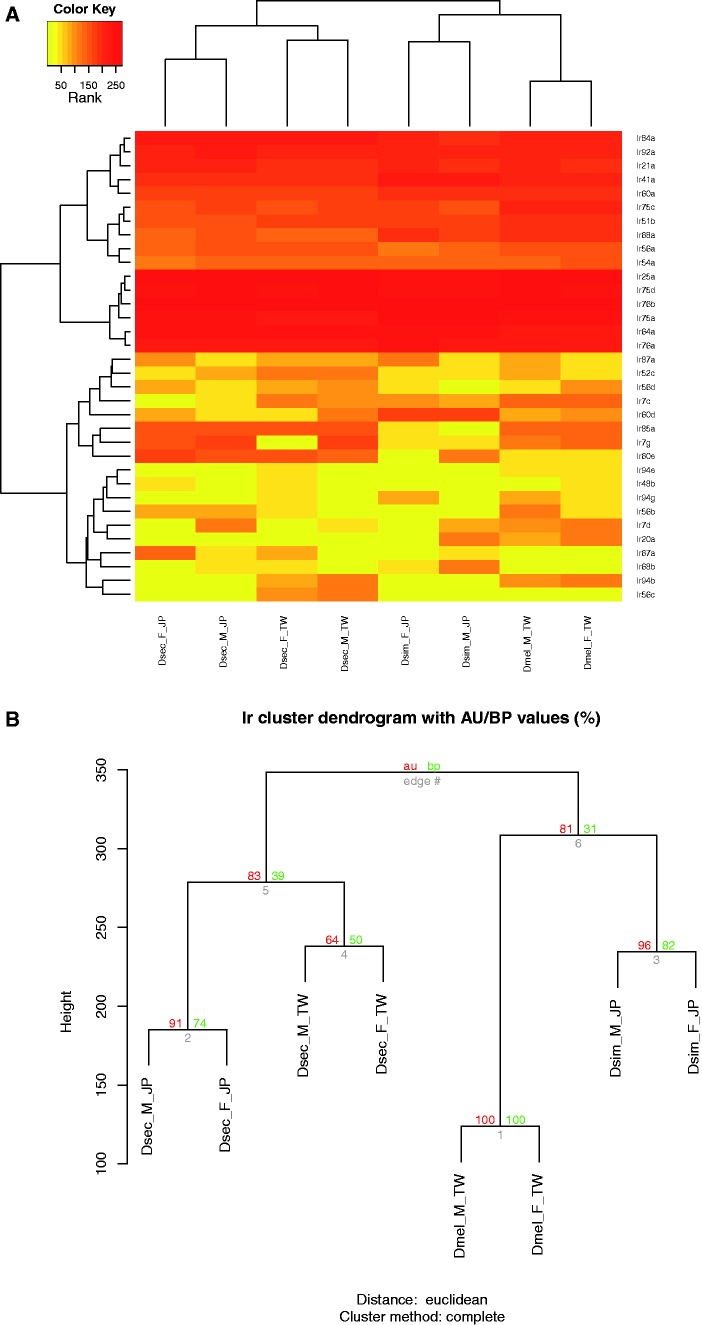
Fig. 3.—
Heat maps of expression profiles of Ir genes. (A) Heat maps of ranked expression levels of Ir genes and (B) statistical significance of clustering results among three species. Color patterns and sample names are as described in figure 2.
