Abstract
Complex carbohydrates perform essential functions in life, including energy storage, cell signaling, protein targeting, quality control, as well as supporting cell structure and stability. Extracellular polysaccharides (EPS) represent mainly structural polymers and are found in essentially all kingdoms of life. For example, EPS are important biofilm and capsule components in bacteria, represent major constituents in cell walls of fungi, algae, arthropods and plants, and modulate the extracellular matrix in vertebrates. Different mechanisms evolved by which EPS are synthesized. Here, we review the structures and functions of membrane-integrated processive glycosyltransferases (GTs) implicated in the synthesis and secretion of chitin, alginate, hyaluronan and poly-N-acetylglucosamine (PNAG).
Introduction
Different mechanisms evolved to polymerize sugar molecules into high molecular weight polysaccharides. Polymer assembly by a GT requires the activation of the monomeric sugar units, often as nucleotide-bound form, and the transfer of these ‘donor’ sugars to an acceptor, a specific hydroxyl group of the growing polysaccharide chain [1••].
GTs come in many forms with finely tuned specificities for different donors and acceptors [1••]. Most GTs catalyze only a single transfer after which the enzyme-product complex dissociates. However, some GTs are highly processive enzymes that do not release the polymer product, thereby achieving astonishing polymerization efficiencies with thousands of sugar units per polymer.
Here we focus on current insights into the mechanisms of chitin, hyaluronan (HA), PNAG and alginate biosyntheses by processive GTs. Cellulose biosynthesis in plants and bacteria has recently been reviewed [2,3] and we use insights gained from bacterial cellulose synthase [4••,5••] to highlight differences and commonalities among processive GTs.
Processive GTs form linear high molecular weight polymers
The GTs discussed below share several traits, Figure 1. The enzymes belong to family-2 of GTs [6], are membrane-integrated, and share a common cytosolic GT domain for donor and acceptor binding [1••]. These GTs transfer sugars from cytosolic nucleotide-activated sugars and generate nucleoside diphosphates (mostly UDP or GDP) as second reaction product [1••], which often competitively inhibit the synthase at elevated concentrations [7•,8•]. Glycosyl transfer is believed to occur via an SN2-like nucleophilic displacement reaction in which the acceptor attacks the donor’s anomeric C1 carbon, thereby inverting its configuration from α to β [1••]. Coupled to polymer synthesis, the enzymes translocate the nascent polysaccharide across the plasma membrane through a pore formed by their transmembrane (TM) region [4••,9–11].
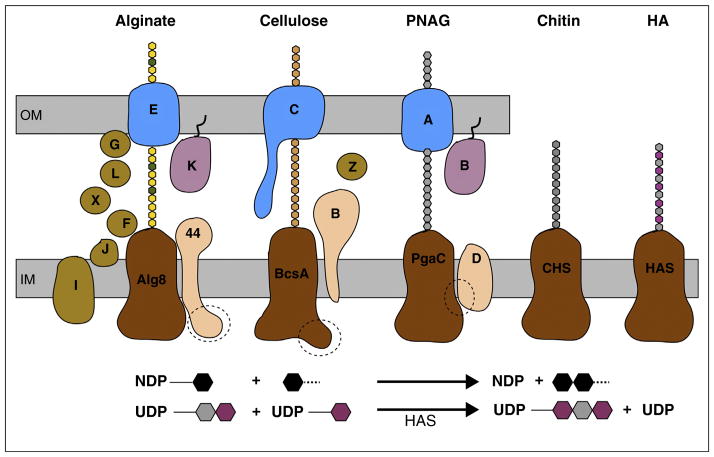
Figure 1.
Membrane-integrated processive GTs synthesize and secrete diverse polysaccharides. The synthases may be part of multi-component complexes or function on their own. The catalytically active subunits (colored brown) share an intracellular GT and a membrane-integrated domain. Alginate consists of mannuronic (yellow) and guluronic acid (green), cellulose of glucose (beige), PNAG of NAG (gray), chitin of NAG, and HA of NAG and GA (magenta) units. A dashed circle indicates the binding site for the signaling molecule cyclic-di-GMP. Lower panel: The enzymes catalyze the transfer of a nucleotide diphosphate (NDP)-activated sugar (black hexagon) to another glycosyl unit, thereby generate NDP as a second reaction product. Among the synthases shown, HAS is the only enzyme that appears to elongate the polymer at its reducing end, thereby generating an UDP-attached polysaccharide. OM, IM: Outer and inner membrane.
