Summary
Nrf2 regulates the expression of numerous anti-oxidant, anti-inflammatory, and metabolic genes. We observed that, paradoxically, Nrf2 protein levels decline in the livers of aged rats despite the inflammatory environment evident in that organ. To examine the cause(s) of this loss, we investigated the age-related changes in Nrf2 protein homeostasis and activation in cultured hepatocytes from young (4-6 months) and old (24-28 months) Fischer 344 rats. While no age-dependent change in Nrf2 mRNA levels was observed (p>0.05), Nrf2 protein content, and the basal and anetholetrithione (A3T)-induced expression of Nrf2-dependent genes were attenuated with age. Conversely, overexpression of Nrf2 in cells from old animals reinstated gene induction. Treatment with A3T, along with bortezomib to inhibit degradation of existing protein, caused Nrf2 to accumulate significantly in cells from young animals (p<0.05), but not old, indicating a lack of new Nrf2 synthesis. We hypothesized that the loss of Nrf2 protein synthesis with age may partly stem from an age-related increase in microRNA inhibition of Nrf2 translation. Microarray analysis revealed that six microRNAs significantly increase >2-fold with age (p<0.05). One of these, miRNA-146a, is predicted to bind Nrf2 mRNA. Transfection of hepatocytes from young rats with a miRNA-146a mimic caused a 55% attenuation of Nrf2 translation that paralleled the age-related loss of Nrf2. Overall, these results provide novel insights for the age-related decline in Nrf2 and identify new targets to maintain Nrf2-dependent detoxification with age.
Keywords: Nrf2, translation, microRNA, aging, antioxidant response element
Introduction
The transcription factor Nuclear Factor, Erythroid Derived 2, Like 2 (NFE2L2 or Nrf2) is a gatekeeper of stress resistance without which cells succumb to both exogenous and endogenous toxins. Over 200 genes responsible for protein homeostasis, antioxidants, and detoxification are under the control of Nrf2 through the Antioxidant Response Element (ARE) binding site. While Nrf2 knockout mice are viable, they display a dramatically lowered ability to detoxify potential carcinogens [1]. Nrf2 is vital for protection against the molecular damage that leads to neoplasia, a risk that increases with age. Unfortunately, Nrf2 protein levels do not appear to be maintained during aging. In fact, we showed previously that Nrf2 levels decline in the aging rat liver [2], which reveals a problematic age-related deficit in the very organ that is responsible for detoxification.
In addition to this loss of detoxification capacity, aging is also characterized by an increase in endogenous reactive oxygen and nitrogen species, and a loss of endogenous antioxidant production [3–5]. It is now recognized that this pro-oxidant cellular environment, which is characterized by chronic inflammation, may be an intrinsic part of the aging process. This so-called “inflamm-aging” contributes to a heightened risk for age-associated diseases (e.g. atherosclerosis, cancer, and dementias) and disuse syndromes (e.g. sarcopenia and frailty) [6]. As Nrf2 plays a key role in coordinating the cellular response to numerous pro-inflammatory insults, electrophilic xenobiotics, and metabolic perturbations, its decline during aging only exacerbates the already heightened risk for chronic pathophysiologies. However, we currently do not know why Nrf2 steady-state levels are lowered with age at the time of life when the need for detoxification is effectively increasing, nor do we know whether its induction with age is compromised.
Nrf2 homeostasis is regulated by an array of interconnected transcriptional, translational, and post-translational mechanisms that allow it to fine-tune gene expression to an array of divergent stress stimuli [7]. Under quiescent conditions, this basic leucine zipper, cap-‘N’-collar transcription factor is normally maintained at very low steady-state levels [8]. The basal content of Nrf2 is sufficient for its binding to cis-acting AREs located in the 5′-flanking regions of target antioxidant and ARE-containing genes. Under acute pro-oxidant conditions or in the presence of electrophilic compounds, Nrf2 rapidly accumulates in the nucleus and enhances expression of detoxification genes. The accumulation of Nrf2 is due in part to its dissociation from Keap1, a Cul3 ubiquitin ligase adaptor protein, thus attenuating its proteasomal degradation. However, this mechanism is only able to preserve existing Nrf2 protein, which is present at very low levels. This seemingly contradictory phenomenon has been explained by several reports showing that increased translation is a critical means to quickly adjust Nrf2 protein levels for adequate cellular response [9–11]. For example, Nrf2 mRNA contains an internal ribosomal entry site (IRES), which allows its translation even during a catabolic state when global cap-dependent translation is reduced [12].
An additional layer of translational control over Nrf2 mRNA may be mediated by microRNAs (miRNAs), a class of small RNAs derived from distinctive hairpin transcripts that are ubiquitous across the plant and animal kingdoms. Once processed, miRNAs average approximately 21-22 nucleotides in length and associate with Argonaute proteins as part of an RNA-induced silencing complex. These complexes primarily serve to control gene expression through post-transcriptional repression of translation. Several miRNAs can regulate the translation of Nrf2 mRNA and therefore lower its steady state levels [13,14]. However, it has never been shown that miRNAs are responsible for the age-related loss of Nrf2.
Despite the importance of Nrf2-mediated response with age, and the complexity of Nrf2 proteostasis, there have been few studies to date that have examined the potential mechanism(s) involved. In this study, we sought to elucidate the mechanism(s) that results in the age-related loss of Nrf2 protein levels using hepatocytes from old and young rats. We hypothesized that Nrf2 protein synthesis declines with age, resulting in the loss of basal Nrf2 protein levels and stress response. Herein, we also describe the contribution of an miRNA, miR-146a, that lowers Nrf2 translation.
Materials and methods
Reagents
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Collagenase type IV was purchased from Worthington Biochemical Corporation (Lakewood, NJ). Anti-Nrf2 (sc-13032) and anti-Keap1 (sc-15246) were ordered from Santa Cruz Biotechnology (Santa Cruz, CA). Bortezomib was obtained from Millennium Pharmaceuticals (Cambridge, MA). PVDF transfer membrane was purchased from Millipore (Billerica, MA). Dual Luciferase Assay Kit was purchased from Promega (Madison, WI).
