Abstract
The exploitation of anomalous signals for biological structural solution is maturing. Single-wavelength anomalous diffraction (SAD) is dominant in de novo structure analysis. Nevertheless, for challenging structures where the resolution is low (dmin ≥ 3.5 Å) or where only lighter atoms (Z ≤ 20) are present, as for native macromolecules, solved SAD structures are still scarce. With the recent rapid development in crystal handling, beamline instrumentation, optimization of data collection strategies, use of multiple crystals and structure determination technologies, the weak anomalous diffraction signals are now robustly measured and should be used for routine SAD structure determination. The review covers these recent advances on weak anomalous signals measurement, analysis and utilization.
Introduction
X-ray diffraction from biomolecules produces characteristic patterns that encode information about atomic structures. Due to absence of suitable lens for hard x-rays, however, only the intensities of diffracted x-rays are recorded; the phases are lost in recording the diffraction patterns and these phases need to be evaluated experimentally for novel structures [1,2•]. When incoming x-ray energies approach the intrinsic absorption edges of heavier elements in the crystal, the diffraction intensities comprise both a normal component and a modulated resonance, also called anomalous scattering, which arises from the coupling of x-ray photons with the bound electrons. Those atoms that scatter anomalously are made distinctive, and thereby they become useful for structure determination. Such atoms may be either intrinsic to the macromolecule or externally introduced by chemical derivatization or biosynthetic incorporation as in selenomethionyl (SeMet) substitution [3]. To retrieve phases experimentally, anomalous signals are first used for substructure determination and this is followed by evaluation of the whole-structure phases by single-wavelength anomalous diffraction (SAD) [1,2•,4••].
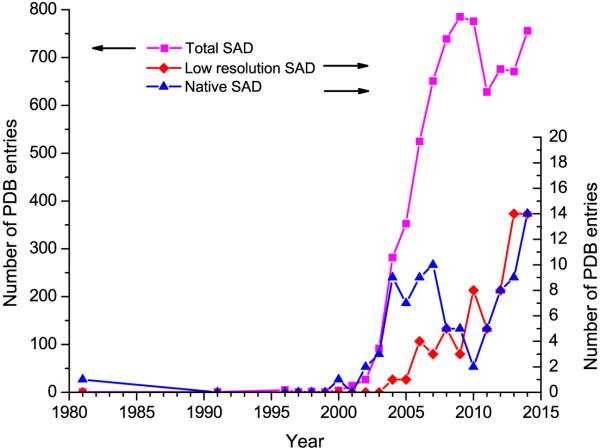
SAD has become the dominant method for de novo structure determination [1,2•]. As of 2014, there were 7163 SAD structures in the PDB (http://www.pdb.org); however, the effectiveness of SAD for more challenging structures is less obvious. For example, relative to over 600 SAD structures each year, there are in total only 67 low-resolution SAD structures (dmin ≥ 3.5 Å) and 91 native-SAD structures (anomalous scatterer Z ≤ 20) (Figure 1). Additionally, SAD is not trivial for membrane proteins, large macromolecular complexes and other challenges. Limited diffraction due to poor intrinsic order, inadequate anomalous scatterers, radiation damage, and other noise-causing factors often militate against sufficient accuracy in measured anomalous signals to support structure determination. Although heavy-atom derivatization and selenomethionyl substitution provide stronger anomalous signals with data collected at appropriate x-ray absorption edges, the challenges associated with poor diffraction and measurement errors may often frustrate structure solution at low resolution (dmin ≥ 3.5 Å), as evidenced by only 14 low-resolution SAD structures in 2014.
Figure 1. SAD structures in the PDB.
PDB entries are as of 31 December 2014. De novo low-resolution SAD structures as defined here have dmin ≥ 3.5 Å. De novo native-SAD structures as defined here have no preceding PDB deposits and have not contained atoms heavier than atomic number 20.
Native biomolecules contain intrinsic light anomalous scatterers, sulfur (Z=16) in proteins and phosphorous (Z=15) in nucleic acids. The resonant edges of these native light elements may not be readily accessible; however, off-resonance anomalous scattering from these elements at lower x-ray energy is weak but measurable, which is very attractive for phasing with no need for heavy-atom derivatization and selenomethionyl substitution, thus promising for automated SAD analysis [5,6,7,8,9,10,11•,12••]. Comparing to heavy atoms, the anomalous signals from light anomalous scatterers are much weaker, limiting their application for routine structure determination by native-SAD. As shown in Figure 1, there were only 91 native-SAD structures in the PDB as of 2014.
