Abstract
The quality of macromolecular crystal structures depends, in part, on the quality and quantity of the data used to produce them. Here, we review recent shifts in our understanding of how to use data quality indicators to select a high resolution cutoff that leads to the best model, and of the potential to greatly increase data quality through the merging of multiple measurements from multiple passes of single crystals or from multiple crystals. Key factors supporting this shift are the introduction of more robust correlation coefficient based indicators of the precision of merged data sets as well as the recognition of the substantial useful information present in extensive amounts of data once considered too weak to be of value.
Introduction
Recent years have seen changes in our understanding of the factors influencing macromolecular crystallographic data quality and in the recommendations for obtaining the highest quality data and selecting an optimal high resolution cutoff for crystallographic refinement. Here, we will focus on three topics related to these changes. First, we discuss the common data quality indicators and their utility. Second, we describe recent results illustrating how high multiplicity1 can improve data quality. Third, we review recent reports providing evidence that extending resolution limits beyond conventional cutoffs to include weaker high resolution data can improve phasing results, electron density maps, and refined models. Understanding of these aspects of data quality is critically important because the observed diffraction data are typically the sole source of experimental information available for supporting a crystallographic structure determination. Strategic considerations regarding other aspects of data collection (e.g. [1,2]) and data reduction (e.g. [3,4••]) are also important, but are beyond the scope of this review.
Common data quality indicators
In Table 1, we list and comment on the utility of eight common statistical indicators reported by current data reduction software, including the new CC1/2 and CC* [5••]. The equations for each are in the literature and are not given here. These indicators all report on data precision, so if substantial systematic errors are present the indicators need not reflect the data accuracy [4••]. We have arranged the data precision indicators into three groups according our view of their utility, and we also specify for each one the crucial distinction of whether it reports on the precision of individual or of merged measurements (Table 1).
Table 1.
Common indicators of data precision and their recommended usage. The ‘type’ defines whether an indicator reports on the precision of individual observations or the final merged data
| Indicator | Type | Recommended usage |
|---|---|---|
| Indicators of first rank | ||
| CC1/2 | Merged | CC between intensity estimates from half data sets. Primary indicator for use for selecting high resolution cutoff for data processing. Is related to the effective signal to noise of the data (see Box 1). |
| CC1/2-anom | Merged | Suggested shorthand for CC between anomalous difference estimates from half data sets. (We suggest CCanom be used if datasets rather than between half data sets are compared [7].) Primary indicator for assessing the resolution limit of useful anomalous signal. Analogous to CC*, a indicator can be calculated from CC1/2-anom. |
| CC* | Merged | Calculated from CC1/2. Indicator useful for comparing data and model quality. Provides the potential for a cross-validation independent indication of overfitting [5••]. CC* is undefined for negative CC1/2. |
| Additional useful indicators | ||
| Rmeas (=Rrim) | Individual | Multiplicity independent replacement of Rmerge and Rsym; Useful for assessing space group symmetry and isomorphism of multiple data sets; Should play no role in determining resolution cutoff. |
| Rpim (~Rmrgd/√2) | Merged | Mainly of value for comparisons with previous practices; Should play no role in determining resolution cutoff, as it rises toward infinity as signal decreases; Also because it is an unweighted sum, if data of varying quality are merged, it will underestimate the quality of the final data. The SFX community's Rsplit = RmrgdI. |
| 〈I/σ〉ind | Individual | Average signal-to-noise ratio of individual observations. The σ for each reflection is calculated according to an ‘error model’ that parameterizes the random and systematic errors. Should play no role in determining resolution cutoff. |
| 〈I/σ〉mrgd | Merged | As 〈I/σ〉ind but for the intensities after a weighted averaging (‘merging’) of equivalent observations. For reflections with multiplicity n, 〈I/σ〉mrgd is at most √n higher than 〈I/σ〉ind, but the increase will be less if reflections to be merged have varying 〈I/σ〉ind. More useful than CC1/2 for assessing quality of low resolution data. If properly estimated data should correlate with CC1/2 values (see Box 1). |
| ISa | Individual | Also 〈I/σ〉asymptotic; theoretical value of 〈I/σ〉ind for an infinitely strong observation of the dataset calculated from coefficients of error model established during scaling [3,17]. Gives insight into the level of fractional error in the dataset. |
| Indicators that should not be used | ||
| Rmerge = Rsym | Individual | Flawed indicators that have been replaced by Rmeas. We recommend these be removed from all data reduction software. |
| Overall value | Both | ‘Overall’ quantities for statistics are not of general value, because they are highly influenced by the distribution of multiplicity. More informative would be reporting ‘low resolution bin’ and ‘high resolution bin’ values. |
With the introduction of CC1/2, all three key indicators we primarily recommend for assessing the precision of the merged data (for both standard and serial crystallography) are Pearson's correlation coefficients (CC) between independent sets of observations characterized as a function of resolution: CC1/2, CC1/2-anom, and CC* (Table 1). CC values range from 1 to –1 for perfectly correlated versus anticorrelated data, but for properly indexed data these indicators should range from near 1 for highly precise data to near 0 for very imprecise data. An advantage of CC-based indicators is that they have well-studied statistical properties so that, for instance, given a CC value and how many observations contributed to it, one can calculate the probability that this value has occurred by chance, i.e. how likely it is that the null hypothesis holds (e.g. http://en.wikipedia.org/wiki/Statistical_significance). CC1/2-anom, our suggested name for the correlation between independent estimates of the anomalous differences from half data sets [6], was the first of these CC-based indicators to be introduced as an extension of work showing that the CC between anomalous differences of two complete data sets helped define which data would be useful for solving anomalous substructures [7].