Animals
Fischer 344 male rats, both young (4-6 months) and old (25-28 months), were purchased from the National Institute on Aging animal colonies. The rats were allowed to acclimatize in the Linus Pauling Institute animal facility for a minimum of 1 week on a 12 hour light cycle (7am to 7pm) and fed standard chow ad libitum. All animal work was approved and in accordance to IACUC guidelines (Assurance Number: A3229-01). The AAALAC-accredited Laboratory Animal Resources Center (LARC) provided management and veterinary care.
Hepatocyte Isolation and Cell Culture
Hepatocyte isolation was performed as described previously [15]. Briefly, the liver was perfused with Hank's balanced salt solution, and disassociated to a single cell suspension using a collagenase solution (1 mg/ml). The resultant cell suspension was filtered through sterile gauze to remove connective tissue and debris. Parenchymal cells were isolated using gravity filtration and washed four times with Krebs– Henseleit solution, pH 7.3. Cell count and viability were assessed using trypan blue exclusion. Freshly isolated hepatocytes were suspended in a modified Williams' E media (5% FBS, 1 mM dexamethasone, 100 ng/ml insulin, 2 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin), dispensed onto collagen coated cell culture plates and incubated for 16 hours at 37 °C in 5% CO2 atmosphere before being used in experiments. Hepatocytes were seeded at 2.5×105 cells/ml of media for regular cell culture, or 1.25×105 cells/ml of media for culture + transfection. We have demonstrated that these outlined culture conditions maintain the age profile of hepatocytes with respect to Nrf2 for at least three days in culture [16].
Transfections
Plasmid transfections of hepatocytes were achieved with the jetPEI-hepatocyte transfection reagent (Polyplus-transfection SA, Illkirch, France). Hepatocytes were incubated with 2 μg plasmid and the recommended amount of jetPEI after hepatocytes had been in culture for 30 hours. Transfection efficiency was assessed to be ≥ 60% by parallel transfection of EGFP vector and cell counting. miRNA transfection of hepatocytes were achieved with mirVana miRNA mimic (Ambion) using RNAiMax (Life Technologies) following the manufacturer's protocol. The miRNA levels were measured using the Taqman MicroRNA Assay (Life Technologies).
Nrf2 Half-life
Hepatocytes were transfected with a human influenza hemagglutinin-tagged Nrf2 (isoform BC061724) expression vector (pHA-Nrf2) with jetPEI as detailed above. After 20 hours, hepatocytes were treated with 100 μM cycloheximide. Hepatocytes were harvested every 20 minutes and the proteins were isolated as described. Immunoblots were used to quantify Nrf2 levels. Statistical analysis was done by a two-way ANOVA.
ARE-gene induction and quantitative PCR
Hepatocytes were transfected with a human influenza hemagglutinin tagged Nrf2 (isoform BC061724) over-expression vector (pHA-Nrf2) or empty vector with jetPEI as detailed above. Cells were harvested 16 hours after transfection, total RNA was collected from hepatocytes with Trizol (Life Technologies, Carlsbad, CA), and reverse transcription was performed using the Retroscript Kit (Life Technologies) following the manufacturer's protocol. The PCR was done on a StepOnePlus PCR machine (Life Technologies) using Taqman Universal Mastermix. Relative quantities were calculated based on ΔΔCt between cells transfected with pHA-Nrf2 and empty vector and assuming an amplification efficiency of 2. All primer-probe mixtures were purchased from Life Technologies except for Nrf2, which were ordered from Eurofins MWG Operon (Huntsville, Alabama) with the following sequence and modifications: TTTTCCAGTGAGGGGATCGATGAG, GTCAGCTACTCCCAGGTTGCCCA, and [6-FAM]ACCACTGTCCCCAGCCCAGAGGCCAC[BHQ1a-Q]. Quantification was normalized to the housekeeping gene EIF2A.
ARE Activity
Hepatocytes in culture were transfected with 1.95 μg ARE-Luc, a Gclc-promoter driven PGL4 luciferase vector, and 0.05 μg of a Renilla expression vector (pRL-CMV). Cells were incubated 18 hours after transfection and then treated with either vehicle control (DMF), 50 μM 5-(4-methoxyphenyl)-3H-1,2-dithiole-3-thione (A3T), or 100 μM R-α-Lipoic Acid (LA) (Makwood). After 16 hours cells were harvested and assayed for luciferase activity using the protocol provided with the Dual Luciferase Assay Kit (Promega).
Lysate preparation and immunoblots
Cultured hepatocytes were washed twice with PBS, pH 7.4, and then collected by scraping. Nuclear and cytoplasmic protein extracts were collected by the use of Sigma-Aldrich CelLytic Nu-CLEAR isolation kit following the manufacturer's protocol. Protein samples were denatured by addition of 2× Laemmli loading buffer and heating at 90 °C for 5 minutes. Protein separation was performed using a neutral Bis-tris acrylamide gel and following the BioRad XT Bis-Tris PAGE XT-Mes protocol. Transfer was accomplished using the Bjerrum Schafer-Nielsen transfer buffer with 20% methanol on a semi-dry transfer apparatus. Proteins were deposited on PVDF transfer membrane. All blots were blocked with a 5% milk (w/v) TBS-T solution overnight at 5°C. Primary and secondary antibodies were each incubated on the membranes for 2 hours. Densitometry, performed with the ImageJ software package (http://imagej.nih.gov/ij/), was used for graphical representation and to perform statistical analysis.
miRNA Microarray
Liver tissue from young and old rats was collected and miRNAs isolated using the PureLink miRNA Isolation Kit (Life Technologies). Samples were frozen in liquid nitrogen and shipped to LC Sciences (Houston, TX) for analysis. Microarray analysis covered the entire Rattus norvegicus miRBase set (miRBase release 12.0). Background subtraction, normalization, and statistical analysis where provided by LC Sciences. Significance was calculated by ANOVA and corrected for multiple comparisons.
