Abstract
Next-generation sequencing (NGS) has transformed our understanding of the dynamics and diversity of virus populations for human pathogens and model systems alike. Due to the sensitivity and depth of coverage in NGS, it is possible to measure the frequency of mutations that may be present even at vanishingly low frequencies within the viral population. Here, we describe a simple bioinformatic pipeline called CoVaMa (Co-Variation Mapper) scripted in Python that detects correlated patterns of mutations in a viral sample. Our algorithm takes NGS alignment data and populates large matrices of contingency tables that correspond to every possible pairwise interaction of nucleotides in the viral genome or amino acids in the chosen open reading frame. These tables are then analysed using classical linkage disequilibrium to detect and report evidence of epistasis. We test our analysis with simulated data and then apply the approach to find epistatically linked loci in Flock House Virus genomic RNA grown under controlled cell culture conditions. We also reanalyze NGS data from a large cohort of HIV infected patients and find correlated amino acid substitution events in the protease gene that have arisen in response to anti-viral therapy. This both confirms previous findings and suggests new pairs of interactions within HIV protease. The script is publically available at http://sourceforge.net/projects/covama
Keywords: Linkage Disequilibrium, Flock House Virus, Human Immunodeficiency Virus Protease, Covariation, RNAseq
1. Introduction
Viral populations are typically very diverse due to the high error-rate of their polymerases. As a result, just a single round of replication can generate many variant species of an original parent virus. The error rate of viral polymerases varies greatly among viral species, but is generally considered to be the highest for positive-strand RNA viruses[1]. This ability of viruses to sample a wide-range of mutational space is what gives rise to their ability to quickly evolve and adapt.
Next-Generation Sequencing (NGS) is well poised to characterize viral intra-host diversity and has been employed during the surveillance of viral epidemics, to monitor the development of drug resistance and importantly to further our fundamental understanding of viral evolution, their lifecycles and the functional components of their genomes. There are many bioinformatic software packages available that enable a user to detect and characterize mutations in viral genome from NGS data[2]. However, it is also important to characterize and understand how these mutations interact with one another and whether they arise independently or in a concerted manner due to a phenotypic co-dependence. There are many methods for haplotype phasing in the genome of higher-eukaryotes (reviewed in [3]). However, haplotype phasing in viruses is considerably more challenging due to the heterogeneous make-up of the viral populations, varying rates of homologous recombination [4], and the requirement for high sequence coverage to detect low-frequency events.
To this end, a number of approaches have been proposed. These generally fall into two categories: direct and indirect approaches. Direct approaches extract information from individually sequenced fragments of DNA or sequence reads. VPhaser[5] and VPhaser 2.0[6] employ a direct approach by measuring the frequency of nucleotide mutations, determining the probability of seeing two mutations together and comparing this prediction to the actual observed read data. This can sensitively infer intrahost diversity, but is limited by the length of the DNA fragments that are sequenced and the underlying error-rates of the sequencing platform. Indirect approaches employ mathematical or statistical models such as mutual information theory [7] and hidden-markov models [4] to infer the association of two mapped mutations from multiple individually sequenced fragments of DNA. However, this imposes certain assumptions about the expected and observed frequencies of distant mutations and the rates of recombination between them (either due sequencing artifacts or the viral replication itself). Indirect approaches also include ‘quasi-species reassembly’ algorithms that aim to reconstruct the multiple variant viral genomes within a population, of which there are multiple strategies (reviewed in [8, 9]).
Here, we describe a simple bioinformatic pipeline called CoVaMa (Co-Variation Mapper) that uses NGS data to search for evidence of covariation by measuring Linkage Disequilibrium (LD) within the mutational landscape of the virus sample. Our approach is ‘direct’ by virtue of counting the frequency of observed mapped nucleotides from the read alignment data and populating large matrices of contingency tables that correspond to every possible pairwise interaction of nucleotides in the viral genome. In this manner, no assumptions are made about the underlying rate or source of single nucleotide differences from the reference genome. Rather, significant epistatic pairs are reported by finding outlying LD values among the entire distribution of all other LD values measured. If provided with an open reading frame, CoVaMa is also able to detect epistatic amino acid pairs. Additionally, CoVaMa can merge the output from multiple read alignments in order to look for evidence of coevolution of alleles within a population. CoVaMa can accept any read length as well as paired-end reads, although the computational demands of the pipeline scale accordingly with much longer reads.
In this manuscript, we use simulated NGS data to verify that expected pairs of mutations can be reliably and sensitively detected. We then demonstrate how co-variance can be found within the genome of Flock House Virus (FHV) when passaged in cell culture and analysed by RNAseq. Finally, we reanalyze RNAseq data from a large cohort of HIV infected patients who have failed to response to anti-viral therapy and find correlated mutations within the protease gene known to confer protease-inhibitor resistance as well as potentially novel associations. While we have applied this pipeline to explore RNA virus epistasis here, CoVaMa may also be applied in a broad range of settings including large DNA viruses, bacteria or even larger organisms but is limited by the computational resources available.
2. Materials and Methods
2.1 Co-Variation Mapper - CoVaMa
A simple program scripted using Python was written to measure linkage disequilibrium in NGS datasets, called Co-Variation Mapper (CoVaMa). The scripts, associated manuals and test data are available at (http://sourceforge.net/projects/covama/). CoVaMa is cross-platform compatible requiring Python version 2.7 and Numpy (http://www.numpy.org/). CoVaMa is computationally intensive and run time can vary dramatically, ranging from a few minutes to a few hours depending upon the complexity, size and length of reads in a NGS dataset. However, we use an Apple Workstation with 16Gb RAM and 8×2 core processors and have found this sufficient for the analyses described in this manuscript. The performance of CoVaMa can be improved considerably (up to 4× improvement) by using the Python-interpreter and JIT compiler, PyPy (http://pypy.org/).
