Abstract
The majority of bacterial proteins are dispensable for growth in the laboratory, but nevertheless play important physiological roles. There are no systematic approaches to identify cell-permeable small molecule inhibitors of these proteins. We demonstrate a strategy to identify such inhibitors that exploits synthetic lethal relationships both for small molecule discovery and for target identification. Applying this strategy in Staphylococcus aureus, we have identified a compound that inhibits DltB, a component of the teichoic acid D-alanylation machinery, which has been implicated in virulence. This D-alanylation inhibitor sensitizes S. aureus to aminoglycosides and cationic peptides and is lethal in combination with a wall teichoic acid inhibitor. We conclude that DltB is a druggable target in the D-alanylation pathway. More broadly, the work described demonstrates a systematic method to identify biologically active inhibitors of important bacterial processes that can be adapted to numerous organisms.
Introduction
Small molecules with specific protein targets are powerful tools to interrogate biological pathways1,2. In bacteria, antibiotics have been used as probes to study divisome assembly and peptidoglycan recycling3–5, to investigate how cytoskeletal dynamics are coupled to cell wall biosynthesis6–9, and to characterize intrinsic resistance mechanisms and stress response pathways, among other processes10. Typical antibiotics, which inhibit targets that are essential for viability under laboratory growth conditions, can be identified in screens for growth inhibition. Unfortunately, the vast majority of proteins in bacteria are dispensable for growth in the laboratory and no systematic approaches for identifying inhibitors of these targets have been established. Here we demonstrate a small molecule discovery strategy that exploits synthetic lethality both to identify bioactive compounds that inhibit physiologically important processes and to identify their targets. Using this strategy, we have identified a compound that inhibits D-alanylation of Staphylococcus aureus teichoic acids.
Synthetic lethality describes a biological interaction in which a given gene is dispensable in a wild-type background, but not in a mutant background in which another gene has been inactivated. The phenomenon implies that the interacting genes have functions that converge on the same essential process11. Large scale deletion and transposon mutant libraries have been used to identify gene-gene synthetic lethal interactions in bacteria and yeast12–15, but a similar principle can be exploited in high throughput screens to discover small molecules that selectively kill a mutant but not a wild-type strain. Such molecules potentially inhibit targets in the synthetic lethal interaction network of the mutant. This screening approach has been used to identify possible anti-cancer therapeutics but has found limited use in bacteria16.
Here, we used a synthetic lethal screening approach to identify compounds that selectively inhibited growth of a S. aureus mutant deficient for synthesis of wall teichoic acids (WTAs). WTAs are anionic polymers that are covalently attached to peptidoglycan in many Gram-positive organisms17. In S. aureus, WTAs comprise up to 50% of the cell wall by mass, making a substantial contribution to cell envelope integrity. Among other functions, they coordinate peptidoglycan synthesis and autolysin activity with cell division, and are also required for host infection18–21. Due to the importance of WTAs in cell envelope physiology, we predicted that they would have numerous synthetic lethal interactions with other cell envelope components15. We confirmed this hypothesis by probing a transposon library with the natural product tunicamycin, a selective inhibitor of TarO, the first enzyme in the WTA biosynthetic pathway20. We found that several genes became indispensible when WTA biosynthesis was inhibited15.
We have now screened a library of small molecules to identify compounds that inhibit proteins in the WTA synthetic lethal network, and have developed a strategy to identify the targets of these small molecules. Like the screening approach, the target identification strategy makes use of synthetic lethality. We report that amsacrine, a small molecule discovered in our chemical screen, inhibits DltB, a membrane-embedded enzyme required for D-alanylation of teichoic acids22. This modification is involved in virulence of S. aureus. We show that amsacrine sensitizes cells to aminoglycosides and cationic antimicrobial peptides23, a phenotype expected for a D-alanylation inhibitor. Hence, our approach can identify biologically active molecules that inhibit pathways implicated in virulence, but which are not essential for survival in vitro. Both the chemical screening approach and the target identification strategy described here can be adapted for other pathways or bacteria to identify inhibitors for use as cellular probes of bacterial physiology and components of synthetic lethal antibacterial compound combinations.
RESULTS
The synthetic lethal network for WTAs
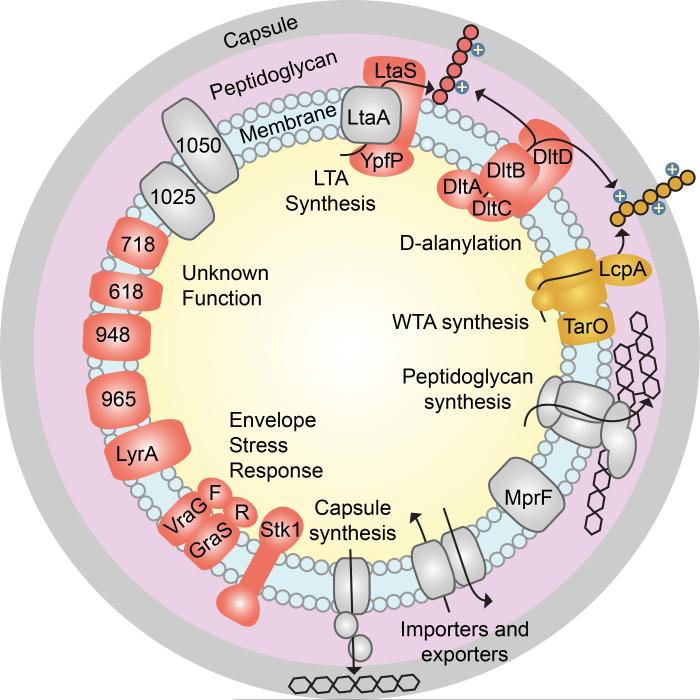
A screen for compounds that inhibit growth of a mutant bacterial strain can be carried out in the absence of prior knowledge of synthetic lethal interactions, but genetic information provides a list of possible targets for any small molecules that are identified. We previously identified a number of synthetic lethal interactions with wall teichoic acids by using tunicamycin to probe a pooled transposon library, and a subset of these interactions were validated15. To develop a comprehensive list of targets that are synthetically lethal with depletion of WTAs, we prepared a larger transposon library by a phage-based mini-Tn delivery mutagenesis method24, subjected it to growth in the presence of tunicamycin, and analyzed the results by Tn-seq25. Transposon insertions in each gene in the presence or absence of tunicamycin were compared to identify genes in which reads were depleted by >80%. These results were combined with those obtained previously to identify potential synthetic lethal partners of WTAs. Null mutants of candidate genes were tested for sensitivity to tunicamycin to confirm synthetic lethality with depletion of WTAs (Supplementary Results, Supplementary Fig. 1). Figure 1 shows a schematic of the S. aureus cell envelope in which all confirmed synthetic lethal targets with respect to WTAs are highlighted in red. The targets are all membrane- or wall-associated proteins, and include components of the lipoteichoic acid biosynthetic pathway26, the four components of the D-alanylation pathway, the cell wall stress response system GraRSVraFG27, and Stk1, a serine/threonine kinase that regulates cell envelope remodeling. Stk1 is known to phosphorylate GraRS, which in turn regulates the expression of dltABCD28. Several membrane proteins of unknown function were also confirmed as synthetically lethal with WTA depletion through this work or the previous study15.
