Abstract
Endosymbiosis is a common phenomenon in nature, especially between bacteria and insects, whose typically unbalanced diets are usually complemented by their obligate endosymbionts. While much interest and focus has been directed toward phloem-feeders like aphids and mealybugs, blood-feeders such as the Lone star tick (Amblyomma americanum), Glossina flies, and the human body louse (Pediculus humanus corporis) depend on obligate endosymbionts which complement their B-vitamin-deficient diets, and thus are required for growth and survival. Glossiphoniid leeches have also been found to harbor distinct endosymbionts housed in specialized organs. Here, we present the genome of the bacterial endosymbiont from Haementeria officinalis, first of a glossiphoniid leech. This as-yet-unnamed endosymbiont belongs to the Gammaproteobacteria, has a pleomorphic shape and is restricted to bacteriocytes. For this bacterial endosymbiont, we propose the name Candidatus Providencia siddallii. This symbiont possesses a highly reduced genome with high A+T content and a reduced set of metabolic capabilities, all of which are common characteristics of ancient obligate endosymbionts of arthropods. Its genome has retained many pathways related to the biosynthesis of B-vitamins, pointing toward a role in supplementing the blood-restricted diet of its host. Through comparative genomics against the endosymbionts of A. americanum, Glossina flies, and P. humanus corporis, we were able to detect a high degree of metabolic convergence among these four very distantly related endosymbiotic bacteria.
Keywords: Haementeria officinalis, Providencia siddallii, leech endosymbiont, blood-feeder, genome reduction, B-vitamin
Introduction
Symbiotic associations between insects and bacteria have been widely studied. The roles these symbionts play in their associations vary greatly from nutrient providing (Akman Gündüz and Douglas 2009), parasitoid and fungal defence (Oliver et al. 2003; Scarborough et al. 2005), cytoplasmic incompatibility (Yen and Barr 1971; Hunter et al. 2003), among others (Chen et al. 2000; Montllor et al. 2002; Nakabachi et al. 2013). Many examples from obligate endosymbionts come from insects feeding on nutrient-deficient diets such as phloem or xylem sap, whose diets are rich in carbohydrates but deficient in most essential amino acids and cofactors (Hansen and Moran 2014). Strict blood-feeders are no strangers to nutritional symbioses, finding obligate endosymbionts harbored in a variety of arthropods such as ticks (Zhong et al. 2007; Liu et al. 2013; Lalzar et al. 2014; Smith et al. 2015), bedbugs (Hosokawa et al. 2010; Nikoh et al. 2014), lice (Allen et al. 2007; Kirkness et al. 2010), Hippoboscoidea flies (Akman et al. 2002; Hosokawa et al. 2012; Rio et al. 2012), and in a particular group of annelids: leeches (Kikuchi and Fukatsu 2002; Siddall et al. 2004; Perkins et al. 2005; Kvist et al. 2011). Analogous to the symbionts from phloem and xylem feeders, nutritional symbionts from strict blood-feeding arthropods have been found to dedicate a part of their reduced gene repertoire to the biosynthesis of B-vitamins (Akman et al. 2002; Rio et al. 2012; Nikoh et al. 2014; Smith et al. 2015). This has been explained through the need to supplement their hosts’ blood-restricted diets, which are poor in these nutrients (Lehane 2005). Even though highly detailed studies of the association between insects and bacteria have been seminal to our current understanding of symbiosis including metabolic complementation, “symbiotic syndrome,” and coevolutionary analyses, little is known about these phenomena outside the class Insecta. Leeches are members of the Phylum Annelida, that together with Mollusca, Platyhelminthes, and other minor phyla form the group Lophotrochozoa, which is by its own, sister group to Ecdysozoa, including Arthropoda and Nematoda, among others (Dunn et al. 2008). Given that leeches and arthropods are distantly related, they represent phylogenetically independent models to study symbiotic relationships and represent a great case to investigate the generality of the phenomena already characterized in Insecta.
Leeches (Hirudinea), together with earthworms (Oligochaeta) form the class Clitellata, a highly derived group of the diverse Phylum Annelida or “segmented worms” (Rousset et al. 2007). Strict blood-feeding leeches are found within all major lineages from the group, and it has been suggested that the last common ancestor of the whole group was a blood-feeder (Apakupakul et al. 1999). Blood-feeding members of the order Arhynchobdellida, including the European medicinal leech Hirudo medicinalis and its North American counterpart Macrobdella decora lack specialized organs to harbor symbiotic bacteria. Nevertheless, leeches from the genus Hirudo form stable and heritable associations with bacteria from the genus Aeromonas and Mucinivorans (Graf 1999; Worthen et al. 2006; Siddall et al. 2011; Nelson et al. 2015). Although the exact method these two bacteria use to be transmitted to offspring in unknown, in Hirudo verbana, it has been noted that Aeromonas veronii is present as soon as cocoons are deposited, whereas Mucinivorans is only detectable at a later time, as identified by diagnostic polymerase chain reactions (PCRs; Rio et al. 2009). The exact contribution of these bacteria to their host is yet to be determined. Strict blood-feeding leeches of the proboscis-bearing order Rhynchobdellida also form apparent vertically transmitted associations with distinct bacteria (Kikuchi et al. 2002; Siddall et al. 2004; Kikuchi and Fukatsu 2005; Goffredi et al. 2012). Within this order, strict blood-feeding leeches from the Glossiphoniidae family have been found to hold intimate relationships with endosymbiotic bacteria harbored in specialized organs called “esophageal glands” or “mycetomes” (hereafter bacteriomes) (Kikuchi and Fukatsu 2002; Siddall et al. 2004; Perkins et al. 2005). Although the way in which these endosymbionts are transmitted to the offspring is not known, two pieces of evidence point toward a vertical transmission from parent to offspring through the egg. First, it has been shown that 100% of Placobdelloides spp. examined eggs were infected with the same bacterial species found in adult bacteriomes (Kikuchi and Fukatsu 2002). Second, juvenile Placobdella parasitica individuals that had never received a blood meal were also found to harbor a large population of bacteria in their bacteriomes (Siddall et al. 2004).
It has been determined that there are at least three independent phylogenetic origins for the bacteriome endosymbionts from glossiphoniid leeches (Siddall et al. 2004; Perkins et al. 2005), one within the Alphaproteobacteria (detected in the leeches of the genus Placobdella) and two within different lineages of Gammaproteobacteria (detected in Placobdelloides and Haementeria species). Previous genomic approaches to the study of the alphaproteobacterial Reichenowia symbiont of P. parasitica, have provided a raw view into this bacterium (Kvist et al. 2011). Nevertheless the low coverage and assembly level greatly impaired further analyses. Regarding the gammaproteobacterial symbionts in the glossiphoniid leech Haementeria ghilianii, the pleomorphic bacterial associate has been found to be embedded in a collagenous extracellular matrix surrounding the mature bacteriomes of the leech (Perkins et al. 2005).