CoVaMa is split into three different scripts, separated to allow storing of data at intermediate stages of the analyses: 1) CoVaMa_Make_Matrices; 2) CoVaMa_Merge_Matrices; and 3) CoVaMa_Analyse_Matrices.
2.1.1 CoVaMa Make Matrices
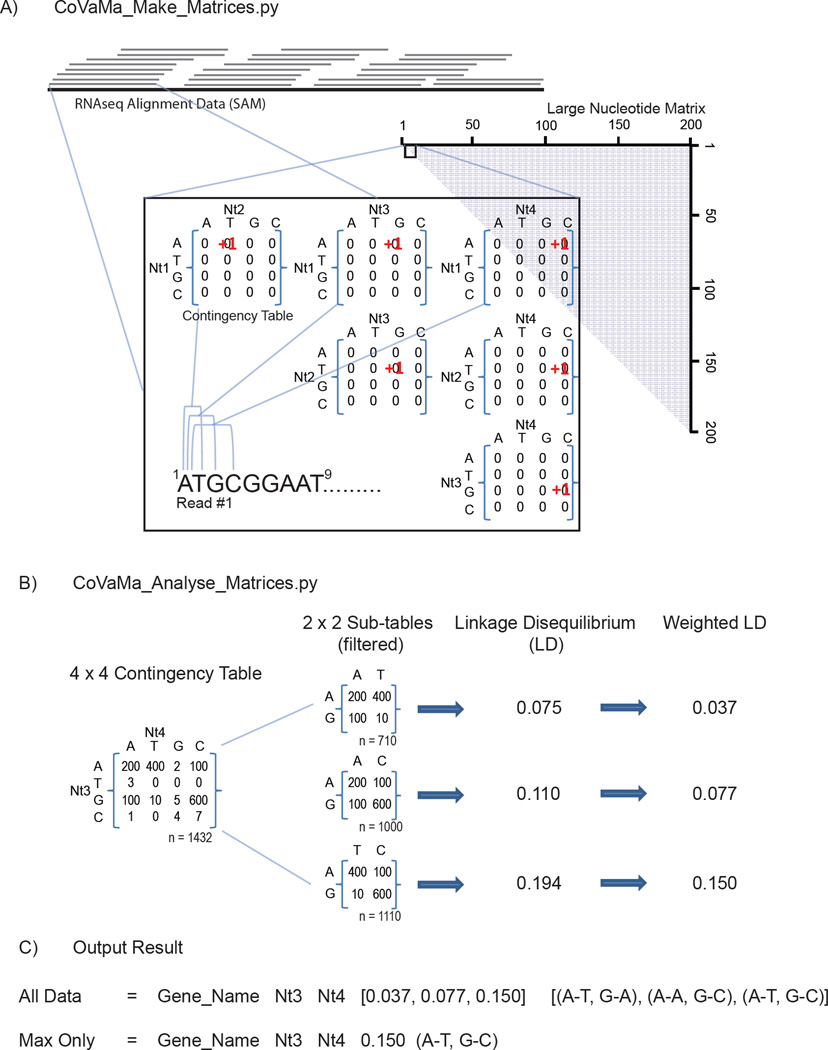
Firstly, CoVaMa generates large matrices corresponding to every possible pair of nucleotides or amino acids in the reference genomic sequence. As illustrated in Figure 1A, each point in these matrices is filled with a contingency table either 4×4 or 20×20 in size for nucleotide matrices and amino acid matrices respectively. The rows of the nucleotide contingency tables correspond to nts (A,T,G,C) at the 5’ nucleotide position of each nucleotide pair and likewise columns of the contingency table correspond to nts (A,T,G,C) at the 3’ nucleotide position. Similarly, each row and column of the 20×20 amino contingency tables corresponds to each of the 20 natural amino acids at the N and C termini respectively. The contingency tables in the matrices are generated using the Python defaultdict subclass in order to avoid building large numbers of unnecessary contingency tables that would otherwise occupy memory and reduce performance.
Figure 1.
CoVaMa generates matrices of pairwise nucleotide or amino acid associations from Sequencing Alignment Mapping (SAM) data from RNAseq. A) CoVaMa_Make_Matrices.py is the first script in CoVaMa that extracts information from each individual aligned read. Every possible pairwise interaction of nucleotides in the aligned read is used to populate 4×4 contingency tables that are components of the large nucleotide matrix. Amino acid matrices containing 20×20 contingency tables are built in an analogous fashion, but by first translating each aligned read in the desired reading frame. B) CoVaMa_Analyse_Matrices.py analyses each individual contingency table in the large matrices for evidence of linkage disequilibrium. All possible combinations of 2×2 sub-tables are extracted from the 4×4 (or 20×20) contingency table provided there are sufficient reads mapped. An LD value is calculated for each 2×2 table and optionally normalized relative to the total number of reads in the 4×4 table. Results are written to an output file as exemplified in C).
Next, CoVaMa extracts the relevant data contained within the Sequence Alignment/Map (SAM) files [10] and stores it in a temporary dictionary. Either single or paired-end alignment data can be used as input data. As it is likely that there are many duplicate reads with identical alignments due to PCR duplication or low-sample degeneracy, identical alignments are processed together as a group rather than individually to optimize performance and run time. An additional option (--Edge) provided in the command-line is to ignore a user-defined number of nucleotides at the 5’ and 3’ end of an aligned read. These portions of an aligned read are usually of the poorest quality and can often contain apparent variances from the reference genome due to the retention of short fragments of the Illumina adaptor sequences. Additionally, if the first or last nucleotides of a sequence read overlap the junction of a recombination event, they may erroneously map to the wild-type reference sequence but be counted as mismatched nucleotides. To mitigate this scenario, we recommend setting the --Edge option to the same value used for mismatch tolerance during the alignment of the sequence data.