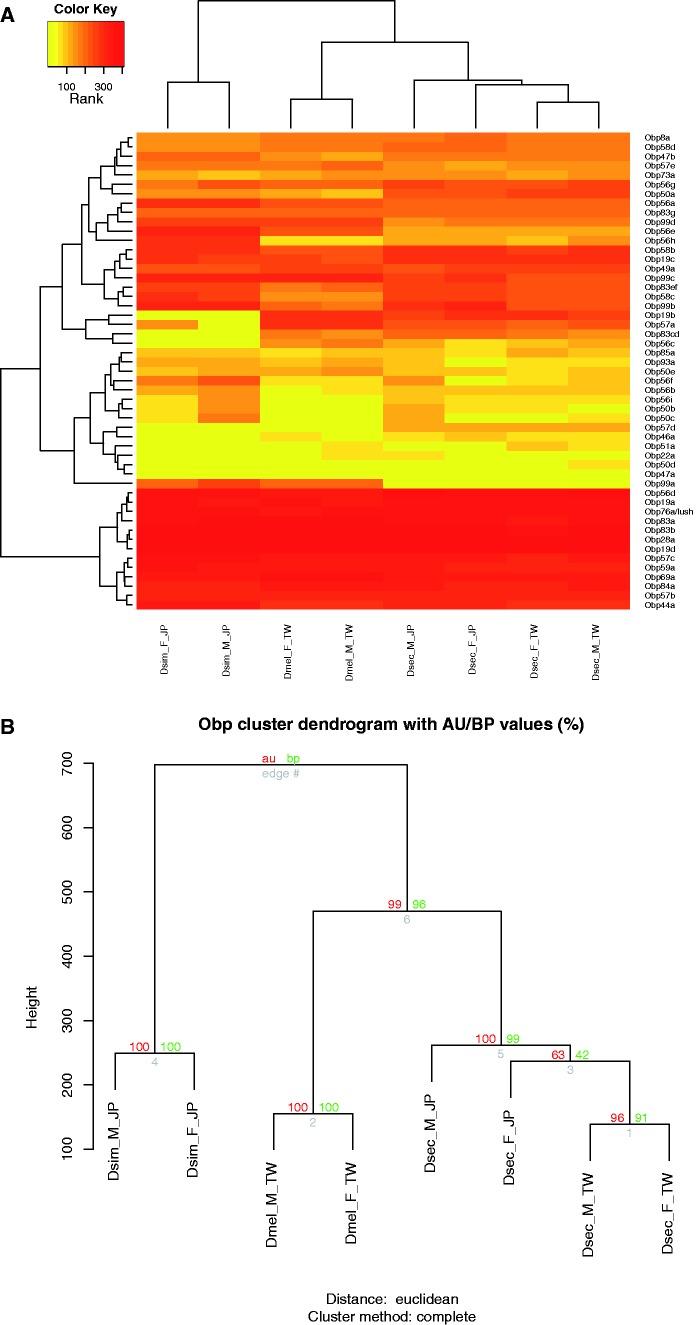
Fig. 4.—
Heat maps of expression profiles of Obp genes. (A) Heat maps of ranked expression levels of Obp genes and (B) statistical significance of clustering results among three species. Color patterns and sample names are as described in figure 2.
The results showed that the expression profiles of Or and Ir genes in D. sechellia antennae were significantly divergent from the two sibling species (figs. 2 and 3, approximately unbiased [au] P value > 0.95). The Obp genes also showed an expression profile divergent from the two sibling species, although D. sechellia was closer to D. melanogaster than to D. simulans (fig. 4). We did not observe any significant divergence in Gr genes or the rest of the transcriptome (supplementary figs. S2–S3, Supplementary Material online). The nonsignificant expression divergence in the Gr genes was likely due to the very low expression level of the gene family in the antennae.
The heat maps showed divergent expression between D. sechellia and the two sibling species for eight Obp and Or genes: Obp99a, Or13a, Or22a, Or22b, Or45a, Or65c, Or67a, and Or83c. Five of them (Obp99a, Or22a, Or22b, Or65c, and Or67a) were not included in OGS because they had undergone recent duplications or deletions in at least one of the three species. The Or13a, Or45a, and Or83c genes were not identified based on the criterion of q ≥ 0.95.
In addition, some of the genes showed differential expression in terms of the FPKM values, but were not seen in the color patterns of heat maps. For instance, Or56a was ranked as one of the highest expressed Or genes in all samples, but the FPKM values were actually significantly different among the three species. We therefore were not able to observe differential expression of this gene from heat map directly. A similar situation was observed in Obp and Ir genes. Five of them (Obp19a, Obp56d, Obp83a, Obp99c, and Ir84a) found to be significantly differentiated were all expressed at very high levels, so they were denoted by the same color patterns.