Statistical Analysis
All statistical analysis was performed using Excel (Microsoft, Inc.) and Prism 7 (GraphPad Software, Inc.) For comparisons between two samples, two-sided Student's t-test was used. Differences between samples that resulted in a p-value of ≤ 0.05 were considered statistically significant. Statistical analysis between multiple endpoints was analyzed by two-way ANOVA and multiple comparisons were evaluated by Tukey's post hoc method.
Results
Nrf2 protein declines with age in rat hepatocytes
We previously showed that constitutive Nrf2 binding to a core consensus ARE sequence declines with age, indicating that basal expression of ARE-mediated genes may be compromised [15]. Figure 1a,b shows a marked decline of Nrf2 protein with age; overall steady-state Nrf2 levels were 42 ± 12% lower in livers of 24-month old rats than in young controls (p<0.05). Moreover, Nrf2 loss was coincident with a significant attenuation of expression of genes that are constitutively regulated by Nrf2. As shown in Figure 1c, mRNA content of Gclc, Gclm, and Gst2a exhibited age-dependent declines of 19.6%, 31.4%, and 40%, respectively. Because Nrf2 mRNA levels vary in tissues [17] and in response to certain toxicological stressors [18–20], we sought to determine whether lowered Nrf2 transcript levels may be responsible for the age-related loss of Nrf2 protein content. On the contrary, hepatic Nrf2 mRNA values exhibited no significant age-dependent changes (Figure 1c; p>0.14), indicating that dysregulation of Nrf2 transcription with age does not account for attenuation of Nrf2 protein levels.
Figure 1.
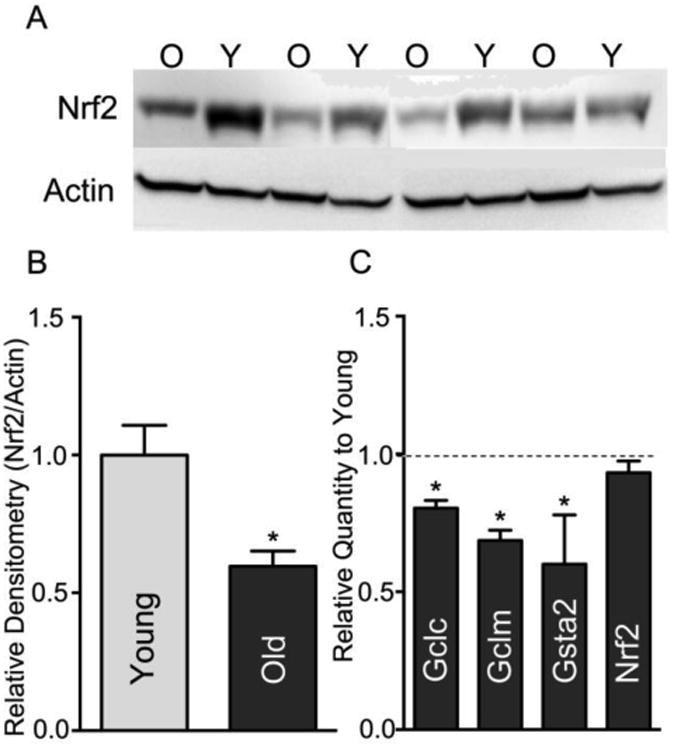
Nrf2 protein levels but not mRNA levels decline with age in rat liver.
(A) Liver tissue lysate from young (Y) and old (O) rats were assayed for Nrf2 protein content by immunoblot. Actin was utilized as a loading control and housekeeping gene. (B) Densitometry graph for the expression of Nrf2 protein, N=4; *p <0.05. (C) QPCR was utilized to assay the relative expression of the Nrf2 regulated genes: gamma glutamyl cysteine ligase catalytic and modulatory subunits (Gclc and Gclm), and glutathione S transferase subunit a2 (Gsta2), as well as Nrf2 itself. Graph shows old relative to young (Y=1.0). Messages that decline significantly with age are noted by * (p<0.05; N=7).
Increasing Nrf2 levels remediates lost ARE gene expression
To determine whether replenishing Nrf2 hepatic protein content may remediate the observed declines in ARE-dependent gene expression, we transfected hepatocytes isolated from young and old rats with a CMV-driven pHA-Nrf2 expression vector. Under Nrf2 overexpression, the decline in ARE-associated target gene expression was reversed, where Gclc and Gclm mRNA content were at least 3-fold higher than observed in hepatocytes transfected with an empty vector (Figure 2; p<0.05), and there were no longer any age-associated differences in their mRNA levels. These results show that the lower steady-state levels of Nrf2 primarily drive the observed loss of ARE-mediated detoxification gene expression with age.
Figure 2.
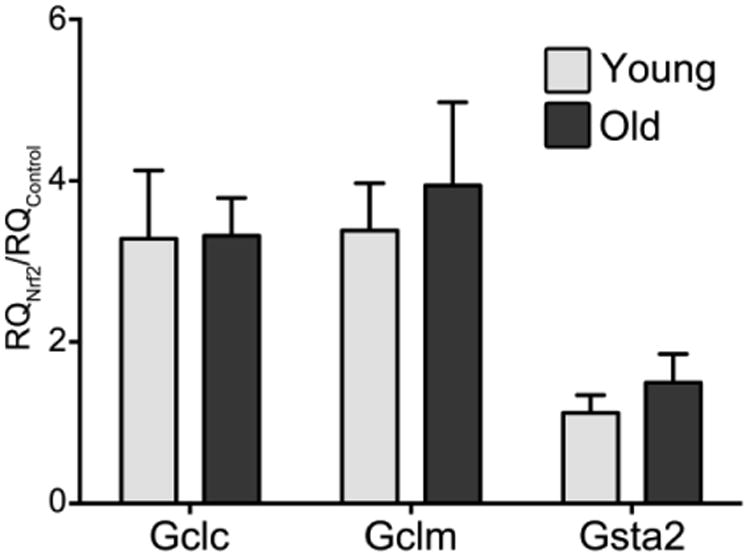
Nrf2 over-expression results in similar induction of ARE regulated genes with age. Cultured hepatocytes from young and old rats were transfected with pHA-Nrf2. After 24 h, qPCR was utilized to assay the relative expression of genes regulated by Nrf2. Analysis shows that there is no significant difference between young and old samples (p>0.05).