Similarly CC1/2, calculated in resolution shells by correlating the intensity values produced from two half data sets [5••], provides a model-free, empirical measure of the level of discernable signal and is equivalent to the Fourier Shell Correlation statistic used to define resolution in cryo-EM studies (e.g. [8]). In fact, a theoretical relationship between CC1/2 and the signal-to-noise of the merged data (〈I/σ〉mrgd) can be derived that helps put CC1/2 on a familiar footing (Box 1). Typically, CC1/2 is near 1.0 (or 100%) at low resolution, and drops smoothly toward 0 as the signal-to-noise ratio decreases. Any deviations from this behavior should be scrutinized as possible indicators of anomalies. Since CC1/2 measures how well one half of the data predicts the other half, it does not directly indicate the quality of the data set after final merging. This, however, is estimated by the quantity CC*.
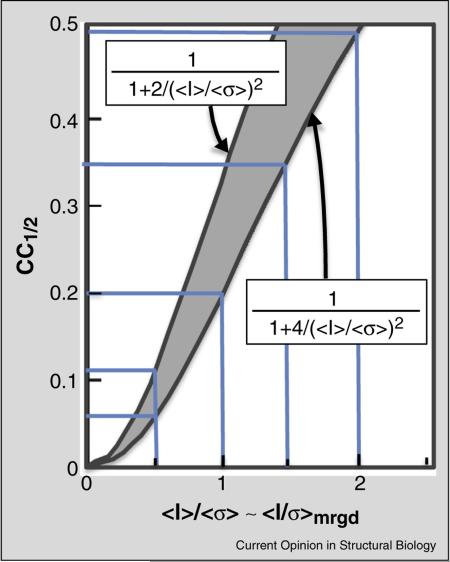
Box 1 Approximate relation between CC1/2 and 〈I/σ〉mrgd.
Assuming that the σ = σmrgd values obtained from data processing are consistent with the spread of observations around their mean, we can derive an approximate expected relationship between CC1/2 and 〈I/σ〉mrgd in a high resolution shell. From the derivation of equation 1 in Karplus and Diederichs [5] we have
where σε denotes the mean error within a half-dataset. Introducing q2 〈I2〉/〈I2〉 – 〈I〉2, we can write
At this point, we note that for acentric reflections following a Wilson distribution q2 = 2 and , which lets us write CC1/2 for acentric reflections as
Then, since 〈I/σ〉 is close to 〈I〉/〈σ〉, in particular at high resolution where σ is approximately the same for all reflections, it is a reasonable approximation that
Two factors that may shift this relationship are (1) that real data may include some centric reflections, for which q2 = 3/2, changing the 4 in the above equations to a 3, and (2) that at very low 〈I/σ〉mrgd the measured intensities are dominated by Gaussian noise and will not follow Wilson statistics and q2 = 1 applies, which changes the 4 in the above equations to a 2. Thus in resolution shells having weak data, the CC1/2 versus 〈I/σ〉mrgd relationship should be fall between the extreme cases of:
And tending to be closer to the first equation for data with ≥1 for which Wilson statistics are still relevant. As seen in the figure, this implies that for accurately estimated σmrgd values, CC1/2 between ~0.1 and ~0.4 can be roughly equated to 〈I/σ〉mrgd values between ~0.5 and ~1.5.
Figure legend: The two curves define limiting relationships for how CC1/2 relates to 〈I〉/〈σ〉 for low signal data. The upper curve is only valid if Gaussian noise dominates the data; so the lower curve should be considered the more relevant above 〈I〉/〈σ〉 ~0.5. The pale blue lines highlight the corresponding values for 〈I〉/〈σ〉 of 0.5, 1.0, 1.5 and 2.0.
CC* is mathematically derived from CC1/2 using the relationship CC* = [2 CC1/2/(1 + CC1/2)]1/2 and provides an estimate of the CC that would be obtained between the final merged data set and the unknown true values that they are representing [5••]. This brings a new ability to compare data and model quality on the same scale because one can compare CC* with a CC between and (i.e. CCwork or CCfree) to discover even without cross-validation if overfitting has occurred during refinement [5••]. The calculations have been built into the PHENIX system [9], and already been used in some reports (e.g. [10•, 11••]). While how to best use this information in guiding and validating refinements is not yet clear, it provides a welcome replacement for the practice of comparing refinement R-factors with data reduction R-factors (e.g. [12•]) that is not correct [5••].
In our view (Table 1), no indicators other than CC1/2 should influence the high-resolution cutoff decisions for data processing. As noted (Box 1), the 〈I/σ〉mrgd statistic (we use subscripts for the two different 〈I/σ〉 values to avoid confusion and emphasize their distinct information content) is related to CC1/2, and so is in principle equally useful for defining a cutoff. However, it is not as useful in practice because the 〈I/σ〉mrgd values obtained during data reduction may not be accurate since they depend on the error model and parameterization used and additional factors such as outlier rejection algorithms and weighting of observations. This causes some irreproducibility across programs as was documented in a report showing that data processed by HKL2000, MOSFLM, and XDS yielded at a certain resolution 〈I/σ〉mrgd values of 2.7, 3.5, and 5.2, respectively [13].
To illustrate why the other common indicators (besides CC1/2 and 〈I/σ〉mrgd) are not useful for guiding the high resolution cutoff decision, we offer the following gedanken experiment. Consider five idealized datasets without radiation damage or systematic errors: ‘Big’ with multiplicity = 2 from a rare large crystal; ‘Tiny’ from a readily grown 100-fold smaller microcrystal; ‘T100’ resulting from the merging of 100 equivalent microcrystal datasets; ‘Big + T100’ resulting from the merging of Big and T100; and ‘Big2’ resulting from the merging of Big with an equivalent dataset from a second large crystal.
