Abstract
Polo-like kinase 1 (Plk1) plays key roles in regulating mitotic processes that are critical for cellular proliferation. Overexpression of Plk1 is tightly associated with the development of certain cancers in humans, and a large body of evidence suggests that Plk1 is an attractive target for anticancer therapeutic development. Drugs targeting Plk1 can potentially be directed at two distinct sites: the N-terminal catalytic domain, which phosphorylates substrates, and the C-terminal polo-box domain, which is essential for protein–protein interactions. In this review, we will summarize recent advances and new challenges in the development of Plk1 inhibitors targeting these two domains. We will also discuss novel strategies for designing and developing next-generation inhibitors to effectively treat Plk1-associated human disorders.
Protein kinases as drug targets
Reversible protein phosphorylation is a fundamental regulatory mechanism underlying diverse biochemical and cellular processes in eukaryotic organisms [1]. Protein kinases catalyze transfer of the gamma phosphate from adenosine triphosphate (ATP) to protein substrates. A plethora of evidence suggests that dysregulation of kinase-dependent processes can contribute to a variety of pathological disorders, including cancers [2]. Therefore, protein kinases are considered to be potential targets for developing anticancer therapeutics. However, generating specific kinase inhibitors was originally thought to be difficult because of high similarities in their overall kinase domain (KD) folds and the inclusion of intrinsically conserved motifs within the KD. Despite this early discouraging perception, as a result of extensive research efforts, protein kinases have now become one of the largest classes of anticancer drug targets [3, 4]. As of today, small-molecule inhibitors have been developed against nearly half of the 518 total cellular protein kinases. Among them, 35 inhibitors are FDA approved for clinical applications and more than 500 inhibitors are currently in clinical trials.
As one of the most attractive anticancer drug targets, polo-like kinase 1 (Plk1) has been the subject of extensive research. As a consequence of these efforts, a large number of Plk1 inhibitors have been developed, with several encouraging agents currently in clinical trials. Structure–activity relationships, biochemical and cellular properties, and the clinical trial status of multiple promising Plk1 inhibitors have been the subject of recent reviews [5–8]. Here, we will focus on discussing the physiological functions and biochemical properties of Plk1, the rationale behind targeting Plk1 for anticancer therapy, and current trends and future directions in developing more efficacious anti-Plk1 therapeutics. Given a high level of interest and multiple layers of ongoing efforts in developing potent and specific Plk1 inhibitors, we will summarize important recent advances and new strategies to timely meet the needs for successful anti-Plk1 drug discovery.
Plks and cancer
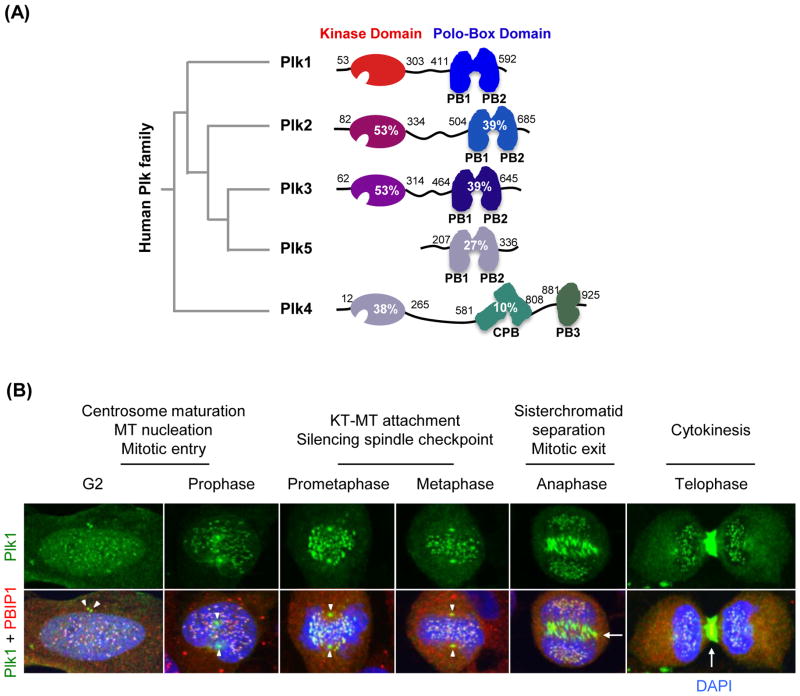
Plk1 belongs to the polo sub-family of Ser/Thr protein kinases (collectively known as Plks), which play critical roles in diverse cellular processes, such as cell cycle progression, differentiation, and survival [9–12]. In mammalian cells, five Plks (Plk1 to 5) that exhibit distinct tissue distributions and physiological functions have been reported to date (Table 1). The Plks are characterized by unique C-terminal non-catalytic protein–protein interaction domains, referred to as “polo-box domains” (PBDs) [13–15] (Figure 1A). With the exception of the recently isolated Plk5, which lacks the N-terminal KD [16], all four Plks (Plk1 to 4) possess a KD that shares varying degrees of sequence homology with Plk1 KD (Figure 1A). Plk4 is more distantly related to Plk1 in having a uniquely structured cryptic polo-box (CPB) and PB3 [17], and it will not be discussed in detail.