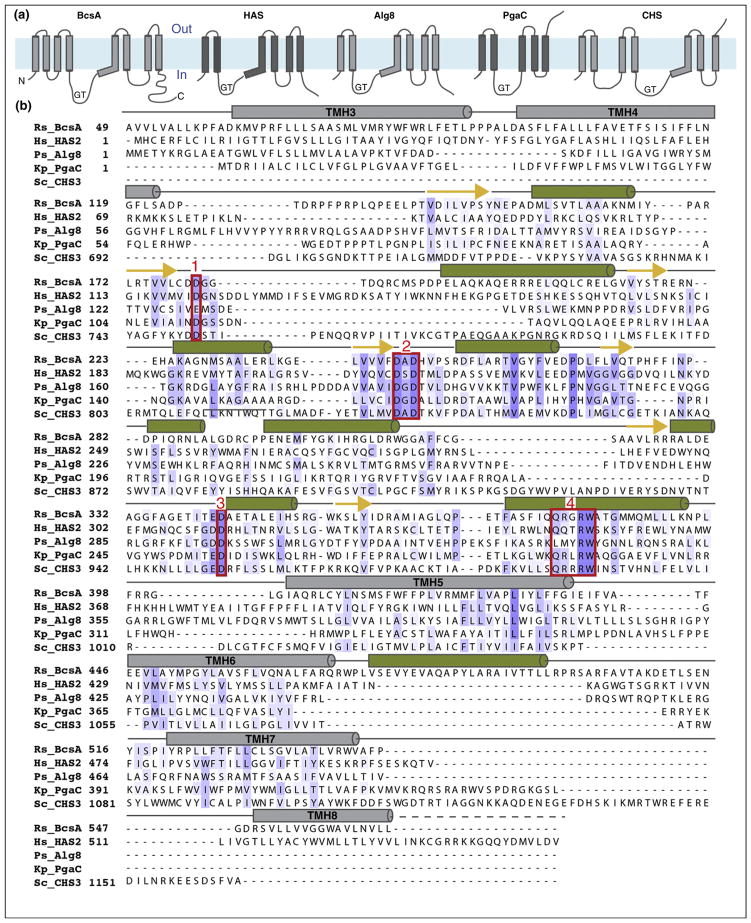
The GT domain contains several sequence motifs that are required for donor and acceptor binding (reviewed in detail in Ref. [3]). Three variably spaced aspartates are crucial for catalytic activity, Figure 2 [12]. The first Asp, frequently belonging to a ‘DDG’ motif (text box 1 in Figure 2), contributes to nucleotide binding [4••]. The second Asp, located in the consensus ‘DxD’ motif (text box 2 in Figure 2), coordinates a Mg2+ or Mn2+ required for GT activity [4••]. The third Asp is also part of a tripeptide motif (‘TED’ in cellulose and ‘GDD’ in HA synthase, text box 3 in Figure 2) and probably functions as the general base that facilitates acceptor deprotonation during glycosyl transfer [4••,5••]. A fourth sequence motif particularly characteristic of processive GTs is a ‘Q/ LxxRW’ pentapeptide (text box 4 in Figure 2) [13•]. On the basis of the cellulose synthase structure, the Trp residue forms vander-Waals contacts with the polymer’s acceptor glucose unit while the preceding Arg residue contacts the substrate’s pyrophosphate group [5••].
Figure 2.
Sequence alignment and predicted secondary structure of selected family-2 GTs. (a) Predicted TM topology of Rhodobacter sphaeroides BcsA, Homo sapiens HAS2, Pseudomonas aeruginosa Alg8, Klebsiella pneumoniae PgaC, and Saccharomyces cerevisiae CHS3. Topology diagrams are shown from the N to the C terminus, labeled N and C for BcsA. GT: glycosyltransferase domain. The membrane region is shown as a blue rectangle. Topologies were predicted with TOPCONS [71]. (b) Multiple sequence alignment of the sequences used in (a). The sequences were aligned in CLUSTALW [72] and predicted TM topologies and secondary structures were used to manually refine the alignment. Predicted TM helices and secondary structure elements are shown as gray bars (TM helices) and green columns and yellow arrows for α-helices and β-strands, respectively. Conserved sequence motifs are framed with a red box and numbered 1–4.
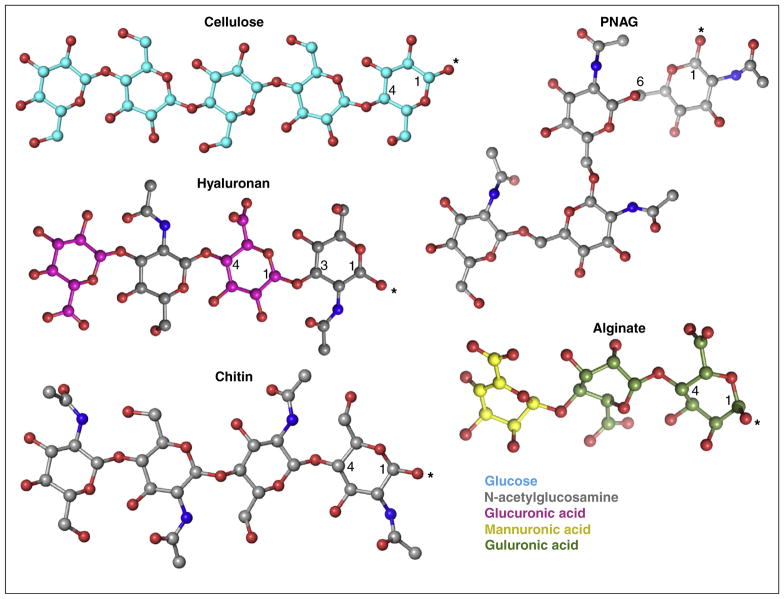
Linear polysaccharides carry reducing and non-reducing ends, referring to the termini with an unmodified and linked hydroxyl group at the anomeric carbon, respectively. All processive GT-2 enzymes characterized to date synthesize linear polymers, Figure 3, which can be modified by soluble proteins after translocation across the plasma membrane. With only a few known exceptions, the enzymes form homo-polysaccharides in which all sugar units are connected by the same glycosidic bond.