Assuming 〈I/σ〉ind = 2 in the highest resolution bin of Big, and that Rmeas ~ 0.8/〈I/σ〉ind) [4••], that CC1/2 is related to 〈I/σ〉mrgd as shown in Box 1, and that n-fold repetition of a measurement reduces its σ by √n, we can generate the following idealized high resolution bin statistics for the five datasets:
| Dataset | Big | Tiny | T100 | Big + T100 | Big2 |
|---|---|---|---|---|---|
| Multiplicity | 2 | 2 | 200 | 202 | 4 |
| 〈I/σ〉ind | 2.0 | 0.2 | 0.2 | 0.22 | 2.0 |
| Rmerge | 28% | 280% | 399% | 395% | 35% |
| Rmeas | 40% | 400% | 400% | 396% | 40% |
| Rpim | 28% | 280% | 28% | 28% | 20% |
| 〈I/σ〉mrgd | 2.8 | 0.28 | 2.8 | 4.0 | 4.0 |
| CC1/2 | 0.66 | 0.04 | 0.66 | 0.80 | 0.80 |
Readily apparent is that according to CC1/2 and 〈I/σ〉mrgd, Big and T100 are of equivalent quality as are Big + T100 and Big2. The huge differences in the values of 〈I/σ〉ind, Rmerge, and Rmeas within these pairs shows why indicators of the precision of individual measurements should never be used for guiding cutoff decisions. Furthermore, a comparison of Big + T100 vs. Big2 shows that even Rpim does not reflect their equivalence. This is because when data of different precision are merged, Rpim and all R-factor based indicators lose relevance because each reflection is weighted equally rather than (as for 〈I/σ〉mrgd) according to its reliability. The impact of this is even more dramatically seen in the 10-fold different Rmeas values of Big + T100 vs. Big2. Also worth noting is that T100 has quite respectable signal in the highest resolution bin even though Tiny does not, emphasizing that high resolution cutoff decisions should only be made after all relevant data have been merged. Finally, the large increase in Rmerge for Tiny vs. T100 reveals how tremendously misleading is the overestimation by Rmerge of precision at low multiplicity [14,15] — making it appear that the datasets merged in T100 were not isomorphous and should not be merged, even while Rmeas correctly indicates the merged data were isomorphous. This is why we recommend (Table 1) that Rmerge never be used.
Further, we suggest that publication standards be changed to require low and high resolution shell data quality statistics rather than ‘overall’ and high resolution shell values (Table 1). It has been shown that ‘overall’ statistics are weighted by multiplicity [14], and so depending on how multiplicity varies with resolution, the ‘overall’ number can take on any value from that of the strongest data to that of the weakest data. Selecting a more generous high resolution cutoff (e.g. [11••, 16•]) and/or increasing the multiplicity of the high resolution data (e.g. [14]), makes the ‘overall’ statistics become worse even though the resulting data are better.
Finally, one uncommon indicator we recommend be reported by data reduction programs (but not in structure reports) is 〈I/σ〉asymptotic or ‘ISa’ ([17]; Table 1). Importantly, 1/ISa provides an estimate of the level of experiment/hardware related systematic (i.e. fractional) error in the data set that limits the precision of strong reflections. For instance, an ISa of near 30, about as high as can be achieved for CCD detector data sets [4••], indicates about a 3.3% (i.e. ~1/30) systematic error. ISa thus has utility as a diagnostic for guiding efforts to improve experimental setups as well as data processing.
Multiplicity can powerfully enhance measurable signal through decreasing noise
A crucial distinction to make regarding data quality is the difference between the level of signal that is measured in a particular data set versus the level of signal that could in principle be measured from that sample (e.g. Figure 1a). Given only random errors, the standard error σ in a measurement is reduced by √n if the measurement is repeated n times. The utility of high multiplicity data sets from single crystals to improve the accuracy of anomalous signal measurements and enable phasing has been powerfully demonstrated many times (e.g. [18,19,20,21]), and it has been recognized that for success sufficient data must be collected before radiation decay degrades the signal (e.g. Figure 1a; [22,23]). Furthermore, theory and practice agree that for a given total crystal exposure time, fractional errors associated with data collection can be minimized and better data produced by collecting higher multiplicity data using shorter exposures [24].
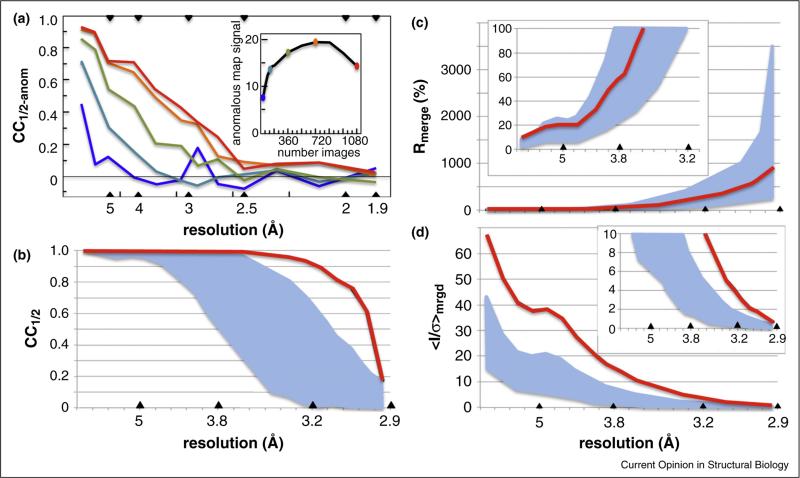
Figure 1.