Figure 1. The S. aureus cell envelope is a complex system that includes numerous components and interactions that are poorly understood.
Wall teichoic acids (pathway and polymer highlighted in gold) are synthetically lethal with the proteins shown in red. Selected proteins that are not synthetically lethal with WTA synthesis are shown in gray.
Prioritizing hits in screens of multiple bacterial strains
We developed a growth inhibition screen to identify inhibitors of targets in the WTA interaction network. We did this using three different S. aureus strains: wild-type Newman, an isogenic WTA-deficient strain (ΔtarO), and an isogenic D-alanylation-deficient strain (ΔdltA). While only a wild-type and one mutant strain are required for a synthetic lethal screen, we included the ΔdltA strain for two reasons. First, we sought an inhibitor of the D-alanylation pathway and screening this strain allowed us to filter out compounds that prevented growth of both the ΔtarO and ΔdltA strains. Second, we hoped to identify inhibitors of pathways that interact with the D-alanylation pathway, but not the WTA pathway, among the hits that inhibited growth of only the ΔdltA strain.
We screened 28,157 small molecules comprising both known bioactives and other commercially available compounds in duplicate against each of the three strains in 384-well plates. Plates were incubated for 16-18 h at 30 °C and growth was assessed by optical density at 600 nm (OD600, Supplementary Fig. 2). We found that the standard method of identifying hits by setting cutoffs based on percent growth inhibition did not work well because the stationary phase densities of the mutant and wild-type strains were not identical. For example, the ΔtarO strain typically grew to an OD600 of 50% of the wild-type or ΔdltA strains. Therefore, a TarO inhibitor, while not lethal, would affect the apparent growth of the wild-type strain substantially. Inhibitors of other unknown targets could also affect stationary phase density, making them difficult to distinguish from compounds that have some toxicity. Compensating for stationary phase defects by loosening cutoff constraints would generate bins containing large numbers of unranked compounds. To focus follow-up efforts, we needed an approach to rank order all hits based on differential OD600.
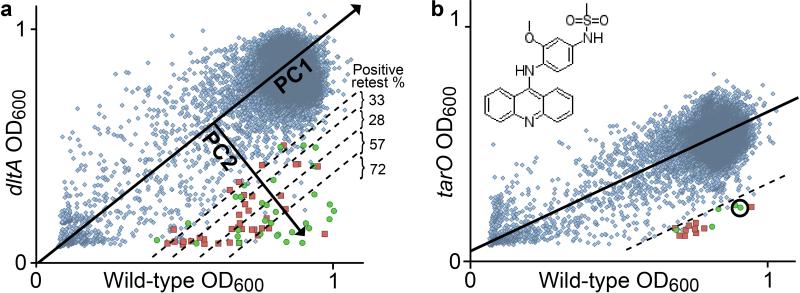
We therefore developed an alternative approach that uses principal component analysis (PCA). In this approach, compounds are plotted according to non-normalized OD600 values against each strain. The controls for no growth (treated with erythromycin for wild-type and ΔtarO or tunicamycin for ΔdltA) and positive growth (treated with DMSO only) were used to define a measure of growth. A line of best fit was drawn through the positive and negative growth controls as the first principal component (PC1). The line of best fit extended from a cluster of compounds near the origin, representing compounds that inhibit growth of both strains, through a dense cloud around the maximum OD600 for each strain, representing compounds that had no effect on growth of either strain. The second principal component (PC2) was drawn perpendicular to PC1 and quantified the difference in growth between each strain for a given compound. Compounds were ranked by PC2 for follow-up.
A comparison of the scatter plots of the data for the mutant versus wild-type strains showed that many more compounds selectively affected growth of the ΔdltA strain compared with the ΔtarO strain (Fig. 2a). Therefore, we used the top ΔdltA hits to assess whether retest rates were higher for compounds with a higher PC2 ranking. The top 100 compounds were retested and compared in groups of twenty. The positive retest rate decreased from >70% for the top twenty ranked compounds to 30% for the bottom sixty compounds, confirming the utility of an approach based on ranking hits by PC2 (Fig. 2a). This method may also be useful for prioritizing hits for strains grown under different conditions.
Figure 2. Principal component analysis (PCA) of high-throughput screening data enables rapid identification of strain-selective growth inhibitors.
(a) Wild-type OD600 plotted against ΔdltA OD600 for 28,157 compounds. The line of best fit through the positive and negative controls is shown as the first principal component (PC1). An arrow perpendicular to the line of best fit is the second principal component (PC2). Dashed lines separate groups of 20 compounds. Confirmed strain-selective inhibitors are shown as green circles. False positives are shown as red squares. (a, inset) Compounds ranked according to PC2 were retested in dose-response assays, and retest rates were calculated as the number of confirmed hits within a group divided by the number of retested compounds. (b) Wild-type OD600 is plotted against ΔtarO OD600 for 28,157 compounds. The top 20 hits by PC2 rank were tested and 5 confirmed. The circled hit is amsacrine, the structure of which is shown in the inset.
ΔdltA hits were enriched in cationic molecules
The majority of the top hit compounds against the ΔdltA strain proved to be positively charged growth inhibitory compounds such as aminoglycosides and the DNA intercalator mitoxantrone. D-alanylation of teichoic acids is thought to play a major role in limiting penetration of positively charged molecules through the cell envelope22, and the ΔdltA mutant appeared highly susceptible to positively charged compounds with intracellular targets that are not in a synthetic lethal interaction network. We tested the relationship between D-alanylation and charge by measuring the minimum inhibitory concentration (MIC) for aminoglycosides in wild-type and ΔdltA strains. We found that the ratio of MICWT to MICΔdltA directly correlated with the number of positive charges (Supplementary Fig. 3). Hence, our screening results confirmed the importance of D-alanylation in establishing a robust barrier to cationic toxins. While there may be inhibitors of synthetic lethal targets among the numerous ΔdltA screening hits, it was problematic to distinguish them from lethal compounds that simply penetrated the cell envelope of the ΔdltA mutant readily. Instead we turned our attention compounds that selectively inhibited growth of the ΔtarO strain.