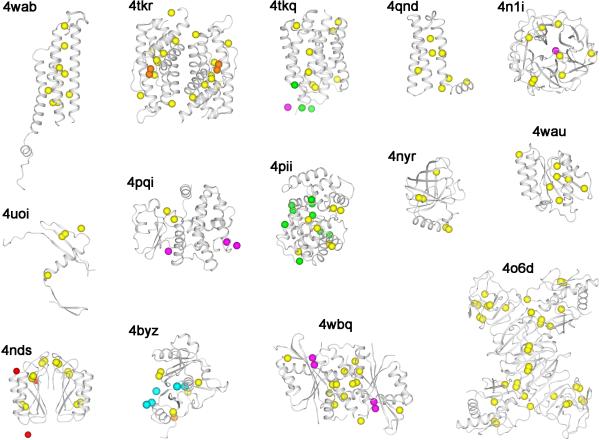
With recent developments in crystal handling, beamline instrumentation, data collection strategies, use of multiple crystals and advances in structure determination, the use of weak anomalous signals for structure determination is now feasible and should be routinely applicable for challenging structure determinations. As in Figure 1, within the recent few years, the number of challenging structures has increased for both low-resolution and native-SAD cases, each associated with use of weak anomalous diffraction signals. There is a clear trend of pushing SAD phasing to more difficult structures. Figure 2 are the ribbon diagrams of 14 native-SAD structure solved in 2014. Relative to previous years, this is a dramatic increment. We expect that more structures to come from weak anomalous signals.
Figure 2. De novo native-SAD structures in 2014.
Structures are identified by associated PDB codes and the contents of the asymmetric unit are shown for each. Protein backbones are represented as ribbon drawings and anomalous scatterers are shown as spheres. Yellow: sulfur; orange: phosphorus; magenta: calcium; green: chloride; cyan: potassium, and red: sodium.
Crystal handling
Biomolecular crystals are made from solutions. For a typical cryocrystallography experiment, crystal handling includes the transfer of crystals by a support from solution into cryoprotectant followed by flash cooling by cryogenic liquid or cryogenic gas. Crystal handling is important for phasing from weak anomalous signals in particular when the crystals are small and fragile. When crystals are of special shapes such as thin plates and long needles, they may be stressed due to humidity changes, surface tension forces, mechanical disturbance, and cryogenic shock. When crystals are small, solutions around crystals can be a problem because of the larger surface-to-volume ratio relative to larger crystals. The increased ratio leads to a greater impact of adherent solvent and increased background noise, which is especially harmful for measuring weak anomalous signals.
There are multiple ways for removing solution from around crystals. Perhaps the simplest of these is manual removal of solution from a micromount-loaded crystal with a filter paper before flash cooling for cryocrystallography [13]. To facilitate removal of solution attached to crystals, Kitago et al. [9] developed a semi-automated loopless capillary top mounting method. They attached a polyimide loop to the tip of a capillary and used the surface tension between the loop and crystal for initial crystal mounting. Before cryocooling, solution was aspirated through the capillary, resulting in only dehydrated crystals. By removing solution around crystals, it may be possible to freeze naked crystals without cryoprotectant, thus alleviating any potential cryoprotectant-caused damage [14]. To facilitate manipulation of crystals grown in viscous solution, Sugahara [11•] developed a rigid polyester-based fiber tool. The tool contains one, two or three fiber fingers that can hold crystals as small as 20 μm at the tip. In addition to fiber fingers, an adhesive crystal holder, commercially known as Crystal Catcher (Kyodo International Inc.), was also developed. Adhesive holders are either metallic microbuses or capillaries that can extrude adhesive materials to attach protein crystals [15••,16,17]. Such crystal supports and holders are attractive for further optimization and development; by reducing crystal shock and adherent solution, they should allow better measurement of weak anomalous signals.
In addition to crystal handling tools that require direct contact with crystals, as above, emerging tools for touchless manipulation of crystals have been developed. These include mounting crystals by acoustic waves known as ADE technology [18,19•], magnetically controlled microrobot, known as RodBot [20], and optical laser tweezers [21]. The combined use of these touchless tools may be promising for automated crystal handing with no sacrifice to the diffraction quality in measuring weak anomalous signals from tiny crystals.
Beamline instrumentation and data collection
Synchrotron beamlines and associated crystallography have deeply influenced the way that biological crystal structures are determined. The implementation of microdiffraction beamlines may push the limit of using tiny crystals for routine structure determination [22]. For an optimal beamline, the instrumentation noise should be minimized and the highest achievable I/σ(I) for the strongest reflections should be a good indicator for quality control of a specific beamline [23,24].
Each biological crystal is an optical device, but one that is subject to damage by x-rays during anomalous diffraction experiments. In general, data collection strategies need to take account of crystal geometry and predicted damage for optimization, and this is particularly true for weak anomalous signals. Therefore, it is generally advisable to deliver x-rays to match the size and shape of the crystal or of an optimal region found by raster scanning [25]. Larger beams contribute unnecessary background noise and smaller beams may produce inadequate diffraction. When using a beam that is smaller than the crystal, offsetting of the rotation axis from the beam axis may be effective for obtaining more diffracted x-ray intensities with the fixed absorbed dose [26].