Table 1.
Expression, subcellular localization, and physiological function of Plks
| Expression | Localization | Function | |
|---|---|---|---|
| Plk1 | Actively dividing cells | Centrosomes, kinetochores, midbody | G2/M transition, mitotic progression, cytokinesis |
| Plk2 | Less-dividing cells | Centrosomes | Stress-response, centriole duplication |
| Plk3 | Less-dividing cells | Centrosomes, midbody, nucleolus | Genotoxic, hypoxic & oxidative stress response, p53 activation |
| Plk5 | Non-dividing cells | Cytoplasm | Neuritic process |
| Plk4 | Actively dividing cells | Centrosomes | Centriole biogenesis |
Figure 1.
A schematic diagram illustrating the structures of the human Plk family and subcellular localization of polo-like kinase 1 (Plk1) during the cell cycle. (A) A Phylogenetic tree is shown at left. Amino acid residues and sequence identities of Plk2–5 compared to Plk1 are indicated. PB1, polo-box motif 1; PB2, polo-box motif 2; CPB, cryptic polo-box; PB3, polo-box motif 3. (B) Subcellular localization pattern of Plk1 in HeLa cells is shown with a centromere/kinetochore-associating protein, PBIP1 [151]. DNA is stained with DAPI. Arrowheads, centrosomes; arrows, midzone and midbody.
A large body of evidence suggests that Plk1 is highly expressed in mitotically active cells and tissues, and that it plays critical roles at multiple stages of M-phase, including mitotic entry, metaphase/anaphase transition, and cytokinesis (Figure 1B). In line with the multitude of Plk1 functions, Plk1 has been shown to localize to centrosomes, kinetochores, and midzones/midbodies in a temporally and spatially regulated manner [15, 18–21] (Figure 1B). Hence, Plk1 interacts with diverse targets at discrete subcellular locations to promote proper M-phase progression.
In contrast to Plk1’s role in cell proliferation, other Plk members appear to play distinct roles that have little functional overlap with one another (Table 1). Although the in vivo functions of Plk2 and Plk3 are yet to be further investigated, a flurry of data suggests that Plk2 mediates cellular stress-induced checkpoint arrest for survival [22, 23], while Plk3 regulates genotoxic stress-response pathways [24–29] (Table 1). In addition, Plk2, Plk3, and Plk5 are required for proper neuronal function and/or differentiation [16, 30–32], while Plk4 plays a key role in centriole biogenesis [33, 34].
In mouse models, a homozygous deletion of Plk1 or Plk4 is lethal to embryos [14, 35], whereas a homozygous deletion of Plk2 is viable but exhibits retarded growth and skeletal development late in gestation [36] (Table 2). Homozygous Plk3-null mice develop spontaneous tumors in various organs at advanced ages, suggesting a role of Plk3 as a tumor suppressor [37]. Interestingly, Plk1- or Plk4-heterozygous mice also develop spontaneous tumors at advanced ages [14, 35]. Given that defects in the function of Plk1 or Plk4 lead to abnormal bipolar spindle formation and chromosome missegregation [9, 10, 12, 38], improper levels of these kinase activities may lead to chromosome instability and mitotic errors that could promote tumorigenesis.
Table 2.
Plks in the development of mouse and human cancers
| Role in tumor | Phenotypes in mouse models | Human cancer association | ||
|---|---|---|---|---|
| Knock out (−/−) | Haploinsufficient(+/−) | |||
| Plk1 | Promoter | Lethal at 4 or 8 cell stage [35] | Develop lymphomas, lung carcinomas, squamous cell carcinomas, and ovarian sarcomas [35] Increased tumor frequency in PLK1+/− TP53−/− cells [35] | Upregulated in breast, oesophageal, lung, and colorectal cancer, gastric, anaplastic thyroid, hepatocellular carcinoma, melanomas and gliomas, non-small cell lung, ovarian, prostate, pancreatic, head and neck cancers [39,40] |
| Plk2 | Suppressor? | Retarded embryonic growth [36] | No apparent phenotype | Downregulated in B cell neoplasias and heptocellular carcinoma [41] |
| Plk3 | Suppressor | Highly vascularized tumors in lung, kidney, liver, and uterus [37] | No apparent phenotype | Downregulated in lung carcinoma, head and neck squamous cell carcinoma, heptocellular carcinoma [41–43] |
| Plk5 | Suppressor? | N/A | N/A | Downregulated in primary brain tumor (astrocytoma and glioblastoma multiforme) [16] |
| Plk4 | Suppressor/promoter | Lethal at E7.5 [14] | Developed in liver and lung cancers [44] | Upregulated in colorectal cancer and poor prognosis breast cancer [45,46] Downregulated in heptocellular carcinoma [41] |
Biochemical properties of Plk1
The Plks constitute a class of enzymes whose catalytic function is intimately related to protein–protein interactions (PPIs) mediated by its C-terminal PBD (or CPB in the case of Plk4; see Figure 1). Detailed biochemical studies have demonstrated that the N-terminal catalytic KD of Plk1 functions in concert with its cis-acting, C-terminal PBD, which interacts with phosphorylated motifs to bring the KD into close proximity with its substrates [47–49]. This cooperative action between the KD and PBD allows Plk1 to achieve rapid amplification of Plk1-dependent biochemical steps [50, 51]. Determination of the crystal structures of the KD, the PBD, and a KD-PBD complex has revealed that KD and PBD reciprocally interact with each other [47, 52–54] to mediate diverse biological processes in a highly regulated fashion [55].