Confirmed ΔtarO hits included amsacrine
The scatter plot comparing growth of the ΔtarO and wild-type strains for each compound showed a shallower line of best fit (Fig. 2b), reflecting the stationary phase defect of the ΔtarO strain. The plot also showed a narrower PC2 distribution for ΔtarO than the ΔdltA vs. wild-type plot. This narrow distribution suggested that the ΔtarO strain, unlike the ΔdltA strain, was not substantially more permeable than the wild-type. We selected the top twenty compounds based on PC2 rank for retesting, and five of these reproduced strain-specific growth inhibition in a dose-dependent manner. One compound that attracted our attention was amsacrine (4’-[9-acridinylamino]-methanesulfon-m-anisidide, Fig. 2b inset, Supplementary Fig. 4), an anticancer agent that inhibits eukaryotic topoisomerases29. Though amsacrine has a net charge of +1, it was not a hit against the ΔdltA strain, and a growth curve did not show increased sensitivity of this strain compared with wild-type.
Amsacrine exhibited an MIC value of 40-80 μg/ml against wild-type S. aureus, but only 5 μg/ml against the ΔtarO strain. The wild-type strain treated with the TarO inhibitor tunicamycin was also sensitive to 5 μg/ml amsacrine. To test whether the WTA-deficient strains are simply more susceptible to topoisomerase inhibition than the wild-type strain, we tested two other topoisomerase inhibitors for activity against the ΔtarO mutant. The eukaryotic topoisomerase inhibitor etoposide and the bacterial topoisomerase inhibitor ciprofloxacin had identical MICs against the wild-type and ΔtarO strains (Supplementary Fig. 5). We also tested amsacrine for inhibition of E. coli DNA gyrase. Unlike ciprofloxacin, amsacrine did not act as a topoisomerase poison under the conditions tested (Supplementary Fig. 6). Finally, we synthesized and tested the amsacrine analog o-AMSA, an amsacrine analog known not to inhibit eukaryotic topoisomerases30. The MIC for o-AMSA against the ΔtarO strain was 10 μg/ml, similar to that of amsacrine; however, o-AMSA was not lethal against the wild-type strain up to the highest concentration tested (320 μg/ml, Supplementary Fig. 7). Hence, we concluded that amsacrine and o-AMSA likely inhibited a target in the WTA synthetic lethal network.
Identification of DltB as the target of amsacrine
A common method for identifying targets of compounds that affect bacterial viability is to raise resistant mutants and locate the mutations responsible for resistance, usually through whole-genome sequencing. This approach does not work for compounds that inhibit dispensable targets, unless one can identify conditions or strain backgrounds in which the compounds are lethal. We had a susceptible strain background in the ΔtarO mutant because screening of that mutant identified amsacrine; however, efforts to raise resistant mutants in the ΔtarO background were unsuccessful.
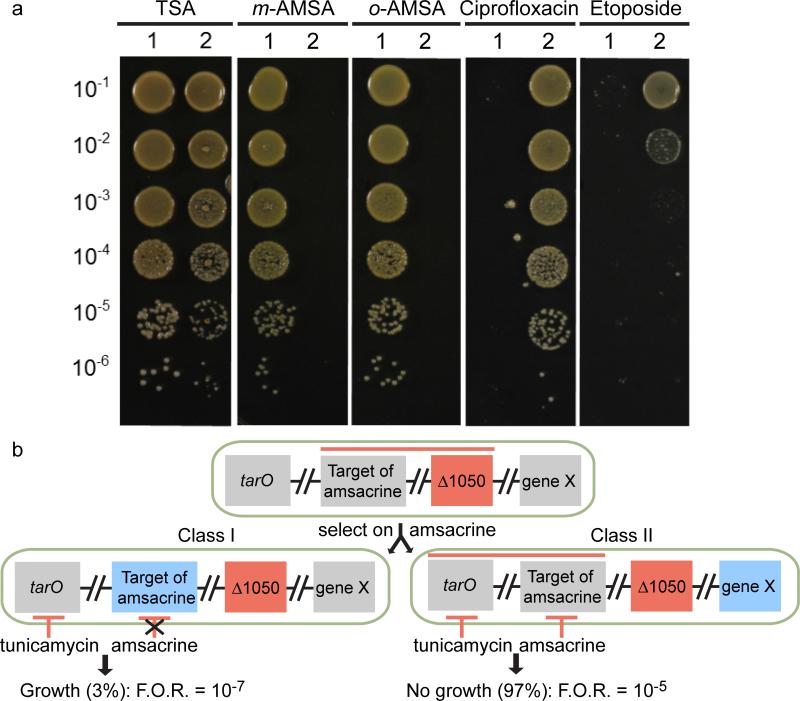
In order to identify amsacrine-susceptible mutants, we grew our existing transposon library in the presence of 10 μg/ml amsacrine and used Tn-seq to identify genes that became essential in its presence. This concentration of amsacrine had no effect on growth of wild-type S. aureus but was above the concentration required to kill the ΔtarO strain. Mutants with transposon insertions in SAOUHSC_01050 were depleted from the transposon library grown in amsacrine. This gene encodes a protein with three predicted membrane-spanning helices and an extracellular domain of unknown function. We obtained a transposon insertion mutant of SAOUHSC_01050, here designated as tn::1050, from the Nebraska MRSA transposon library (mutant NE1420)31, and verified that this mutant did not grow on amsacrine or o-AMSA (Fig. 3a). MICs for amsacrine and o-AMSA were 5 and 10 μg/ml respectively, identical to their MICs against the ΔtarO strain (Supplementary Fig. 7). We also tested the tn::1050 strain for increased sensitivity to topoisomerase inhibition. In contrast to amsacrine and o-AMSA, tn::1050 is resistant to both ciprofloxacin and etoposide at concentrations lethal to wild-type (Fig. 3a). These results provided additional evidence that the target of amsacrine and o-AMSA, the inhibition of which is lethal in the tn::1050 and ΔtarO backgrounds, is not topoisomerase/gyrase.
Figure 3. The target of amsacrine was identified by exploiting synthetic lethality.