Averaging multiple measurements can substantially enhance data quality. (a) CCanom is plotted as a function of resolution for a data set of 1080 18 images in a sulfur-SAD phasing case study [23]. Statistics for data merged from 30 (blue), 120 (cyan), 360 (green), 720 (orange), and 1080 (red) images are shown. Based on 30 images (3.5 fold multiplicity), there is no apparent anomalous signal beyond 4 Å, but with 720 images (75-fold multiplicity) the apparent signal extends beyond 3 Å resolution. Inset shows the quality of the anomalous difference map (maximal rrms) increases substantially and then, as radiation damage systematically alters the structure, decreases even while CCanom stays high. (b–d) Behavior of CC1/2, Rmerge, and 〈I/σ〉mrgd as a function of resolution for individual crystals (breadth of values indicated by cyan swaths) and for a set of data merged from 18 crystals (red traces) and successfully used for sulfur-SAD phasing and refinement at 2.9 Å resolution [11••]. Insets show close-ups of the low or high resolution regions. According to the authors, the best individual crystal would only have been useful to ca. 3.2 Å resolution, and by the panel C inset, the averaged data would have been truncated at near 3.8 Å based on an Rmerge ~ 60% cutoff criterion.
An important recent advance has been the (re)-discovery that high multiplicity can improve signal strength, even for anomalous signal, through combining of data from multiple crystals [25••, 26•, 27] as long as the individual data sets are tested for isomorphism. Building on this work, Akey et al. ([11••]) merged data from 18 crystals to generate anomalous data with 50-fold multiplicity and data for refinement with 100-fold multiplicity thus enabling phasing and a higher resolution refinement than could be accomplished using data from any single crystal. These data illustrate how during merging, the final Rmeas becomes roughly the average of the individual Rmeas values but the final 〈I/σ〉mrgd and CC1/2 values can improve substantially (Figure 1b–d). Recent serial femtosecond crystallography (SFX) results, for which each crystal only provides a single image [28], provide further examples of the power of enhancing data quality through merging data from multiple crystals. In a time-resolved SFX study of the photoactive yellow protein, to obtain sufficient quality difference electron density maps, the workers aimed for a multiplicity of ~1500 in the highest resolution bin [29•].
Evidences that data beyond Rmeas ~ 60% and〈I/σ〉mrgd ~ 2 contain useful information
Until recently, a common and recommended practice has been truncating data at the resolution at which Rmeas remains below ~60% and 〈I/σ〉mrgd is ~2 or higher ([30] and Figure S1 of [5••]). Our report [5••] introducing CC1/2 also introduced paired refinement tests and showed that, for our test cases, including data out to a CC1/2 value of between 0.1 and 0.2 led to an improved refined model even though the data at that resolution had Rmeas ~ 450% and 〈I/σ〉mrgd ~ 0.3. We also showed that these weak data improved the quality of difference maps (see Figure S2 of [5••]). This reinforced earlier evidence for the value in refinement of data having 〈I/σ〉mrgd ~ 0.5 [31,32]. The damage caused by using an Rmeas ~ 60% cutoff criterion grows with increasing multiplicity, because the excluded data have a higher and higher 〈I/σ〉mrgd. For instance, for the 100-fold multiplicity data set of Akey et al. [11••], an Rmeas ~ 60% cutoff corresponds to ~3.7Å resolution at which 〈I/σ〉mrgd is ~ 12 (Figure 2c and d). In another study, extending the resolution from 2.85 Å (Rmeas ~ 60%) to 2.1 Å (Rmeas ~ 680%; CC1/2 = 0.22, 〈I/σ〉mrgd = 0.9) improved the MR-Rosetta [33] solution to a challenging molecular replacement problem from Rfree ~ 40% to Rfree ~ 31% [34].
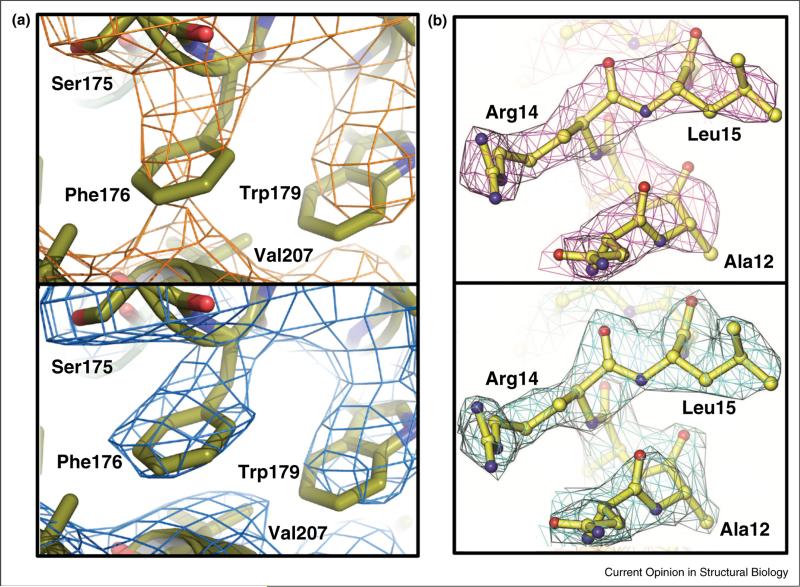
Figure 2.