Figure 3.
Chemical diversity of polysaccharides. Coordinates for the shown oligosaccharides were obtained and adjusted from pdb entries 4P02 (cellulose), 3AFL (alginate), 2JCQ (HA), 4P7R (PNAG), and 3WH1 (chitin). The carbon atoms of the individual sugars are colored as in Figure 1, oxygen and nitrogen atoms are colored red and blue, respectively. The polymer’s reducing ends are labeled with an asterisk.
Chitin synthase
Chitin is one of the most abundant biopolymers and occurs in various contexts across a broad range of species. It is a linear, β-1,4-linked polymer of N-acetylglucosamine (NAG), Figure 3, that is best known for its strengthening and protective role in the body wall cuticles of arthropods and several other invertebrates.
Insect chitin synthases (CHSs) have recently been reviewed in detail by Merzendorfer [9]. The enzymes can be divided into three distinguishable domains termed A, B and C. Domain A is located at the N-terminus and has limited sequence conservation among different species. In fungal class I–III and VI CHSs, the A domains do not contain any transmembrane helices (TMHs), whereas class IV + V and VII enzymes contain 2–3 TMHs [14]. The B domain forms a central cytosolic GT domain that is followed by the C domain containing 3–7 TMHs [9].
Substrate, chain elongation and primer hypothesis
Biochemical data on CHSs and related GTs, including the bacterial lipochito-oligosaccharide synthase NodC [12], led to a general reaction scheme for chitin biosynthesis. The polymer is formed from UDP-activated NAG and is extended at its non-reducing end [15,16]. Similar to the arrangement of glucose units in cellulose, the individual NAG units of chitin are rotated by approximately 180° relative to their neighbors [17]. Initial mechanistic models thus assumed that CHS binds its substrate in alternating orientations [18], resembling discussions on cellulose biosynthesis [19]. Recent insights into cellulose biosynthesis, however, explained that this alternating arrangement can be established by a simple rotation of the terminal sugar unit around the glycosidic bond, eliminating the need for a dual substrate binding site [4••].
Chitin biosynthesis has primarily been studied in Saccharomyces cerevisiae, which differentially expresses three chS genes [20]. CHS1 is responsible for only 10% of the in vivo chitin pool, but accounts for most of the chitin synthase activity recovered in vitro [21]. Interestingly, this activity is enhanced in the presence of NAG as well as chitooligosaccharides, which may prime chitin biosynthesis [22].
Regulation of chitin biosynthesis
Chitin biosynthesis is regulated on multiple levels. Regulation can directly affect CHS, its cellular localization, or the activities of enzymes involved in substrate biosynthesis [18].
The intracellular localization and stability of CHS can be controlled by phosphorylation. S. cerevisiae CHS2 synthesizes chitin for the primary septum [23] and is phosphorylated by Cdk1 at four positions near its N-terminus. Phosphorylation leads to the ER retention of CHS2 until mitotic kinases are degraded during cell cycle progression [24–26]. As observed for some fungal and insect CHSs, chitin biosynthesis may be stimulated by endopeptidases, such as trypsin [25]. This observation led to the suggestion that CHS enzymes exist in two states, either zymogenic or active. Yet, the biological significance of these observations is still unclear [18].
HA synthase
HA is ubiquitously expressed among vertebrates and is one of the most abundant glucosaminoglycans in the human body where it represents a major component of the extracellular matrix [27•,28•]. HA is particularly enriched in soft connective tissues aiding in osmo-regulation, cell differentiation, and cell adhesion and migration, among other functions [27•]. It is a linear polysaccharide of strictly alternating NAG and glucuronic acid (GA) units linked via β-1,3 and β-1,4 glycosidic linkages, respectively, Figure 3. The polymer can reach astonishing lengths, ranging from several hundred to tens of thousand disaccharide units. Importantly, the physiological effects of HA are modulated by its length. It has recently been reviewed [27•,29] that low and high molecular weight HA exerts opposing physiological effects, including pro or anti-inflammatory and angiogenic properties. However, it is also possible that low molecular weight HA is a consequence, rather than the cause, of inflammatory processes as it can be chemically degraded by, for example, reactive oxygen species [30].
HA is also produced by some bacteria and even viruses [31,32•]. The identification of bacterial hyaluronan synthases (HAS) by Weigel and colleagues paved the way for a detailed biochemical characterization [33,34,35••]. While bacterial HA (primarily produced by vertebrate pathogens) forms a non-immunogenic capsule component probably to avoid host immune responses, the biological significance of viral HA is currently unclear. Although most bacterial HASs are membrane-integrated enzymes like their eukaryotic counterparts, a second class of bacterial HAS has been identified in Pasteurella multocida, that is non-processive and not membrane-integrated [36].