Discussion
We speculate that the adaptation of D. sechellia to the new host was under positive selection. This view was supported by the enrichment of differentially expressed chemosensory genes, particularly upregulated chemosensory genes, in the D. sechellia lineage. We also showed that chemosensory genes expressed in different developmental stages or responsible for courtship behavior in one of the sibling species might have evolved new function and contributed to the host shift of D. sechellia. In addition, Ir, Csp, and Che genes might also have played a role in the host shift of D. sechellia.
Dekker et al. (2006) proposed that the host shift in D. sechellia resulted from great differences in the numbers of sensilla in antennae. According to their results, the number of ab3 sensilla increased 2.5- to 3-fold in D. sechellia and a reduction of 93% to 100% of ab2 sensilla. Based on this observation, we expected to see upregulation of ab3-specific genes but downregulation of ab2-specific genes in D. sechellia. However, among ab3-specific genes, we found upregulation of only Or85b but not Or22a and Or22b in D. sechellia. Moreover, genes expressed on the ab2 sensilla, such as Or59b and Or85a, were not expressed consistently lower in D. sechellia than in the other two species as D. sechellia has fewer ab2 sensilla than the other two species. Therefore, the host shift may not be related to different numbers of sensilla in the antennae among the three species.
In this study, we included two strains of D. sechellia from two different labs, which were cultured in similar conditions. The two strains were derived from the same strain that has been widely used in fly laboratories. We did an analysis to compare expression levels of genes between the two strains of D. sechellia. There were only 131 genes differentially expressed (with the criterion of q ≥ 0.95) between the two strains. Thus, our data from the two D. sechellia strains may be taken as two biological replicates.
Csp Genes
Compared with other chemosensory gene families, the Csp gene family has the smallest number of genes: Only four genes in the Drosophila genome including a10 (also known as Os-D), CG30172, PebIII, and Phk-3 (Zhou et al. 2006). Among these four genes, only a10 is antenna-specific and expressed 6-fold higher than the other Csp genes in D. melanogaster (Zhou et al. 2006; Foret et al. 2007). Our data confirmed that a10 was expressed much higher than other Csp genes in all three species.
If q = 0.8 was used to determine DEGs (supplementary tables S5–S8, Supplementary Material online), then Phk-3 was expressed significantly higher in D. sechellia than in the two sibling species (q = 0.85–1.00 in the comparisons). Gene a10 and PebIII were expressed higher in D. sechellia than in D. melanogaster but lower in D. sechellia than in D. simulans. CG30172 showed higher expression in D. simulans than in D. sechellia, but not higher than in D. melanogaster. Based on these observations, we suggest that only Phk-3 might be associated with the host shift.
Ir Genes
Benton et al. (2009) identified 61 Ir genes in Drosophila. Using qRT-PCR or RNA in situ hybridization, the authors showed that 15 Ir genes were expressed in fly antennae. No expression was detected for the other Ir genes in any of the tissues studied. It remains unclear whether these genes were not expressed, expressed at different life stages, or expressed at levels below the detection threshold of the two techniques. In addition, 13 of the 15 Ir genes did not overlap in expression with the Or genes or Orco in adult antennae. The RNA-seq data in this study confirmed the expression of 10 of the 15 Ir genes reported in Benton et al. (2009) (supplementary table S4, Supplementary Material online). The other five genes (Ir8a, Ir31a, Ir40a, Ir75b, and Ir93a) were not included in the OGS, possibly due to incomplete gene annotation in the D. simulans and D. sechellia genomes.