For standard crystallographic experiments, only a single rotation axis, referred as phi or omega dependent on beamline configuration, is commonly used for data collection. Multi-axis data collection has been used to reduce the blind region for more complete data measurement [27]. Recently, Brockhauser et al. [28•] developed control software that can accurately align prospective mirror planes relative to the beam. With a crystal so aligned, Bijvoet mates can be collected from the same diffraction patterns as was achieved earlier by manual alignment [29,30,31]. With the automated kappa-geometry setup, weak anomalous signals from sulfur atoms were obtained from two paths, one alignment path for accurate Friedel pairs and one random pass for data completeness. Mirror-symmetry alignment eliminates the need for inverse-beam data collection when applicable. Separately, a precision multi-axis goniometer, called PRIGo, was developed at Swiss Light Source (SLS) [32•]. Integrated at SLS beamline X06DA, PRIGo can be used for accurate positioning of crystals for multi-axis data collection. By collecting highly redundant data from multiple crystal orientations, weak anomalous signals were measured from which eleven native-SAD structures were solved, including a 260 kDa multiprotein-ligand tubulin complex solved from data obtained at four orientations of a single crystal [33••]. By using multiple orientations, some systematic errors can be reduced. This is of special advantage for measuring weak anomalous signals, as demonstrated by direct comparison with single-axis data from the tubulin example. Multi-axis data collection is attractive and can suffice with a single crystal for measuring weak anomalous signals provided that radiation damage is not limiting.
For weak anomalous signals measurement, detector noise also needs to be considered. Although CCD-based detectors have been a big success at synchrotron beamlines, emerging and rapidly developing pixel array detectors (PADs), for example PILATUS (Dectris), are transforming modern synchrotron crystallography [34]. Relative to CCD detectors, photon-counting pixel array detectors are advantageous in having zero readout and dark-current noise, a sharp point-spread function, fast readout time and high dynamic range [35]. These physical specifications allow shutterless, fine phi-slicing diffraction data collection for improved data quality including weak anomalous signals [12••,36,37]. In addition, the sensitivity and energy threshold of individual detector pixels may be tuned to filter out additional noises for a specific experiment, thus further enhancing the weak anomalous signals.
Multiple crystals
Before the coming of cryogenic data collection, it was typically necessary to use many crystals to obtain complete data because of radiation-sensitivity of macromolecular crystals in sealed capillaries. The lifetime of crystals was greatly extended with cryo-crystallography, and single-crystal cryo-crystallography became routine [1,38], although multiple crystals were still required for challenging problems with relatively weak diffraction [31]. With frozen crystals, however, lattice uniformity and isomorphism often suffered [39•]. To achieve completeness when precluded by radiation damage or to reach high multiplicity to enhance accuracy, it was more acceptable to merge data from different parts of a single crystal [40] or from perfectly isomorphous crystals such as naturally formed baculovirus polyhedra crystals [41]. For crystals with extreme radiation sensitivity, particularly very small crystals as from membrane proteins in lipidic cubic phase [25], data were merged from multiple crystals but without obtaining anomalous signals for use in phase evaluation.
Multi-crystal SAD at low resolution
In attempting to solve the crystal structure of HK3S, a large histidine-kinase sensor domain (1456 residues in the asymmetric unit), by SeMet-SAD phasing, we collected diffraction data from eight different crystals and were able to extract weak anomalous signals to support SAD phasing at 3.5 Å resolution. Even though the eight crystals were not identical, phasing efficacy improved with progressive addition of crystals [42•]. With this success, we then solved the crystal structure of a bacterial sodium-dependent dicarboxylate transporter based on weak anomalous signals from three different crystals [43]. Here, the merged data allowed unambiguous determination of the structure at 3.5 Å resolution based on 92 Se atoms in a monoclinic lattice; eventual refinement was extended to 3.2 Å resolution.
Several other recently reported challenging structures are from low-resolution SAD analyses that used multiple crystals. These include the first complete structure of an eukaryotic ribosome, which was solved at 3.0 Å resolution from eleven crystals and ~1400 osmium heximide sites [44]; sodium/proton antiporter PaNhaP, which was solved by SeMet SAD at 3.2 Å resolution from two crystals [45]; the RAG1-RAG2 recombinase complex, which was solved by SeMet SAD (54 ordered sites) at 3.7 Å resolution from five crystals and then extended to 3.2 Å resolution [46••]; and an eukaryotic origin replication complex, which was solved at 3.5 Å resolution based on a four-crystal Gd-SAD experimental map at 4.0 Å resolution and aided by sulfur sites from an S-SAD experiment [47]. We expect that the use of multiple crystals for low-resolution SAD phasing will become a desirable option for challenging problems.