(a) The USA300 wild-type strain (1) and tn::1050 strain (2) were grown on TSA or amsacrine (m-AMSA), o-AMSA, or the topoisomerase inhibitors ciprofloxacin and etoposide. The strains exhibit opposite sensitivities to the AMSA analogs as compared to the topoisomerase inhibitors. (b) Synthetic lethality between 1050 and the target of amsacrine (denoted by red line, top) was exploited in a selection on amsacrine for resistant mutants. A Δ1050 strain (tn::1050) was plated on 10 μg/ml amsacrine. Class I mutants with alterations in the target of amsacrine (left) were distinguished from class II mutants containing extragenic suppressors (right, denoted by blue “gene X”) by testing for growth in a combination of tunicamycin (1 μg/ml) and amsacrine (10 μg/ml). Extragenic suppressors preserve the synthetic lethality between WTAs and the target of amsacrine, while mutations in the target of amsacrine that prevent inhibition disrupt this interaction.
We selected for resistant mutants by plating the tn::1050 mutant on solid media containing 10 μg/ml amsacrine. After overnight incubation at 30 °C, the frequency of resistance was approximately 1×10−5. We expected the resistant colonies to comprise two classes of mutants (Fig. 3b), one of which would suppress the action of amsacrine through target mutation. The second class would comprise other, extragenic suppressors of lethality. To identify target suppressors, we exploited the synthetic lethal interaction between tunicamycin and amsacrine. This was possible because this gene is not in the synthetic lethal network for WTAs and the tn::1050 mutant is not sensitive to tunicamycin (Fig. 1). Therefore, we sorted the mutants based on their ability to grow in a combination of amsacrine and tunicamycin (Fig. 3b). We expected mutants with mutations in the target of amsacrine to grow in the presence of a tunicamycin-amsacrine combination. Mutants that suppressed the tn::1050 defect were expected to remain sensitive to this compound combination. We selected 400 amsacrine-resistant colonies and tested them for the ability to grow in the presence of tunicamycin plus amsacrine. 97% of the mutants were susceptible to the compound combination, implying that they did not contain mutations in the target of amsacrine, but rather suppressed the disruption of SAOUHSC_01050. These mutants were set aside. The remaining 3% of resistant mutants with possible target mutations occurred with an overall frequency of resistance of ~10−7, consistent with point mutations in a single gene. Seven mutants obtained from plating independent cultures were submitted for whole genome sequencing. Six of these contained point mutations in the gene encoding DltB, and four of these six contained mutations that resulted in substitution of Phe255 for either leucine or isoleucine. Missense mutations in two other codons in the dltB gene were also identified (Ser175Thr, Ala219Asp). Although all strains also contained mutations in other genes, there was no overlap in these additional mutated genes (Supplementary Fig. 8). All amsacrine-resistant mutants were cross-resistant to o-AMSA.
To confirm that the dltB point mutants were responsible for amsacrine or o-AMSA resistance, we expressed all four mutant alleles in trans. Expression of the DltB variants S175T, A219D, F255L, or F255I rendered wild-type S. aureus resistant to the combination of tunicamycin and amsacrine, while expression of wild-type DltB in trans did not confer resistance (Supplementary Fig. 9). These data strongly suggested that DltB is the primary target of amsacrine in S. aureus. DltB was a plausible target because we have previously shown that D-alanylation of lipoteichoic acids is required for viability when wall teichoic acids are absent15.
Amsacrine inhibits lipoteichoic acid D-alanylation
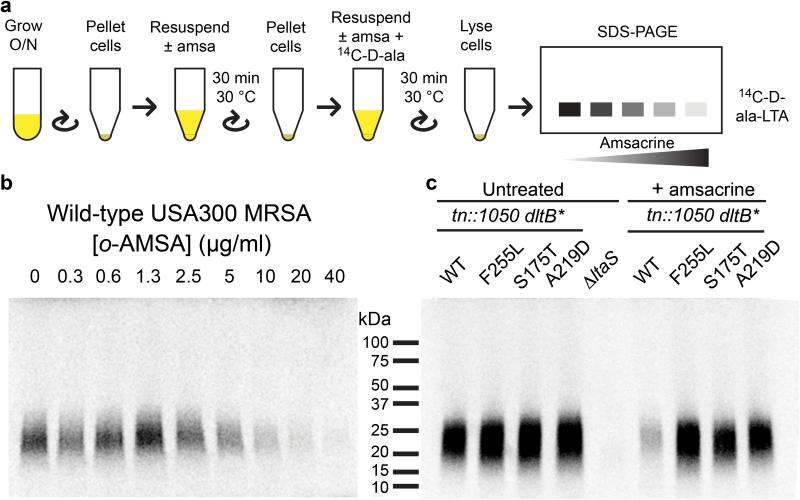
We next tested whether amsacrine and o-AMSA inhibit D-alanylation of lipoteichoic acids, as predicted from the genetic results. Wild-type S. aureus was treated with increasing concentrations of amsacrine or o-AMSA in the presence of 14C-D-alanine and the lipoteichoic acids were then separated by SDS-PAGE and visualized by autoradiography (Fig. 4a). Incorporated radiolabel decreased in a dose-dependent manner for both compounds (Fig. 4b, Supplementary Fig. 10). Inhibition of D-alanylation was also observed in the tn::1050 background, but the dltB mutants were able to incorporate D-alanine into LTA in the presence of amsacrine as efficiently as the untreated controls. (Fig. 4c). Based on the combined genetic and biochemical results, we have concluded that amsacrine inhibits D-alanylation in S. aureus by inhibiting DltB.
Figure 4. Amsacrine inhibits incorporation of 14C-D-alanine into LTA in wild-type S. aureus but not in mutants dltB mutants.
(a) Schematic of assay for D-alanine incorporation into LTAs to assess DltABCD activity. Samples were normalized based on OD during initial pellet and resuspension. For full details, see Supplementary Methods. (b) 14C-D-alanine labeling of LTA in S. aureus USA300 decreases with increasing amounts of o-AMSA. (c) 14C-D-alanine is incorporated into LTA produced in tn::1050 dltB* mutants even in the presence of amsacrine (15 μg/ml). Amino acid changes due to the dltB mutations are indicated. Though amsacrine is lethal to the parental tn::1050 strain, brief treatment is possible.