Grosjean et al. (2011) proposed that Ir84a, a food-derived receptor, promotes male courtship in D. melanogaster. The authors showed that Ir84a was activated by food-derived aromatic odors, phenylacetic acid and phenylacetaldehyde, but not by fly-derived chemicals. The two chemicals are common in plants and fruits used as food sources and oviposition sites by Drosophila. This receptor was expressed in the OSNs that also expressed Ir75d, or Ir76a and Ir76b (Benton et al. 2009). Interestingly, we found that Ir84a and Ir76a were differentially expressed in D. sechellia. Ir84a was overexpressed in D. sechellia compared with the two sibling species. Ir76a was expressed significantly higher in D. sechellia than in D. melanogaster in both sexes, and was expressed significantly higher in females, though significantly lower in males compared with D. simulans (supplementary table S8, Supplementary Material online). Thus, Ir84a and Ir76a could have been associated with the host shift of D. sechellia.
Other Genes
We now discuss the genes previously proposed to be associated with the host shift in D. sechellia. The FPKM values of these genes are summarized in table 2.
Table 2.
Expression Levels of Genes Proposed to be Associated with Host-Plant Adaptation in Drosophila sechellia in Literature
| Taiwan |
Japan |
||||||
|---|---|---|---|---|---|---|---|
| Gene Name | Function/Sensilla Types | References | Sex | Drosophila melanogaster | Drosophila sechellia | Drosophila simulans | Drosophila sechellia |
| Orco | Universal OR | Males | 1,860.84 | 1,418.23 | 1,587.59 | 1,156.52 | |
| Females | 1,996.82 | 1,400.83 | 1,856.26 | 1,100.28 | |||
| Or19a | Sensing citrus | Dweck et al. (2013) | Males | 2.00 | 14.07 | 7.64 | 13.32 |
| Females | 2.16 | 16.01 | 9.73 | 14.84 | |||
| Or22a | Ab3 sensilla | Hallem and Carlson (2006) | Males | 11.67 | 1.06 | 75.07 | 1.09 |
| Kopp et al. (2008) | Females | 15.04 | 0.29 | 108.11 | 0.74 | ||
| Dekker et al. (2006) | |||||||
| Or22b | Ab3 sensilla | Kopp et al. (2008) | Males | 71.97 | 0.00 | 61.53 | 0.00 |
| Dekker et al. (2006) | Females | 82.33 | 0.00 | 85.56 | 0.00 | ||
| Or59b | Ab2 sesilla | Dekker et al. (2006) | Males | 73.95 | 106.80 | 192.70 | 104.11 |
| Females | 120.07 | 151.79 | 303.92 | 142.07 | |||
| Or85a | Ab2 sesilla | Dekker et al. (2006) | Males | 16.32 | 45.68 | 31.47 | 22.87 |
| Females | 21.92 | 74.15 | 51.18 | 35.98 | |||
| Or98a | Sensing MeHex | Hallem and Carlson (2006) | Males | 34.96 | 50.92 | 5.62 | 29.05 |
| Females | 39.94 | 58.93 | 7.96 | 29.32 | |||
| Or85b | Sensing MeHex | Hallem and Carlson (2006) | Males | 42.97 | 156.59 | 17.20 | 128.78 |
| Ab3 sensilla | Kopp et al. (2008) | Females | 50.11 | 151.82 | 20.00 | 107.25 | |
| Dekker et al. (2006) | |||||||
| Or47b | Sensing MeHex | Hallem and Carlson (2006) | Males | 89.18 | 66.00 | 62.48 | 119.29 |
| Females | 51.51 | 39.30 | 41.11 | 65.91 | |||
| Obp57d | Host-plant preference | Higa and Fuyama (1993) | Males | 0.00 | 0.53 | 0.00 | 0.56 |
| Matsuo et al. (2007) | Females | 0.00 | 0.37 | 0.00 | 0.63 | ||
| Obp57e | Host-plant preference | Higa and Fuyama (1993) | Males | 6.45 | 2.43 | 2.66 | 0.98 |
| Matsuo et al. (2007) | Females | 3.27 | 1.11 | 3.79 | 0.74 | ||
Note.—Expression levels (FPKMs) in bold indicate that the gene was differentially expressed between the two species. The FPKM values for some of the genes, such as Or22a and Or22b, were not normalized because they were not included in the OGS. Obp57d overlapped with Cpr57A based on gene annotation file, resulting in no read mapped to Obp57d in D. melanogaster and D. simulans.