Multi-crystal native SAD
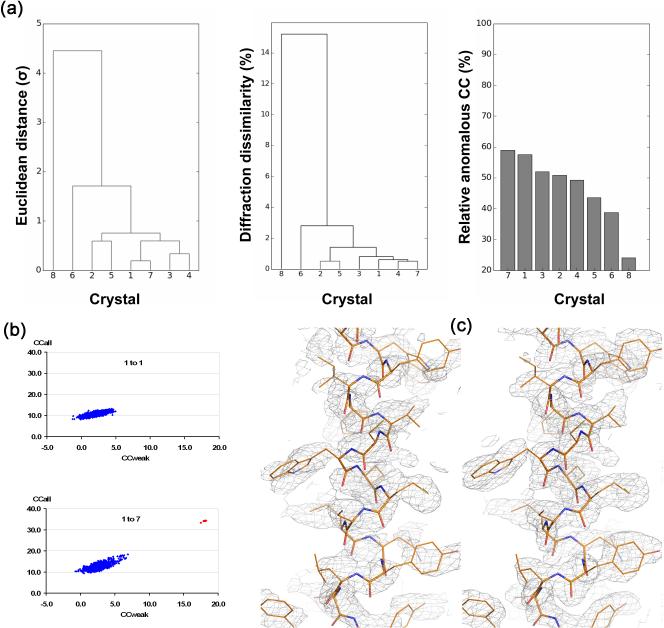
With success in enhancing weak anomalous signals at low resolution, we hypothesized that the merged data could be useful as well for enhancing intrinsic anomalous signals from sulfur and other light anomalous scatterers (Z≤20) in protein. To make this useful, a key challenge would be to make sure that only compatible single-crystal data sets are combined. Therefore, we collectively used unit-cell variation, diffraction dissimilarity and relative anomalous correlation coefficient (RACC) as the criteria for statistical equivalence [48••,49]. The native-SAD procedure that we devised can be illustrated by its application to CysZ, a unique membrane protein that facilitates sulfate uptake into cells. Anomalous diffraction data were collected from eight different CysZ crystals with an x-ray energy near 7 keV (Fe K-edge, 1.743 Å). Statistical analyses on unit cell variations, diffraction dissimilarity and RACC identified crystal 8 as a non-compatible outlier (Figure 3a). After rejecting crystal 8, the remaining seven data sets were merged for SAD phasing. Although each of the single-crystal data sets failed for substructure determination, including the best one, the merged 7-crystal data set led to 34 correct solutions from 1000 SHELXD tries (Figure 3b) and these refined to the 22 methionine and cysteine sulfur atoms of CysZ plus four chloride ions and one sulfate ion. Moreover, this substructure produced a readily interpretable SAD-phased experimental map, which was fitted automatically to give a model for 408 of 453 ordered residues; the refined substructure also produced interpretable maps when used with single-crystal data sets, but these maps were inferior to that from 7-crystal data set (Figure 3c).
Figure 3. Multiple crystals in SAD phasing.
Data are from studies on CysZ from Idiomarina loihiensis, a transmembrane sulfate permease [48]. (a) Clustering analysis of crystal variations. Crystal 8 is an outlier. (left) Unit-cell variation, (middle) diffraction dissimilarity, and (right) relative anomalous correlation coefficient (RACC). (b) Substructure determination analyses by SHELXD. (top) Analysis from data of only the best crystal (1 to 1), and (bottom) analysis for data as merged from all seven statistically equivalent crystals (1 to 7). (c) SAD electron densities for a representative helix after density modification. (left) The map based on the substructure model deduced from the 1-to-7 data set but applied to the data from Crystal 1 alone, and (right) the map after SAD phasing of the 1-to-7 data set.
Other real-life applications have been made in determining various structures at resolutions between 2.3 Å to 2.8 Å with unique protein sizes of between 144 and 1200 amino acid residues. Recently, we systematically compared the phasing efficacy of weak anomalous signals at 6 keV relative to 7 keV, and we were able to solve three native-SAD structures at around 3.0 Å resolution, including two novel membrane proteins [50,51]. In addition to our own successes, other native-SAD structures based on data from multiple crystals are also emerging. Examples include the flavivirus NS1 protein [52,53••], the N-terminal domain of HCV E1 [54••,55], dimeric PPR-protein complex [56], and a folate receptor [57].
Various advances are contributing to native SAD studies that benefit from multiple crystals. To aid the selection of compatible data sets for scaling and merging, program BLEND has been available which uses various clustering methods for crystal rejections [58•]. To reduce the changes of unit cell variations during crystal manipulation [39•], a controlled dehydrated technique was proposed to assist cryocrystallography [59]. Finally, when radiation damage is not a problem, weak anomalous signals may be enhanced additionally or alternatively by collecting multiple-orientation data sets from a single crystal [33••].
As a reflection of recent advance in measuring weak anomalous signals, 14 native-SAD structures were deposited into the PDB in 2014 (Figure 1 and 2). These structures are of different sizes, different lattice and different resolutions. From a retrospective view, in light of recent successes, more than 94% of crystal structures in the PDB would be candidates for contemporary native-SAD analysis. With x-ray energy going lower in conjunction with developments in crystal handling, beamline instrumentation, data collection strategies, and data analysis of multi-crystal data sets, we expect that native-SAD structure determinations will grow increasingly popular.
Besides structure determination, enhanced weak anomalous signals from multiple crystals may help element identification of previously unknown anomalous scatterers [49]. This technique has been used for identification of anomalous elements in a number of structures, including Mg2+ in DnaK [60], an Hsp70 chaperone, and phosphorus in ThiT [50], a thiamine transporter. At low resolution, the enhanced anomalous signals are also effective for residue registration as recently used for tracing an eukaryotic origin replication complex [47]. Therefore, even though the initial phases may be obtained from other resources such as molecular replacement, enhanced weak anomalous signals from multiple crystals may still help the structural analysis process and should be utilized when available.