Although the primary sequence of Plk1 exhibits a high level of homology with closely related Plk2 and Plk3 (Figure 1), in vitro biochemical analyses have shown that the three Plks have dissimilar substrate specificities [56, 57]. Furthermore, the PBDs of each Plk interact with specific phosphoepitopes on their binding targets [47, 58, 59]. Thus, the specificity of Plk1-dependent biochemical reactions is regulated at two consecutive steps: i) PBD-dependent substrate binding and ii) KD-dependent substrate phosphorylation. Consistent with the cooperative nature of the KD and PBD of Plk1, inhibition of either one of these two domains is sufficient for abrogating Plk1 function in vivo [50, 51, 55, 57, 60].
Plk1 addiction in cancer
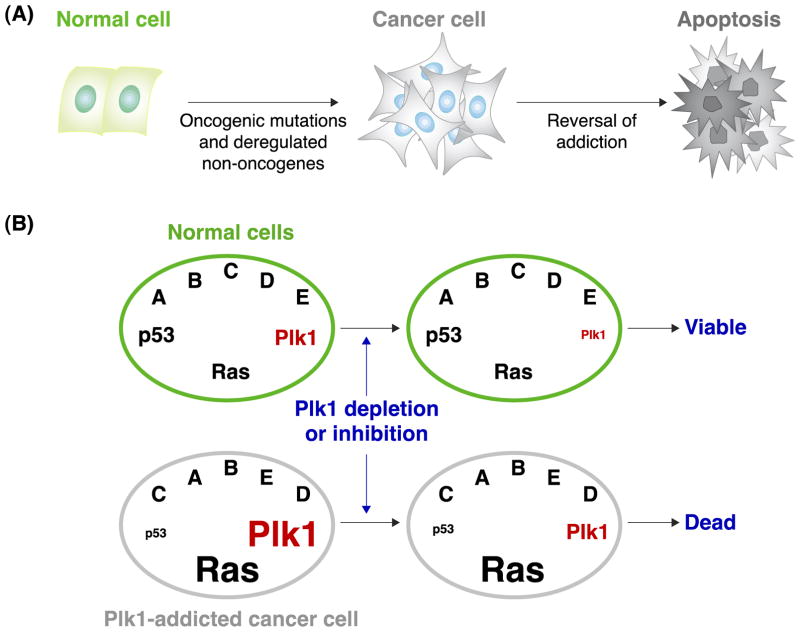
Plk1 has drawn a lot of attention because its overexpression is tightly associated with neoplastic transformation of human cells. Plk1 overexpression is thought to promote tumorigenesis by overriding cellular checkpoints and inducing genetic instability [40, 61, 62]. Interestingly, while Plk1 inhibition leads to mitotic arrest and apoptotic cell death in a wide variety of human cancer cells, Plk1 reduction by up to ~80% did not have a significant detrimental effect in primary human cells and various organs of adult Plk1 RNAi mice [63]. Cancer cells are known to be addicted to oncogenes and many non-oncogenes, such as Plk1 [64, 65]. Reversing oncogene addictions in cancer cells has been shown to induce apoptotic cell death [64, 66]. Therefore, increased sensitivity of cancer cells to Plk1 interrogation may likely stem from their altered signaling pathways and biochemical steps in Plk1-addicted cancer cells (Figure 2A).
Figure 2.
A schematic diagram illustrating cancer cell–selective killing by the reversal of addictions to oncogenes and non-oncogenes. (A) Cancer cells arise as a consequence of mutations in oncogenes or deregulation of non-oncogenes. Reversal of this addiction can induce apoptotic cell death in cancer cells. (B) In a hypothetical cancer cell, where p53 is downregulated and Ras is mutationally activated, Plk1 is overexpressed. Because of cancer cells’ addiction to Plk1, depletion or inhibition of Plk1 can induce selective killing of cancer cells, but not normal cells.