Haementeria species are geographically restricted to the Americas, with the bulk of species in South America and a few in Mexico (Oceguera-Figueroa 2012). These leeches feed on vertebrates’ blood, mainly mammals, and therefore it is expected their endosymbiont could be complementing its B-vitamin deficient diet. In this study, we have sequenced the genome of the bacterial endosymbiont of the Mexican leech Haementeria officinalis, the first whole-genome from an obligate endosymbiont from a leech. We have analyzed its genomic characteristics, and through phylogenomic methods we were able to confidently determine the free-living bacterial clade more closely related to this bacterium. Additionally, we have reconstructed and analyzed the metabolic capabilities of the symbiont and propose a nutrient-provisioning basis for the bacterial-leech symbiosis. Finally, we used the genomic data available from various distinct and phylogenetically distantly related obligate endosymbiotic bacteria with putative similar ecological roles (coming also from distantly related hosts) to perform broad comparative genomic analyses on the biosynthetic capabilities of B-vitamins. Through this, we gained further insight into the underlying requirements from these nutrients from the endosymbionts’ hosts. Also, we propose some ways in which the different hosts could control the production of these vitamins and we hypothesize on the different pathways particular blood-feeders’ endosymbionts are able to biosynthesize these essential compounds.
Materials and Methods
Leech Collection, DNA Extraction, and Sequencing
Haementeria officinalis individuals were collected on November 1, 2011, by Alejandro Oceguera-Figueroa and Javier Vargas Sánchez. Leeches were collected by immersing legs into the water in the edges of the pond, waiting for about 1 min and then examining for leeches attached to the skin. The collection site is located at the municipality of Coroneo, Guanajuato (20°17′56ʺ N 100°25′44ʺ W, Altitude 2270). Dissection was performed on 35 individuals to retrieve the four bacteriomes of each specimen. DNA extraction was performed with the JETFLEX Genmic DNA Purification Kit (GENOMED 600100). The extracted DNA was then sequenced at the sequencing facility at the FISABIO (http://fisabio.san.gva.es/en/secuenciacion-masiva-y-bioinformatica, last accessed January 6, 2015) on the 454 platform (1 plate) using the FLX+ system. Other Haementeria species were collected from different locations (supplementary table S1, Supplementary Material online) and, after bacteriome dissection, DNA was extracted using the DNeasy extraction kit (QIAGEN). Bacterial 16S rRNA gene sequences were amplified using bacterial universal primers BSF8 and BSR1541 in 25 µl volumes. PCR conditions were set as Perkins et al. (2005).
Preassembly
For all 454 reads, we first performed an extraction of the RAW reads using the program sff_extract v0.3.0 (http://bioinf.comav.upv.es/sff_extract/download.html, last accessed January 6, 2015), developed by the COMAV Institute (http://bioinf.comav.upv.es/index.html, last accessed January 6, 2015). We discarded reads shorter than 100 bp, longer than 1,000 bp, those without a single base pair with quality higher than 35 and those with undefined nucleotides (“N”) using Pyrocleaner v1.3 (Jérôme et al. 2011). The remaining reads were taxonomically assigned using PhymmBL v4.0 (Brady and Salzberg 2011) with custom-added genomes of various representatives from the class Insecta (Atta cephalotes, Acyrthosiphon pisum, Drosophila melanogaster, and Tribolium castaneum), human Homo sapiens GRCh37.p5, and the leech Helobdella robusta, along with their corresponding mitochondrial genomes. We determined that around 53% of the 646,927 reads corresponded to the Gammaproteobacteria class (344,438 reads), as visualized using Krona v 2.4 (Ondov et al. 2011).
Genome Assembly
The 454 reads were assembled using MIRA v4.0.2 (Chevreux et al. 1999). Manual editing in Gap4 from the Staden package (Staden et al. 1999) followed the automatic assembly resulting in four contigs with an average coverage of 253× and a total joined length of 843,823 bp. We then tried remapping unused reads to the assembled contigs, but these were not extended. Reads that closed the gaps were manually searched for, but no matches were found. wgs-assembler v7.0 (Myers et al. 2000) and gsAssembler v2.8 (ROCHE) were also tested in an attempt to close this four gaps, but same results were obtained. As an alternative, primers were designed on each 5′- and 3′-ends of the contigs and attempts to amplify every combination possible were unsuccessful.
Genome Annotation and Metabolic Reconstruction
The four contigs underwent a first round of open reading frame (ORF) prediction using Prodigal v2.5 (Hyatt et al. 2010) and were annotated using BASys server v1.0 (Van Domselaar et al. 2005). tRNAs were annotated using the standalone version of tRNAscan-SE v1.3.1 (Lowe and Eddy 1997) (COVE-only) and checked using TFAM v1.4 (Tåquist et al. 2007). rRNAs, regulatory RNA structures, and other ncRNAs were annotated using Infernal v1.1.1 (Nawrocki and Eddy 2013) and the Rfam database v12.0 (Burge et al. 2013) with a step of manual curation for the 16S and 23S ribosomal genes to correct boundaries. RBSfinder (Suzek et al. 2001) was used to both correct start codons and to predict putative ribosome-binding sites of coding sequences (CDSs). Manual curation on the annotation of genes and search for pseudogenes and other features was done on UGENE (Okonechnikov et al. 2012) using NCBI’s BLASTx, BLASTp (Altschul et al. 1997), and delta-BLASTp (Boratyn et al. 2012) against NCBI’s nr and nt databases when needed. Priority for these searches was as following: 1) against Escherichia coli K-12 substrain MG1655 and 2) against the whole nr database. CDSs were considered functional if they presented no frameshifts disrupting the CDS or if they preserved all the regions and domains that have previously been identified by experimental evidence as essential for gene function, based on information available at EcoCyc (Keseler et al. 2013) or from other published experiments. If they did not complied to the criteria explained before, they were considered pseudogenes. A search of all the CDS’s protein sequence was done using the standalone version of InterProScan v5.8 (Hunter et al. 2012) against the database v48.0 to infer GO terms, Pfam, InterPro motifs, etc.
The four fully annotated contigs were then submitted to the metabolic annotation process implemented in Pathway Tools v18.5 (Karp et al. 2010) against BioCyc and MetaCyc databases (Caspi et al. 2014). After the automatic reconstruction, manual curation of the database was done comparing to known reactions and complexes present in BioCyc.
COG Profiles
COG categories were assigned using various ad hoc perl scripts to find nonoverlapping hits against the COG database using BLASTp with an e-value cutoff of 1e-03. The COG profile displays and clustering were made using the heatmap2 function from the R (R Core Team 2015) package gplots. Absolute COG category frequencies were divided by the strains total number of COG assigned CDSs. For identifying the niche-specific differences between the phloem and blood-feeders’ metabolic profiles, we subtracted all the COG assignments belonging to core proteins. For assessing functional divergence of the leech gammaproteobacterial symbiont from the free-living Providencia strains, a mean of per COG category frequency was calculated for the latter and subtracted from the given category for both the leech symbiont and Providencia strains as in (Manzano-Marín et al. 2012).
Phylogenetic Analyses of Symbionts of Haementeria spp. and Ortholog Groups of Proteins
Bayesian phylogenetic placement of the endosymbionts identified from the different Haementeria species was done using MrBayes v3.2.4 (Ronquist et al. 2012) under the general time reversible GTR+I+G model (two independent runs, four chains each). Sequences were aligned along with other bacterial species retrieved from the SILVA database (Quast et al. 2013) (supplementary table S2, Supplementary Material online) using ssu-aligner v0.1 (Available from: http://selab.janelia.org/software/ssu-align/, last accessed March 13, 2015). Manual editing was performed to the alignment, and then Gblocks v0.91 (Talavera and Castresana 2007) was used to produce final alignment for phylogenetic inference (supplementary file S2, Supplementary Material online).