HAS are small membrane-integrated GTs
Compared to cellulose, chitin and alginate synthases, HASs are remarkable enzymes with unique properties. First, HAS recognizes two different substrates, UDP-NAG and UDP-GA; second, the enzyme catalysis the formation of β-1,3 and β-1,4 linkages; third, HA has a strictly alternating NAG-GA sequence; and fourth, HAS is necessary and sufficient to catalyze the synthesis and membrane translocation of HA [10••].
Bacterial and vertebrate HAS differ with respect to their predicted TM topologies, containing 4–5 and 6–7 TM regions, respectively. The enzymes share a conserved cytosolic GT domain, usually located between TMHs 2 and 3. Radiation inactivation analyses on Streptococcal HAS showed that the enzyme functions as a monomer [34], however, recent immuno-precipitation analyses on human HAS suggest homo-oligomerization of the enzyme [36].
Studies on bacterial HAS provided the first biochemical evidence for the dual functionality of processive GTs. Hubbard and colleagues reconstituted purified Streptococcal HAS into proteoliposomes for HA translocation studies [10••]. By initiating HA biosynthesis with substrates added to the outside of the lipid vesicles, this study demonstrated that the newly synthesized HA indeed accumulated in the vesicle lumen, thereby corroborating that bacterial HAS is necessary and sufficient for the synthesis and translocation of HA [10••,11].
In contrast to most other processive family-2 GTs characterized to date, HAS appears to elongate the reducing end of HA. This was demonstrated using primarily pulse-chase labeling of HA produced by bacterial and vertebrate enzymes and requires that the growing HA polymer is attached to UDP [37–41]. Hence, UDP-activated GA and NAG would function as the acceptors during glycosyl transfer and not as donors [10••]. Further structural and functional analyses are required to corroborate this mechanism and to delineate architectural adaptations of HAS that distinguish it from other processive family-2 GTs.
Regulation of HAS activity
Human HAS isoforms 1, 2 and 3 have been reported to produce HA polymers of different molecular weight [42], while evidence for length regulation of bacterial HA is sparse. Transcriptional control of haS genes and the availability of substrates might be one mechanism by which HA polymerization is controlled [43]. The biosyntheses of UDP-activated GA and NAG depend on the cytosolic glucose concentration as well as the regulation of enzymes required for their biosyntheses. For example, over-expression of S. equisimilis HAS together with UDP-glucose 6-dehygrogenase, glucose-1-P uridyltransferase, and NAG-uridyltransferase in E. coli led to the accumulation of high titers of HA in the growth medium [44]. In addition, Kumari and colleagues identified two conserved polar amino acids in Streptococcal HAS TMH2 and TMH4 that are crucial for the biosynthesis of high MW HA [45] and Pummill and DeAngelis identified Ser77 in Xenopus HAS1 as a crucial residue for HA length regulation [46]. HAS1 mutants carrying a point mutation at position 77 either produced lower or higher MW HA, depending on the amino acid introduced.
Posttranslational modifications
In vivo and vitro studies on vertebrate HAS have shown that the enzymes can be post-translationally phosphorylated, glycosylated and ubiquitinated [47–49]. For example, human HAS2 is glycosylated on Ser221 with NAG (O-GlcNAc), which increases the enzyme’s activity and stability [50]. Human HAS3 expressed in COS-7 cells is phosphorylated at one or more serine positions with unknown impact on enzymatic function [49]. In another study, mouse HAS2, which is essential for normal fetal development, was shown to be mono-ubiquitinated on Lys190 [48]. Replacing Lys190 with Arg abolished the in vitro activity of the enzyme. Lys190 belongs to a ‘GKR’ motif of the GT domain that is conserved among bacterial and eukaryotic HAS, supporting its putative regulatory role, Figure 2.
Alginate synthase
Alginate is a major component of the cell wall in brown algae but is also produced by some bacteria as a biofilm component. In particular, alginate production by Pseudomonas aeruginosa in cystic fibrosis patients correlates with poor prognosis and increased morbidity.
Alginate is a hetero-polysaccharide of β-1,4 linked D-mannuronic acid interspersed with L-guluronic acid units [51], Figure 3. Guluronic acid is the C5 epimer of mannuronic acid. In bacteria and algae, it is introduced into the polymer by epimerization of mannuronic acid units by a soluble epimerase after the secretion of the polymer across the plasma membrane [52,53]. The distribution of these two units in the polymer varies between species [54], but usually consists of homogenic regions of the same sugar joined by heterogenic regions. The polymer can further be acetylated in the periplasm on the mannuronic acid’s C2 and C3 hydroxyl groups [55].
Alginate biosynthesis in gram-negative bacteria requires a multi-component complex that probably spans the inner and outer membrane (recently reviewed by Whitney and Howell [56]). At the inner membrane, a complex of the GT Alg8 and the non-catalytic Alg44 subunits forms the active synthase that binds the substrate GDP-mannuronic acid and appears to be sufficient for alginate synthesis and membrane translocation [57•]. Transport across the outer membrane is achieved by a complex of the outer membrane porin AlgE and periplasmic AlgK subunits [58]. It is currently unknown whether the Alg8/Alg44 complex directly interacts with the outer membrane AlgK/E components [59]. An attractive model would be that alginate interacts with an ensemble of modifying enzymes within the periplasm and that these subunits chaperone the polymer from the inner to the outer membrane [56,60].