Once the alignment data has been extracted, CoVaMa populates the contingency tables within the larger matrices. The alignment data provides the position in the reference genome that the reads map as well as any point mutations that may have occurred. Ambiguous (‘N’) nucleotides are ignored. Every nucleotide within an alignment is paired with every other nucleotide and the corresponding contingency table is populated according to which nucleotides are at found at each pair of loci. For example, for an alignment containing a ‘T’ at nt 20 and a ‘C’ at nt 40, CoVaMa will go to contingency table with the coordinates 20:40 in the nucleotide matrix and add 1 successful mapping to this table at coordinates corresponding to T:C, which is row 2, column 4 under the protocol used here. Pairs are assessed only once (i.e. nt 20 is paired with nt 40, but nt 40 is not paired with nt 20 as this would duplicate information) and a nucleotide cannot pair with itself. Therefore the number of nucleotide pairs in an alignment is given by: n(n−1)/2. Each read provided in the alignment data is also translated into an amino acid sequence in an open reading frame if provided by the user in the command line, up until a ‘STOP’ codon is found or until the end of the read segment. In this manner, every amino acid is paired and this information is used to populate a 20×20 contingency table.
After CoVaMa has populated the contingency tables for all of the read data, the data is written out as a pickle file. The purpose of this is so that the contingencies tables can be analysed repeatedly using different parameters without having to regenerate the matrix as it is this stage of the analysis that is most computationally intensive. Only contingency tables represented by a minimum number of reads (as specified in the command-line) are written out to prevent expending memory on tables that have too few data points to be statistically significant.
2.1.2 CoVaMa Merge Matrices
In some situations, it may be appropriate to merge individual matrices together, for example, when multiple datasets are obtained for multiple replicate samples. As each dataset may contain a different level of sequencing coverage or depth, it would be inappropriate to directly add the matrices to one another as this would weigh the contingency tables in favor of which ever dataset had the most reads. Therefore, when contingency tables are merged, the number of reads mapping at each nucleotide or amino acid pair is normalized relative to the total number of reads that mapped in the local contingency table. After normalization, the contents of equivalent tables in the individual matrices are averaged together to generate a single matrix.
2.1.3 CoVaMa Analyse Matrices
To test for evidence of mutational correlation, each of the contingency tables generated in the first half of the CoVaMa script are analysed for evidence of linkage disequilibrium (LD). LD is a measure of the non-random association of two alleles within a population of genomes and determines whether pairs of alleles tend to occur together (disequilibrium or co-variance) or occur independently of one another (equilibrium) [11]. Linkage disequilibrium between two alleles can be present for either of two reasons: 1) variant alleles have been inherited together and so are associated by mutual descent; or 2) the two variant alleles are epistatically linked either via positive-selection of linked loci or the elimination of unlinked loci. While usually applied in the setting of population genetics, this approach can also be straight-forwardly applied to study sequence diversity within a single viral population as the frequency of alleles can be measured using NGS. However, NGS provides a complex description of the allelic composition of a viral population by assessing the frequency of all possible nucleotides at a given position. Therefore, to measure LD in a viral population, every nucleotide can be considered to be an allele and every single nucleotide position within a viral genome can be assessed for linkage to every other nucleotide position.
To measure LD, the allele frequency is measured at each locus (‘Aa’ and ‘Bb’) within the population and the linkage disequilibrium for alleles Aa and Bb is determined using: LD = (pAB * pab) − (pAb * paB). LD can be either positive or negative, depending on the arbitrary assignment of the alleles as ‘A’ or ‘a’ and ‘B’ or ‘b’. Two loci that are in complete equilibrium have a LD value of 0.0, whereas two loci in disequilibrium will have an LD maximum value of +/− 0.25.
Linkage disequilibrium calculations require a 2×2 contingency table (‘A’ and ‘a’ vs ‘B’ and ‘b’). However, we are generating 4×4 contingency tables to compare the frequencies of all possible nucleotides at pairs of loci and 20×20 contingency tables for amino acid pairs. Consequently, we must extract 2×2 tables one by one from every contingency table and evaluate every 2×2 table for evidence of linkage disequilibrium as illustrated in Figure 1B. There are 36 possible 2×2 unique tables in a single 4×4 table and 36100 in a single 20×20 table.
CoVaMa analyzes each possible 2×2 table for evidence of LD, but first applies a simple filter to each 2×2 table to ensure that each of the alleles are sufficiently well represented to provide a meaningful analysis and to ensure that 2×2 tables do not report variance that could be accounted for by sequencing errors in the NGS platform. Firstly, each 2×2 table must have a minimum number of mapped reads (--Min_Coverage), and secondly, each allele must be present within the total population above a certain fraction, as chosen in the command line (--Min_Pop_Fraction). These parameters have default values of 1000 and 0.1% respectively, but can be adjusted taking into account the known error rate of the sequencing platform/technique used and the depth of the sequence coverage. The typical error-rate in the Illumina technologies is primarily due to base-calling errors and mutations that arise during the RT-PCR and PCR steps of library generation. After appropriate quality filtering, this rate is typically found to be in the order of 10−2 to 10−3 errors per nucleotide [12] but can be improved via a number of strategies including PrimerID [13], CircSeq [14, 15], and by merging together the overlapping portions of paired-end reads.
CoVaMa includes an option to normalize each recorded LD value based on the ratio of the number of reads used to calculate the LD for an individual 2×2 table to the total number of reads in the whole 4×4 or 20×20 contingency table. The purpose of this is to prevent skewing the data in favor of infrequently occurring pairs of mutations that are nonetheless in strong disequilibrium. Without this weighting, rare pairs of correlated mutations may appear to be highly significant in terms of their LD value, but in fact only make up a very small portion of the total population. Finally, any contingency table displaying evidence for linkage disequilibrium is written to an output file. An example of the output for a single contingency table is shown in Figure 1C.