Amsacrine sensitizes S. aureus to cationic compounds
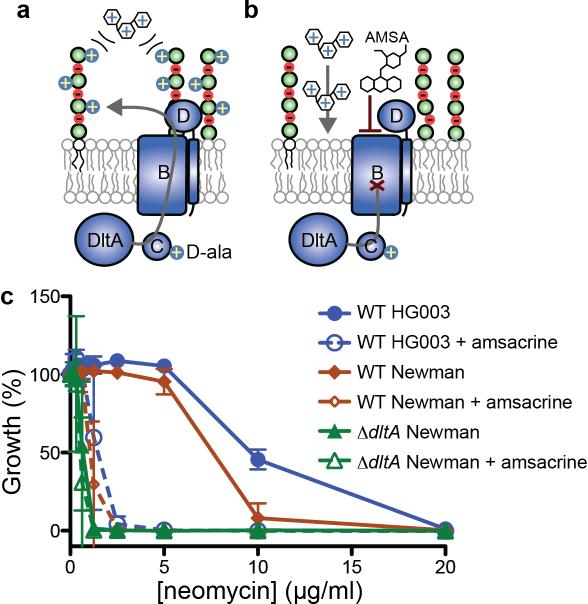
D-alanylation of teichoic acids is important for S. aureus defense against positively charged defensins, antimicrobial peptides, and antibiotics22. D-alanylation neutralizes the negative charges of the backbone phosphates of teichoic acids, providing a barrier that repels positively-charged molecules such as aminoglycosides (Fig. 5a). D-alanine-deficient teichoic acids no longer provide this barrier, sensitizing S. aureus to these molecules (Fig. 5b). Amsacrine and o-AMSA, as DltB inhibitors, were predicted to increase susceptibility of wild-type S. aureus to cationic antibiotics and antimicrobial peptides. We tested this by measuring the MICs of neomycin and gentamicin in the presence of amsacrine or o-AMSA. The MICs of these cationic antibiotics decreased 5- to 15-fold, depending on the antibiotic and were identical to their MICs against the ΔdltA strain (Fig. 5c, Supplementary Fig. 11). We conclude that amsacrine treatment phenocopies genetic deletion of the D-alanylation pathway with respect to potentiation of cationic antibiotics.
Figure 5. Amsacrine potentiates activity of aminoglycosides.
(a) Lipoteichoic acid D-alanylation repels positively-charged compounds such as aminoglycoside antibiotics. (b) Inhibiting D-alanylation potentiates entry of aminoglycosides into cells. (c) Treatment with Amsacrine (10 μg/ml) to inhibit D-alanylation reduced the MIC of neomycin against S. aureus. The reduced MIC was comparable to the MIC of neomycin against a ΔdltA strain.
Discussion
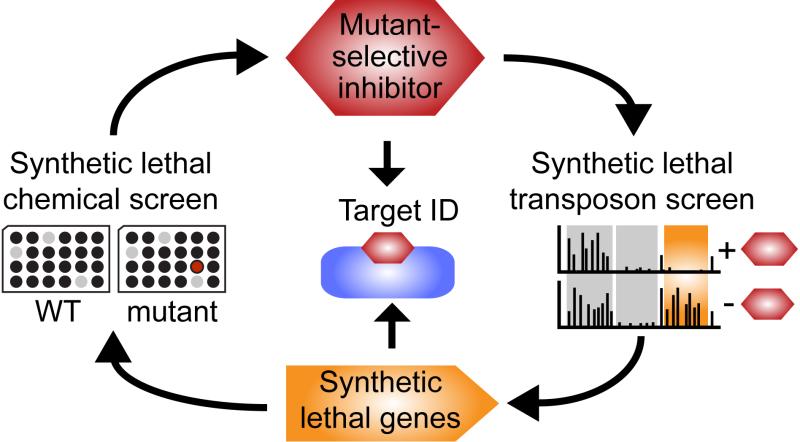
We have established a general approach to identify small molecules that inhibit pathways that are dispensable for in vitro growth but play important roles in physiology (Fig. 6). The approach exploits synthetic lethal interactions for both compound discovery and target identification. To demonstrate the approach, we focused on WTAs, cell wall polymers implicated in fundamental aspects of S. aureus physiology,17 which have a rich network of synthetic lethal interactions15. We screened wild-type S. aureus and a mutant lacking the first gene in the WTA biosynthetic pathway against a small molecule library in order to identify compounds that selectively inhibited growth of the mutant. These compounds were predicted to inhibit targets in the WTA synthetic lethal network. Amsacrine was chosen as a promising compound for follow-up.
Figure 6. Synthetic lethal chemical genetic discovery cycle identifies mutant-selective inhibitors and their targets.
Starting with a mutant of interest, the discovery cycle begins with a pathway-directed screen for compounds that inhibit mutant, but not wild-type growth. Confirmed mutant-selective inhibitors are then used to probe a transposon library for Tn-seq to identify genes with insertions that drop out of the library. Strains lacking these genes are susceptible to compound and can be used to select mutants for target identification. Target mutations can be identified if an inhibitor of an orthogonal synthetic lethal interaction is available (see text). Alternatively, common mutations that occur in different susceptible backgrounds can reveal the target. The cycle not only provides compounds and their targets but also genetic information on pathway interactions to elucidate biological function.
Identifying targets of compounds that do not inhibit growth of wild-type bacteria presents special challenges. It is possible to raise resistant mutants in a susceptible mutant strain, but if mutant selection fails for the screening strain used to identify the compound one must have a strategy to identify other susceptible mutants. Furthermore, one should anticipate mutants that suppress compound toxicity via target mutation and mutants that suppress the defect in the susceptible strain background. Here we have outlined a systematic approach to identifying the target of amsacrine that can be applied to other compounds that do not inhibit growth of wild-type bacteria. This approach involved probing a pooled transposon library with compound and using Tn-seq to identify genes that became essential when the target of the compound was inhibited. We focused on a gene, SAOUHSC_01050, which was not in the synthetic lethal network for WTAs. After confirming that a tn::1050 mutant was sensitive to amsacrine, we selected resistant mutants in this background.
To differentiate between resistant mutants with alterations in the target of amsacrine from extragenic suppressors of the tn::1050 defect, we tested mutants for growth in the presence of a combination of amsacrine and tunicamycin. Only mutants that directly suppressed amsacrine activity were expected to grow. Seven mutants that grew in the compound combination were sequenced and six were found to encode DltB variants with amino acid substitutions, suggesting that amsacrine blocked D-alanylation of teichoic acids by inhibiting DltB. We confirmed that amsacrine and o-AMSA inhibited LTA D-alanylation in whole cells fed with 14C-D-Ala and that D-alanylation was restored in mutants expressing the resistant DltB alleles, leading us to conclude that DltB is the target of amsacrine. Hence, after first identifying amsacrine in a synthetic lethal screen against a WTA-deficient strain, we exploited a different synthetic lethal interaction to select resistant mutants, and we then sorted the mutants based on whether synthetic lethality of amsacrine and tunicamycin, which inhibits WTA synthesis, was suppressed. When a synthetic lethal pair of compounds is not available for sorting mutants, one can identify likely target mutations for one compound by raising resistant mutants in two or more susceptible strain backgrounds and sequencing them to identify the common mutations. The ability to use Tn-seq to identify synthetic lethal interactions with unknown targets is thus crucial to this approach.