Regulation of alginate biosynthesis
Alg8 contains 5 predicted TMHs with the GT domain between TMH2 and 3 [57•]. The subunit is catalytically inactive in the absence of Alg44 [61], which contains a single TMH framed by an intracellular N-terminal PilZ domain and a periplasmic C-terminal domain. PilZ domains, first identified as regulatory components of cell motility, bind the bacterial signaling molecule cyclic-di-GMP, which affects many cellular processes, including biofilm formation [62•]. Similar to bacterial alginate production, synthesis of bacterial cellulose is allosterically activated by cyclic-di-GMP [7•,63••]. Here, the activator binds to a PilZ domain located at the C terminus of the catalytic cellulose synthase subunit BcsA. Cyclic-di-GMP binding affects the accessibility of BcsA’s active site via a ‘gating loop’ [3,4••,5••]. How cyclic-di-GMP regulates alginate biosynthesis is currently unknown but is particularly important for the development of novel antimicrobial therapeutics.
PNAG synthase
Poly-NAG (PNAG) is an extracellular homo-polysaccharide frequently found in bacterial biofilms [64,65]. It is produced by the Pga machinery consisting of the PgaABCD proteins [66,67•] and is formed from UDP-activated NAG that is polymerized via β-1,6 linkages, Figure 3.
Similar to bacterial alginate and cellulose synthases, the Pga complex probably spans the inner and outer bacterial membrane [56]. PgaC and PgaD form an inner membrane-integrated complex. PgaC is the catalytic GT containing 4–5 predicted TMHs. PgaD probably contains two TMHs with a short periplasmic loop and small cytosolic C terminus. PgaA is an outer membrane porin similar to AlgE [58] and might associate with PgaB, a periplasmic lipoprotein that is probably anchored to the outer membrane. PgaB deacetylates a small fraction of the polymer’s NAG units [68].
Production of PNAG strongly depends on the presence of cyclic-di-GMP, similar to alginate and cellulose biosynthesis [69••]; however, PgaC or D lack classical cyclic-di-GMP-binding domains [62•]. With a clever cross-linking and mutagenesis approach, Steiner and colleagues demonstrated that cyclic-di-GMP interacts with and stabilizes a PgaCD complex [69••]. In the absence of cyclic-di-GMP, PgaCD dissociates, rendering PgaD prone to proteolysis. It is thus likely that allosteric activation of PNAG biosynthesis by cyclic-di-GMP is based on stabilization of the PgaCD interaction [5••].
BcsA as a model for processive GTs?
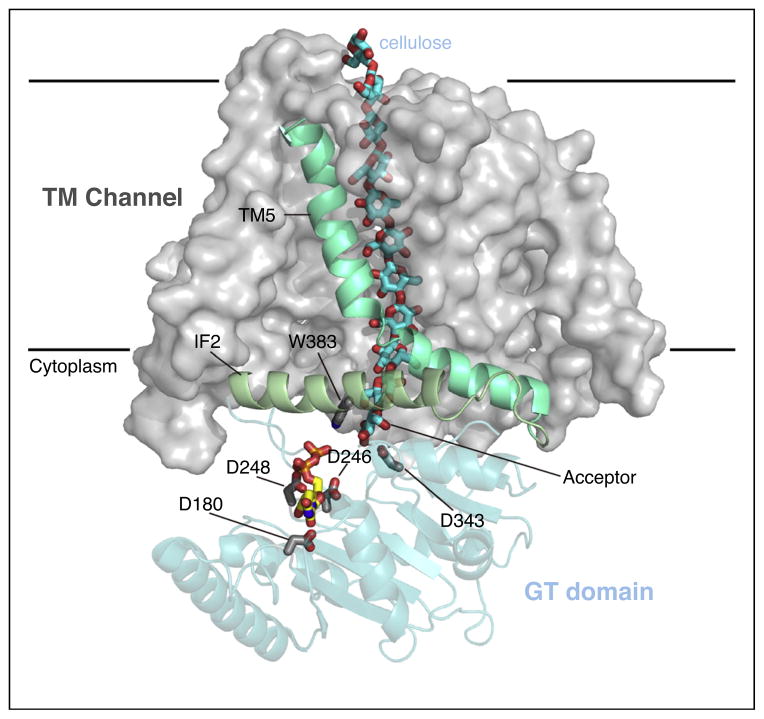
Biochemical and structural analyses suggest that processive family-2 GTs directly couple polysaccharide synthesis and secretion, Figure 1. Structural analyses on bacterial cellulose synthase revealed that this is achieved by a tight association of the GT-domain with pore forming TMHs [3]. The cellulose synthase structure demonstrates that the binding site for the acceptor (i.e. the cellulose’s non-reducing terminal glucose unit) is formed directly at the entrance to the TM-channel, such that the elongated polysaccharide translocates into the TM pore rather than dissociates from the enzyme [5••], Figure 4. The interface between the GT domain and TM channel is formed by amphipathic interface helices, one of which contains the Q/ LxxRW motif implicated in acceptor binding [3].
Figure 4.