2.2 Statistical Analysis
A very large number of LD values are generated during the analysis of even a very short fragment of RNA/DNA (potentially millions of data points). Hence it is important to follow a robust methodology for selecting those LD values that are significantly different. Here, we employ the 3σ rule for comparing a single data point to a very large distribution of other data points [16]. After CoVaMa has completed its analysis, it finds the mean and standard deviation of all the LD values and reports the 3σ critical value (μ+3σ) and 5σ critical value (μ+5σ). The output data can be filtered to remove points falling below this cut-off to reveal only the significant data points. It should be noted that this process is conservative, as the distribution of LD values tends to be heavy-tailed rather than normally distributed. This is due to the fact that LD values are confined to limits between 0 and 0.25 and that the majority of background noise is assumed to occur randomly and thus contain no mutual information; i.e. they will be in equilibrium and therefore tend towards an LD value of 0.
3. Results
3.1 Simulated Data
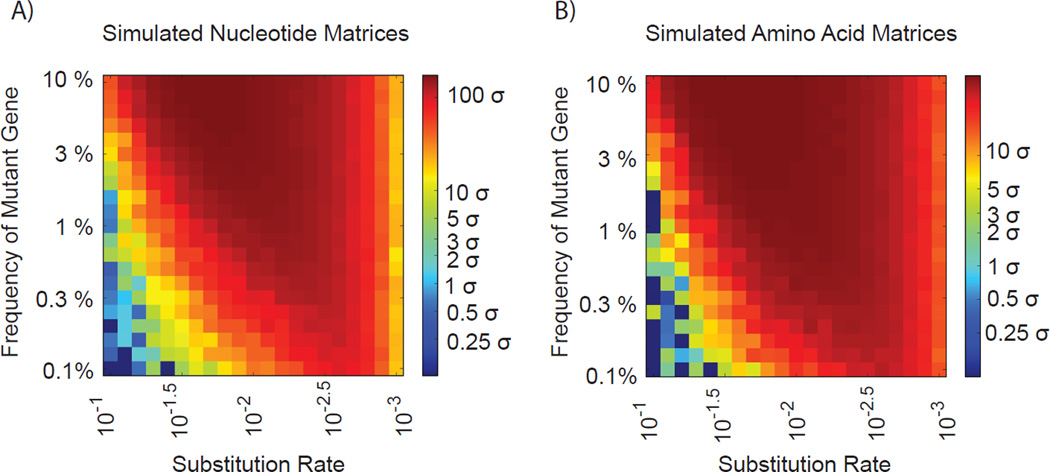
To test the CoVaMa pipeline and determine whether epistatic mutations can be reported faithfully, simulated NGS datasets were generated mimicking an RNAseq experiment analyzing Flock House Virus (FHV) RNA3, a subgenomic RNA 387 nts in length that expresses protein B2. Two reference RNAs were generated, a wild type version and a second version containing two nucleotide variants at nt 81 and nt 133 that would translate into two amino acid residue variants in the correct reading frame. 400 simulated datasets were generated covering a range of random nucleotide substitution rates in the raw reads and a range of frequencies of the mutant RNA relative to the wild type RNA. For each dataset, 100’000 100 nt reads were extracted from a random position in the reference RNAs in the same fashion described in our previous studies[17]. This analysis was designed to determine how sensitive CoVaMa would be to rare mutant RNAs across a range of error rates.
Each simulated dataset was initially aligned to the wild type reference RNA3 end-to-end using Bowtie[18]. The output SAM files were then passed to the CoVaMa pipeline and the linkage disequilibrium was reported for all pairs of mapped nucleotides as well as their corresponding pairs of amino acid residues. Then, for each dataset the LD for the known mutant pair was compared to the distribution of LD values for all of the other mapped nucleotide or amino acid pairs. Figures 2A and 2B show the number of standard deviations (σ) that the expected mutant pair (nt 81 to nt 133) was separated from the mean of the entire distribution of LD values for each dataset for both nucleotide pairs and amino acid pairs respectively.
Figure 2.
Simulated datasets with varying degrees of random substitution rates (x-axis) was generated for a hypothetical RNAseq experiment covering a range of ratios (y-axis) of wild-type to mutant Flock House Virus RNA 3 and passed through the CoVaMa pipeline. Each point in the heatmaps represents the output from an individual simulated dataset for A) the known mutated nucleotide pair and B) the known mutated amino acid pair. The colour of the heatmap reflects the number of standard deviations (σ) that the expected mutant pair was distinct from the mean of the entire distribution of LD values.
As expected, these plots indicate that confidence in identifying the mutant pair is poor when the simulated substitution rate is high but the mutant pair frequency is low. Conversely, with a low mutation rate and higher abundance the mutant pair is readily detected with very high significance. Importantly, at the range of errors commonly found for the Illumina platform, the expected mutant pair is correctly identified with a confidence greater than 3σ even when the mutant pair is only present with a 0.1% frequency.
3.2 Flock House Virus – Nucleotide Matrices
We have extensively studied FHV as a model system for positive sense RNA virus evolution and recombination[17, 19]. FHV provides a well-characterized and simple viral system by virtue of only comprising two RNA genes encoding three gene products. Nonetheless, in spite of such simplicity the mutational spectrum of FHV has proven to be highly dynamic with single nucleotide polymorphisms rapidly arising in cell culture and prolific RNA recombination resulting in the evolution of defective RNA species. FHV can be passaged in a straight-forward manner in Drosophila melanogaster S2 cells in culture and particles purified from the lysed cells [20]. As such, FHV provides an ideal model system to validate the computational approaches described here and explore intra-host diversity under limited conditions.