D-alanylation is important for a variety of functions in vitro and during infection. A functional dltABCD operon is required for virulence of several pathogens, including Listeria monocytogenes, Streptococcus pneumoniae, and Staphylococcus aureus23,32. One mechanism by which D-alanylation of teichoic acids may contribute to virulence is by conferring resistance to cationic molecules including cationic antimicrobial peptides such as host defensins33. S. aureus mutants lacking dltA are also very sensitive to neutrophil killing and the action of host-produced phospholipases that degrade bacterial membrane lipids34. Because D-alanylation has been implicated in S. aureus virulence as well as intrinsic resistance to antibiotics, including aminoglycosides and vancomycin35, it has been speculated that the D-alanylation pathway could be a target for treating bacterial infections36.
Small molecules that inhibit DltA in vitro have been reported previously36, but amsacrine is, to our knowledge, the only molecule shown to inhibit D-alanylation in cells. Therefore, we used it to investigate whether cells treated with amsacrine resembled a dlt null strain with respect to susceptibility to cationic compounds. Amsacrine and o-AMSA sensitized wild-type S. aureus to aminoglycosides and cationic antimicrobial peptides as much as 15-fold, on par with D-alanylation-deficient mutants. Furthermore, LTA D-alanylation was inhibited by amsacrine in whole cells fed radiolabeled D-alanine. The discovery of this D-alanylation inhibitor shows that DltB is “druggable” and demonstrates a cell-based strategy that should be useful for identifying more potent D-alanylation inhibitors better suited for use in animals.
DltB is a poorly characterized polytopic membrane protein that belongs to the mBOAT family of enzymes that transfer acyl groups from a CoA donor to a peptide or lipid acceptor. DltB is speculated to transfer D-alanine from a carrier protein to a membrane lipid to produce an intermediate that serves as an acyl donor for transfer of D-ala to lipoteichoic acids37. The amino acid substitutions in DltB that confer amsacrine resistance are found in adjacent transmembrane helices and a cytoplasmic loop that may partially insert into the membrane (Supplementary Fig. 8b). These features in DltB are conserved in other MBOAT family members, including ghrelin o-acyl transferase GOAT38. The DltB mutations identified here suggest that amsacrine blocks D-alanylation by binding to a conserved region of the protein important for catalytic function.
In closing, we point out that small molecules that inhibit physiologically important pathways in bacteria have many applications. As we have shown, they are useful for probing transposon libraries in order to map synthetic lethal interactions between pathways. They may also be useful for revealing functions of unknown proteins by enabling selection for suppressors of genetic defects. Finally, by creating a condition in which another protein is essential such inhibitors can enable identification of functionally important residues in that protein. The utility of molecules that make genes conditionally essential has been appreciated by bacterial geneticists for some time, but a systematic approach to discover such molecules and identify their targets has not previously been available39. The approach to compound and target discovery described here can be used repeatedly to assemble collections of small molecules useful for elucidating interaction networks and illuminating gene function in a wide variety of bacteria.
Methods
Reagents and General Methods
S. aureus was grown in tryptic soy broth (TSB) or on TSB with 1.5% agar at 30 °C. Transduction was performed using Φ85 as described40. Antibiotic concentrations used were 5 μg/ml erythromycin (Erm). Amsacrine was purchased from Abcam (ab142742). E. coli DNA gyrase (TG2000G) and DNA gyrase assay kit (TG1003) was purchased from TopoGEN.
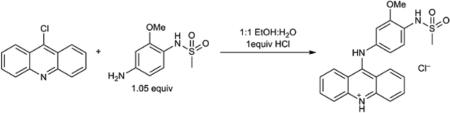
Chemical Synthesis of o-AMSA hydrochloride
All chemicals were purchased from commercial sources as noted below and used as received. NMR spectra were recorded on a Varian Inova 500 (500 MHz for 1H and 125 MHz for 13C) instrument in CD3OD and were recorded at ambient temperatures. Chemical shifts are reported in units of parts per million (ppm). 1H NMR were calibrated using the residual protio-solvent as a standard. 13C NMR spectra are calibrated using the deutero-solvent as a standard. High-resolution mass spectra (HRMS) was obtained on a Bruker micrOTOFQ-II mass spectrometer.
o-AMSA hydrochloride was prepared from 9-chloroacridine (Aldrich) and N-(4-amino-2-methoxyphenyl)methanesulfonamide (ChemBridge) according to a literature procedure41. 1H NMR (500 MHz, CD3OD): δ 8.23 (ddd, J = 8.8, 1.3, 0.6 Hz, 2H), 8.03 – 7.98 (m, 2H), 7.95 (ddd, J = 8.6, 1.3, 0.6 Hz, 2H), 7.55 (d, J = 8.4 Hz, 1H), 7.48 (ddd, J = 8.8, 6.7, 1.3 Hz, 2H), 7.17 (d, J = 2.3 Hz, 1H), 7.02 (dd, J = 8.4, 2.3 Hz, 1H), 3.84 (s, 3H), 3.02 (s, 3H); 13C NMR (126 MHz, CD3OD): δ 157.3, 154.1, 141.8, 139.7, 136.9, 127.6, 126.8, 125.9, 125.4, 120.2, 118.1, 115.2, 109.5, 56.8, 39.9; HRMS (m/z): [M]+ calcd. for [C21H20N3O3S]+, 394.1220; found, 394.1224. see Supplementary Fig. 12 for NMR spectra.
Transposon sequencing
Transposon sequencing was performed as described previously42. Briefly, a pooled transposon library was grown in the presence or absence of 1 μg/ml tunicamycin at 30 °C for 5 h. Genomic DNA was then harvested and prepared for Illumina high-throughput sequencing for identification of the genomic location of the transposon in the surviving mutant. Sequencing data was processed using the Tufts University Genomics Core Facility Galaxy server43–45 and mapped to the genome of NCTC832 using Bowtie software46. Mapped reads were processed to identify depleted genes using the Mann-Whitney U test. Genes that had a >5-fold change in the number of reads between control and treated samples were considered enriched or depleted, and the significance threshold was set to P < 0.05 after correcting for false discovery rate.
All genes that were considered a hit in any comparison were then compiled. The depletion ratio for each gene in each sample was then combined by geometric averaging (taking the sixth root of the product of all six depletion ratios). P values were combined by using the Chi Squared statistic with six degrees of freedom.