Pore organization of bacterial cellulose synthase subunit BcsA. BcsA’s TMHs are shown as a gray surface with the exception of TMH5 and IF2, which contains the ‘QxxRW’ motif. The GT domain is shown as a blue cartoon and cellulose and UDP are shown as sticks with blue and yellow carbon atoms, respectively. Selected residues belonging to the conserved DDG (D180), DxD (D246 and 248), TED (D343), and Q/ LxxRW (W383) motifs are shown as sticks. Cellulose synthase elongates the polymer at the non-reducing end, which is stabilized at the acceptor site within the catalytic pocket.
A sequence comparison of cellulose, chitin, HA, alginate and PNAG synthases is difficult based on primary sequences alone. However, considering the sequence conservation of the GT domains, predicted TM topologies and secondary structures, as well as experimental topology studies [57•,70], we aligned the sequences of selected GT-2 family members, Figure 2.
Interestingly, TMH predictions [70] of the region directly C-terminal of the GT domains of HA, alginate and chitin synthases predict either a long TMH or two TMHs linked via a tight 2–3 residue long extracellular loop. This region corresponds to TMH5 in BcsA [4••], which forms a cytosolic interface helix over its N terminal half and inserts into the membrane past a conserved Pro residue. The amphipathic portion of the TMH is part of the TM channel entrance. Thus, it is possible that a curved TMH is a common feature of processive GTs. The sequence alignment shown in Figure 2 can only be a cautious attempt to compare BcsA with other processive enzymes but should aid in the design of biochemical experiments to further characterize these exceedingly important enzymes.
The limited space available precluded discussing many other polysaccharides formed by processive GTs, including bacterial acetylated cellulose, mixed linkage glucans in plants and bacteria, as well as curdlan.
Acknowledgments
We apologize to all colleagues whose work was not cited in this review, due to space limitations. PP and JZ are supported by the Center for LignoCellulose Structure and Formation, Energy Frontier Research Center, US Department of Energy, Office of Science, Grant DE-SC0001090. YB is supported by the National Institutes of Health, Grant 1R01GM110143 and CH received financial support from a Cell and Molecular Biology Training Grant, National Institutes of Health, T32 GM008136.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1••.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. A comprehensive review of the structures, functions and reaction mechanisms of glycosyltransferases. [DOI] [PubMed] [Google Scholar]
- 2.McFarlane HE, Döring A, Persson S. The cell biology of cellulose synthesis. Annu Rev Plant Biol. 2014;65:69–94. doi: 10.1146/annurev-arplant-050213-040240. [DOI] [PubMed] [Google Scholar]
- 3.McNamara J, Morgan JLW, Zimmer J. A molecular description of cellulose biosynthesis. Annu Rev Biochem. 2015;84:17.11–17.27. doi: 10.1146/annurev-biochem-060614-033930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Morgan J, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. The crystal structure of bacterial cellulose synthase provides the first insights into the mechanism of processive GTs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Morgan JLW, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014;21:489–496. doi: 10.1038/nsmb.2803. The structure of cyclic-di-GMP bound to bacterial cellulose synthase reveals how cyclic-di-GMP allosterically activates cellulose biosynthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantarel B, Coutinho P, Rancurel C, Bernard T. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Omadjela O, Narahari A, Strumillo J, Mélida H, Mazur O, Bulone V, Zimmer J. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci U S A. 2013;110:17856–17861. doi: 10.1073/pnas.1314063110. A detailed in vitro biochemical analysis of bacterial cellulose biosynthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Tlapak-Simmons VL, Baron CA, Weigel PH. Characterization of the purified hyaluronan synthase from Streptococcus equisimilis. Biochemistry. 2004;43:9234–9242. doi: 10.1021/bi049468v. A detailed characterization of purified bacterial HAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- 10••.Hubbard C, McNamara J, Azumaya C, Patel M, Zimmer J. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. A HA biosynthesis assay is described that reveals that HAS is necessary and sufficient for HA synthesis and membrane translocation. [DOI] [PubMed] [Google Scholar]
- 11.Tlapak-Simmons VL, Baggenstoss BA, Clyne T, Weigel PH. Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis. J Biol Chem. 1999;274:4239–4245. doi: 10.1074/jbc.274.7.4239. [DOI] [PubMed] [Google Scholar]