FHV particles were grown in cell culture for a total of 48 hours, purified over a series of ultracentrifugation steps and treated with nucleases to remove non-encapsidated RNAs, as per our previous analyses [17, 19]. The encapsidated genomic RNA was then extracted and prepared for RNAseq using ClickSeq (a cDNA library generation method we recently developed that considerably reduces artifactual recombination[21]). Final cDNA sequencing libraries were prepared for paired-end sequencing with an average fragment length of 150–200bps and sequenced on an Illumina NextSeq giving 150bp for each read. Overlaps on the paired read data were exploited to reconstruct longer single reads and correct sequencing errors using BBmerge from the BBmap suite (http://sourceforge.net/projects/bbmap/). The raw data was adaptor-trimmed and quality-filtered with cutadapt[22] and the fastx_toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) using the commands as shown in Box 1, Step 1.
BOX 1. FHV mapping commands.
Command line entries to generate the data presented in Table 1.
A brief description of the parameters in each command followed by the command itself is given for the analysis performed to find LD in the FHV genome.
-
1)Overlaps in the paired-end reads were exploited to reconstruct longer single reads using BBmerge:
- bbmap/bbmerge.sh in1= R1_raw.fastq in2=R2_raw.fastq out=Merged.fastq
-
2)Adaptor trimming and quality filtering: Reads must be a minimum of 70 nts in length after adaptor trimming. The first and last ten nucleotides are removed as these correspond to random nucleotides included in the ClickSeq adaptors. Finally, the reads were quality filtered requiring 98% of each read to contain base-calling PHRED scores of no less than 20.
- python cutadapt -b agatcggaagagc -m 70 Merged.fastq | fastx_trimmer -Q33 -f 10 | fastx_trimmer -Q33 -t 10 | fastq_quality_filter -Q33 -p 98 -q 20 -o Prep.txt
-
2)Alignment of read data to reference FHV genome: End-to-end mapping (v-mode) allowing only 3 mismatches per aligned read. Only the best alignment is reported to the output in SAM format.
- bowtie –v 3 --best –S FHV_Genome_Index Prep.txt FHV_mapping.sam
-
4)Generation and population of matrices containing nucleotide contingency tables: The ‘Directory’ is the chosen directory for the output data and the reference genome is in fasta format. The input file is in SAM format. 5 nucleotides were trimmed from the 5’ and 3’ extremities of each aligned read to prevent mismatches being called in these regions. Only contingency tables containing 100 or more reads were written out to a pickle file called ‘Total_Matrices.py.pi’.
- pypy CoVaMa_Make_Matrices.py ./Directory/ FHV_Genome.fasta --SAM1 FHV_mapping.sam --Ends 5 --Min_Coverage_Output 100
-
5)Analysis of nucleotide contingency tables for evidence of linkage disequilibrium: The input file is the output pickle file from the previous step. The output data is a written to a text file in the chosen ‘Directory’. Only contingency tables where each allele is represented by 100 or more reads were evaluated for LD and were weighted as described.
- pypy CoVaMa_Analyse_Matrices.py Total_Matrices.py.pi FHV_NtLD.txt ./Directory/ --Min_Coverage 100 -Weighted
-
6)Extraction of significant data: The fourth data field in the output data gives the LD value. 3σ is 0.0021 for the described FHV RNAseq dataset.
- awk ‘{if($4>0.0021)print;}’ FHV_Nt-LD.txt > FHV_Nt-LD_Significant-Results.txt
The raw data was aligned to the FHV reference genomic RNAs using an end-to-end alignment in Bowtie[18] allowing a maximum of 3 mismatches per read. The output SAM file was then passed to the CoVaMa_Make_Matrices script to generate contingency tables of nucleotide pairwise associations. Only contingency tables that were occupied by at least 100 reads were retained and these were passed to the CoVaMa_Analyse_Matrices script to find evidence of linkage disequilibrium. The only contingency tables analysed were those where variant alleles at each loci were represented by at least 100 reads and were present at a frequency no less than 0.1% of the total reads in the contingency table. LD values for each variant pair were normalized relative to the number of reads that occupied each contingency table, as described above and in Figure 1B. The command line entries used for each of these steps (nucleotide matrices only) are shown in Box 1, Steps 2–6.
In this analysis, a total of 551’381 tables were evaluated with 10’862 passing the coverage filters and returning an LD value. The mean and standard deviation of these values was 2.3×10−4 and 6.8×10−4 respectively, yielding a 3σ critical value of 2.1×10−3. Only 10 pairs of nucleotides were found with significant (>3σ) linkage disequilibrium (Table 1). Of these 10 pairs, 7 of them are found in FHV RNA 1 and 3 in FHV RNA 2. We also looked for evidence of linkage disequilibrium between pairs of amino acids but found no significant LD values in the FHV RNA 1, RNA 2 or RNA 3 open reading frames.
Table 1.