It has been demonstrated that non-oncogenic Plk1 inhibition is particularly vulnerable in cancer cells bearing an inactivating TP53 mutation, in comparison to their isogenic TP53 wild-type (WT) cells [67–71]. In a genome-wide study, Plk1 was isolated as one of several mitotic regulators whose overexpression is selectively required for the viability of activated Ras-bearing cancer cells, but less so for the viability of their respective normal cells [72]. Other related mitotic kinases, such as Cdc2, AurA, and AurB, have not been identified from the same screen. Since a large fraction of human cancers bear inactivated TP53 and/or activated RAS mutations, antagonizing Plk1 function may have a broad applicability in selectively interrogating cancer cells (Figure 2B).
Validation of Plk1 as a drug target
Data obtained from multiple studies showed that patients with high Plk1 expression in their tumors exhibit a poorer survival rate than those with low Plk1 expression [73–76], suggesting that Plk1 overexpression is closely associated with cancer aggression. Intriguingly, interference with Plk1 function by various experimental means efficiently induces mitotic block and apoptotic cell death in most cancer cells, and is sufficient to prompt tumor regression in mouse xenograft models [77, 78]. In contrast, a similar level of Plk1 depletion or inhibition fails to induce a severe effect in normal or primary cells, even though it results in a somewhat reduced rate of cell division [67, 68, 72, 79, 80]. The differential levels of Plk1 required for the viability of cancer vs. normal cells makes Plk1 a particularly attractive target for anticancer drug discovery (Figure 2B).
Strategies for targeting Plk1
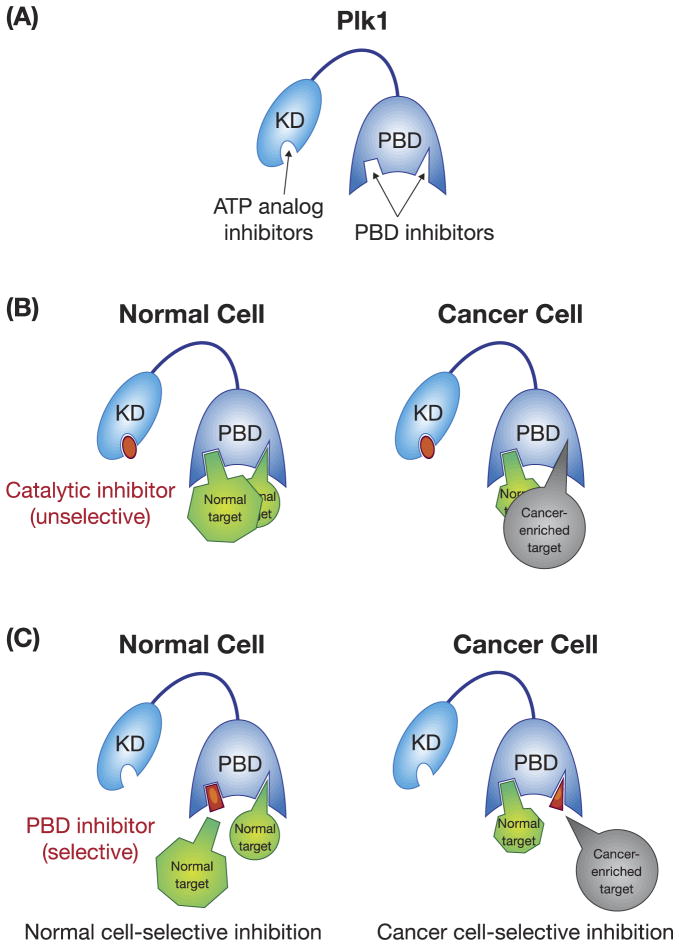
The functionally essential KD and PBD of Plk1 represent two distinct drug targets available within one molecule (Figure 3A). Over the years, a lot of effort has been made to target the catalytic activity of Plk1, yielding a large number of ATP-competitive inhibitors with varying degrees of success (Table 3). While most of these inhibitors are at the preclinical stage of development, several are currently in clinical trials with favorable results. However, mainly due to the large number of protein kinases in mammalian cells and the high degree of structural conservation among ATP-binding pockets, these inhibitors exhibit a significant level of cross-reactivity with other functionally unrelated kinases. Moreover, since closely related Plk2 and Plk3 play a role in checkpoint-mediated cell-cycle arrest in response to cellular or genotoxic stress [22, 28], and Plk3 is proposed to function as a tumor suppressor [29, 37, 81], development of Plk1-specific ATP-competitive inhibitors is likely important for selectively interrogating Plk1-addicted cancer cells.
Figure 3.