Construction of the ortholog groups of proteins was done using OrthoMCL v2.0.9 (Chen et al. 2007) as in (Manzano-Marín et al. 2012) using genomes available for representatives from different genera of bacteria belonging to different Gammaproteobacteria (supplementary table S3, Supplementary Material online), mainly Enterobacteriaceae, to which a close relative from the leech H. ghilianii endosymbiont was phylogenetically placed through phylogenetic reconstruction using the 16S rRNA gene sequence (Perkins et al. 2005). We identified 55 out of the 69 single-copy core genes used for phylogenetic analysis (Bayesian inference) of endosymbiotic bacteria in (Husník et al. 2011). Then, both alignment (supplementary file S3, Supplementary Material online) and phylogenetic reconstruction were conducted as in (Husník et al. 2011) using Dayhoff6 recoded amino acid alignments as implemented in Phylobayes v3.3f (Lartillot et al. 2009) under the CAT+GTR model. We ran six independent chains for 9,666 generations and a burn-in of 3,000 was chosen. Both bipartition maxdiff and summary variables were less than 0.07 and all effective sizes of all summary variables were higher than 173. Visual display of the summary tree was done using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed January 13, 2015) and edited in Inkscape v0.91 (http://www.inkscape.org/en/, last accessed January 13, 2015).
Results
Candidatus Providencia siddallii sp. nov.
We propose the name Candidatus Providencia siddallii strain officinalis-GTOCOR (hereafter referred to as Providencia siddallii Off) for the gammaproteobacterial symbiont living in the bacteriomes of the leech Haementeria officinalis. Our current study has positioned this endosymbionts nested within the Providencia clade (supplementary fig. S1, Supplementary Material online), having Providencia stuartii strain MRSN 2154 as its closest relative by 94% 16S nucleotide identity. Using a generic name for this newly characterized Gammaproteobacteria would render Providencia paraphyletic, which is not desirable given the fact that only monophyletic groups should be recognized and named. Previous phylogenetic analyses conducted on the endosymbiont from H. ghilianii, a close relative of H. officinalis, have also positioned this relative of Pr. siddallii Off as a close relative of Pr. stuartii (Perkins et al. 2005). The species name siddallii refers to Mark E. Siddall, Curator of Invertebrates at the American Museum of Natural History (NY), and Leech expert who, together with Susan Perkins, was the scientist that identified the bacterial symbionts of Haementeria for the first time. The prefix strain name “officinalis,” refers to the Haementeria leech species host of this Gammaproteobacteria. The use of the species name Pr. siddallii is recommended for all bacterial endosymbionts from Haementeria leeches, forming a monophyletic clade, closely related to Providencia.
Providencia siddallii’s Bacteriome Localization and Monophyly of Haementeria Endosymbionts
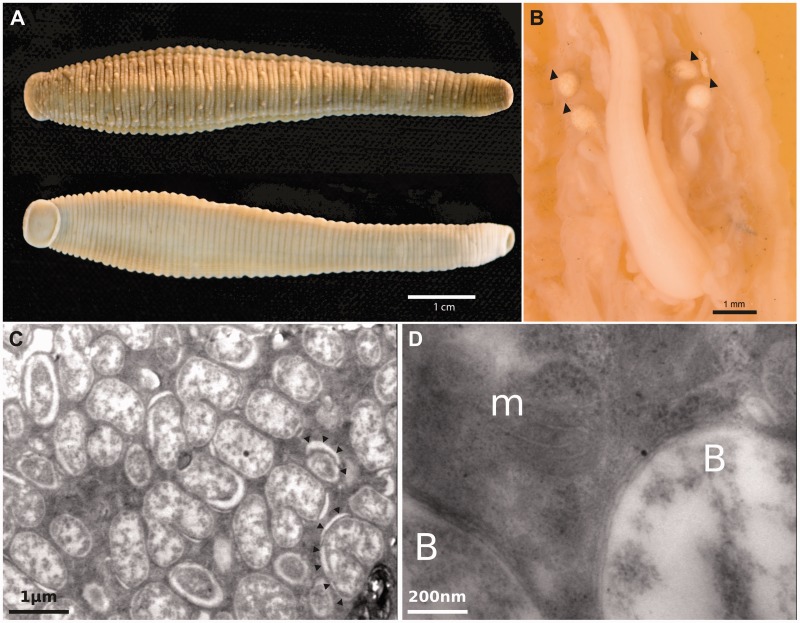
The Mexican leech H. officinalis (de Filippi 1849) (fig. 1A) has a bacteriome, which consists of four large globular sacks which join to the esophagus via thin ducts (fig. 1B), similar to the bacteriome of H. ghilianii (Perkins et al. 2005). The bacteriome of both species is populated by pleomorphic bacterial cells (Perkins et al. 2005 and fig. 1C). The symbiont of H. officinalis is located intracellularly and seems to be contained in a symbiosome (fig. 1C and D). The 16S rRNA gene sequence retrieved from Pr. siddallii Off has a 95% identity to the partial sequence published of the bacterial endosymbiont of H. ghilianii (GenBank accession number AY999969). Hereafter, we will refer to this relative of Pr. siddallii Off as simply Pr. siddallii Ghi (strain ghilianii). Providencia siddallii Ghi differs from Pr. siddallii Off in that this has a “CCAGCGACTTTAGTCGGGA” insertion relative to Pr. siddallii Ghi’s 16S rRNA gene (positions 1114–1132 from Pr. siddallii Off’s 16S rRNA). Also, we were able to determine using various 16S rRNA gene sequences retrieved from five different species of Haementeria leeches, that they formed a monophyletic cluster which probably originated from within the Providencia clade (supplementary fig. S1, Supplementary Material online). Also, from this phylogeny, It is evident the general congruency of the endosymbionts’ phylogeny with that of its hosts (Oceguera-Figueroa 2012), possibly indicating parallel evolution.
Fig. 1.—
Haementeria officinalis, bacteriome, and resident Pr. siddallii Off bacteria. (A) View of an H. officinalis leech. (B) Bacteriomes of a dissected H. officinalis leech indicated by black arrowheads. (C) Providencia siddallii Off’s pleomorphic cells present in the bacteriomes of H. officinalis. Putative symbiosomes boundaries are marked with black arrowheads. (D) Magnification showing bacterial cells (B) and mitochondrion (m), evidencing the intracellular localization of Pr. siddallii Off. Scale-bars are presented in different colors to be easily distinguished.
The Pr. siddallii Off Genome
The genome of Pr. siddallii Off has been assembled to four nonoverlapping contigs spanning 843,824 bp with a 454 average coverage of 253×. The sequences have been deposited at DDBJ/EMBL/GenBank under the project number PRJEB6644.