Examples of tangible electron density map improvement enabled by extending resolution cutoffs. (a) Comparison of the 2Fo–Fc electron density (contoured at 1 ρrms) for a region of the prokaryotic sodium channel pore using an 〈I/σ〉mrgd ~2 cutoff (Rpim = 47%, 〈I/σ〉mrgd = 1.9, CC1/2 = 0.78) 4.0 Å resolution (upper panel) versus a more generous CC1/2 ~ 0.1 based cutoff (Rpim = 213%, 〈I/σ〉mrgd = 0.3, CC1/2 = 0.14) 3.46 Å resolution (lower panel). The 4 Å resolution cutoff was already somewhat generous as the Rpim of 47% with a multiplicity of 12 would be expected to correspond to an Rmeas value of above 150% (47%*√12). Used with permission from Figure S1 of [48•]. (b). Comparison of the 2Fo–Fc electron density (contoured at 1 ρrms) for a region of the E. coli YfbU protein using for the phase extension a fairly conventional cutoff (Rmeas = 77%, 〈I/σ〉mrgd = 3.5, CC1/2 = 0.85) of 3.1Å resolution (upper panel) versus a more generous 〈I/σ〉mrgd ~ 0.5 or CC1/2 ~ 0.1 cutoff Rmeas = 302%, 〈I/σ〉mrgd = 0.5, CC1/2 = 0.14) of 2.5 Å resolution (lower panel). The additional weak data did not just extend the resolution of the map, but improved the quality of the phases obtained at 3.1 Å resolution. Images used with permission from the International Union of Crystallography from Figure 3 of [40••] (http://dx.doi.org/10.1107/S1399004714005318).
In terms of the value of using data beyond 〈I/σ〉mrgd ~ 2, one set of systematic refinement tests showed small improvements with no negative impacts by including data out to CC1/2 between 0.2 and 0.4 corresponding to 〈I/σ〉mrgd between 0.5 and 1.5 [35••]. Interestingly this correspondence between 〈I/σ〉mrgd and CC1/2 roughly matches that expected from theory (Box 1). Another study using distinct tests similarly concluded that useful information is present in reflections out to CC1/2 between 0.1 and 0.5, and that extending the resolution by ~0.2 Å beyond an 〈I/σ〉mrgd ~ 2 cutoff provided a marginal benefit and no adverse effects [36••]. A third study showed that the practice of selectively removing weak reflections within a given resolution bin introduced systematic errors into the data and leads to worse refined models [37••].
Also, many analyses are now using the more generous CC1/2-based cutoffs (with high resolution Rmeas values as high as ~1000% and 〈I/σ〉mrgd values as low as ~0.3) and authors comment on the benefits (e.g. [11••, 16•,38,39•]). One striking example is shown in Figure 2a. Weak data have further been shown to improve the phasing of a crystal with 16-fold non-crystallographic symmetry. Phase extension and automated modeling using data truncated per conventional criteria at 3.1 Å resolution stalled at Rfree ~ 35%, whereas using an extended 2.5 Å resolution cutoff produced an excellent model with Rfree ~ 24.5% and improved electron density maps (Figure 2b; [40••]). Although the signal per reflection is rather weak for the extended data, the tangible impact on phase extension, refinement, and map quality can be rationalized in that the numbers of added reflections are very large, in some cases doubling the data available and they help to minimize series termination error. Wang [41•] describes a perverse incentive researchers have to truncate datasets to obtain more attractive R/Rfree values for any given model, and proposes an intriguing modified Rfactor that emphasizes the value of using more data.
As there is no single ‘correct’ cutoff for every case, using paired refinements [5••] provides a controlled approach to decide for any dataset what resolution cutoff yields the best model. And the PDB-REDO server is now available as a refinement tool that includes a paired refinement option [42•]. Another conservative approach to the cutoff question is to process one's data out to CC1/2 ~ 0.1, but carry out initial refinements using a self-selected conservative resolution limit until the residual Fo–Fc difference map has no interpretable peaks. Then, one can recalculate the difference map using an extended resolution cutoff, and any interpretable peaks provide evidence of tangible information brought by the newly included weak data. In one project for which this was done, the extended difference map was highly informative, and further refinement improved our 2.6 Å resolution ‘final’ model with R/Rfree = 18.9/23.2% to a lower R/Rfree = 17.4/22.0% even at the extended 2.3 Å resolution [43•].
Conclusions and outlook
When we pointed out the flawed multiplicity dependence of Rmerge and recommended making resolution cutoff decisions based on precision of the data after merging [14], we predicted that this ‘should stimulate a shift in data collection strategies, so that the current bias toward using single crystals for complete data sets whenever possible will shift to favor multiple crystal data sets which have increased multiplicity and hence more accurate reduced structure factors.’ The continued use of Rmerge (or even Rmeas) to define cutoffs hindered this from occurring, but now, with the introduction of CC1/2 as a statistically robust indicator of the precision of merged data and with definitive evidence that the inclusion of weak data improves models and that merging data from multiple crystals can be highly beneficial, practices are changing in this direction. Increasing multiplicity is a reasonable strategy to pursue not just to enhance anomalous signals for phasing, but also for obtaining the best high resolution data set for refinement. Important to note, though, is increasing the signal-to-noise of high resolution data occurs by decreasing noise rather than increasing the intensity, so it does not increase the relative contribution of these structure factors to the electron density map. Also worth noting is that obtaining the best high resolution data does not guarantee the best model will be obtained; that depends on care being taken by the crystallographer during model building and refinement.
There obviously need to be changes to what journals require for Table 1 statistics, and we think a useful set would simply be the high resolution bin CC1/2 and number of reflections it is based on (since the low resolution bin CC1/2 is always ~1 and not very informative) and 〈I/σ〉mrgd in the low and high resolution bins, and potentially (for calibration with the past) the resolution at which 〈I/σ〉mrgd ~ 2. CC1/2 also needs to be added to the PDB deposition form. In terms of what this does to the meaning of resolution, we are in agreement with Phil Evans [44] that the nominal resolution of a structure has always referred to which reflections are included in the Fourier summation rather than guaranteeing a certain quality in terms of the apparent resolution of the resulting electron density maps.