A schematic diagram showing two strategies that target either the catalytic activity or the PBD of Plk1. (A) The catalytic activity of Plk1 is mostly targeted by ATP-competitive inhibitors, while the PBD is inhibited by PBD-binding antagonists. K.D., kinase domain; PBD, polo-box domain. (B) Catalytic inhibitors are unselective because they can abolish the catalytic activity of Plk1 in both normal and cancer cells. (C) PBD inhibitors can be designed in such a way that they can selectively inhibit cancer cell–enriched PBD-dependent interactions. The normal and cancer-enriched targets are drawn in different sizes in (B) and (C) to indicate altered levels of PBD-dependent interactions between normal and cancer cells.
Table 3.
Plk1 kinase domain inhibitors
| Compound (chemical class) | Structure | Mode of action | IC50/Selectivity | References |
|---|---|---|---|---|
| PRECLINICAL | ||||
| MLN0905 (Benzolactam derivative) | ATP-competitive inhibitor | Plk1 IC50 = 2 nM Plk2 & Plk3 IC50 = ND |
[83] | |
| RO3280 | ATP-competitive inhibitor | Plk1 IC50 = 3 nM Plk2 & Plk3 IC50 = ND |
[84] | |
| ZK-thiazolidinone | ATP-competitive inhibitor | Plk1 IC50 = 19 ± 12 nM Plk2 & Plk3 IC50 < 100nM |
[85] | |
| SBE13 | Non ATP-competitive inhibitor | Plk1 IC50 = 0.2 nM Plk2 IC50 > 66 μM Plk3 IC50 = 875 nM >4000-fold selectivity over Aurora A |
[86] | |
| Cyclapolin 1 (benzothiazole N-oxide) | Putative covalent binding with a residue in the ATP-binding cleft | Plk1 IC50 < 20 nM Plk2 & Plk3 IC50 = ND |
[87] | |
| DAP-81 (diaminopyrimidine (DAP) derivative) | Predicted to target the nucleotide pocket | Plk1 IC50 = 900 nM Plk2 & Plk3 IC50 = ND |
[88] | |
| Compound 36 (imidazopyridine derivative) | Not determined | Plk1 IC50 = 9.8 nM Plk2 IC50 = 21 nM Plk3 IC50 = 178 nM |
[89] | |
| CLINICAL | ||||
| GSK461364 (thiophene derivative) | ATP-competitive inhibitor | Plk1 Kiapp < 0.5 nM Plk2 Kiapp = 860 nM Plk3 Kiapp = 1000 nM |
[90] | |
| NMS-P937 (pyrazoloquinazoline scaffold) | ATP-competitive inhibitor | Plk1 IC50 = 2 nM Plk2 IC50 > 10,000 nM Plk3 IC50 > 10,000 nM Selectivity against a panel of > 250 kinases |
[91,92] | |
| TAK-960 | ATP-competitive inhibitor | Plk1 IC50 = 0.8 nM Plk2 IC50 = 16.9 nM Plk3 IC50 = 50.2 nM |
[93] | |
| BI 2536 (dihydropteridinone derivative) | ATP-competitive inhibitor | Plk1 IC50 = 0.83 nM Plk2 IC50 = 3.5 nM Plk3 IC50 = 9.0 nM ~1000-fold selectivity against Tyr & Ser/Thr kinases |
[78] | |
| BI 6727 (dihydropteridinone derivative) | ATP-competitive inhibitor | Plk1 IC50 = 0.87 nM Plk2 IC50 = 5 nM Plk3 IC50 = 56 nM No inhibition against > 50 kinases |
[94] | |
| ON 01910 (benzylstyryl sulphone) | Affects microtubule dynamics | Plk1 IC50 = 9 nM Plk2 IC50 = 260 nM Plk3 > 10,000 nM |
[95] | |
The PBD has drawn a lot of attention as an alternative target for anti-Plk1 drug discovery. The early finding that a five-residue-long phosphopeptide binds to the Plk1 PBD with high affinity and specificity [82] clearly demonstrated that the Plk1 PBD contains a small and unique binding cleft that can be probed for drug discovery. Consistent with this finding, several small-molecule and peptide-derived inhibitors have been developed and examined in various preclinical studies (Table 4). Although PBD inhibitors are at the early stages of development, the specific nature of PPIs allow them to be highly specific and, therefore, more amenable for combination therapy with less toxicological problems. Unlike the ATP-binding site of Plk1 KD that binds to ATP as the only ligand, the Plk1 PBD binds to a large spectrum of target proteins with different degrees of binding affinities. These differences make it feasible to develop PBD inhibitors that can be selective against a set of PBD-binding targets. ATP analog inhibitors impede Plk1 catalytic activity equally in both normal and cancer cells (Figure 3B and 3C), although the outcome of the same degree of Plk1 inhibition could be more severe in cancer cells than normal cells because of the cancer cells’ addiction to Plk1 (Figure 2). Since PBD-mediated biochemical pathways could be altered in cancer cells as a result of their addiction to Plk1, selectively interfering with cancer cell–enriched PBD-binding target(s) could serve as a promising strategy for imposing an additional layer of cancer cell–selective killing effect (Figure 3C).