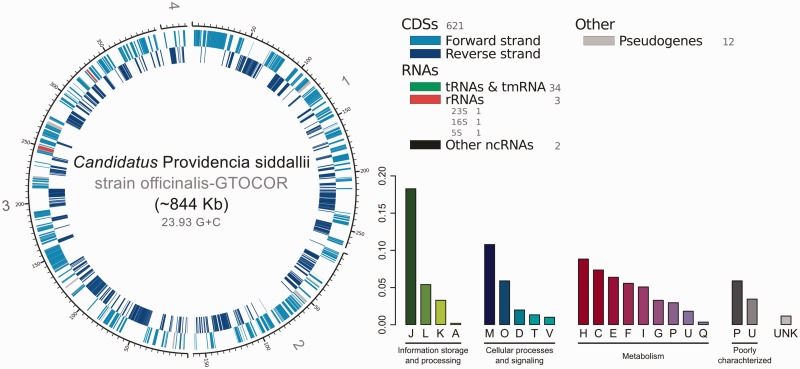
Providencia siddallii Off possesses a small genome with a very low average G+C content of 23.93% (fig. 2: left) and a coding density of only 73.48%. It presents 26 pseudogenes and 15 intact genes that are putatively recent events of gene shrinkage (nonessential protein components missing or recognizably present in another reading frame or after an interrupting stop codon, see Materials and Methods). These last group, still preserve distinguishable DNA sequence belonging to the original gene, but that are now not part of the CDS as a result of a frameshift or an early stop codon. The genome presents no gene duplication, and as other endosymbionts from blood-sucking insects, has a good part of its metabolic genetic repertoire dedicated to the metabolism of coenzymes (fig. 2: barplot).
Fig. 2.—
Providencia siddallii strain officinalis-GTOCOR genome. Left: Pr. siddallii Off genome plot. Bottom right: Barplot of COG categories relative abundance. The highest amount of genes involved in metabolism are the ones present in category H (coenzyme metabolism), to which B-vitamin biosynthetic genes belong to.
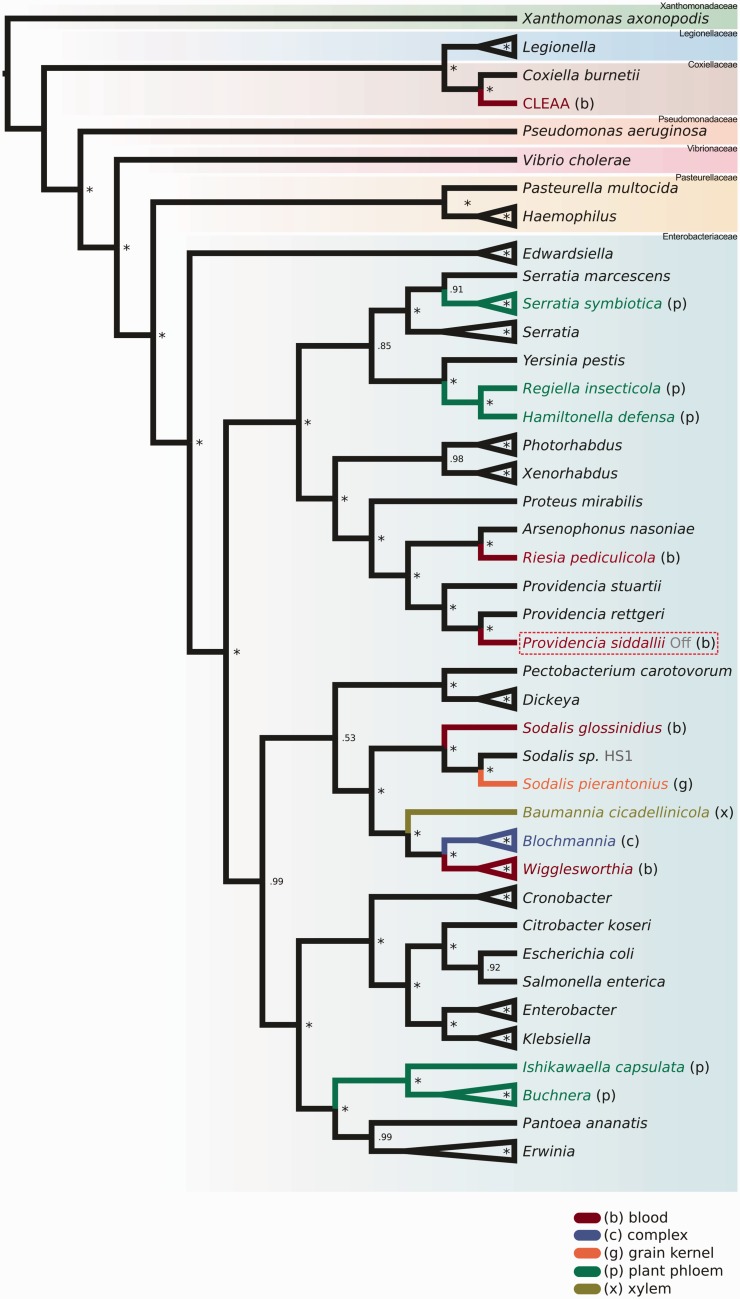
Applying the same method described by Husník et al. (2011) for studying the phylogenetic affinities of the endosymbiotic bacteria, we were able to confidently assign Pr. siddallii Off as an endosymbiont nested within the Providencia clade (fig. 3), corroborating the preliminary results obtained from the 16S rRNA gene phylogeny. From this concatenated gene phylogeny, we recovered four independent lineages of blood-feeders’ obligate endosymbionts, two of them associated to arthropods: the Coxiela-like endosymbiont of the Lone star tick A. americanum (hereafter CLEAA), nested within the Coxiellaceae (Smith et al. 2015), and Riesia pediculicola, which stands as a sister clade to Arsenophonus (Nováková et al. 2009; Kirkness et al. 2010); a third one, consisting of Pr. siddallii Off, nested within the Providencia genus. The phylogenetic affinities of Glossinia flies obligate endosymbiont Wigglesworthia could not be confidently assigned, as it clustered within a symbiont clade including Sodalis, Baumannia, and Blochmannia, a group that could be influenced by phylogenetic artefacts and/or lack of full genomes from free-living relatives (Husník et al. 2011). However, it is clear that it represents without doubts a fourth independent lineage of Enterobacteriaceae associated with a blood-feeder.
Fig. 3.—
Providencia siddallii Off phylogenetic positioning. Providencia siddallii Off phylogenetic positioning in a cladogram according to a Bayesian reconstruction done using 55 concatenated single-copy core genes present in diverse Gammaproteobacteria using phylobayes. Providencia siddallii Off is identified as arising from within the Providencia clade. Also, four different origins for obligate endosymbionts from blood-suckers are evident. Sodalis glossinidius is considered as facultative. The full phylogeny can be found as supplementary fig. S2, Supplementary Material online. Red dotted box highlights the position of Pr. siddallii Off. Asterisks denote a posterior probability of 1.