Aging compromises Nrf2 activation of ARE-mediated gene expression
Our results point to a connection between lower Nrf2 steady-state levels and decrements in stress response genes with age. However, it is not known whether Nrf2 levels are still sufficient to initiate a proper response to stress stimuli. To determine the extent of any age-dependent changes in Nrf2 stress response, we transfected hepatocytes from young and old rats with an ARE-containing luciferase reporter (ARE-luc), then treated the cells with known inducers of Nrf2/ARE-mediated gene expression. Treating cells from young animals with 100 μM R-α-lipoic acid (LA) [2] or 50 μM anethole trithione (5-[4-methoxyphenyl]-3H-1,2-dithiole-3-thione; A3T) [23], yielded a robust activation of the ARE-luc reporter vector, indicating a strong induction of the Nrf2-mediated stress resistance response in these cells. However, hepatocytes derived from old animals showed only a small induction in luciferase activity with either LA or A3T, which was 6.5- and 7.0-fold lower, respectively, than what was observed in cells from young rats (p<0.05) (Figure 3). These results show that the age-related loss of Nrf2 not only compromises basal expression of certain ARE-mediated genes, but also has the potential for limiting the induction of ARE-mediated stress response from xenobiotic and stress stimuli.
Figure 3.
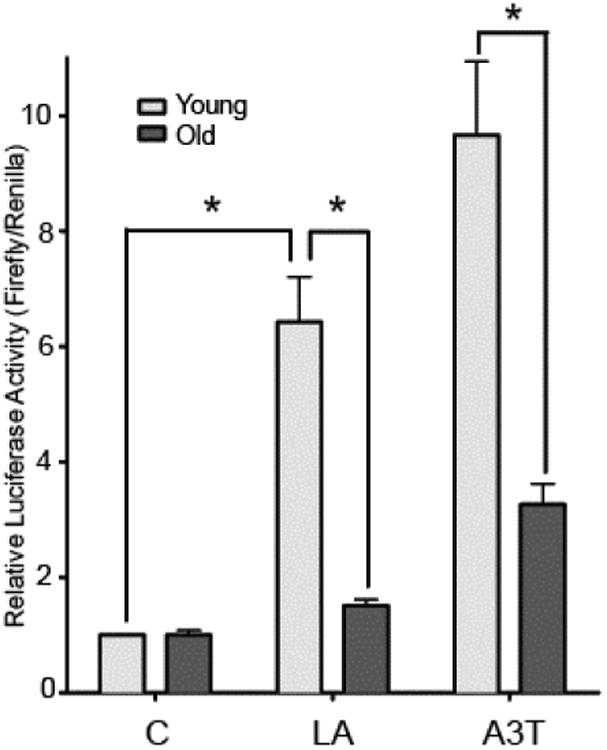
Stress response of the ARE is attenuated with age.
Hepatocytes isolated from young and old rats were transfected with the ARE-Firefly luciferase and Renilla-PLG4 (control) luciferase reporter vectors. After 24 h, cells were treated with vehicle control (C), 100 μM lipoic acid (LA), or 50 μM A3T for 16 h, then lysed and assayed for luciferase activities. (N=3 *p<0.05). Control values were normalized to 1.0 for both young and old in order to show the relative increase.
Nrf2 steady-state levels are an interplay between its rates of synthesis and degradation. Considering that Figure 1c shows no decrements in Nrf2 mRNA levels with age, we next examined the accumulation of the Nrf2 protein itself under stress stimuli in cells pretreated with bortezomib (BTZ), an inhibitor of the 26S proteasome, to prevent Nrf2 turnover. While Nrf2 levels accumulated in cells from young animals both basally and with LA or A3T stress induction, the same was not true in cells from old animals (Fig. 4A,B). In fact, no significant increase in Nrf2 occurred in cells from old animals, and the use of BTZ shows that this was not due to increased Nrf2 proteasomal degradation. In a follow-up experiment, we measured the levels of Keap1 by immunoblot. As Keap1 is a Cul3 ubiquitin ligase that facilitates Nrf2 degradation by the 26S proteasome, a significant increase in Keap1 in cells from old animals vs young would suggest that more Nrf2 degradation occurs with age. Contrary to this, we found that Keap1 levels declined 42% on an age basis (Figure 4C) (p<0.05). This surprising result prompted us to measure Nrf2 half-life in both sets of hepatocytes. We overexpressed Nrf2 in hepatocytes from young and old rats and directly quantified the rate of Nrf2 turnover using cycloheximide as a means to block protein degradation. Results showed marked differences in Nrf2 turnover rates between young and old rat hepatocytes. A single-phase decay model to fit the data revealed that Nrf2 half-life increased >5-fold with age (p<0.05; Figure 4D), with T1/2 estimates of 21.6 ± 3.0 and 119.8 ± 21.5 minutes in young and old hepatocytes, respectively. These results, along with the age-related loss of Keap1, suggest a markedly slower rate of Nrf2 degradation, showing that Nrf2 degradation is not responsible for the steady-state decline in hepatic Nrf2 levels with age, and may even be a mechanism used by the cell to compensate for Nrf2 loss. All of these results are in accordance with our previous data in immortalized cells [10] showing that Nrf2 mRNA translation is an important aspect of its ability to respond quickly to oxidative or xenobiotic stress, whereas it was formerly supposed that all Nrf2 entering the nucleus was being rescued from Keap1-mediated degradation. However, our current data suggests for the first time that the age-related loss of Nrf2 may be due to a change in its translation.