Significant LD values (>3σ) between pairs of nucleotides found by CoVaMa using RNAseq data of Flock House Virus grown in cell culture.
| Reference | Nt 1 | Nt 2 | Change | LD Value |
|---|---|---|---|---|
| RNA1 | 31 | 36 | A-C to C-A | 0.0099 |
| 35 | 36 | A-C to C-A | 0.0113 | |
| 36 | 54 | A-C to C-T | 0.0187 | |
| 187 | 240 | T-G to G-A | 0.0037 | |
| 192 | 240 | T-G to G-A | 0.0040 | |
| 240 | 259 | A-C to G-T | 0.0052 | |
| 1044 | 1101 | T-C to C-T | 0.0066 | |
| RNA2 | 130 | 226 | T-G to C-A | 0.0102 |
| 130 | 247 | T-C to C-T | 0.0043 | |
| 226 | 247 | A-T to G-C | 0.0725 |
It is not clear why these loci are linked from our analysis alone. It is interesting to note, however, that all of the detected loci are found in regions that are conserved in Defective-Interfering RNAs [17] that arise during FHV passage in cell culture and so it is possible that these loci contribute to the formation of functional or structural motifs. Moreover, the four nucleotides in disequilibrium with one another at FHV RNA 1 at nts 187, 192, 240 and 259 (Table 1) fall within a well-characterized cis-acting response element (cis-RE) from nt 40 to nt 290 that is required for the replication of FHV RNA 1 [23]. Fine-grain deletion analyses in this cis-RE indicate that nts 187, 192, 240 and 259 are in regions required for FHV RNA 1 replication. Both nt 187 and 192 form part of a stem of predicted RNA secondary structure in this region that is also required for the recruitment of FHV RNA 1 to the mitochondrial membrane (the site of RNA replication) in a manner dependent upon FHV RNA-dependent RNA polymerase and so may act in a concerted fashion. It would be interesting to confirm with experimental analyses whether these mutations promote or attenuate viral fitness in cell culture and whether they have a cooperative or co-dependent effect, as would be suggested by the data presented here.
3.3 HIV Protease – Amino Acid Matrices
The development of resistance to HIV protease inhibitors has contributed to the persistence of the HIV/AIDS epidemic and so the evolution of HIV protease in response to anti-retroviral treatment has been well-studied. There are a number of characterised amino acid substitutions known to affect protease activity and rescue wild-type proteolysis in the presence of protease inhibitors[24]. Often, multiple mutations occur in a correlated fashion. Previous analyses have applied linkage disequilibrium studies to detect correlated evolution by comparing consensus viral genome sequences isolated from drug-treated versus drug naive patients[25]. However, this approach is insensitive to the mutations found within the diverse viral population found within each patient. Here, we use CoVaMa to take into account the mutant spectra within each patient and subsequently measure linkage disequilibrium within the protease gene within a large patient cohort.
In a previous paired-end sequencing analysis of 93 patients from the US military HIV Natural History Study, we described the co-evolution of distant parts of the HIV genome in protease and gag through mutual information theory[7, 26]. In this study, viral RNA was obtained from the serum or plasma of patients after therapy had failed to adequately suppress viral replication (1,000 copies/mL) with multiple samples taken for some patients. For paired-end sequencing, cDNA amplicons were generating by RT-PCR amplifying two 1-kilobase segments of the viral genome that spanned the protease and gag polyprotein, as described [26]. These amplicons were prepared for paired-end sequencing, yielding ~5M paired reads per patient sample with fragment sizes averaging 275bp. This fragment size is sufficient to cover the protease gene in its entirety.
Here, we reanalyzed the RNAseq data to find evidence of LD in the amino acid sequence of protease. The consensus genome for each patient was generated as part of our previous investigation [7, 26] and the paired-end reads for each individual dataset were aligned end-to-end to the viral genome using bowtie [18]. Using patient-specific consensus genomes rather than the reference HX2B genome for alignment is very important as it ensures that the maximum number of reads will align successfully. After the final alignment to the consensus genome, the output SAM files were passed to the CoVaMa pipeline to generate contingency tables. This process was performed individually for every dataset in the patient cohort using the command line entries shown in Box 2 steps 1–3.
BOX 2. HIV mapping commands.
Command line entries to generate the data presented in Table 2.
A brief description of the parameters in each command followed by the command itself is given for the analysis performed to find LD in HIV protease.
- Adaptor trimming and quality filtering for each read pair: Reads must be a minimum of 50 nts in length after adaptor trimming. The reads were quality filtered requiring 98% of each read to contain base-calling PHRED scores of no less than 20.
- python cutadapt -b agatcggaagagc -m 50 Raw_read_R1.fastq | fastq_quality_filter -Q33 -p 98 -q 20 -o HIV_reads_R1.fastq
- python cutadapt -b agatcggaagagc -m 50 Raw_read_R2.fastq | fastq_quality_filter -Q33 -p 98 -q 20 -o HIV_reads_R2.fastq
- Alignment of read data to the reference HIV genome: End-to-end mapping (v-mode) allowing only 3 mismatches per aligned read. Only the best alignment is reported to the output in SAM format.
- bowtie -v 3 --best -S HIV_index HIV_reads_R1.fastq HIV_align_R1.sam
- bowtie -v 3 --best -S HIV_index HIV_reads_R2.fastq HIV_align_R2.sam
- Generation and population of matrices containing amino acid contingency tables: Only contingency tables containing 100 or more reads were written out to a pickle file called ‘Total_AA_Matrices.py.pi’. The ‘Directory’ is the chosen directory for the output data for each patient. The open reading frame for HIV protease is found between nucleotides 2253 and 2549.
- pypy CoVaMa_Make_Matrices_wAA.py --SAM1 HIV_align_R1.sam --SAM2 HIV_align_R2.sam --Min_Fusion_Coverage 100 ./Directory1/ HIV_genome.fasta 2253 2549
- The amino-acid matrices generated for each patient in the cohort were merged into one single matrix: Each Directory and Matrix for each patient are listed and output file is written into the current working directory.
- pypy CoVaMa_Merge_Matrices.py ‘Directory1/Matrix1 Directory2/Matrix2 Directory3/Matrix3…’ HIV-PR_Merged_LD.py.pi ./Directory/
- Analysis of amino acid contingency tables for evidence of linkage disequilibrium: The input is a pickle-file and the output is written as a text file in the specified directory. The input matrix is a merged one, and only contingency tables that have generated by merging together 50 or more individual tables are evaluated. Only contingency tables where each allele is represented by 100 or more reads were evaluated for LD and were weighted as described.