Genetic Reduction in Pr. siddallii Off
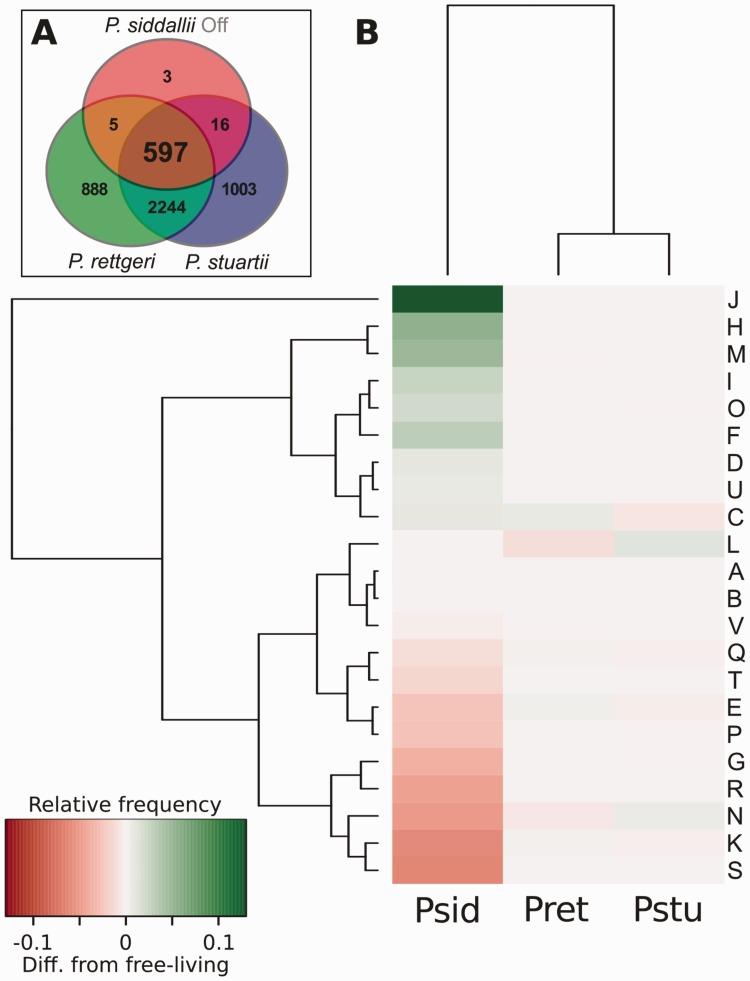
Given that we identified Pr. siddallii as an endosymbiont nested within the Providencia clade, we collected the full genomes from both Providencia rettgeri and Pr. stuartii (INSDC project numbers PRJNA162193 and PRJNA181279, respectively) and analyzed the genetic-repertoire reduction and functional profile divergence of Pr. siddallii Off relative to its free-living providencia relatives (fig. 4; see Materials and Methods: COG Profiles). First, it is evident that Pr. siddallii Off’s genetic repertoire consists mainly of a subset of that of Providencia, with the exception of three genes (fig. 4A). These consist of one 31-residue hypothetical protein and two genes (ybdM and ybdN) whose specific function remains unknown. Looking at the functional profile, we can see that while the free-living Providencia show a very similar COG functional profile between them two, Pr. siddallii Off has a greatly disturbed one (fig. 4B). It shows a great reduction in poorly characterized genes (R and S), transcription (K), cell motility (N), carbohydrate transport and metabolism (P and G), and a moderate one in secondary metabolites biosynthesis, transport and catabolism (Q), signal transduction mechanisms (T), and inorganic ion and amino acid transport and metabolism (P and E). This last one, contrasts the trend seen in phloem-feeders obligate endosymbionts, where this category is greatly preserved to supplement the hosts’ essential-amino-acids-poor diet (Hansen and Moran 2014). In contrast to these reduced categories, and apart from the typical ones that highly reduced genomes tend to preserve (J, M, I, O), we found coenzyme transport and metabolism genes (H) to be highly retained. This is in congruence with the preservation of many genes dedicated to these pathways in all blood-feeders’ endosymbionts sequenced so far (Akman et al. 2002; Rio et al. 2012; Boyd et al. 2014; Nikoh et al. 2014; Smith et al. 2015). Additionally, we were able to detect through a COG profile comparison of blood-feeders’ endosymbionts vs. phloem and grain-feeders’ (see Materials and Methods: COG Profiles), that the relative amount of genes in category H is the most disparate of them all (supplementary fig. S3, Supplementary Material online), clearly pointing toward the importance in retention of genes in this functional group. Also, category E was very dissimilar between blood and phloem-feeders’ endosymbionts. The last ones, preserving a contrastingly higher relative amount of genes devoted to the synthesis of essential amino acids.
Fig. 4.—
Providencia siddallii Off genetic reduction from a free-living Providencia strain. (A) Venn diagram of the pangenome of free-living Providencia strains and Pr. siddallii Off as calculated by OrthoMCL. It is evident Pr. siddallii Off’s genetic repertoire mainly represents a subset of Providencia’s. (B) Functional profile divergence of Pr. siddallii Off compared with free-living Providencia strains. The most disparate categories are located on the top and bottom rows, going to the most similar in the central rows. Psid: Pr. siddallii Off; Pret: Providencia rettgeri; Pstu: Pr. stuartii.
Metabolic Capabilities
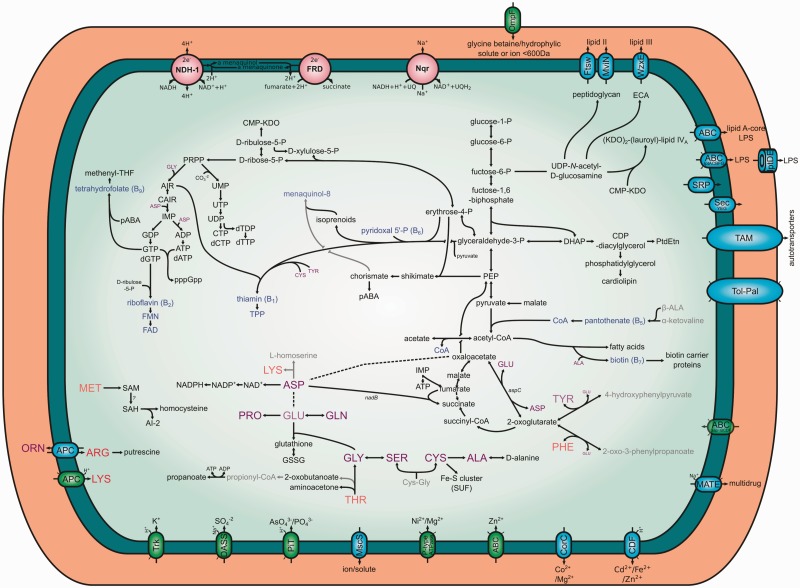
With the help of Pathway Tools and the BioCyc and MetaCyc databases, we reconstructed the metabolic pathways for Pr. siddallii Off (fig. 5). It is an obligate anaerobe, having a complete NADH to fumarate electron transfer respiratory chain. It possesses a Sodium(+)-translocating NADH-quinone reductase, which would act as the primary Na+ pump, while the NADH-ubiquinone reductase I (NDH-1) would act as the primary H+ one. It retains some genes involved in the tricarboxylic acid cycle which could still synthesize malate, fumarate, succinate and 2-oxoglutarate from oxaloacetate through the transamination of glutamic acid. It also preserves complete pathways for the synthesis of both purines and pyrimidines from PRPP and can perform glycolysis from glucose-1-P, although we were unable to find specific transport systems for any sugar. It can produce UDP-N-acetyl-D-glucosamine from fructose-6-P and in turn biosynthesize peptidoglycan and (KDO)2-(lauroyl)-lipid IVA. It also codes for a complete pathway for enterobacterial common antigen.