The major volatile odors of M. citrifolia fruits are contributed by a variety of compounds, including HA, OA, methyl octanoate (MeOct), methyl hexanoate (MeHex), and 3-methyl-3-butenyl octanoate (Legal et al. 1994; Dekker et al. 2006; Wei et al. 2011). Although the acids HA and OA comprise the vast majority of the volatiles in M. citrifolia, a previous study showed that D. sechellia is more sensitive to the ester hexanoate than to acids, and is particularly sensitive to the hexanoate MeHex (Dekker et al. 2006). Hallem and Carlson (2006) showed that the two ester compounds, MeHex and MeOct, mostly stimulate Or22a, Or85b and Or98a, whereas they mostly inhibit Or47b in D. melanogaster (fig. 2). Using microarray expression analysis, Kopp et al. (2008) showed that Or22a, Or22b, and Or85b were significantly overexpressed in D. sechellia. However, Or22a and Or22b were also differentially expressed between D. simulans and D. melanogaster, and Or22a showed significant differences between two strains of D. melanogaster. For these two genes, our data are inconclusive as there are duplicated genes in one of the genomes, so that Or22a and Or22b were not included in OGS. In our data, Or85b was expressed at a significantly higher level in D. sechellia than in D. melanogaster and D. simulans (all the q values > 0.97). Also, Or98a was expressed at significant higher levels in both sexes in D. sechellia. Or47b, a receptor inhibited in D. melanogaster, showed no significant difference between D. melanogaster and D. sechellia, although a significantly higher expression level was detected in D. sechellia than in D. simulans. Therefore, only Or85b might be involved in the host shift of D. sechellia.
Dweck et al. (2013) proposed that Or19a is very important for the preference of citrus fruits as egg-laying substrate in flies. Our data are consistent with this hypothesis, because we found Or19a significantly upregulated in D. sechellia compared with the other two species.
Matsuo and his coworkers suggested that Obp57d and Obp57e are associated with the host-plant preference of D. sechellia (Higa and Fuyama 1993; Matsuo et al. 2007). Knocking out Obp57d and Obp57e in D. melanogaster led to a preference to the odorants emitted by the host-plant of D. sechellia. Using qRT-PCR, the authors found that Obp57d and Obp57e were expressed mainly in the legs of all three species. Obp57d was not included in our OGS because it completely overlaps with a cuticular protein 57A (Cpr57A) gene in D. melanogaster, making it impossible to do an informative differential expression analysis with D. melanogaster. Moreover, Obp57d was expressed at a very low level in the antennae of D. sechellia (FPKM < 1 in all D. sechellia samples). As for Obp57e, when lowering the criterion to q = 0.8, we found it to be expressed at a significantly lower level in D. sechellia (FPKM = 0.74 in Taiwan and 2.43 in Japan) than in D. melanogaster (FPKM = 6.45, P = 0.82) and D. simulans (FPKM = 3.79, P = 0.84).
Hungate et al. (2013) identified a genomic locus spanning approximately 170 KB comprising 18 genes, which are related to the OA tolerance in D. sechellia. These 18 genes included two major gene clusters belonging to the Obp gene family and the Osi gene family. The Obp gene cluster includes three Obp genes: Obp83cd, Obp83ef, and Obp83g. In our data, Obp83ef was upregulated in D. sechellia compared with D. melanogaster, but not D. simulans. The reason is that Obp83ef was expressed at very low levels in D. melanogaster compared with the other two species. Thus, it is unlikely that Obp83ef was involved in the host shift of D. sechellia. Obp83cd was not included in OGS and Obp83g showed an expression level of FPKM < 10 across all the data sets. All the genes (nine genes) in the Osi gene cluster with the exception of Osi8 and Gasp were expressed at very low levels in the antennae of all three species. Taken together, these observations suggest that this genomic locus probably does not play an important role in antenna-mediated adaptation, though they may be important in OA tolerance in other body parts.