- pypy CoVaMa_Analyse_Matrices_wAA.py HIV-PR_Merged_LD.py.pi HIV-PR_LD_m50.txt ./Directory/ -Merge --Min_Merged_Matrices 50 --Min_Coverage 500 –Weighted
- Extraction of significant data: The fourth data field in the output data gives the LD value. 3σ is 0.013 for the described HIV merged RNAseq dataset.
- awk ‘{if($4>0.013)print;}’ HIV-PR_LD_m50.txt > HIV-PR_Significant_Results.txt
Next, all the contingency tables for each individual were ‘merged’ together into a single table that represented the mutant spectra found within the total cohort, rather than just the individuals. As linkage between alleles can occur due to both epistatic coevolution or due to co-inheritance, averaging together the contingency tables for multiple patients will remove any evidence of linkage due to inheritance. Any remaining linkage is thus likely to occur due to correlated evolution of the protease in response to drug treatment. The merged matrices were analysed using CoVaMa, yielding a large number of correlated mutations in the drug-resistant variant species. The command line entries used for each of the steps are shown in Box 2 steps 4–6.
With this analysis, we found a large number of correlated mutations in the amino acid sequence of HIV protease. In total, 414 pairs of mutations were found with LD values above the 3σ critical value and 162 above 5σ. The top 25 pairs are shown in Table 2. Importantly, we find many pairs of mutations that have been previously characterized as giving rise to protease inhibitor resistance both independently [24] and in a coordinated fashion [7, 27–30] (21 out of the 25 pairs shown in Table 2). This gives us confidence in the ability of the methodology described here to detect covariation in NGS datasets and cohorts. Interestingly, single amino acid loci often displayed high levels of LD with multiple other amino acid loci, indicating that amino acid substitutions may cluster together into networks that have evolved collectively [28]. As we have merged the alignment data from multiple patient samples, it is highly unlikely that these networks are present due to a common inheritance (identity-by-descent), but rather reflect a functional cooperativity of these residues in the evolution of drug resistance. Additionally, we find highly significant events that have not been previously described. Further experimentation will determine the validity and nature of these correlated mutations.
Table 2.
Twenty-five highest correlated pairs of amino acid residues in the protease gene of HIV found by CoVaMa in an RNAseq study of a large cohort of drug-resistant HIV patients.
| Pair 1 | Pair 2 | LD | References of known associations |
|---|---|---|---|
| 41K-93L | 41R-93I | 0.063462 | [7, 30] |
| 77I-93L | 77V-93I | 0.056952 | [27–30] |
| 46L-82A | 46M-82V | 0.056426 | [7, 27, 28] |
| 30D-88N | 30N-88D | 0.055541 | [7, 27, 28] |
| 10I-93L | 10L-93I | 0.052307 | [7, 27–29] |
| 35D-36I | 35E-36M | 0.051156 | [7, 27–29] |
| 63L-93I | 63P-93L | 0.051149 | [30] |
| 10I-62V | 10L-62I | 0.050766 | [27] |
| 63L-90L | 63P-90M | 0.04806 | [27, 28] |
| 10I-90M | 10L-90L | 0.047771 | [27, 28] |
| 36I-77V | 36M-77I | 0.042961 | [27, 29, 30] |
| 62I-71A | 62V-71V | 0.040321 | [29] |
| 46I-90M | 46M-90L | 0.040264 | |
| 41K-63P | 41R-63L | 0.03999 | |
| 54I-82V | 54V-82A | 0.039912 | [27] |
| 62I-93I | 62V-93L | 0.039224 | [27, 30] |
| 10I-82A | 10L-82V | 0.037066 | [27, 28] |
| 10I-35D | 10L-35E | 0.036709 | |
| 36I-62V | 36M-62I | 0.036042 | [27–29] |
| 10I-36I | 10L-36M | 0.034866 | |
| 62I-63L | 62V-63P | 0.034603 | [30] |
| 30D-35E | 30N-35D | 0.034588 | [7] |
| 71A-93I | 71V-93L | 0.034398 | [29, 30] |
| 10I-71V | 10L-71A | 0.034022 | [27–29] |
| 62I-90L | 62V-90M | 0.033936 | [27] |
4. Discussion
In this manuscript, we described how data generated from NGS studies can be used to measure linkage disequilibrium among variant nucleotides and amino acids in the viral mutational landscape. Our script, CoVaMa, provides a simple tool to perform this analysis. The studies illustrated here could easily be applied using our scripts to other individual or cohort studies investigating viral diversity by NGS or of other more complex organisms.
One major limitation of NGS is the length of the sequence reads that can be generated. NGS platforms and chemistries are currently limited in the length to only a few hundred nucleotides. As a result of extracting information directly from reads alignment data and making no inferences as to how these reads might themselves be linked to one another, CoVaMa is limited to finding correlated pairs of mutations that are separated by a distance no longer than the read length. Nonetheless, CoVaMa is still capable of revealing important correlated mutations and as new technologies and methods arise that extend the length of high-throughput sequencing reads and improve long-read error-rates, the strength of approach described here will scale accordingly.
Our first analysis using simulated data demonstrated that linkage disequilibrium can successfully and sensitivity extract coordinated patterns of mutations from NGS sequence alignment. Of course, this represented an idealized scenario. In reality the distribution of LD values may be affected by multiple simultaneous mutations and the raw data may contain many other types of errors such as artifactual recombination events, homo-polymer slipping, micro-deletions, and PCR duplications. Nonetheless, in the range of errors commonly found for the Illumina platform, the expected mutant pair is correctly identified with a confidence greater than 3σ even when this mutant pair is present with only a 0.1% frequency within the population. This gives us confidence in the ability of CoVaMa to sensitively and reliably detect co-variation in NGS datasets.