Fig. 5.—
Metabolic reconstruction of Pr. siddallii Off. Metabolic reconstruction of Pr. siddallii Off as done by PathwayTools. Intact pathways are shown in solid black lines, while almost-complete ones are shown in gray. Compounds whose biosynthesis is not explained by any intact routes present in the organism are shown in faded coloring. Importers are shown using green ovals, whereas exporters and exporters/importers are shown in blue. Essential amino acids and nonessential ones are shown in red and purple coloring, respectively. Cofactors and B-vitamins are shown in blue coloring. The three cellular compartments typical of a gram-negative bacterium are represented in different coloring.
Regarding the biosynthesis of amino acids, it has lost the capability of synthesizing all essential amino acids through canonical routes. This comes as expected, since after water, proteins are the most abundant compounds in blood (Lehane 2005), which could serve as a source of amino acids. Nevertheless, in the case of phenylalanine, it could be synthesized by the action of the aspC gene product, provided the input of 2-oxo-3-phenylpropanoate. Also, in regards to lysine, it preserves an almost-complete pathway missing only the last step, catalyzed by the action of the lysA gene product (diaminopimelate decarboxylase). This “missing-of-the-last-step” is a common feature in highly reduced insect endosymbionts, where it has been proposed, for the pea aphid, that the host might control these last steps through enzymes of its own (Hansen and Moran 2011). In addition, it also preserves specific transporters for importing arginine and lysine, this last one further supporting the hypothesis that the last step might be done by the host. As for nonessential amino acids, it could de novo synthesize proline and glutamine, given the input of either aspartic acid or glutamic acid. It can also de novo synthesize serine and glycine from D-glyceraldehyde-3-P, and alanine and cysteine provided an L-cystenyl-glycine input. Also, tyrosine could be produced from 4-hydroxyphenylpyruvate and glutamic acid through the action of the aspC gene product.
Finally, with respect to the biosynthesis of co-enzymes and vitamins, It retains the capability of completely synthesizing thiamin, thiamin diphosphate (TPP), riboflavin, flavin mononucleotide, flavin adenine dinucleotide (FAD), pantothenate (provided β-alanine and α-ketovaline), coenzyme A (CoA), pyridoxal-5′-P (PLP), biotin, and tetrahydrofolate. The pathway for menaquinol-8 is incomplete, lacking the genes menH, menI, and ubiE.
B-Vitamin and Coenzyme Pathway Convergence among Blood-Feeders’ Gammaproteobacterial Endosymbionts
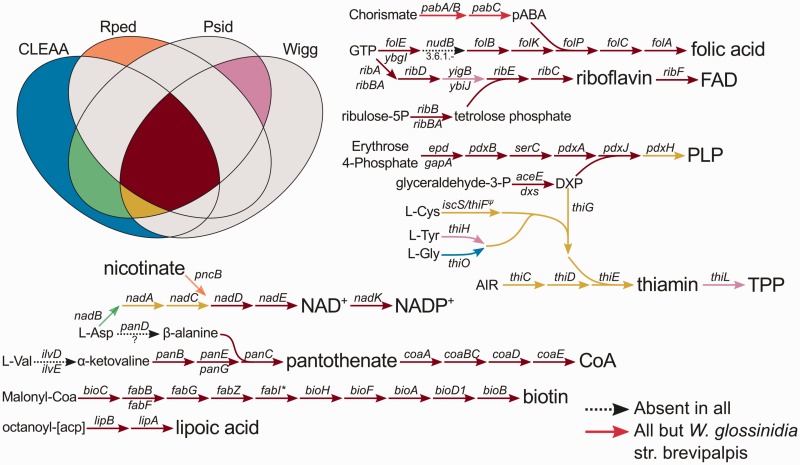
Strict blood-feeders, such as some arthropods and leeches, face a deficiency in B-vitamins from their diets, so it is expected that their bacterial endosymbionts might supplement the host’s nutritionally unbalanced diet. We therefore decided to analyse the different obligate endosymbionts from strict blood feeders and their metabolic capabilities for synthesizing different cofactors and B-vitamins. We found evidence of the biosynthetic routes for folic acid (B9), riboflavin (B2), FAD, PLP (B6), thiamin (B1), TPP, lipoic acid, biotin (B7), pantothenate (B5), CoA, NAD+, and NADP+ to be greatly preserved among all blood-feeders (fig. 6).
Fig. 6.—
B-vitamin biosynthetic pathways retained by gammaproteobacterial endosymbionts from blood-feeders. Pathways representing the conserved production of diverse B-vitamins and cofactors by blood-feeders endosymbionts. Arrows represent reactions catalyzed by the enzymes represented by the names above and below them. A dash in the names of the enzymes represent that both enzymes are involved in the reaction(s). Dotted lines represent absent reactions in all organisms. Bright red lines indicate pathways present in all organisms but W. glossinidia strain brevipalpis. The rest of the color codes are represented in the Venn-like diagram on the left. Rped: R. pediculicola; Psid: Pr. siddallii Off; Wigg: Wigglesworthia endosymbionts.
Regarding the synthesis of para-aminobenzoic acid, the only endosymbiont which would not be able to produce this compound would be Wigglesworthia glossinidia strain brevipalpis. This endosymbiont has been found to have this metabolic difference with its close relative W. glossinidia strain morsitans. This disparity has been postulated to contribute to the higher parasite susceptibility of the host species (Glossina morsitans morsitans) (Rio et al. 2012), given that African trypanosomes have been found to be auxotrophs for folic acid (which they salvage exogenously) (Berriman et al. 2005). All endosymbionts lack the nudB gene (coding for a dihydroneopterin triphosphate pyrophosphohydrolase). However, in E. coli, a deletion mutant shows no detectable growth defect, although the mutant cells contain significantly reduced levels of B9 (Gabelli et al. 2007). Additionally, work done in Bacillus subtilis has suggested that there is no enzymatic requirement of this dephosphorylation (De Saizieu et al. 1995). With reference to the biosynthesis of B2, a lack of yigB/ybiJ (coding both for 5-amino-6-(5-phospho-D-ribitylamino) uracil phosphatases) by both CLEAA and R. pediculicola was observed. Both of these functions could be possibly substituted by other nonspecific phosphatase(s).
Concerning the biosynthesis of PLP, only R. pediculicola lacks a complete pathway, It misses the pdxH gene (coding for a pyridoxine 5′-phosphate oxidase). Nevertheless, this step could be carried out by the host (Pediculus humanus corporis), which codes for a gene transcript putatively coding for this enzyme (VectorBase accession: PHUM170130), therefore controlling the production of this compound. In addition, R. pediculicola is the only analyzed endosymbiont from blood-feeders who is not able to synthesize thiamin but preserves a thiamin import system (thiamin/TPP ABC transporter coded by the genes tbpA, thiP, and thiQ). Additionally, P. humanus corporis codes for a thiamin-pyrophosphokinase (VectorBase accession: PHUM522680), which could indicate that thiamin is acquired through another source and then converted into TPP, to then be imported by R. pediculicola. This other source could be either another associate or a yet-unknown way of using the low concentrations of thiamin that are present in the human blood (Kimura et al. 1982). It is also important to note that CLEAA uses L-glycine to produce 2-iminoacetate, through the action of the gene product of the thiO gene (Nishiya and Imanaka 1998; Settembre et al. 2003), rather than using L-tyrosine, as is the case for both Pr. siddallii Off and Wigglesworthia endosymbionts.