- 12.Dorfmueller HC, Ferenbach AT, Borodkin VS, van Aalten DM. A structural and biochemical model of processive chitin synthesis. J Biol Chem. 2014;289:23020–23028. doi: 10.1074/jbc.M114.563353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Saxena IM, Brown RM, Dandekar T. Structure–function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry. 2001;57:1135–1148. doi: 10.1016/s0031-9422(01)00048-6. A primary sequence comparison reveals commonalities and differences between processive GTs and highlights the conservation of the Q/LxxRW motif. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Herrera J, Gonzalez-Prieto JM, Ruiz-Medrano R. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 2002;1:247–256. doi: 10.1111/j.1567-1364.2002.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 15.Kamst E, Bakkers J, Quaedvlieg NE, Pilling J, Kijne JW, Lugtenberg BJ, Spaink HP. Chitin oligosaccharide synthesis by rhizobia and zebrafish embryos starts by glycosyl transfer to O4 of the reducing-terminal residue. Biochemistry. 1999;38:4045–4052. doi: 10.1021/bi982531u. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama J, Boisset C, Hashimoto M, Watanabe T. Molecular directionality of beta-chitin biosynthesis. J Mol Biol. 1999;286:247–255. doi: 10.1006/jmbi.1998.2458. [DOI] [PubMed] [Google Scholar]
- 17.Minke R, Blackwell J. The structure of alpha-chitin. J Mol Biol. 1978;120:167–181. doi: 10.1016/0022-2836(78)90063-3. [DOI] [PubMed] [Google Scholar]
- 18.Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur J Cell Biol. 2011;90:759–769. doi: 10.1016/j.ejcb.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Carpita NC. Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1 → 4)-beta-D-glycans. Plant Physiol. 2011;155:171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- 21.Silverman SJ. Similar and different domains of chitin synthases 1 and 2 of S. cerevisiae: two isozymes with distinct functions. Yeast. 1989;5:459–467. doi: 10.1002/yea.320050605. [DOI] [PubMed] [Google Scholar]
- 22.Becker HF, Piffeteau A, Thellend A. Saccharomyces cerevisiae chitin biosynthesis activation by N-acetylchitooses depends on size and structure of chito-oligosaccharides. BMC Res Notes. 2011;4:454. doi: 10.1186/1756-0500-4-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teh EM, Chai CC, Yeong FM. Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle. 2009;8:2964–2974. [PubMed] [Google Scholar]
- 25.Martinez-Rucobo FW, Eckhardt-Strelau L, Terwisscha van Scheltinga AC. Yeast chitin synthase 2 activity is modulated by proteolysis and phosphorylation. Biochem J. 2009;417:547–554. doi: 10.1042/BJ20081475. [DOI] [PubMed] [Google Scholar]
- 26.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 27•.Girish KS, Kemparaju K. The magic glue hyaluronan and its eraser hyaluronidase: a biological overview. Life Sci. 2007;80:1921–1943. doi: 10.1016/j.lfs.2007.02.037. A detailed review of HA’s many physiological functions and its implication in diseases. [DOI] [PubMed] [Google Scholar]
- 28•.Weigel PH, Deangelis PL. Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem. 2007;282:36777–36781. doi: 10.1074/jbc.R700036200. A comprehensive review of HA biosynthesis. [DOI] [PubMed] [Google Scholar]
- 29.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 30.Menzel EJ, Farr C. Hyaluronidase and its substrate hyaluronan: biochemistry, biological activities and therapeutic uses. Cancer Lett. 1998;131:3–11. doi: 10.1016/s0304-3835(98)00195-5. [DOI] [PubMed] [Google Scholar]
- 31.DeAngelis PL. Microbial glycosaminoglycan glycosyltransferases. Glycobiology. 2002;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- 32•.DeAngelis P, Jing W, Graves M, Burbank D, Van Etten J. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. The first biochemical characterization of viral HA biosynthesis highlights differences and commonalities with bacterial and eukaryotic HASs. [DOI] [PubMed] [Google Scholar]
- 33.Kumari K, Weigel PH. Molecular cloning, expression, and characterization of the authentic hyaluronan synthase from group C Streptococcus equisimilis. J Biol Chem. 1997;272:32539–32546. doi: 10.1074/jbc.272.51.32539. [DOI] [PubMed] [Google Scholar]
- 34.Tlapak-Simmons VL, Kempner ES, Baggenstoss BA, Weigel PH. The active streptococcal hyaluronan synthases (HASs) contain a single HAS monomer and multiple cardiolipin molecules. J Biol Chem. 1998;273:26100–26109. doi: 10.1074/jbc.273.40.26100. [DOI] [PubMed] [Google Scholar]
- 35••.DeAngelis PL, Papaconstantinou J, Weigel PH. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993;268:19181–19184. The identifcation and biochemical characterization of bacterial HAS is described, representing the first characterization of a HAS overall. [PubMed] [Google Scholar]
- 36.DeAngelis PL, Jing W, Drake RR, Achyuthan AM. Identification and molecular cloning of a unique hyaluronan synthase from Pasteurella multocida. J Biol Chem. 1998;273:8454–8458. doi: 10.1074/jbc.273.14.8454. [DOI] [PubMed] [Google Scholar]
- 37.Prehm P. Synthesis of hyaluronate in differentiated teratocarcinoma cells. Mechanism of chain growth. Biochem J. 1983;211:191–198. doi: 10.1042/bj2110191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tlapak-Simmons VL, Baron CA, Gotschall R, Haque D, Canfield WM, Weigel PH. Hyaluronan biosynthesis by class I streptococcal hyaluronan synthases occurs at the reducing end. J Biol Chem. 2005;280:13012–13018. doi: 10.1074/jbc.M409788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prehm P. Biosynthesis of hyaluronan: direction of chain elongation. Biochem J. 2006;398:469–473. doi: 10.1042/BJ20060431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigel PH, West CM, Zhao P, Wells L, Baggenstoss BA, Washburn JL. Hyaluronan synthase assembles chitin oligomers with -GlcNAc(alpha1→)UDP at the reducing end. Glycobiology. 2015;25:632–643. doi: 10.1093/glycob/cwv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodevin-Authelet S, Kusche-Gullberg M, Pummill PE, DeAngelis PL, Lindahl U. Biosynthesis of hyaluronan: direction of chain elongation. J Biol Chem. 2005;280:8813–8818. doi: 10.1074/jbc.M412803200. [DOI] [PubMed] [Google Scholar]