By applying this approach to real RNAseq data from a cell-culture based study of Flock House Virus, we demonstrated that correlated mutations arise even within the limited environment of cell culture. Under these conditions, there are very few barriers to viral progression and so concerted viral adaptations seem less likely than when compared to viral passaging in the wild. Nonetheless, we observed a few modifications to the RNA sequences that may affect local RNA motifs. Finally, we demonstrate that CoVaMa can find correlated mutations in the protease gene within a large patient cohort. Most of the correlated mutations found here have been previously found in a number of studies including our previous analysis of the same patient cohort. This demonstrates that the methodology applied here has been successful and can robustly detect epistatic co-evolution. Having identified these correlated pairs of mutations, experiments can be designed to measure the effect that they may have on viral fitness and the viral lifecycle.
Highlights.
Mutations in populations of RNA viruses often evolve in a correlated manner
CoVaMa is a Python script that detects Linkage Disequilibrium in RNAseq data
We find correlated mutations in simulated and real RNAseq of Flock House Virus
Correlated mutations are also found in protease genes from an HIV patient cohort
Acknowledgements
This work was funded by a seed grant from the Scripps Translational Science Institute 5UL1TR001114 to B.E.T, by the HIVE center grant 1P50GM103368 to B.E.T. and by The National Institutes of Health 5R01-GM54076 to J.E.J. M.W.C. was supported by The National Institutes of Health T32AI007354.
Support for this work (IDCRP-000-03) was also provided by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed through the Uniformed Services University of the Health Sciences. This project has been funded in whole, or in part, with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under Inter-Agency Agreement Y1-AI-5072.
The Views expressed are those of the author(s) and do not necessarily reflect the official views of the Uniformed Services University of the Health Sciences, the United States Air Force, or the Department of Defense.
Abbreviations
- FHV
Flock House Virus
- NGS
Next-Generation Sequencing
- LD
Linkage Disequilibrium
- HIV
Human Immunodeficiency Virus
- SAM
Sequence Alignment Map
- cis-RE
cis-acting response element
- CoVaMa
Co-Variation Mapper
- Nt
Nucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 2.Sharma D, Priyadarshini P, Vrati S. J Virol. 2015;89:1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browning SR, Browning BL. Nat Rev Genet. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. Journal of computational biology : a journal of computational molecular cell biology. 2013;20:113–123. doi: 10.1089/cmb.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macalalad AR, Zody MC, Charlebois P, Lennon NJ, Newman RM, Malboeuf CM, Ryan EM, Boutwell CL, Power KA, Brackney DE, Pesko KN, Levin JZ, Ebel GD, Allen TM, Birren BW, Henn MR. PLoS computational biology. 2012;8:e1002417. doi: 10.1371/journal.pcbi.1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Charlebois P, Macalalad A, Henn MR, Zody MC. BMC Genomics. 2013;14:674. doi: 10.1186/1471-2164-14-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn WF, Chang MW, Tan Z, Oliveira G, Yuan J, Okulicz JF, Torbett BE, Levy RM. PLoS computational biology. 2015;11:e1004249. doi: 10.1371/journal.pcbi.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prosperi MC, Yin L, Nolan DJ, Lowe AD, Goodenow MM, Salemi M. Scientific reports. 2013;3:2837. doi: 10.1038/srep02837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerenwinkel N, Gunthard HF, Roth V, Metzner KJ. Frontiers in microbiology. 2012;3:329. doi: 10.3389/fmicb.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slatkin M. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minoche AE, Dohm JC, Himmelbauer H. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Proc Natl Acad Sci U S A. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou DI, Hussmann JA, McBee RM, Acevedo A, Andino R, Press WH, Sawyer SL. Proc Natl Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acevedo A, Brodsky L, Andino R. Nature. 2014;505:686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnov NVe, Dunin-Barkovskiĭ IV. Mathematische Statistik in der Technik: Kurzer Lehrgang, Deutscher Verlag der Wissenschaften. 1969 [Google Scholar]
- 17.Routh A, Johnson JE. Nucleic Acids Res. 2014;42:e11. doi: 10.1093/nar/gkt916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Routh A, Ordoukhanian P, Johnson JE. J Mol Biol. 2012;424:257–269. doi: 10.1016/j.jmb.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneemann A, Marshall D. J Virol. 1998;72:8738–8746. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Routh A, Head SR, Ordoukhanian P, Johnson JE. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 23.Van Wynsberghe PM, Ahlquist P. J Virol. 2009;83:2976–2988. doi: 10.1128/JVI.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. Topics in antiviral medicine. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Lee C. PLoS One. 2007;2:e814. doi: 10.1371/journal.pone.0000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang MW, Oliveira G, Yuan J, Okulicz JF, Levy S, Torbett BE. J Virol Methods. 2013;189:232–234. doi: 10.1016/j.jviromet.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee SY, Liu TF, Holmes SP, Shafer RW. PLoS computational biology. 2007;3:e87. doi: 10.1371/journal.pcbi.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu TD, Schiffer CA, Gonzales MJ, Taylor J, Kantor R, Chou S, Israelski D, Zolopa AR, Fessel WJ, Shafer RW. J Virol. 2003;77:4836–4847. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman NG, Schiffer CA, Swanstrom R. Virology. 2003;314:536–548. doi: 10.1016/s0042-6822(03)00484-7. [DOI] [PubMed] [Google Scholar]
- 30.Counts CJ, Ho PS, Donlin MJ, Tavis JE, Chen C. PLoS One. 2015;10:e0123561. doi: 10.1371/journal.pone.0123561. [DOI] [PMC free article] [PubMed] [Google Scholar]