As for the biosynthesis of CoA and B5, none of the endosymbionts analyzed code for the steps transforming L-valine and L-aspartate into α-ketovaline and β-alanine, respectively. Two possibilities could take place: either these reactions could be taken over by other transaminase(s) and decarboxylase(s), respectively, or both α-ketovaline and β-alanine would have to be imported into the endosymbionts. Additionally, CLEAA does not present a panE gene (which codes for a 2-dehydropantoate 2-reductase). However, in a recent study in Francisella tularensis subsp. tularensis, a hypothetical protein which can convert 2-dehyropantoate to pantoate (locus tag: FTT_1388), for whose gene they propose the name panG, was identified (Miller et al. 2013). Therefore, panG could substitute the function of the panE gene in B5 biosynthesis in CLEAA, which presents and homologue of panG. It is also important to mention that the panE gene is also missing in Coxiella burnetii, but it too preserves a homologous gene to panG.
Also, on the biosynthesis of NAD+, only R. pediculicola and both Wigglesworthia have incomplete pathways. The former, could bypass the lack of the nadB, nadA, and nadC gene products by importing nicotinate, as it is the only endosymbiont analyzed here that preserves the gene pncB (coding for a nicotinate phosphoribosyltransferase). This gene product would transform nicotinate to β-nicotinate D-ribonucleotide (NaMN) to then be utilized by the nadD gene product to continue the synthesis of NAD+. Wigglesworthia endosymbionts lack the nadB gene (coding for an L-aspartate oxidase). This would in turn mean that they both either need to import α-iminosuccinate or that another closely related protein, such as that coded by the sdhA gene (succinate dehydrogenase), could have lost specificity for its substrate and can now also act on L-aspartate. It is important to note that SdhA shares an unusual tertiary structure within the substrate biding site with both the nadB and frdA gene products (Mattevi et al. 1999; Bossi et al. 2002).
Finally, in relation to the biosynthesis of lipoic acid and B7, all endosymbionts analyzed preserved complete pathways for the production of these compounds. Nevertheless, it is important to remark that both CLEAA and Pr. siddallii Off, instead of using the fabI gene product (enoyl-[acp] reductase), would be putatively using a trans-2-enoyl-CoA reductase, similar to that recently described in the facultatively anaerobic Euglena gracilis mitochondrion and that has been found to be widely distributed in prokaryotic genomes (Hoffmeister et al. 2005), to perform the fatty acid synthesis steps involved in the metabolic pathway leading to B7.
Discussion
Even though insect endosymbionts from sap-feeding hosts have received much attention, little has been paid on other groups of hosts with equally intimate associations such as blood-feeding leeches and their endosymbionts. While for the sap-feeders, the endosymbiotic partners are mainly specialized on the biosynthesis of essential amino acids, vitamins and cofactors, for the blood-feeders it has been shown that they preserve various metabolic pathways dedicated to the biosynthesis of B-vitamins and some cofactors. This has been explained through the need of blood-suckers to complement their B-vitamin-deficient diet through symbiosis. Blood-feeding leeches are no exception to the rule, and have been shown to form different apparently vertically transmitted associations with a variety of bacteria. Glossiphoniid leeches present an especially interesting case, since they possess specialized organs in which they harbor specific apparently obligate bacteria. As we have shown, Haementeria leeches form tight associations with members of the Pr. siddallii species, housed in bacteriomes. Given that bacterial samples obtained from different species of Haementeria from disparate geographical areas form a monophyletic and well supported group, it is apparent that a single infection from a Providencia-like bacterium must have occurred to the last common ancestor of Haementeria species, followed by subsequent speciation events as evidenced by general congruence of the phylogeny of their endosymbionts with that of their hosts. Providencia siddallii Off displays many of the characteristics of settled obligate endosymbionts from insects, including a small genome with a high A+T content and a limited metabolic repertoire. Nevertheless, it preserves almost-complete pathways for the production of various B-vitamins and some cofactors, while having lost almost all pathways for producing essential amino acids.
Additionally, through the genomic analysis of Pr. siddallii Off, a more interesting opportunity arises, the possibility of comparing distantly related endosymbionts in charge of B-vitamin production for their equally distant blood-sucking hosts. The comparison of these independently established associations, is expected to shed light into the core B-vitamin production pathways that are retained among these bacterial associates and to give clues into the ways in which these organisms’ nutrient production could be controlled. We have found an impressive level of functional convergence in the retention of most B-vitamin biosynthetic pathways, with the exception of that of thiamin in R. pediculicola. This last could be a species-specific adaptation to an external source of this nutrient, for which we have found a specific transporter, unlike for the rest of blood-sucking endosymbionts. The production of at least four compounds (thiamin, NADP+, pantothenate, and CoA) could be regulated by the amount of available amino acids. These last, could provide clues into a way the host could regulate its endosymbiont’s nutrient production. Some of these amino acids could come from the proteins that are present in the host diet (Lehane 2005).
It is evident the importance of the expansion of strict blood-feeders’ endosymbionts studies. Leeches provide a very attractive symbiotic model for these relationships (Graf et al. 2006), and their study provides distant, phylogenetically independent and interesting comparisons with blood-sucker endosymbionts from arthropods. The further analyses of these systems and their genomic and metabolic characteristics will continue to provide clues into the adaptations these endosymbiotic bacteria have suffered to accommodate to their hosts’ peculiar diets.