High-throughput chemical screening
Overnight cultures of the wild type and mutant strains were grown in tryptic soy broth (TSB) at 30 °C overnight until stationary phase. 30 μl of TSB were dispensed into a 384-well plate (Corning® 3009) using a Matrix WellMate® plate filler. DMSO alone was added to column 23 as a negative control for full growth in the absence of any inhibitors, and column 24 contained final concentrations of 0.4 μg/ml tunicamycin47,48 for the ΔdltA strain and 10 μg/ml erythromycin for the WT and ΔtarO strains, to serve as positive controls for the growth inhibitory activity of synthetic lethal inhibitors. Stock solutions of library compounds (5-25 μg/ml final) in DMSO were then pin-transferred into lanes 1–22 of duplicate plates. Final compound concentration depended on library source concentration, as all were diluted 300 nl into 80 μl. An additional 50 μl of TSB containing approximately 2 × 106 CFU/ml of S. aureus Newman (1000-fold dilution of overnight culture) were then dispensed into all wells of each plate. Plates were incubated at 30 °C for 16 hours. The optical density at 600 nm (OD600) of each well was read on a PerkinElmer Envision plate reader.
Principal Component Analysis
Principal component analysis (PCA) yielded the line of best fit through the origin as the first principle component and the distance from that line as the second component. In order to perform Principal Component Analysis on two-dimensional data, the line of best fit was first determined. This was performed by creating an X-Y scatterplot of compound data with growth (optical density) for each strain along each axis. A trend line (line of best fit) was then generated, passing through the positive and negative controls.
Next, we determined the line perpendicular to the trend line that includes a given data point (point X_value, Y_value). To do this, we first calculated the slope of the trend line and the y-axis intercept (b). The slope of the perpendicular line is the negative reciprocal. Using this slope and the data point, we then calculated the Y intercept (Y_intercept) of the perpendicular line that includes that point. In Excel, the calculation is:
Next, we calculated the X and Y values of the point along the line of best fit that is closest to each individual data point. This is calculated by finding where the line of best fit and the altitude intercept and setting their respective X and Y values equal to each other (X_on_line, Y_on_line). In Excel, these calculations are:
Given these X and Y coordinates, we then can calculated the distance between two points (Abs_distance) using the formula:
This yields the absolute distance but does not indicate a direction. It is critical to account for whether a point is above or below the line of best fit to determine which strain the compound specifically inhibits. We assumed any point above the line had a positive direction while any point below the line had a negative direction. In Excel this can be accomplished by a simple If statement:
To ensure that distances have been corrected properly, we confirmed t the average of all Distance values is very close to zero (<0.001). Compound ranks can then be determined based on this Distance value.
For further analysis, the second principle component exhibited a nearly normal distribution, allowing the use of standard statistics. The standard deviation was calculated by using the formula:
It was then possible to calculate the Z score by dividing each Distance by the Standard_Deviation.
Dose-response MIC determination
Hits were retested for effects on growth of a larger subset of strains in a dose-dependent format. Aliquots (1.5 μL) were then transferred to 96-well plates so that compound concentrations varied along the rows. Stationary phase cultures of bacteria were diluted to OD600 = 1.0 and then 1/1000 in fresh TSB and 150 μL aliquots were dispensed into all wells. The plates were then covered and incubated at 30 °C for 16–18 h.
In vitro inhibition of bacterial topoisomerase
To assay the effects of amsacrine on topoisomerase activity, purified E. coli gyrase and the topoisomerase activity kit from TopoGEN Inc. were used. The topoisomerase assay buffer consisted of 35 mM Tris-HCl, pH 7.5, 24 mM KCl, 4 mM MgCl2, 1 mM ATP, 2 mM DTT, 1.8 mM spermidine, 6.5% glycerol, and 100 μg/mL BSA. To this was added 0, 5, or 100 μg/mL m-AMSA as well as 200 ng concatenated, kinetoplast DNA (kDNA) and 8 units of E. coli gyrase. The reaction was incubated at 37 °C for 1 h before addition of loading dye (final concentrations were 1% sarkosyl, 0.025% bromophenol blue, and 5% glycerol) and electrophoresis in a 1% agarose gel containing 0.5 μg/mL ethidium bromide.
The fluoroquinolone antibiotic ciprofloxacin was used as a positive control for induction of double-strand DNA breaks by E. coli topoisomerase II in the absence of ATP49. For cleavable complex formation, the assay was incubated for 30 min at 37 °C before addition of 0.15% SDS and 75 μg/mL proteinase K (final concentrations), then incubated an additional 30 min at 37 °C.
Whole Genome Sequencing
The USA300 JE2 SAOUHSC_01050::tn(ermR) S. aureus mutant was grown on tryptic soy agar (TSA) with 10 μg/ml of amsacrine overnight at 30 °C. Resistant colonies were re-streaked onto TSA with compound for confirmation and to obtain single colonies. After sorting for target mutants based on growth in 10 μg/mL of amsacrine and 0.4 μg/mL of tunicamycin, resistant mutants and parental strains were then grown in liquid culture for genomic DNA extraction. DNA was processed using the Nextera XT kit and sequenced at the Tufts University Core Facility for Genomics using an Illumina MiSeq in paired end 250 nucleotide runs.
Whole genome fastq sequencing data was analyzed using Geneious v7.1 software50. The parental strain was assembled de novo, and contigs were aligned to the sequenced grandparent strains to generate a consensus sequence. Resistant mutant strains were aligned to the appropriate consensus sequence with default parameters and up to five iterations of remapping. Single nucleotide polymorphisms (SNPs) and insertion/deletions (indels) were identified with default parameters and a minimum variant frequency (i.e. the number of reads sharing a mutation) of 75%.
Detection of D-alanylation
S. aureus USA300 tn::SAOUHSC_01050 and several amsacrine-resistant mutants were tested for 14C-D-alanine incorporation into LTA in the presence and absence of amsacrine and o-AMSA. An ΔltaS strain from the laboratory of Angelika Gründling51 and several other wild-type strains were included as controls. Cultures were normalized prior to cycloserine and amsacrine treatment to ensure equal amounts of lipoteichoic acid in each culture. Normalized aliquots of bacteria were grown to an OD600 of 0.6 in TSB and were then treated with 0, 0.3, 0.6, 1.3, 2.5, 5, 10, 20, 40, 80, or 160 μg/ml amsacrine or o-AMSA in 0.25× TSB, pH 6.0 containing 200 μg/ml D-cycloserine for 1 h at 30 °C. D-cycloserine was included to prevent the rapid consumption of label via D-alanine-D-alanine ligase of peptidoglycan biosynthesis. 14C-D-alanine was added after the first 30 min.