We also support the ongoing efforts to archive raw diffraction data to maximize the potential benefit of research funds invested by providing maximal flexibility for correcting mistakes and improving existing structures as technologies improve [45–47]. In the meantime, we encourage users to process data out to CC1/2 ~ 0.1 (after merging of crystals!) even if one is not planning to use it, and to deposit unmerged intensity values together with the merged values. While much is still to be learned about how to obtain the best data, we hope the examples provided here will help crystallographers collect better data and determine better structures.
Highlights.
Common data quality indicators grouped as primary, secondary, or wrong/misleading.
A gedanken experiment reveals shortcomings of some common indicators.
Reviews evidence that massive multiplicity improves data for phasing and refinement.
Cites examples showing value of extending data past conventional resolution cutoffs.
A derived relationship between CC1/2 and 〈I/σ〉mrgd makes CC1/2 more intuitive.
Acknowledgements
This work was supported in part by NIH grant GM083136 (to PAK). We thank Vladimir Lunin for discussions regarding the relationship between CC1/2 and 〈I/σ〉mrgd.
Footnotes
We prefer the term multiplicity to the more common redundancy as it emphasizes that the multiple observations of a reflection are not actually redundant with each other, but together provide more information than any of the individual observations.
Conflict of interest statement
The authors have no conflicts of interest in preparing this work.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Krojer T, Pike AC, von Delft F. Squeezing the most from every crystal: the fine details of data collection. Acta Crystallogr D: Biol Crystallogr. 2013;69(Pt 7):1303–1313. doi: 10.1107/S0907444913013280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeldin OB, Brockhauser S, Bremridge J, Holton JM, Garman EF. Predicting the X-ray lifetime of protein crystals. Proc Natl Acad Sci U S A. 2013;110:20551–20556. doi: 10.1073/pnas.1315879110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D: Biol Crystallogr. 2011;67(Pt 4):282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Diederichs K. Crystallographic data and model quality. In: Ennifar, editor. Nucleic Acids Crystallography: Methods and Protocols. Vol. 320. Springer Science + Business Media; New York: 2015. (in press) [Provides a rather in-depth discussion of random and systematic errors that impact crystallographic data quality, the distinction between precision and accuracy, and the use of data quality indicators. Then walks readers through some lesser known tools that can be used to troubleshoot and optimize data reduction using XDS.] [Google Scholar]
- 5••.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [Introduces paired refinement concept, CC1/2 and CC* indicators. Uses these and difference map analyses to prove that conventional high-resolution cutoff criteria discard useful data. Further shows how data quality R-factors are not comparable to refinement R-factors and that their values should not be used in defining a high-resolution cutoff. That the new indicators are being used and practices are changing is indicated by the over 400 citations already garnered by the work.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans P. Scaling and assessment of data quality. Acta Crystallogr D: Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 7.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D: Biol Crystallogr. 2002;58(Pt 10 Pt 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 8.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D: Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Bae B, Davis E, Brown D, Campbell EA, Wigneshweraraj S, Darst SA. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of σ70 domain 1.1. Proc Natl Acad Sci U S A. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [Early example of low resolution (~4Å) structure analyses using extended CC1/2-based resolution limits and including supplementary plots showing CCwork and CCfree behavior along with Rwork and Rfree showing that despite the use of data with, in one case, Rmeas values over 400% the refinements behave well.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Akey DL, Brown WC, Konwerski JR, Ogata CM, Smith JL. Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus ns1. Acta Crystallogr D: Biol Crystallogr. 2014;70(Pt 10):2719–2729. doi: 10.1107/S1399004714017556. [Along with the de novo solution of a large protein structure by S-SAD phasing enabled by merging data from many crystals, the authors provide a retrospective analysis of how the high multiplicity improved the data, how excluding data from certain crystals improved the results, and how using data well-beyond conventional resolution limits immensely improved the structure determination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Holton JM, Classen S, Frankel KA, Tainer JA. The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures. FEBS J. 2014;281:4046–4060. doi: 10.1111/febs.12922. [A valuable study using real and simulated to show that substantial information exists in diffraction data that are not accounted for by refined protein models, and suggests these are related to inadequately modeled aspects of protein dynamics and the solvent organization. Relevant to this review, is the evidence it provides of the deeply engrained nature of certain misconceptions regarding data quality indicators. These include the use of Rmerge much more than Rmeas, the incorrect consideration of these values rather than Rpim — as being relevant to the quality of the merged data, and finally as treating intensity-based data quality R-factors of any kind as being quantitatively comparable with the crystallographic R-factors from refinement. We think that the conclusions of the work would still hold up using more appropriate comparators, and may in fact even become stronger.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krojer T, von Delft F. Assessment of radiation damage behaviour in a large collection of empirically optimized datasets highlights the importance of unmeasured complicating effects. J Synchrotron Radiat. 2011;18(Pt 3):387–397. doi: 10.1107/S0909049511008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 15.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Cryst. 1997;30(Pt 2):203–205. [Google Scholar]