Supplementary Material
Supplementary files S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors would like to acknowledge Diego Santos-Garcia (Spain) for his advice in genomic DNA extraction. Javier Vargas (Mexico), Rodrigo Ponce de León, and Odile Volonterio (Uruguay) provided assistance in the collection of leeches. Mark E. Siddall from the American Museum of Natural History is an inspiration for those interested on leeches and fueled our interest in the relationships of leeches and bacteria from the beginning. And finally, Lourdes Agredano, Susana Gómez, and Aldo Merlo from the UNAM provided generous assistance in the photo documentation of the bacteriomes and leeches. This work was supported by the Ministerio de Economía y Competitividad (Spain) co-financed by FEDER funds [BFU2012-39816-C02-01 to A.L.]; Generalitat Valenciana (Spain) [PrometeoII/2014/065 to A.M.]; the European Commission [Marie Curie FP7-PEOPLE-2010-ITN SYMBIOMICS 264774 to A.M.-M.]; the Consejo Nacional de Ciencia y Tecnología (Mexico) [Doctoral scholarship CONACYT 327211/381508 to A.M.-M.]; the Consejo Nacional de Ciencia y Tecnología (Mexico) [Postdoctoral scholarship CONACYT 165414 to A.O.-F.]; and the UNAM (Mexico) [Programa de Apoyo a Proyectos de Investigación e innovación Tecnológica IA204114 to A.O.-F.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Akman Gündüz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 276:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman L, et al. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia . Nat Genet. 32:402–407. [DOI] [PubMed] [Google Scholar]
- Allen JM, Reed DL, Perotti MA, Braig HR. 2007. Evolutionary relationships of ‘Candidatus Riesia spp.,’ endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 73:1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apakupakul K, Siddall ME, Burreson EM. 1999. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Mol Phylogenet Evol. 12:350–359. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. 2005. The Genome of the African Trypanosome Trypanosoma brucei. Science 309:416–422. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, et al. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct. 7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi RT, Negri A, Tedeschi G, Mattevi A. 2002. Structure of FAD-bound l -aspartate oxidase: insight into substrate specificity and catalysis. Biochemistry 41:3018–3024. [DOI] [PubMed] [Google Scholar]
- Boyd BM, Allen JM, de Crécy-Lagard V, Reed DL. 2014. Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee lice. G3 4:2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A, Salzberg S. 2011. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nat Methods. 8:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge SW, et al. 2013. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41:D226–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, et al. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42:D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D-Q, Montllor CB, Purcell AH. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl. 95:315–323. [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. 2007. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2:e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. In: Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB). Heidelberg (Germany) p. 45–46. Available from: http://www.bioinfo.de/isb/gcb99/talks/chevreux/. [Google Scholar]
- De Saizieu A, Vankan P, van Loon AP. 1995. Enzymic characterization of Bacillus subtilis GTP cyclohydrolase I. Evidence for a chemical dephosphorylation of dihydroneopterin triphosphate. Biochem J. 306 (Pt 2):371–377. [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749. [DOI] [PubMed] [Google Scholar]
- Gabelli SB, et al. 2007. Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis. Structure 15:1014–1122. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Morella NM, Pulcrano ME. 2012. Affiliations between bacteria and marine fish leeches (Piscicolidae), with emphasis on a deep-sea species from Monterey Canyon, CA. Environ Microbiol. 14:2429–2444. [DOI] [PubMed] [Google Scholar]
- Graf J. 1999. Symbiosis of Aeromonas veronii Biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect Immun. 67:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Kikuchi Y, Rio RVM. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365–371. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 108:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2014. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 23:1473–1496. [DOI] [PubMed] [Google Scholar]
- Hoffmeister M, Piotrowski M, Nowitzki U, Martin W. 2005. Mitochondrial trans-2-enoyl-CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J Biol Chem. 280:4329–4338. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, et al. 2012. Reductive genome evolution, host-symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J. 6:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 107:769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MS, Perlman SJ, Kelly SE. 2003. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc Biol Sci. 270:2185–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, et al. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40:D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jérôme M, Noirot C, Klopp C. 2011. Assessment of replicate bias in 454 pyrosequencing and a multi-purpose read-filtering tool. BMC Res Notes. 4:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. 2010. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 11:40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, et al. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Fukatsu T. 2002. Endosymbiotic bacteria in the esophageal organ of glossiphoniid leeches. Appl Environ Microbiol. 68:4637–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Fukatsu T. 2005. Rickettsia infection in natural leech populations. Microb Ecol. 49:265–271. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Sameshima S, Kitade O, Kojima J, Fukatsu T. 2002. Novel clade of Rickettsia spp. from leeches. Appl Environ Microbiol. 68:999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Fujita T, Itokawa Y. 1982. Liquid-chromatographic determination of the total thiamin content of blood. Clin Chem. 28:29–31. [PubMed] [Google Scholar]
- Kirkness EF, et al. 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 107:12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist S, Narechania A, Oceguera-Figueroa A, Fuks B, Siddall ME. 2011. Phylogenomics of Reichenowia parasitica, an alphaproteobacterial endosymbiont of the freshwater leech Placobdella parasitica. PLoS One 6:e28192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalzar I, Friedmann Y, Gottlieb Y. 2014. Tissue tropism and vertical transmission of Coxiella in Rhipicephalus sanguineus and Rhipicephalus turanicus ticks. Environ Microbiol. 16:3657–3668. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Lehane MJ. 2005. Managing the blood meal. In: The biology of blood-sucking in insects. Cambridge University Press; p. 84–115. Available from: http://dx.doi.org/10.1017/CBO9780511610493.007. [Google Scholar]
- Liu L, et al. 2013. Coinfection of Dermacentor silvarum olenev (acari: ixodidae) by Coxiella-like, Arsenophonus-like, and Rickettsia-like symbionts. Appl Environ Microbiol. 79:2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Lamelas A, Moya A, Latorre A. 2012. Comparative genomics of Serratia spp.: two paths towards endosymbiotic life horn. PLoS One 7:e47274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattevi A, et al. 1999. Structure ofL-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure 7:745–756. [DOI] [PubMed] [Google Scholar]
- Miller CN, et al. 2013. PanG, a new ketopantoate reductase involved in pantothenate synthesis. J Bacteriol. 195:965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montllor CB, et al. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 27:189–195. [Google Scholar]
- Myers EW, et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 23:1478–1484. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MC, Bomar L, Maltz M, Graf J. 2015. Mucinivorans hirudinis gen. nov., sp. nov., an anaerobic, mucin-degrading bacterium isolated from the digestive tract of the medicinal leech Hirudo verbana. Int J Syst Evol Microbiol. 65:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2014. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 111:10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya Y, Imanaka T. 1998. Purification and characterization of a novel glycine oxidase from Bacillus subtilis. FEBS Lett. 438:263–266. [DOI] [PubMed] [Google Scholar]
- Nováková E, Hypsa V, Moran NA. 2009. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oceguera-Figueroa A. 2012. Molecular phylogeny of the New World bloodfeeding leeches of the genus Haementeria and reconsideration of the biannulate genus Oligobdella. Mol Phylogenet Evol. 62:508–514. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A. 100:1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Bergman NH, Phillippy AM. 2011. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SSL, Budinoff RBR, Siddall MEM. 2005. New Gammaproteobacteria associated with blood-feeding leeches and a broad phylogenetic analysis of leech endosymbionts. Appl Environ Microbiol. 71:5219–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, et al. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Available from: http://www.r-project.org/. [Google Scholar]
- Rio RVM, et al. 2012. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont wigglesworthia. MBio. 3(1):e00240–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio RVM, Maltz M, McCormick B, Reiss A, Graf J. 2009. Symbiont succession during embryonic development of the European medicinal leech, Hirudo verbana. Appl Environ Microbiol. 75:6890–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset V, Pleijel F, Rouse GW, Erséus C, Siddall ME. 2007. A molecular phylogeny of annelids. Cladistics 23:41–63. [DOI] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HCJ. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. [DOI] [PubMed] [Google Scholar]
- Settembre EC, et al. 2003. Structural and mechanistic studies on ThiO, a glycine oxidase essential for thiamin biosynthesis in Bacillus subtilis. Biochemistry 42:2971–2981. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Min G-S, Fontanella FM, Phillips AJ, Watson SC. 2011. Bacterial symbiont and salivary peptide evolution in the context of leech phylogeny. Parasitology 138:1815–1827. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Perkins SL, Desser SS. 2004. Leech mycetome endosymbionts are a new lineage of alphaproteobacteria related to the Rhizobiaceae. Mol Phylogenet Evol. 30:178–186. [DOI] [PubMed] [Google Scholar]
- Smith TA, Driscoll T, Gillespie JJ, Raghavan R. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol. 7:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. 1999. The Staden Package, 1998. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. Vol. 132. Totowa (NJ): Springer; p. 115–130. [DOI] [PubMed] [Google Scholar]
- Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123–1130. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tåquist H, Cui Y, Ardell DH. 2007. TFAM 1.0: an online tRNA function classifier. Nucleic Acids Res. 35:W350–W353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33:W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthen PL, Gode CJ, Graf J. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol. 72:4775–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JH, Barr AR. 1971. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232:657–658. [DOI] [PubMed] [Google Scholar]
- Zhong J, Jasinskas A, Barbour AG. 2007. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One 2:e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.