Structure determination
When anomalous signals are weak, the structure determination can be challenging even with best efforts at signal-to-noise enhancement. Traditional SAD structure determination first involves the determination of the substructure by dual-space direct method with or without seeding from anomalous-difference Patterson, as implemented in programs such as SHELXD [61], HySS [62] and SnB [63]. With a complete or partial substructure available, SAD phasing programs refine and complete the substructure of anomalous scatterers through maximum-likelihood algorithm as originally implemented in SHARP [64] and adopted by PHASER [65] and CRANK [66]. To extend the substructure determination from weak anomalous signals, likelihood-based methods were recently introduced for substructure determination in combination with the anomalous-difference Patterson and implemented in latest version of HySS [67••]. Through further optimization of the method and incorporation of direct method, more effective structure determination is expected from weak anomalous signals.
With the initial SAD phases from substructure, density modifications are employed to break the phase ambiguity as well as for selection of the correct enantiomorph. For weak anomalous signals, the initial phases may not produce electron density maps of sufficient quality for model building and refinement. Therefore, combining phases from different resources, for example from a partial molecular replacement or from a partially built model, could be useful for phase improvement. To make use of weak signals for phasing, a new likelihood-based phase probability function was devised to push the structure determination of SAD structures by combining phases from SAD substructure, density modification and partial model [68••]. The combined approach allowed automatic solution of SAD structures when the currently existing approaches had failed. The combined approach has been implemented in program CRANK2 through CCP4.
Other procedures for improving structure determination from weak signals include automatic sharpening B factors at middle to low resolutions, automatic anisotropic scaling and corrections, using multiple-models in density modifications and refinement, using the prior knowledge in automated model building and new iterative ways for model building from partial models [62,69,70]. Nevertheless, at low resolution where models cannot be built automatically, new algorithms are still needed and perhaps those being developed for cryo-electron microscopy may also work for interpretation of low resolution SAD electron density maps [71]. At low resolution, where there are many fewer reflections than atomic parameters, deformed elastic network (DEN) refinement in CNS [62] and phenix.refine [72] and jelly-body refinement in REFMAC5 [73] are very useful for introducing additional restraints and constraints for high-quality refinements. To further improve the parameter-to-observation ratios during structural refinement, SAD amplitudes (|F+(obs)| and |F−(obs)|) have been incorporated for refinements by programs such as REFMAC5 [74,75••] and phenix.refine [62,72]. With anomalous signals available, they should always be used in refinement programs for improved structures and statistics.
Conclusion
Weakness of anomalous signals has often prevented robust and routine SAD structure determination. Recent advances in experimental tools and associated methods are now making structure solutions more routine for two typically challenging subjects characterized by weak anomalous signals, namely, low resolution structures and native structures having only light-atom anomalous scatterers. The optimization of these methods and their combination will ensure the applicability of weak anomalous signals for challenging structure determination.
Highlights.
Crystal handling to reduce damage, variations and background.
Beamline instrumentation and data collection strategies for weak anomalous signals.
High multiplicity strategies to enhance signal-to-noise from weak anomalous signals.
Structure determination algorithms for use of weak anomalous signals.
Acknowledgments
This work was supported in part by NIH grants GM095315 and GM107462.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Garman EF. Developments in x-ray crystallographic structure determination of biological macromolecules. Science. 2014;343:1102–1108. doi: 10.1126/science.1247829. [DOI] [PubMed] [Google Scholar]
- 2•.Hendrickson WA. Anomalous diffraction in crystallographic phase evaluation. Q Rev Biophys. 2014;47:49–93. doi: 10.1017/S0033583514000018. [This is a comprehensive review of phase evaluation from anomalous diffraction measurements, including discussion of analyses from weak anomalous signals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Hendrickson WA, Teeter MM. Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulfur. Nature. 1981;290:107–113. doi: 10.1038/290107a0. [This is the first structure solved by native-SAD phasing although it was not called SAD by that time.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang BC. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 6.Dauter Z, Dauter M, de La Fortelle E, Bricogne G, Sheldrick GM. Can anomalous signal of sulfur become a tool for solving protein crystal structures? J Mol Biol. 1999;289:83–92. doi: 10.1006/jmbi.1999.2743. [DOI] [PubMed] [Google Scholar]
- 7.Dauter Z, Adamiak DA. Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy. Acta Crystallogr D Biol Crystallogr. 2001;57:990–995. doi: 10.1107/s0907444901006382. [DOI] [PubMed] [Google Scholar]