Figure 4.
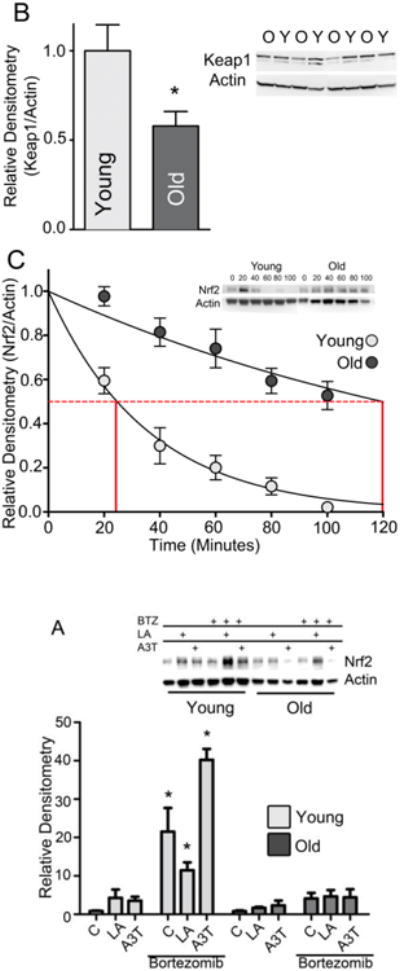
Both Nrf2 synthesis and degradation decline with age.
(A) Hepatocytes from young and old rats were treated with 50 μM A3T, 100 μM lipoic acid (LA), and/or 100 nM bortezomib for 6 h. Nuclear fractions were isolated from the harvested samples and assayed for Nrf2 content. Samples were quantitated by densitometry relative to the loading control Actin. Statistical analysis was done by two-way ANOVA and Tukey's post-hoc method. *p<0.05 vs young control without BTZ. (N=3). (B) Graphic representation of Keap1 immunoblots using liver tissue from young and old rats N=4; *p <0.05. Actin loading control is from the same western blot membrane shown in Figure 1A. (C) Half-life of Nrf2 protein was determined by transfecting primary hepatocytes from young and old rats with the pHA-Nrf2 expression vector, then adding 100 μM cycloheximide 20 h post-transfection. Samples were taken every 20 minutes and protein was extracted. Nrf2 protein levels were determined by immunoblot, quantified by ImageJ, and used to create the graph. N=3.
MicroRNA 146a targets Nrf2 translation and is increased with age
While Nrf2 translation is lower with age, the partial induction of Nrf2 with A3T shows that new synthesis of Nrf2 is still functional, albeit at a markedly lower rate. Because of growing evidence that mRNA translation in general and Nrf2 translation in particular are fine-tuned to environmental conditions via short non-coding interfering RNA (miRNA) [14], we hypothesized that the sharp attenuation of hepatic Nrf2 mRNA with age stemmed from an increase in miRNA-dependent inhibition. Accordingly, a top-down analysis of age-dependent changes to microRNA was performed using tissue extracts from young and old rat liver. Of the microRNA analyzed, only thirteen transcripts met both the 2-fold and statistical threshold for significance. Most of these miRNA species exhibited an age-associated decline (data not shown), which is in agreement with other reports showing dicer-dependent decline in miRNA maturation with age [24]. However, a small cadre of miRNA transcripts significantly increased on an age-related basis (Fig. 5a). While there are currently few reports identifying genes associated with many of these miRNAs, it does appear that most if not all of the miRNA species that become elevated with age (34a, 146a, 28, 101a) are also known to be induced under chronic inflammatory conditions [25–28]. The oxidant-enriched milieu common in aging tissues has been hypothesized to contribute to a sterile chronic necro-inflammatory environment that goes unresolved by cellular stress response pathways [29,30].
Figure 5.
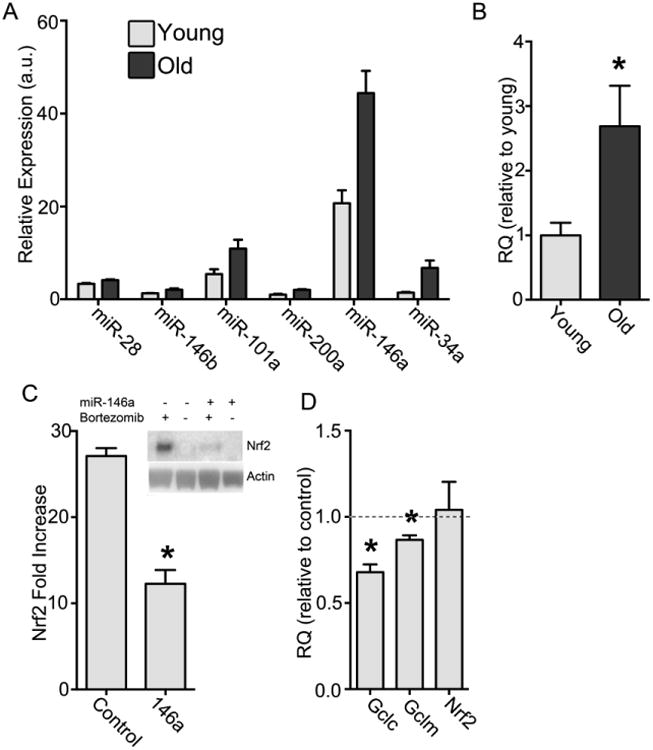
Increased miR-146a with age reduces Nrf2 protein levels.