To analyze by SDS-PAGE, the cell pellets were freeze/thawed before resuspending in 30 uL SDS loading buffer, boiled for 15 min, centrifuged at 21,000 × g for 5 min, and 15 μL were loaded onto a 4-20% gradient Tris/glycine gel (Biorad). The dried gel was exposed to a phosphor storage screen for ~67 hr before imaging.
Supplementary Material
Acknowledgments
The authors thank Dr. Ting Pang for providing plasmids and other reagents, Dr. Caroline Shamu, Dr. Jennifer Smith, and the staff at the ICCB-Longwood Screening Facility for compound screening, and the Harvard Medical School Biopolymers Facility and the Tufts University Core Facility for sequencing. This work was funded by NIH grants U19AI109764 to SW and TCM, U54AI057159 to the ICCB-L Screening Facility, P01AI083214 and R01AI099144 to SW, and F32AI118160 to SHM.
Footnotes
Author Contributions
L.P. and J.P.S.M. performed the chemical screen; T.C.M and M.S. prepared the transposon library, and L.P. T.C.M., and M.S. performed the TnSeq experiments; W.L. constructed the ΔypfP mutant used for validation; L.P. selected for resistance mutations and analyzed whole genome sequences; B.M.W. performed LTA D-alanylation and gyrase assays; S.H.M. constructed complementation mutants; L.M. carried out MIC and spot dilution assays; and S.E.S.M. synthesized O-amsa. S.W. designed and supervised the project, and figure design, preparation, and writing was primarily done by L.P. and S.W with important contributions from L.M., J.S.M., and B.M.W. All authors edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hung DT, Rubin EJ. Chemical biology and bacteria: not simply a matter of life or death. Curr. Opin. Chem. Biol. 2006;10:321–326. doi: 10.1016/j.cbpa.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison TJ. Towards a pharmacological genetics. Chem. Biol. 1994;1:3–6. doi: 10.1016/1074-5521(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 3.Chung HS, et al. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitano K, Tomasz A. Triggering of autolytic cell wall degradation in Escherichia coli by beta-lactam antibiotics. Antimicrob. Agents Chemother. 1979;16:838–848. doi: 10.1128/aac.16.6.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H, Uehara T, Bernhardt TG. Article Beta-Lactam Antibiotics Induce a Lethal Malfunctioning of the Bacterial Cell Wall Synthesis Machinery. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garner EC, et al. Coupled, Circumferential Motions of the Cell Wall Synthesis Machinery and MreB Filaments in B. Subtilis. Science (80−. ) 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirner K, et al. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat. Chem. Biol. 2014;11:38–45. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domínguez-Escobar J, et al. Processive Movement of MreB-Associated Cell Wall Biosynthetic Complexes in Bacteria. Science (80−. ) 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 10.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 11.Le Meur N, Gentleman R. Modeling synthetic lethality. Genome Biol. 2008;9:R135. doi: 10.1186/gb-2008-9-9-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Typas A, et al. A tool-kit for high-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butland G, et al. eSGA: E. coli synthetic genetic array analysis. Nat. Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 14.Tong AHY, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 15.Santa Maria JP, et al. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc. Natl. Acad. Sci. 2014;111:12510–12515. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLornan DP, List A, Mufti GJ. Applying Synthetic Lethality for the Selective Targeting of Cancer. N. Engl. J. Med. 2014;371:1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 17.Brown S, Santa Maria JP, Walker S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlag M, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 2010;75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 19.Atilano ML, et al. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell J, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 2011;6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidenmaier C, et al. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 2004;10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 22.Neuhaus FC, Baddiley J. A Continuum of Anionic Charge: Structures and Functions of D-Alanyl-Teichoic Acids in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins LV, et al. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 24.Santiago M, et al. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genomics. 2015;16:252. doi: 10.1186/s12864-015-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems- level analysis of microorganisms. Nat. Rev. Microbiol. 2013;11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Percy MG, Gründling A. Lipoteichoic Acid Synthesis and Function in Gram-Positive Bacteria. Annu. Rev. Microbiol. 2014:81–100. doi: 10.1146/annurev-micro-091213-112949. doi:10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 27.Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012;56:1047–1058. doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fridman M, et al. Two unique phosphorylation-driven signaling pathways crosstalk in staphylococcus aureus to modulate the cell-wall charge: Stk1/Stp1 meets GraSR. Biochemistry. 2013;52:7975–7986. doi: 10.1021/bi401177n. [DOI] [PubMed] [Google Scholar]
- 29.Ketron AC, Denny WA, Graves DE, Osheroff N. Amsacrine as a Topoisomerase II Poison: Importance of Drug-DNA Interactions. Biochemistry. 2012 doi: 10.1021/bi201159b. doi:dx.doi.org/10.1021/bi201159b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson EM, Tewey KM, Liu LF. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4’-(9-acridinylamino)-methanesulfon-m anisidide. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:1–8. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyart C, et al. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 2003;49:1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x. [DOI] [PubMed] [Google Scholar]
- 33.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 34.Hunt CL, Nauseef WM, Weiss JP. Effect of D-alanylation of (lipo)teichoic acids of Staphylococcus aureus on host secretory phospholipase A2 action before and after phagocytosis by human neutrophils. J. Immunol. 2006;176:4987–4994. doi: 10.4049/jimmunol.176.8.4987. [DOI] [PubMed] [Google Scholar]
- 35.Peschel A, Vuong C, Otto M, Götz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 2000;44:2845–7. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.May JJ, et al. Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall. FEBS J. 2005;272:2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 37.Reichmann NT, Picarra Cassona C, Gründling A. Revised mechanism of D-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 44. 2013 doi: 10.1099/mic.0.069898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MS, et al. Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase. J. Biol. Chem. 2013;288:32211–32228. doi: 10.1074/jbc.M113.510313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Santa Maria JP, et al. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc. Natl. Acad. Sci. 2014;111:12510–12515. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cain BF, Seelye RN, Atwell GJ. Potential Antitumor Agents 14. Acridylmethanesulfonanilides. J. Med. Chem. 1974;17:922–930. doi: 10.1021/jm00255a003. [DOI] [PubMed] [Google Scholar]
- 42.Santiago M, et al. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genomics. 2015;16:252. doi: 10.1186/s12864-015-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blankenberg D, et al. Galaxy: A web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010:1–21. doi: 10.1002/0471142727.mb1910s89. doi:10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giardine B, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell J, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 2011;6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. (Tokyo) 1971;24:215. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 49.Pan XS, Fisher LM. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: Overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.