- 8.Ramagopal UA, Dauter M, Dauter Z, Sulfur SAD. What is the limit? Biophys J. 2003;84:355a–356a. [Google Scholar]
- 9.Kitago Y, Watanabe N, Tanaka I. Semi-automated protein crystal mounting device for the sulfur single-wavelength anomalous diffraction method. J Appl Crystallogr. 2010;43:341–346. [Google Scholar]
- 10.Yang C, Pflugrath JW, Courville DA, Stence CN, Ferrara JD. Away from the edge: SAD phasing from the sulfur anomalous signal measured in-house with chromium radiation. Acta Crystallogr D Biol Crystallogr. 2003;59:1943–1957. doi: 10.1107/s0907444903018547. [DOI] [PubMed] [Google Scholar]
- 11•.Sugahara M. A fibre-based crystal mounting technique for protein cryocrystallography. J Appl Crystallogr. 2012;45:362–366. [New fiber finger tools were development for crystal handling. The tools can be used reduced solution around crystals to support measurement of weak anomalous signals.] [Google Scholar]
- 12••.Mueller M, Wang M, Schulze-Briese C. Optimal fine phi-slicing for single-photon-counting pixel detectors. Acta Crystallogr D Biol Crystallogr. 2012;68:42–56. doi: 10.1107/S0907444911049833. [Pixel array detectors, for example PILATUS 6M, have fundamentally different physics in measuring x-ray diffraction intentieis compared to CCD detectors. The research show that fine phi-slicing allowed optimal quality of anomalous signals to be measured when the oscillation angle is comparable to the crystal mosaicity as reported by program XDS.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorne R, Stum Z, Kmetko J, O'Neill K, Gillilan R. Microfabricated mounts for high-throughput macromolecular cryocrystallography. J Appl Crystallogr. 2003;36:1455–1460. [Google Scholar]
- 14.Pellegrini E, Piano D, Bowler MW. Direct cryocooling of naked crystals: are cryoprotection agents always necessary? Acta Crystallogr D Biol Crystallogr. 2011;67:902–906. doi: 10.1107/S0907444911031210. [DOI] [PubMed] [Google Scholar]
- 15••.Mazzorana M, Sanchez-Weatherby J, Sandy J, Lobley CM, Sorensen T. An evaluation of adhesive sample holders for advanced crystallographic experiments. Acta Crystallogr D Biol Crystallogr. 2014;70:2390–2400. doi: 10.1107/S1399004714014370. [A new method for catching crystals in their crystallization drops through extrusion of an adhesive reagent.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitatani T, Adachi H, Sugiyama S, Matsumura H, Murai R, Takahashi Y, Murakami S, Inoue T, Mori Y, Takano K. A manipulating tool for protein microcrystals in solution using adhesive materials. Jap J Appl Phys. 2009;48:118001. [Google Scholar]
- 17.Kitatani T, Sugiyama S, Matsumura H, Adachi H, Yoshikawa HY, Maki S, Murakami S, Inoue T, Mori Y, Takano K. New technique of manipulating a protein crystal using adhesive material. Appl Phys Expr. 2008;1:037002. [Google Scholar]
- 18.Guo F, Zhou W, Li P, Mao Z, Yennawar NH, French JB, Huang TJ. Precise manipulation and patterning of protein crystals for macromolecular crystallography using surface acoustic waves. Small. 2015;11:2733–2737. doi: 10.1002/smll.201403262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Roessler CG, Kuczewski A, Stearns R, Ellson R, Olechno J, Orville AM, Allaire M, Soares AS, Héroux A. Acoustic methods for high-throughput protein crystal mounting at next-generation macromolecular crystallographic beamlines. J Synchrotron Radiat. 2013;20:805–808. doi: 10.1107/S0909049513020372. [Acounstic waves were used to eject crystals on to a x-ray transparent polyimide support belt for high-throughput data collection and analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tung H-W, Sargent DF, Nelson BJ. Protein crystal harvesting using the RodBot: a wireless mobile microrobot. J Appl Crystallogr. 2014;47:692–700. [Google Scholar]
- 21.Wagner A, Duman R, Stevens B, Ward A. Microcrystal manipulation with laser tweezers. Acta Crystallogr D Biol Crystallogr. 2013;69:1297–1302. doi: 10.1107/S090744491300958X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JL, Fischetti RF, Yamamoto M. Micro-crystallography comes of age. Curr Opin in Struct Biol. 2012;22:602–612. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebschner D, Dauter M, Rosenbaum G, Dauter Z. How good can our beamlines be? Acta Crystallogr D Biol Crystallogr. 2012;68:1430–1436. doi: 10.1107/S0907444912034658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diederichs K. Quantifying instrument errors in macromolecular X-ray data sets. Acta Crystallogr D Biol Crystallogr. 2010;66:733–740. doi: 10.1107/S0907444910014836. [DOI] [PubMed] [Google Scholar]
- 25.Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 μm size X-ray synchrotron beam. J Roy Soc Interface. 2009;6(Suppl 5):S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeldin OB, Brockhauser S, Bremridge J, Holton JM, Garman EF. Predicting the X-ray lifetime of protein crystals. Proc Natl Acad Sci U S A. 2013;110:20551–20556. doi: 10.1073/pnas.1315879110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dauter Z. Carrying out an optimal experiment. Acta Crystallogr D Biol Crystallogr. 2010;66:389–392. doi: 10.1107/S0907444909038578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Brockhauser S, Ravelli RB, McCarthy AA. The use of a mini-kappa goniometer head in macromolecular crystallography diffraction experiments. Acta Crystallogr D Biol Crystallogr. 2013;69:1241–1251. doi: 10.1107/S0907444913003880. [A reliable alignment method and associated control and analysis software were developed for improved measurement of weak anomalous signals when an even-fold axis is present in the crystal.