Table 4.
Plk1 PBD domain inhibitors
| Compound (chemical class) | Structure | Mode of action | IC50/Selectivity | References |
|---|---|---|---|---|
| PRECLINICAL | ||||
| Thymoquinone | Inhibits PBD-dependent binding & subcellular localization | Plk1 IC50 = 1.14 ± 0.04 μM Plk2 IC50 = 1.9 ± 0.1 μM Plk3 IC50 = 22.4 ± 0.8 μM |
[109] | |
| Poloxin (thymoquinone derivative) | Inhibits in vitro PBD-dependent binding & subcellular localization | Plk1 IC50 = 4.8 ± 1.3 μM Plk2 IC50 = 18.7 ± 1.8 μM Plk3 IC50 = 53.9 ± 8.5 μM |
[110] | |
| Poloxipan | Pan inhibitor of Plk1–3 PBDs | Plk1 IC50 = 3.2 ± 0.3 μM Plk2 IC50 = 1.7 ±0.2 μM Plk3 IC50 = 3.0 ± 0.1 μM |
[110] | |
| Purpurogallin (benzotropolone-containing compound) | Inhibits PBD-dependent binding in vitro and in vivo | Plk1 IC50 < 0.3 μM inhibits PBD of PLK2 but not of PLK3 |
[111] | |
| 4j derivatives | Inhibits PBD-dependent binding in vitro | Plk1 IC50 4j = 3 nM (320 μM)† 4j* = 3 nM (85 μM)† 3c = 1 nM (85 μM)† 4c = 2 nM (55 μM) |
[106,112] | |
ND, Not determined; EC50, effector concentration for half-maximum response; IC50, half-maximal inhibitory concentration; PBD, polo box domain; PLK, polo-like kinase.
Extracellular concentration required to achieve 50% mitotic block in cultured HeLa cells.
Targeting the catalytic domain of Plk1
Targeting the catalytic activity of a protein kinase has long been the prevailing approach to generating kinase inhibitors. Since the catalytic domain furnishes a defined target site, such as an ATP-binding pocket, and inhibitors are designed to annihilate the catalytic activity, examining biochemical and biological effects of this class of inhibitors is relatively straightforward, thus making it possible to put the inhibitors forward quickly for advanced preclinical and/or clinical studies.
Molecular architecture of the ATP-binding sites of Plks
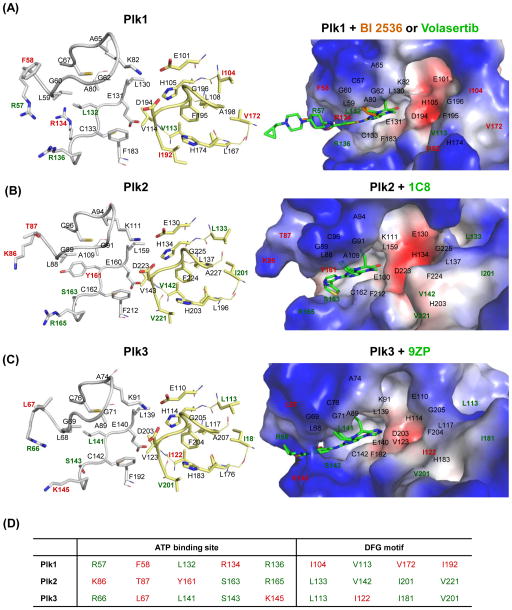
ATP-binding pockets found in various kinases are shown to be highly conserved [1]. Not surprisingly, analyses of the crystal structures of the catalytic domains for Plk1–3 show that the overall architectures of their ATP-binding sites are similar to one another (Figure 4A–C; gray stick models). However, among the residues lining the ATP pockets of the Plk1, F58, and R134 residues are unique to Plk1, while the R57, L132, and R136 residues are found only in two of the ATP pockets of Plk1–3 (Figure 4D). The hinge region residue, L132, is generally conserved as either a Tyr or Phe residue, and R136 is frequently a Gly residue in non-Plks [53, 96]. Moreover, in non-Plks, C67 in the roof of the ATP-binding pocket is commonly replaced with Val, while F183 at the bottom of the pocket is replaced with either Leu or Met at its respective position [53, 96, 97].
Figure 4.