In conclusion, our study suggests that all chemosensory gene families might have contributed to the host shift of D. sechellia, likely through upregulation of some chemosensory genes. That is, besides the genes proposed by previous studies, some additional chemosensory genes might have contributed to the host shift of D. sechellia.
Although we have shown that there was enrichment of differentially expressed chemosensory genes in D. sechellia, functional tests are needed to check whether any of the candidate genes contributed to the host shift of D. sechellia. Two approaches could be used to study the function of candidate chemosensory genes in the future. First, we could apply the Escherichia coli-based cell free expression system developed by Tegler et al. (2015) to express olfactory receptor genes of D. melanogaster to test the ligand binding affinity. This system would be able to help us confirm the binding affinity of candidate genes to the chemicals emitted by the host plant. Second, we could apply the CRISPR/Cas9 system to generate loss-of-function mutations in a specific gene or introduce a desired mutation by homologous repair in Drosophila (the review by Bassett and Liu 2014). With this technique, plasmids carrying guiding DNA of candidate chemosensory genes could be injected into embryos of D. sechellia and crossed to Cas9 strains. We can evaluate the function of mutated candidate genes by observing the behavior of mutated D. sechellia in terms of moving toward or avoiding chemicals emitted by the host plant.
Data Accessibility
The raw sequencing data reported in this work have been deposited in the NCBI GEO with accession numbers GSE67587, GSE67861 and GSE67862 for D. sechellia (Taiwan), D. sechellia (Japan) and D. simulans (Japan), respectively.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S15 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by Postdoctoral Fellows Program, Academia Sinica, Taiwan (to M.-S. S.); Plan Nacional (BFU2011–28575 to C.N. and J.-M.C.); Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013–2017” (SEV- 2012–0208 to C.N. and J.-M.C.); “Fundacio’ Obra Social la Caixa” (to J.-M.C.); and European Research Council (ERC- 2008-AdG No 232947 to J.-M.C.). The authors thank Dr Carol Lee, University of Wisconsin-Madison for valuable comments on the manuscript.
Literature Cited
- Arya GH, et al. 2010. Natural variation, functional pleiotropy and transcriptional contexts of odorant binding protein genes in Drosophila melanogaster. Genetics 186:1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Liu J-LL. 2014. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics. 41:7–19. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, et al. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22:327–338. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. 2000. Candidate taste receptors in Drosophila. Science 287:1830–1834. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 15:1535–1547. [DOI] [PubMed] [Google Scholar]
- Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. 2006. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 16:101–109. [DOI] [PubMed] [Google Scholar]
- Dweck HK, et al. 2013. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23:2472–2480. [DOI] [PubMed] [Google Scholar]
- Dworkin I, Jones CD. 2009. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics 181:721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Wanner KW, Maleszka R. 2007. Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 37:19–28. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159:1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, et al. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22:1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y, et al. 2011. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478:236–240. [DOI] [PubMed] [Google Scholar]
- Ha TS, Smith DP. 2006. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 26:8727–8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. 2006. Coding of odors by a receptor repertoire. Cell 125:143–160. [DOI] [PubMed] [Google Scholar]
- Higa I, Fuyama Y. 1993. Genetics of food preference in Drosophila sechellia. Genetica 88:129–136. [DOI] [PubMed] [Google Scholar]
- Hungate EA, et al. 2013. A locus in Drosophila sechellia affecting tolerance of a host plant toxin. Genetics 195:1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Ritchie MG. 2012. The genomic response to courtship song stimulation in female Drosophila melanogaster. Proc Biol Sci. 279:1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445:86–90. [DOI] [PubMed] [Google Scholar]