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickson WA, Pahler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc Natl Acad Sci U S A. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemons WM, Brodersen DE, McCutcheon JP, May JLC, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J Mol Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- 32•.Waltersperger S, Olieric V, Pradervand C, Glettig W, Salathe M, Fuchs MR, Curtin A, Wang X, Ebner S, Panepucci E. PRIGo: a new multi-axis goniometer for macromolecular crystallography. J Synchrotron Radiat. 2015;22:895–900. doi: 10.1107/S1600577515005354. [This device provides a solution for multi-axis diffraction that is proving to be effective.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Weinert T, Olieric V, Waltersperger S, Panepucci E, Chen L, Zhang H, Zhou D, Rose J, Ebihara A, Kuramitsu S. Fast native-SAD phasing for routine macromolecular structure determination. Nat Methods. 2015;12:131–133. doi: 10.1038/nmeth.3211. [This makes an important advance in showing the value of high multiplicitiy for the accrurate measurement of weak anomalous signals with reduction of systematic errors from having multiple orientations. With a PILATUS pixel array detector, a PRIGo multi-axis goniometer and optimized exposure time per frame, weak anomalous signals were readily measured for successful determination of eleven native-SAD structures, each from a single crystal.] [DOI] [PubMed] [Google Scholar]
- 34.Broennimann C, Eikenberry E, Henrich B, Horisberger R, Huelsen G, Pohl E, Schmitt B, Schulze-Briese C, Suzuki M, Tomizaki T. The PILATUS 1M detector. J Synchrotron Radiat. 2006;13:120–130. doi: 10.1107/S0909049505038665. [DOI] [PubMed] [Google Scholar]
- 35.Gruner S, Eikenberry E, Tate M. Comparison of X-ray detectors. In: Arnold E, Himmel DM, Rossmann MG, editors. International Tables for Crystallography Volume F: Crystallography of biological macromolecules. 177-182. International Union of Crystallography; 2012. [Google Scholar]
- 36.Henrich B, Bergamaschi A, Broennimann C, Dinapoli R, Eikenberry EF, Johnson I, Kobas M, Kraft P, Mozzanica A, Schmitt B. PILATUS: A single photon counting pixel detector for X-ray applications. Nucl Instrum Meth A. 2009;607:247–249. [Google Scholar]
- 37.Loeliger T, Bronnimann C, Donath T, Schneebeli M, Schnyder R, Trub P. Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) 610-615. IEEE; 2012. The new PILATUS3 ASIC with instant retrigger capability. [Google Scholar]
- 38.Garman EF, Weik M. Radiation damage to macromolecules: kill or cure? J Synchrotron Radiat. 2015;22:195–200. doi: 10.1107/S160057751500380X. [DOI] [PubMed] [Google Scholar]
- 39•.Giordano R, Leal RM, Bourenkov GP, McSweeney S, Popov AN. The application of hierarchical cluster analysis to the selection of isomorphous crystals. Acta Crystallogr D Biol Crystallogr. 2012;68:649–658. doi: 10.1107/S0907444912006841. [The study used clutering based on correlation coefficients of diffraction intensities among crystals for selection of compatible data sets for structural analysis by SAD.] [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen SG, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 41.Ji XY, Sutton G, Evans G, Axford D, Owen R, Stuart DI. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 2010;29:505–514. doi: 10.1038/emboj.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Liu Q, Zhang Z, Hendrickson WA. Multi-crystal anomalous diffraction for low-resolution macromolecular phasing. Acta Crystallogr D Biol Crystallogr. 2011;67:45–59. doi: 10.1107/S0907444910046573. [Low-resolution SAD phasing is challenging due to poor diffraction patterson. With an example of a relatively large structure (~1500 residues) in the asymmetric unit, the utility of using anomalous signals from multiple crystals to support both substructure determination and phasing at low resolution is demonstrated for cases when a single-crystal routine fails.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancusso R, Gregorio GG, Liu Q, Wang DN. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature. 2012;491:622–626. doi: 10.1038/nature11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 45.Wohlert D, Kuhlbrandt W, Yildiz O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. Elife. 2014;3:e03579. doi: 10.7554/eLife.03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [This paper describes the use of multiple crystals in the analysis of a challenging and novel structure at low resolution.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, Shapiro L, Mancia F, Hendrickson WA. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [Native-SAD analysis of native biological macromolecules is very attractive; while this is far from routine due to associated weak anomalous signals. This study provided a robust method for enhancing weak anomalous signals by using sensitive merger of lower-energy data from multiple crystals. The method may allow routine native-SAD analysis with no need of heavy-atom derivatization, thus promising for automated SAD structure analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Liu Q, Hendrickson WA. Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr D Biol Crystallogr. 2013;69:1314–1332. doi: 10.1107/S0907444913001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, Guo Y, Chang Y, Cai Z, Assur Z, Mancia F, Greene MI, Hendrickson WA. Multi-crystal native SAD analysis at 6 keV. Acta Crystallogr D Biol Crystallogr. 