(A) Liver tissue from 6 young (6 months) and 6 old (24 months) rats were analyzed on a miRNA array. Results were limited to p values of less than 0.05 and corrected for multiple comparisons. The miRNAs predicted and/or experimentally determined to modulate Nrf2 are shown. (B) Liver tissue from 6 young and 6 old rats were analyzed by RT-qPCR for levels of miR-146a. The difference between young and old is significant by Student's t-test *p<0.05. (C) Hepatocytes isolated from young rats were transfected with rno-miR-146a miRNA mimic or scrambled RNA oligomer, treated with BTZ for 6 h, and compared to vehicle-treated hepatocytes (N=3 for each condition). Nrf2 protein levels and loading control Actin were measured by immunoblot analysis and quantitated by densitometry. Graph shows Nrf2 fold increase over non-BTZ-treated cells (= 1) (*p<0.05). (D) mRNA levels were measured and compared to hepatocytes transfected with a scrambled RNA oligomer (= 1.0). Although no significant difference was seen in the RQ of the Nrf2 mRNA, both Gclc and Gclm decrease significantly, *p<0.05.
Using mirSVR sequencing analysis and available literature reports, six miRNAs were identified [12,30–32] that increased significantly with age and/or could theoretically bind to the 3′ region of Nrf2 mRNA. A preliminary test was performed by transfecting chemically modified double-stranded RNAs that mimic these miRNAs into the rat hepatoma cell line H4IIE to determine their potential for inhibiting Nrf2 translation (data not shown). While the other miRNAs failed to inhibit Nrf2 expression in this immortalized cell line, the rno-miR-146a mimic (rno = Rattus norvegicus) markedly reduced Nrf2 protein and thus was selected for further experimentation. In order to confirm the array results, liver tissue from young and old rats was harvested, microRNA isolated, and RT-qPCR was performed (Fig. 5b). This analysis revealed a 2.7 ± 0.3-fold increase in miR-146a in aging rat liver versus young controls, providing strong evidence this is an age-affected miRNA in the liver.
In order to investigate whether miR-146a influences the expression of Nrf2, hepatocytes from young rats were transfected with the mimic of the endogenous version, as used above in immortalized cells. Immunoblot analysis showed that Nrf2 protein levels were significantly lower in mimic-transfected cells versus those transfected with a scrambled control (Fig. 5c). As miRNAs either affect mRNA translation or bind and elicit message degradation, additional experiments examined whether rno-miR-146a altered Nrf2 mRNA levels and Nrf2-controlled genes (Fig. 5d). qPCR analysis revealed that rno-miR-146a treatment resulted in no significant difference in Nrf2 mRNA levels between the control and rno-miR-146a-treated samples. This is despite the observed change in its protein accumulation, which suggests that rno-miR-146a inhibits ribosomal association of Nrf2 mRNA but does not induce its degradation. However, mRNA levels of the Nrf2-mediated genes, Gclc and Gclm, were lowered by the mimic treatment, 32.1 ±4.5% and 13.4 ±2.6% respectively. These data reflect the loss of Nrf2 protein synthesis in spite of preserved mRNA levels seen in liver tissues and isolated hepatocytes from old rats. Thus, the elevated level of miR-146a with age is consistent with a role in the attenuated translation of Nrf2.
Discussion
Nrf2 is not only involved in detoxification, but is increasingly recognized as an important longevity-assurance transcription factor because it regulates expression of numerous genes involved in stress resistance and also metabolism genes associated with longevity and health. Even some of the longevity enhancing effects of dietary restriction may stem from Nrf2-mediated gene regulation [34]. In accordance with our previous work [10] showing complex control of Nrf2 levels through mRNA translation, we found that the age-related lesion in Nrf2 proteostasis and inducibility is attenuated Nrf2 translation. Given the attenuated response to LA and A3T (Fig. 3), our data clearly point to a deficit in synthesis of Nrf2 protein with aging.
Decline in protein translation appears to be a trait of aging, and our data indicates that the decline in Nrf2 synthesis is in keeping with this general trait [35]. However, because Nrf2 has such a short half-life, Nrf2 levels and the genes that it regulates may be more adversely affected than other proteins with longer half-lives. We show that steady-state Nrf2 levels fall by approximately 40% with age despite a significantly slower turnover rate. Thus, adaptive mechanisms to maintain steady-state amounts of Nrf2 are not adequate as the aging process gathers pace.
The precise mechanism(s) that lower overall steady-state Nrf2 with age are yet to be completely defined. Our current data indicates that miRs which arise during pro-inflammatory conditions may be at least one cause for attenuated Nrf2 translation. We show that miR-146a reduces Nrf2 translation directly or indirectly due to its own increase with age, though it is likely one of several miRNAs with this influence. Herein, we showed that only miRNAs associated with inflammation increase with age in rat liver, while miRNA species connected to other metabolic pathways actually decline. miR-146a was the most abundant inflammation-induced miRNA that increased with age, and has a predicted binding sequence in the 3′-flanking region of Nrf2. Moreover, mimicking the age-related increase in miR-146a in hepatocytes from young animals partially recapitulates the loss of Nrf2 seen with age. While these results do not rise to the level of a cause-and-effect relationship, it does suggest the possibility that the pro-inflammatory environment of the aging rat liver chronically induces expression of miR-146a, which in turn, adversely affects Nrf2 translation.