- Kim MS, Repp A, Smith DP. 1998. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 150:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman RM, et al. 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156:1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, et al. 2008. Evolution of gene expression in the Drosophila olfactory system. Mol Biol Evol. 25:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. 2005. The molecular basis of odor coding in the Drosophila larva. Neuron 46:445–456. [DOI] [PubMed] [Google Scholar]
- Lachaise D, et al. 1988. Historical biogeography of the drosophila-melanogaster species subgroup. Evol Biol. 22:159–225. [Google Scholar]
- Larsson MC, et al. 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43:703–714. [DOI] [PubMed] [Google Scholar]
- Legal L, Chappe B, Jallon JM. 1994. Molecular-basis of Morinda citrifolia (L)—toxicity on Drosophila. J Chem Ecol. 20:1931–1943. [DOI] [PubMed] [Google Scholar]
- Louis J, David JR. 1986. Ecological specialization in the Drosophila melanogaster species subgroup—a case-study of Drosophila sechellia. Acta Oecol Oecol Generalis. 7:215–229. [Google Scholar]
- Mathew D, et al. 2013. Functional diversity among sensory receptors in a Drosophila olfactory circuit. Proc Natl Acad Sci U S A. 110:E2134–E2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. 2007. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR. 2007. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177:1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SR, Kliman RM. 2008. Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data. PLoS one 3:e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio B, Couturier G, Lemeunier F, Lachaise D. 1983. Evolution d’une specialisation saisonniere chez Drosophila erecta (Diptera, Drosophilidae). Ann Entomol Soc Fr. 19:235–248. [Google Scholar]
- Rkha S, Capy P, David JR. 1991. Host plant specialization in the Drosophila melanogaster species complex—a physiological, behavioral, and genetic-analysis. Proc Natl Acad Sci U S A. 88:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao MS, et al. 2012. Transcriptomes of mouse olfactory epithelium reveal sexual differences in odorant detection. Genome Biol Evol. 4:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao MS, et al. 2013. Transcriptional profiling of adult Drosophila antennae by high-throughput sequencing. Zool Stud. 52:42–51. [Google Scholar]
- Starostina E, Xu A, Lin H, Pikielny CW. 2009. A Drosophila protein family implicated in pheromone perception is related to Tay-Sachs GM2-activator protein. J Biol Chem. 284:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengl M, Funk NW. 2013. The role of the coreceptor Orco in insect olfactory transduction. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 199:897–909. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Dekker T, Hansson BS. 2003. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc Biol Sci. 270:2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- Swarup S, Morozova TV, Sridhar S, Nokes M, Anholt RR. 2014. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem Senses. 39:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, et al. 2012. NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet 17:18–19. [Google Scholar]
- Tegler LT, et al. 2015. Cell-free expression, purification, and ligand-binding analysis of Drosophila melanogaster olfactory receptors DmOR67a, DmOR85b and DmORCO. Sci Rep. 5:7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacas L, Bachli G. 1981. Drosophila sechellia n. sp., huiteme espece du sous-group melanogaster des iles Sechelles (Diptera, Drosophilidae). Rev Fr Entomol. 3:146–150. [Google Scholar]
- Vieira FG, Rozas J. 2011. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96:725–736. [DOI] [PubMed] [Google Scholar]
- Wei GJ, Ho CT, Huang AS. 2011. Analysis of volatile compounds in noni fruit (Morinda citrifolia L.) juice by steam distillation-extraction and solid phase microextraction coupled with GC/AED and GC/MS. J Food Drug Anal. 19:33–39. [Google Scholar]
- Xu A, et al. 2002. Novel genes expressed in subsets of chemosensory sensilla on the front legs of male Drosophila melanogaster. Cell Tissue Res. 307:381–392. [DOI] [PubMed] [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. 2005. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45:193–200. [DOI] [PubMed] [Google Scholar]
- Younus F, et al. 2014. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem Mol Biol. 53:30–43. [DOI] [PubMed] [Google Scholar]
- Zhang YV, Ni J, Montell C. 2013. The molecular basis for attractive salt-taste coding in Drosophila. Science 340:1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Kan Y, Antoniw J, Pickett JA, Field LM. 2006. Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem Senses. 31:453–465 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data reported in this work have been deposited in the NCBI GEO with accession numbers GSE67587, GSE67861 and GSE67862 for D. sechellia (Taiwan), D. sechellia (Japan) and D. simulans (Japan), respectively.