2014;70:2544–2557. doi: 10.1107/S1399004714013376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang Y, Bruni R, Kloss B, Assur Z, Kloppmann E, Rost B, Hendrickson WA, Liu Q. Structural basis for a pH-sensitive calcium leak across membranes. Science. 2014;344:1131–1135. doi: 10.1126/science.1252043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akey DL, Brown WC, Dutta S, Konwerski J, Jose J, Jurkiw TJ, DelProposto J, Ogata CM, Skiniotis G, Kuhn RJ, et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343:881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Akey DL, Brown WC, Konwerski JR, Ogata CM, Smith JL. Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1. Acta Crystallogr D Biol Crystallogr. 2014;70:2719–2729. doi: 10.1107/S1399004714017556. [This paper describes methodology used in determining the structure of flavivirus structural protein NS1 at low resolution from multiple native-SAD data sets measured at 7.1 keV.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.El Omari K, Iourin O, Kadlec J, Fearn R, Hall DR, Harlos K, Grimes JM, Stuart DI. Pushing the limits of sulfur SAD phasing: de novo structure solution of the N-terminal domain of the ectodomain of HCV E1. Acta Crystallogr D Biol Crystallogr. 2014;70:2197–2203. doi: 10.1107/S139900471401339X. [A challenging native-SAD structure of HCV E1 was solved at low resolution with anomalous signals merged from 32 crystals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun. 2014;5:4874. doi: 10.1038/ncomms5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ke J, Chen RZ, Ban T, Zhou XE, Gu X, Tan MH, Chen C, Kang Y, Brunzelle JS, Zhu JK, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20:1377–1382. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Ke J, Zhou XE, Yi W, Brunzelle JS, Li J, Yong EL, Xu HE, Melcher K. Structural basis for molecular recognition of folic acid by folate receptors. Nature. 2013;500:486–489. doi: 10.1038/nature12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Foadi J, Aller P, Alguel Y, Cameron A, Axford D, Owen RL, Armour W, Waterman DG, Iwata S, Evans G. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2013;69:1617–1632. doi: 10.1107/S0907444913012274. [A program for automated analysis and merging of diffraction data sets from multiple crystals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farley C, Burks G, Siegert T, Juers DH. Improved reproducibility of unit-cell parameters in macromolecular cryocrystallography by limiting dehydration during crystal mounting. Acta Crystallogr D Biol Crystallogr. 2014;70:2111–2124. doi: 10.1107/S1399004714012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, Yang J, Wong JL, Vorvis C, Hendrickson WA. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Baker D, Brunger AT, Das R, DiMaio F, Read RJ, Richardson DC, Richardson JS, Terwilliger TC. Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Ann Rev Biophys. 2013;42:265–287. doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weeks CM, Miller R. The design and implementation of SnB version 2.0. J Appl Crystallogr. 1999;32:120–124. [Google Scholar]
- 64.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 65.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr D Biol Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pannu NS, Waterreus WJ, Skubak P, Sikharulidze I, Abrahams JP, de Graaff RA. Recent advances in the CRANK software suite for experimental phasing. Acta Crystallogr D Biol Crystallogr. 2011;67:331–337. doi: 10.1107/S0907444910052224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Bunkóczi G, McCoy AJ, Echols N, Grosse-Kunstleve RW, Adams PD, Holton JM, Read RJ, Terwilliger TC. Macromolecular X-ray structure determination using weak, single-wavelength anomalous data. Nat Methods. 2015;12:127–130. doi: 10.1038/nmeth.3212. [Use of weak anomalous signals for SAD structure anlaysis is challenging for both substructure determination and phasing. The research combines the anomalous difference Patterson with the likelihood methods for substructure determination and completion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Skubak P, Pannu NS. Automatic protein structure solution from weak X-ray data. Nat Commun. 2013;4:2777. doi: 10.1038/ncomms3777. [Use of weak anomalous signals for structure determination is challenging. The study describes a new likelihood-based phase combination method of including density modification and partial model information. The cycles between phase combination, density modification and model building and refinement may result in structure from poor initial phases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, Valkov E, Alon A, Fass D, Axelrod HL. Improved molecular replacement by density-and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terwilliger TC, DiMaio F, Read RJ, Baker D, Bunkóczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, Baker D. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicholls RA, Long F, Murshudov GN. Recent advances in low resolution refinement tools in REFMAC5. In: Read R, Urzhumtsev AG, Lunin VY, editors. Advancing Methods for Biomolecular Crystallography. 231-258. Springer; 2013. [Google Scholar]
- 74.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75••.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68:404–417. doi: 10.1107/S090744491105606X. [Low resolution structural refinement is challenging due to inadquate observations to refinement parameters. Recent developmen in REFMAC5 included external structural information as well as B-factor-based map sharpening for low resolution structure refinement and map interpretation.] [DOI] [PMC free article] [PubMed] [Google Scholar]