The loss of steady-state Nrf2 with age may be a compensatory mechanism for the increased risk of neoplasia. Although Nrf2 is responsible for xenobiotic detoxification and is therefore protective against potentially DNA-damaging compounds [21], the end result of Nrf2 activation is cytoprotection and, in many cases, escape from apoptosis [22]. Increased Nrf2 has been found in some cancers, and Nrf2 has also been implicated in cancer progression via its activation of metabolic enzymes in the pentose phosphate pathways [1]. Thus, lower Nrf2 at a basal level would not be detrimental as long as the system is still inducible. Unfortunately, this is not the case; when we induced Nrf2-regulated gene expression with A3T or LA, Nrf2-dependent stress response was severely attenuated compared to that in young animals (Figs. 3,4). While it remains to be fully elucidated the extent to which Nrf2 translation is activated under stress stimuli, these results do indicate that the attenuated Nrf2 translation of aging may be an underlying factor in the well-known age-dependent loss of xenobiotic and oxidant-induced stress response. In agreement with this, we previously showed that old animals are more vulnerable to exposure to lipid hydroperoxides, which are detoxified in a glutathione- and Nrf2-mediated manner [36]. Other studies have also demonstrated an age-related vulnerability to various drugs including H2-receptor antagonists (antacids), anesthetics, acetaminophen, and alcohol. All of these drugs are detoxified via Nrf2-dependent genes [37–39]. Thus, the age-related loss of Nrf2 translation may play a pivotal role in vulnerability to a variety of xenobiotic insults, further justifying the growing public health concerns associated with polypharmacy in the elderly [40,41]. As definitive examination of age-related changes in detoxification capacity, and its potential consequences on the therapeutic threshold of pharmacological agents in older individuals are scant, further work will be necessary to elucidate the extent and precise nature that the loss of Nrf2 protein homeostasis plays in increased susceptibility to environmental and pharmacological insults with age.
Outside of its involvement in susceptibility to acute toxicological insults, attenuated Nrf2 translation may be a factor in increasing the risk for chronic age-dependent pathophysiologies. For example, we previously demonstrated that significant age-related changes occur to hepatic gene expression where an ontological analysis of transcript changes revealed enrichment of immune response, immune cell infiltration, pro-inflammatory, and tissue remodeling genes [29]. Most importantly, there was no apparent coincident increase in expression of antioxidant/detoxification genes. Therefore, the loss of Nrf2 could directly contribute to perpetuation of the low-grade inflammation associated with aging. Nrf2 directly regulates numerous genes that act as negative feedback inhibitors to down-regulate cytokine and NFKB-mediated inflammation [42], which suggests that loss of Nrf2-mediated target genes may play a critical role for numerous pathophysiologies where chronic inflammation is part of the underlying etiology (e.g. dyslipidemia, fibrosis, cirrhosis, and cancer). Given the prevalence of miR-146a in inflammation, it is also interesting to note that the loss of Nrf2 translation may not only potentiate chronic inflammation, but may be perpetuated by the necro-inflammatory environment of the aging liver. The advantages of “cross-regulation” between inflammation and Nrf2, via miRNA, are presently unclear; however, it may be an example of antagonistic pleiotropy. Elevated miR-146a is a normal response to inflammation and appears to be a feedback mechanism to lower inflammatory response. Normally, inflammation is a transient condition and therefore so is the increased expression of miR-146a; however, in the aged animals inflammation persists. The age-related loss in Nrf2 translation may thus be an indirect consequence of persistent upregulation of pro-inflammatory cytokines. The resulting attenuation of Nrf2 would ironically limit the means to resolve the inflammatory response.
In summary, we have identified, for the first time, an age-related change that results in the attenuation of Nrf2 protein levels in the liver of old rats. Chiefly, Nrf2 protein synthesis declines with age. This observation can, in part, explain why Nrf2 levels decline despite the age-related increase in ROS. This results in the increased susceptibility to acute stressors that rely on basal levels of Nrf2 for detoxification and likely chronic challenges as well. Further research will be required to characterize the consequence of the loss of Nrf2 translation in regards to chronic inflammatory conditions. Finally, restoration of Nrf2 synthesis, perhaps via disruption of miR-146a, and the effects of this reversal continue to be investigated.
Highlights.
- Nrf2 protein levels and activation are attenuated in aging rat liver.
- Nrf2 translation is lowered with age, while its transcription is maintained.
- MicroRNA 146a is increased with age and contributes to loss of Nrf2 translation.
Acknowledgments
This work was supported by NIH Grant P01AT002034 and a grant from the Medical Research Foundation of Oregon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moon EJ, Giaccia A. Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radic Biol Med. 2015;79:292–299. doi: 10.1016/j.freeradbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suh JH, Shenvi SV, Dixon BM, Liu H, Jaiswal AK, Liu RM, Hagen TM. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci U S A. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nohl H. Involvement of free radicals in ageing: a consequence or cause of senescence. Br Med Bull. 1993;49:653–667. doi: 10.1093/oxfordjournals.bmb.a072638. [DOI] [PubMed] [Google Scholar]
- 4.Schöneich C. Reactive oxygen species and biological aging: a mechanistic approach. Exp Gerontol. 1999;34:19–34. doi: 10.1016/s0531-5565(98)00066-7. [DOI] [PubMed] [Google Scholar]