- 16•.Bunker RD, Bulloch EM, Dickson JM, Loomes KM, Baker EN. Structure and function of human xylulokinase, an enzyme with important roles in carbohydrate metabolism. J Biol Chem. 2013;288:1643–1652. doi: 10.1074/jbc.M112.427997. [Early example using CC1/2 ~ 0.1–0.2 to define the resolution cutoff for refinement despite Rmeas values ~200–600% and 〈I/σ〉mrgd values of ~0.5. State that ‘The inclusion of the weak data . . . significantly improved the quality of the refined models.’.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diederichs K. Quantifying instrument errors in macromolecular X-ray data sets. Acta Crystallogr D: Biol Crystallogr. 2010;66(Pt 6):733–740. doi: 10.1107/S0907444910014836. [DOI] [PubMed] [Google Scholar]
- 18.Dauter Z, Adamiak DA. Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy. Acta Crystallogr D: Biol Crystallogr. 2001;57(Pt 7):990–995. doi: 10.1107/s0907444901006382. [DOI] [PubMed] [Google Scholar]
- 19.Debreczeni JE, Bunkoczi G, Girmann B, Sheldrick GM. In-house phase determination of the lima bean trypsin inhibitor: a low-resolution sulfur-SAD case. Acta Crystallogr D: Biol Crystallogr. 2003;59(Pt 2):393–395. doi: 10.1107/s0907444902020917. [DOI] [PubMed] [Google Scholar]
- 20.Debreczeni JE, Bunkoczi G, Ma Q, Blaser H, Sheldrick GM. In-house measurement of the sulfur anomalous signal and its use for phasing. Acta Crystallogr D: Biol Crystallogr. 2003;59(Pt 4):688–696. doi: 10.1107/s0907444903002646. [DOI] [PubMed] [Google Scholar]
- 21.Weiss MS. Global indicators of X-ray data quality. J Appl Cryst. 2001;34(Pt 2):130–135. [Google Scholar]
- 22.Cianci M, Helliwell JR, Suzuki A. The interdependence of wavelength, redundancy and dose in sulfur SAD experiments. Acta Crystallogr D: Biol Crystallogr. 2008;64(Pt 12):1196–1209. doi: 10.1107/S0907444908030503. [DOI] [PubMed] [Google Scholar]
- 23.Sarma GN, Karplus PA. In-house sulfur SAD phasing: a case study of the effects of data quality and resolution cutoffs. Acta Crystallogr D: Biol Crystallogr. 2006;62(Pt 7):707–716. doi: 10.1107/S0907444906014946. [DOI] [PubMed] [Google Scholar]
- 24.Liu ZJ, Chen L, Wu D, Ding W, Zhang H, Zhou W, Fu ZQ, Wang BC. A multi-dataset data-collection strategy produces better diffraction data. Acta Crystallogr A. 2011;67(Pt 6):544–549. doi: 10.1107/S0108767311037469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, Shapiro L, Mancia F, Hendrickson WA. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [Extension of Liu et al. [27] work to show that with filtering for nonisomorphous crystals, improved anomalous signals achieved by merging the data from multiple crystals to give a multiplicity of 100–150 allows SSAD phasing for five proteins. Even individual data sets with no apparent signal on their own (i.e. CC1/2-anom ~ 0) could be combined to provide a merged data set with substantial signal emphasing that ‘no apparent signal present’ does not mean ‘no signal present.’.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Liu Q, Liu Q, Hendrickson WA. Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr D: Biol Crystallogr. 2013;69(Pt 7):1314–1332. doi: 10.1107/S0907444913001479. [Further details of the multi-crystal native SAD phasing approach reported in [25••], through a focus on one of the five proteins in that study — the DnaK:ATP complex. Includes a discussion of broader aspects of such analyses supporting the conclusion that the appropriate merging of data from multiple statistically equivalent crystals should be generally applicable.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Zhang Z, Hendrickson WA. Multi-crystal anomalous diffraction for low-resolution macromolecular phasing. Acta Crystallogr D: Biol Crystallogr. 2011;67(Pt 1):45–59. doi: 10.1107/S0907444910046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boutet S, Lomb L, Williams GJ, Barends TR, Aquila A, Doak RB, Weierstall U, DePonte DP, Steinbrener J, Shoeman RL, Messerschmidt M, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, Oberthuer D, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014346:1242–1246. doi: 10.1126/science.1259357. [Serial femtosecond crystallography study combining single shots from 30 000–100 000 crystals per state to generate individual data sets with multiplicity of 1000–2000. The value of the multiplicity is seen in that for one data set (in their Figure S4a), merging 2000 images led to a low resolution Rsplit (equivalent to Rpim) of ~30% and the extension to 64 000 images dropped that value to under 5%.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275:1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J. Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure. Acta Crystallogr D: Biol Crystallogr. 2010;66(Pt 9):988–1000. doi: 10.1107/S0907444910029938. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Boisvert DC. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)14 at 2.0 Å resolution. J Mol Biol. 2003;327:843–855. doi: 10.1016/s0022-2836(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger TC, Dimaio F, Read RJ, Baker D, Bunkoczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. Phenix.MR_Rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kean KM, Codding SJ, Asamizu S, Mahmud T, Karplus PA. Structure of a sedoheptulose 7-phosphate cyclase: vala from Streptomyces hygroscopicus. Biochemistry. 2014;53:4250–4260. doi: 10.1021/bi5003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D: Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [Introduces the new scaling program AIMLESS and has an extensive, thoughtful discussion of how to choose a high resolution cutoff. Included are tests comparing the impact on refinement of including real versus fake weak high resolution data that lead to the conclusion that ‘by the time CC1/2 has fallen to around 0.2–0.4, or 〈I/σ〉 to around 0.5–1.5, there is little information remaining.’ Appendix 1 shows that for a refinment against random data, Rfree approaches 42% rather than a value around 55–60% as is commonly assumed.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Luo Z, Rajashankar K, Dauter Z. Weak data do not make a free lunch, only a cheap meal. Acta Crystallogr D: Biol Crystallogr. 