- 42.Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem. 1999;274:25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson A, Brinck J, Briskin MJ, Spicer AP, Heldin P. Expression of human hyaluronan synthases in response to external stimuli. Biochem J. 2000;348(Pt 1):29–35. [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, Stephanopoulos G. Metabolic engineering of Escherichia coli for biosynthesis of hyaluronic acid. Metab Eng. 2008;10:24–32. doi: 10.1016/j.ymben.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Kumari K, Baggenstoss BA, Parker AL, Weigel PH. Mutation of two intramembrane polar residues conserved within the hyaluronan synthase family alters hyaluronan product size. J Biol Chem. 2006;281:11755–11760. doi: 10.1074/jbc.M600727200. [DOI] [PubMed] [Google Scholar]
- 46.Pummill PE, DeAngelis PL. Alteration of polysaccharide size distribution of a vertebrate hyaluronan synthase by mutation. J Biol Chem. 2003;278:19808–19814. doi: 10.1074/jbc.M301097200. [DOI] [PubMed] [Google Scholar]
- 47.Tammi RH, Passi AG, Rilla K, Karousou E, Vigetti D, Makkonen K, Tammi MI. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 2011;278:1419–1428. doi: 10.1111/j.1742-4658.2011.08070.x. [DOI] [PubMed] [Google Scholar]
- 48.Karousou E, Kamiryo M, Skandalis SS, Ruusala A, Asteriou T, Passi A, Yamashita H, Hellman U, Heldin C-H, Heldin P. The activity of hyaluronan synthase 2 is regulated by dimerization and ubiquitination. J Biol Chem. 2010;285:23647–23654. doi: 10.1074/jbc.M110.127050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goentzel BJ, Weigel PH, Steinberg RA. Recombinant human hyaluronan synthase 3 is phosphorylated in mammalian cells. Biochem J. 2006;396:347–354. doi: 10.1042/BJ20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vigetti D, Deleonibus S, Moretto P, Karousou E, Viola M, Bartolini B, Hascall VC, Tammi M, De Luca G, Passi A. Role of UDP-N-acetylglucosamine (GlcNAc) and O-GlcNAcylation of hyaluronan synthase 2 in the control of chondroitin sulfate and hyaluronan synthesis. J Biol Chem. 2012;287:35544–35555. doi: 10.1074/jbc.M112.402347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haug A, Larsen B. Biosynthesis of alginate. Epimerisation of D-mannuronic to L-guluronic acid residues in the polymer chain. Biochim Biophys Acta. 1969;192:557–559. doi: 10.1016/0304-4165(69)90414-0. [DOI] [PubMed] [Google Scholar]
- 52.Jain S, Franklin MJ, Ertesvåg H, Valla S, Ohman DE. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol. 2003;47:1123–1133. doi: 10.1046/j.1365-2958.2003.03361.x. [DOI] [PubMed] [Google Scholar]
- 53.Nyvall P, Corre E, Boisset C, Barbeyron T, Rousvoal S, Scornet D, Kloareg B, Boyen C. Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 2003;133:726–735. doi: 10.1104/pp.103.025981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haug A, Larsen B, Smidsrød O. Uronic acid sequence in alginate from different sources. Carbohydr Res. 1974;32:217–225. [Google Scholar]
- 55.Davidson IW, Sutherland IW, Lawson CJ. Localization of O-acetyl groups of bacterial alginate. J Gen Microbiol. 1977;98:603–606. doi: 10.1099/00221287-98-1-223. [DOI] [PubMed] [Google Scholar]
- 56.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. PhoA fusion constructs are used to determine the TM topology of Alg8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2011;108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain S, Ohman DE. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BHA. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol. 2013;79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remminghorst U, Rehm BHA. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 62•.Römling UU, Galperin MYM, Gomelsky MM. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. A comprehensive review of cyclic-di-GMP signaling and its cellular effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. Discovery of cyclic-di-GMP based on its ability to activate bacterial cellulose biosynthesis. [DOI] [PubMed] [Google Scholar]
- 64.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litrán T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Itoh Y, Rice J, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge T, Preston, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67•.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. Identification of the genes involved in PNAG biosynthesis in E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Little DJ, Bamford NC, Pokrovskaya V, Robinson H, Nitz M, Howell PL. Structural basis for the de-N-acetylation of poly-β-1,6-N-acetyl-D-glucosamine in Gram-positive bacteria. J Biol Chem. 2014;289:35907–35917. doi: 10.1074/jbc.M114.611400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein–protein interaction. EMBO J. 2013;32:354–368. doi: 10.1038/emboj.2012.315. Characterization of the activation of PNAG biosynthesis by cyclic-di-GMP and its interaction with and stabilization of the PgaCD complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heldermon C, DeAngelis PL, Weigel PH. Topological organization of the hyaluronan synthase from Streptococcus pyogenes. J Biol Chem. 2001;276:2037–2046. doi: 10.1074/jbc.M002276200. [DOI] [PubMed] [Google Scholar]
- 71.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2. 0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]