- 5.Vasilaki A, Mansouri A, Van Remmen H, van der Meulen JH, Larkin L, Richardson AG, McArdle A, Faulkner JA, Jackson MJ. Free radical generation by skeletal muscle of adult and old mice: effect of contractile activity. Aging Cell. 2006;5:109–117. doi: 10.1111/j.1474-9726.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- 6.Sohal RS, Orr WC. The redox stress hypothesis of aging. Free Radic Biol Med. 2012;52:539–555. doi: 10.1016/j.freeradbiomed.2011.10.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of nrf2 protein in activation of antioxidant response by oxidants. Mol Pharmacol. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 10.Shay KP, Michels AJ, Li W, Kong ANT, Hagen TM. Cap-independent Nrf2 translation is part of a lipoic acid-stimulated detoxification stress response. Biochim Biophys Acta. 2012;1823:1102–1109. doi: 10.1016/j.bbamcr.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu B, Zhang J, Strom J, Lee S, Chen QM. Myocardial ischemic reperfusion induces de novo Nrf2 protein translation. Biochim Biophys Acta. 2014;1842:1638–1647. doi: 10.1016/j.bbadis.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Thakor N, Xu EY, Huang Y, Chen C, Yu R, Holcik M, Kong AN. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010;38:778–788. doi: 10.1093/nar/gkp1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N, Muthusamy S, Liang R, Sarojini H, Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev. 2011;132:75–85. doi: 10.1016/j.mad.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Ku CH, Siow RCM. Regulation of the Nrf2 antioxidant pathway by microRNAs: New players in micromanaging redox homeostasis. Free Radic Biol Med. 2013;64:4–11. doi: 10.1016/j.freeradbiomed.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Shenvi SV, Smith E, Hagen TM. Identification of age-specific Nrf2 binding to a novel antioxidant response element locus in the Gclc promoter: a compensatory means for the loss of glutathione synthetic capacity in the aging rat liver? Aging Cell. 2012;11:297–304. doi: 10.1111/j.1474-9726.2011.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenvi SV, Dixon BM, Petersen Shay K, Hagen TM. A rat primary hepatocyte culture model for aging studies. Curr Protoc Toxicol. 2008;Chapter 14 doi: 10.1002/0471140856.tx1407s37. Unit 14.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker JR, Su AI, Self DW, Hogenesch JB, Lapp H, Maier R, Hoyer D, Bilbe G. Applications of a rat multiple tissue gene expression data set. Genome Res. 2004;14:742–749. doi: 10.1101/gr.2161804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hur W, Gray NS. Small molecule modulators of antioxidant response pathway. Curr Opin Chem Biol. 2011;15:162–173. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Dhivya Vadhana MS, Siva Arumugam S, Carloni M, Nasuti C, Gabbianelli R. Early life permethrin treatment leads to long-term cardiotoxicity. Chemosphere. 2013;93:1029–1034. doi: 10.1016/j.chemosphere.2013.05.073. [DOI] [PubMed] [Google Scholar]
- 21.Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 22.Geismann C, Arlt A, Sebens S, Schäfer H. Cytoprotection “gone astray”: Nrf2 and its role in cancer. Onco Targets Ther. 2014;7:1497–1518. doi: 10.2147/OTT.S36624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Munday R. Dithiolethiones for cancer chemoprevention: where do we stand? Mol Cancer Ther. 2008;7:3470–3479. doi: 10.1158/1535-7163.MCT-08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, Chu Y. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. 2012;318:1175–1184. doi: 10.1016/j.yexcr.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 27.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 28.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900–904. doi: 10.1016/j.crohns.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Finlay LA, Michels AJ, Butler JA, Smith EJ, Monette JS, Moreau RF, Petersen SK, Frei B, Hagen TM. R-α-lipoic acid does not reverse hepatic inflammation of aging, but lowers lipid anabolism, while accentuating circadian rhythm transcript profiles. Am J Physiol Regul Integr Comp Physiol. 2012;302:R587–R597. doi: 10.1152/ajpregu.00393.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salminen A, Kaarniranta K, Kauppinen A. Inflammaging: disturbed interplay between autophagy and inflammasomes. Aging (Albany NY) 2012;4:166–175. doi: 10.18632/aging.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1 -independent mechanism. Breast Cancer Res Treat. 2011;129:983–991. doi: 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hine CM, Mitchell JR. NRF2 and the Phase II Response in Acute Stress Resistance Induced by Dietary Restriction. J Clin Exp Pathol. 2012;S4 doi: 10.4172/2161-0681.S4-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen TM, Vinarsky V, Wehr CM, Ames BN. (R)-alpha-lipoic acid reverses the age-associated increase in susceptibility of hepatocytes to tert-butylhydroperoxide both in vitro and in vivo. Antioxid Redox Signal. 2000;2:473–483. doi: 10.1089/15230860050192251. [DOI] [PubMed] [Google Scholar]
- 37.Wiberg GS, Trenholm HL, Coldwell BB. Increased ethanol toxicity in old rats: changes in LD50, in vivo and in vitro metabolism, and liver alcohol dehydrogenase activity. Toxicol Appl Pharmacol. 1970;16:718–727. doi: 10.1016/0041-008x(70)90077-3. [DOI] [PubMed] [Google Scholar]
- 38.Richie JP, Lang CA, Chen TS. Acetaminophen-induced depletion of glutathione and cysteine in the aging mouse kidney. Biochem Pharmacol. 1992;44:129–135. doi: 10.1016/0006-2952(92)90046-l. [DOI] [PubMed] [Google Scholar]
- 39.Testa R, Ghia M, Mattioli F, Borzone S, Caglieris S, Mereto E, Giannini E, Risso D. Effects of reduced glutathione and n-acetylcysteine on lidocaine metabolism in cimetidine treated rats. Fundam Clin Pharmacol. 1998;12:220–224. doi: 10.1111/j.1472-8206.1998.tb00945.x. [DOI] [PubMed] [Google Scholar]
- 40.Butler JM, Begg EJ. Free drug metabolic clearance in elderly people. Clin Pharmacokinet. 2008;47:297–321. doi: 10.2165/00003088-200847050-00002. [DOI] [PubMed] [Google Scholar]
- 41.McLachlan AJ, Bath S, Naganathan V, Hilmer SN, Le Couteur DG, Gibson SJ, Blyth FM. Clinical pharmacology of analgesic medicines in older people: impact of frailty and cognitive impairment. Br J Clin Pharmacol. 2011;71:351–364. doi: 10.1111/j.1365-2125.2010.03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, Nel AE. Role of the Nrf2-mediated signaling pathway as a negative regulator of inflammation: implications for the impact of particulate pollutants on asthma. Antioxid Redox Signal. 8:88–98. doi: 10.1089/ars.2006.8.88. [DOI] [PubMed] [Google Scholar]
- 43.Kleene KC, Bagarova J, Hawthorne SK, Catado LM. Quantitative analysis of mRNA translation in mammalian spermatogenic cells with sucrose and Nycodenz gradients. Reprod Biol Endocrinol. 2010;8:155. doi: 10.1186/1477-7827-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]