The mode of small-molecule inhibitors binding to the ATP-binding site of Plk1–3. (A) The Plk1 catalytic domain with BI 2536 (brown; PDB 2RKU) [53] and volasertib (green; PDB 3FC2) [94]; (B) the Plk2 catalytic domain with 1C8 (PDB 4I6H), [152]; and (C) the Plk3 catalytic domain with 9ZP (PDB 4B6L) are shown. (Left) The active sites of Plks are displayed in stick models in the left panel. Residues establishing interactions with the ligands are displayed in gray stick models. Residues at and around the DFG motif are shown in yellow stick models. (Right) The binding modes of specific ligands are shown in electrostatic surface representations. Residues specific to each Plk are indicated in red, while residues present in two of Plk1–3 are shown in green. The catalytic domain of Plk4 is excluded because of a low level of similarity to Plk1–3. (D) Unique residues in each Plk isoform are indicated in red, while residues overlapped in two of Plk1–3 are denoted in green.
Notably, the residues around the DFG motif of Plk1–3 are somewhat less conserved than those around the ATP-binding pocket (Figure 4A–C; yellow stick models). Plk1 is activated by a phosphorylation at T210 in the activation loop [98], present just downstream of the conserved DFG motif (residues 194–196), suggesting that T210 phosphorylation may influence the allosteric switching of the DFG motif. Interestingly, a recent report suggested that Plk1 can be trapped in a DFG-out (i.e., the DFG motif is flipped out, creating an allosteric hydrophobic pocket) and catalytically inactive conformation, [86], which has been observed in many Tyr kinases and a few Ser/Thr kinases [99–102]. On the contrary, the crystal structure of a catalytically inactive Plk1 T210V mutant has been shown to exhibit a DFG-in conformation, in which the F195 is buried in the hydrophobic pocket [96]. Therefore, whether an inactive Plk1 can adopt a DFG-out confirmation under specific conditions and whether a less-conserved, DFG-out allosteric pocket can be utilized for anti-Plk1 drug discovery remain to be investigated (see “New strategies” below).
Currently available inhibitors targeting the catalytic activity of Plk1
Most Plk1 KD inhibitors are targeting the ATP-binding site, while a few are reported to target the allosteric- or substrate-binding site within Plk1 KD [86, 95]. Although most of these inhibitors perform reasonably well in preclinical studies, only a handful of them have reached clinical trials. The list of Plk1 KD inhibitors developed to date is shown in Table 3. To address some of the key issues important for anti-Plk1 drug discovery, here we have chosen some of the advanced Plk1 inhibitors to discuss the current status of their development and to diagnose potential problems that need to be tackled to improve their efficacies.
BI 2536 is a dihydropteridinone-derived ATP-competitive inhibitor [78, 103] that has been widely used in both preclinical and clinical studies [5, 6]. BI 2536 inhibits Plk1 efficiently (IC50 = 0.83 nM), and cross inhibits closely related Plk2 (IC50 = 3.5 nM) and Plk3 (IC50 = 9 nM) somewhat less efficiently [78]. BI 2536 was reported to exhibit a high degree of selectivity against a panel of 63 other kinases tested. Subsequent studies revealed, however, that BI 2536 inhibits several other kinases as well [104] and antagonizes the function of bromodomains present in diverse nuclear proteins regulating transcription and chromatin modulation [105]. Analysis of an X-ray co-crystal structure revealed that the binding mode of BI 2536 to Plk1 KD occurs through multiple hydrogen bonds with R57, L59, and C133 residues [53]. BI 2536 also makes contact with various other surrounding residues, including C67, L130, L132, R136, and F183. These multiple interactions collectively contribute to BI 2536’s binding affinity and specificity to Plk1. Although it exhibits an impressive antitumor efficacy in preclinical studies, BI 2536 monotherapy induced only weak clinical efficacy with significant adverse effects, such as neutropenia and leukopenia. Because of these dose-limiting side effects during clinical trials, monotherapy with BI 2536 was terminated during the Phase II study, although a combinatorial study is still underway for several solid tumors [6, 106, 107].
BI 6727 (volasertib) also belongs to the class of dihydropteridinone derivatives. The mode of interactions between the Plk1 catalytic domain and volasertib appears to be essentially the same as those described above with BI 2536 (Figure 4A). As expected, volasertib exhibits in vitro potency and selectivity, similar to what is exhibited by BI 2536, and also shows excellent antitumor activity in various xenograft models of human cancers [94]. In comparison to BI 2536, however, volasertib exhibits a significantly improved safety and pharmacokinetic profile, and is highly efficacious and potent in multiple preclinical/clinical cancer models [94, 108]. Volasertib has been evaluated under Phase III clinical trials and shows the most promising clinical efficacy against acute myeloid leukemia (AML), for which it received a breakthrough therapy designation from the USA FDA in 2013. Further examination is necessary to determine whether cancers with activated RAS mutations or without functional TP53 alleles are highly sensitive to volasertib treatment, as has been previously demonstrated with BI 2536 [68, 72].