2014;70(Pt 2):253–260. doi: 10.1107/S1399004713026680. [Excellent background discussion regarding resolution cutoffs, and careful studies of four 1.4–2Å resolution test cases. Tests comparing observed vs. randomized weak data imply it is genereally helpful to include data ~0.2Å beyond an 〈I/σ〉mrgd = 2 cutoff; however, a surveyed set of quantitive model quality indicators (Rfree, estimated coordinate error, and maximum likelihood target functions) did not show consistent favorable changes in both Refmac and PHENIX refinements. Interestingly, the optical resolution of electron density maps was shown to increase upon the inclusion of even the weakest data tested (in their Figure 3). Despite the minimal tangible evidences of model improvements, the authors concluded that the ‘extension of the maximum resolution at the stage of data collection and structure refinement is cheap in terms of the required effort and is definitely more advisable than accepting a too conservative resolution cutoff.’.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Diederichs K, Karplus PA. Better models by discarding data? Acta Crystallogr D: Biol Crystallogr. 2013;69(Pt 7):1215–1222. doi: 10.1107/S0907444913001121. [Shows using paired refinements that merging a weaker data set with a stronger one can produce improved models, and that selectively removing weak data from datasets leads to worse models even though data quality R-factors are lower. Also shows that in these tests CC1/2 even more than 〈I/σ〉mrgd predicts which data sets will produce better models systematic error are documented and discussed, presumably because 〈I/σ〉mrgd can increase even when data quality is not improving, whereas the empirical CC1/2 accurately reflects when merging certain data actually degrades the precision. Also includes a theoretical analysis of how some systematic errors (including those introduced by certain data filtering practices) impact CC1/2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Structure of the mediator head module. Nature. 2012;492:448–451. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- 39•.Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JH. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [Using a CC1/2 significant at p < 0.001 cutoff (in this case CC1/2 = 0.14, Rmeas = 205%, 〈I/σ〉mrgd = 0.3) allowed extension of the resolution from 2.4 to 2.1 Å for refinement. Authors conservatively state the resolution as 2.4Å in the abstract, but did their analyses using the more informative 2.1Å resolution cutoff. The authors comment on the enhanced detail visible in the extended resolution maps, and Supplementary Figure 2 contains seven examples showing subtle but real differences that improve the interpretability. The higher resolution allowed them to better define the solvation of a ribosome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Wang J, Wing RA. Diamonds in the rough: a strong case for the inclusion of weak-intensity X-ray diffraction data. Acta Crystallogr D: Biol Crystallogr. 2014;70(Pt 5):1491–1497. doi: 10.1107/S1399004714005318. [A compelling study showing that including weak high resolution data greatly improves the accuracy of phases obtained during phase extension using 16-fold non-crystallographic symmetry averaging. The authors also include additional analyses of their results and extensive discussion of the value of weak data. Images from this paper are included here as Figure 2b.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Wang J. Estimation of the quality of refined protein crystal structures. Protein Sci. 2015;24:661–669. doi: 10.1002/pro.2639. [A report documenting that many PDB entries have been refined at resolutions even lower than conventional criteria would indicate and showing how this may be attractive to crystallographers because it leads to lower R/Rfree values for any given model, even though the resulting model would generally be of worse quality than a model generated using additional higher resolution data. A novel goodness of fit measure is proposed, but not yet validated, that incorporates the number of observations (reflections) used in the refinement and so rewards the use of more data.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1(Pt 4):213–220. doi: 10.1107/S2052252514009324. [Reports updates in the PDB_REDO procedures noting that these include a ‘paired refinements’ test. If the reflection file contains higher resolution data than the recorded resolution of the structure, PDB_REDO tests if including the additional data produces a better model using the Rfree, the weighted Rfree, the CCfree, and the free log likelihood, accepting the higher resolution limit unless two of the indicators are degraded.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Driggers CM, Dayal PV, Ellis HR, Karplus PA. Crystal structure of Escherichia coli SsuE: defining a general catalytic cycle for FMN reductases of the flavodoxin-like superfamily. Biochemistry. 2014;53:3509–3519. doi: 10.1021/bi500314f. [A structure analyses for which refinement had been first completed using data sets with conventionally defined resolution limits of near 2.3Å, but which could then be extended by ~0.3Å based on CC1/2-based limits and lead to improved structures as measured by improved R/Rfree values even at the higher resolution limit so that a paired refinement comparison was not needed to justify the extension.] [DOI] [PubMed] [Google Scholar]
- 44.Evans P. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
- 45.Tanley SW, Diederichs K, Kroon-Batenburg LM, Schreurs AM, Helliwell JR. Experiences with archived raw diffraction images data: capturing cisplatin after chemical conversion of carboplatin in high salt conditions for a protein crystal. J Synchrotron Radiat. 2013;20(Pt 6):880–883. doi: 10.1107/S0909049513020724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terwilliger TC. Archiving raw crystallographic data. Acta Crystallogr D: Biol Crystallogr. 2014;70(Pt 10):2500–2501. doi: 10.1107/S139900471402118X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC, Bricogne G. Continuous mutual improvement of macromolecular structure models in the PDB and of X-ray crystallographic software: the dual role of deposited experimental data. Acta Crystallogr D: Biol Crystallogr. 2014;70(Pt 10):2533–2543. doi: 10.1107/S1399004714017040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL., Jr Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. [Used CC1/2 ~0.1 cutoff criterion to extend the resolution from 4.0 to 3.46 Å and state that ‘the obvious differences in map quality reinforce the assertion that adherence to traditional metrics for defining resolution limits can result in the omission of useful diffraction data.’ Images from this paper are included here as Figure 2a.] [DOI] [PMC free article] [PubMed] [Google Scholar]