GSK461364A is a thiophene-derived compound that is shown to competitively inhibit the ATP-binding site of Plk1 with a high potency [90, 109]. Remarkably, it shows an enhanced antitumor effect in cancer cells where p53 is downregulated or mutated [69]. Given that a large fraction of human cancers bear inactivating mutations in the TP53 gene, this finding is particularly important. Studies with GSK461364A demonstrated that targeting Plk1 is an effective therapeutic approach against undifferentiated thyroid carcinoma cells [110]. In the clinical Phase I study, however, GSK461364A induced dose-limiting toxicities, such as neutropenia, thrombocytopenia, and venous thrombotic emboli [111]. Although toxicities like neutropenia are often seen with anti-mitotic microtubule-targeting agents, such as taxanes or vinca alkaloids, the multiple side effects of GSK461364A hint that it may also inhibit other unintended cellular proteins. For advanced patient studies, the specificity of GSK461364A may have to be improved.
RO3280, a pyrimidodiazepine-derived compound, exhibits high in vitro cellular potency in multiple leukemia cell lines and a significant level of antitumor activity in xenograft mouse models. Although RO3280 is still at the preclinical stage of development, it appears to have a potential therapeutic value for AML [84, 112].
ON01910 is a substituted benzyl styryl sulfone that was initially reported to antagonize Plk1 efficiently (IC50 = 9–10 nM) by inhibiting the substrate-binding site of the enzyme in a non-ATP-competitive manner [95]. However, later studies showed that it possesses only weak anti-Plk1 activity (IC50 = ~30 μM) and inhibits multiple other kinases, including PI3K [78, 88, 113, 114]. The exact mechanism of action for ON01910 remains unknown. Nevertheless, ON01910 exhibits potent tumor inhibitory activity in xenograft nude mouse models, and its efficacy is currently being examined under Phase I clinical trials.
HMN-214 is a prodrug of stilbene-derived HMN-176 and is proposed to alter Plk1 function [115]. HMN-214 is currently under Phase I clinical trials [115, 116]. However, whether it is a bona fide Plk1 inhibitor and, if so, how it inhibits Plk1 need to be clarified.
SBE13 is a dimethoxy-benzeneethanamine derivative that was proposed to lock Plk1 in the inactive DFG-out conformation. SBE13 inhibits Plk1 with an IC50 value of 0.2 nM, whereas it inhibits Plk2 and Plk3 only marginally (IC50 values of 66 μM for Plk2 and 875 nM for Plk3) [86]. It has been speculated that this superb Plk1 selectivity stems from the specific binding of SBE13 to the DFG-out-induced allosteric pocket [86]. However, whether SBE13 indeed binds to Plk1’s DFG-out conformation has not been verified at present.
New strategies
Less than acceptable dose-limiting toxicity appears to be one of the major problems commonly associated with currently available Plk1 inhibitors. Since dose-limiting toxicity is often raised as a consequence of non-specific activity of the inhibitors [117], improving Plk1 specificity is one of the pressing issues to be addressed prior to advanced clinical studies. Notably, Plk1 KD inhibitors commonly exhibit a substantial level of inhibitory activities against Plk2 and Plk3, which are required for stress response and survival [22, 28] and tumor suppression [29, 37, 81], respectively. In addition, it has been shown that BI 2536 and, to a lesser degree, volarsertib inhibit pro-apoptotic death-associated protein kinase 2 (DAPK-2) [104]. Inhibiting DAPKs would negate the effect of Plk1 inhibition in inducing mitotic arrest and apoptotic cell death. Therefore, specific inhibition of Plk1 is necessary for an effective treatment of Plk1-addicted cancers.
Examining the crystal structures of Plk1–3 in complex with their cognate inhibitor compounds (volasertib, a dihydropteridinone derivative IC8, and a pteridin derivative 9ZP, respectively) shows that all three inhibitors bind to their corresponding ATP-binding site in an overlapping manner (Figure 4A–C). Among the residues lining the ATP-binding pockets of Plk1–3, only the F58 and R134 residues (highlighted in red in Figure 4D) are uniquely found in Plk1, thus making it particularly challenging to generate Plk1-specific inhibitors. Analysis of the two co-crystal structures obtained with either BI 2536 [53] or volasertib [94] revealed that, in addition to the R57, L59, and C133 residues that both compounds directly interact with, other residues, such as C67, L130, L132, R136, and F183, are also in contact with these compounds. Among these residues, the R57 and L132 residues are present in both Plk1 and Plk3 (Figure 4D), and C67, L132, and F183 residues are somewhat selective against non-Plks. The F58 and R134 residues specific to the Plk1 ATP-binding site are not involved in the interaction with either BI 2536 or volasertib. These findings partially explain why BI 2536 and volasertib are less discriminatory among Plk1–3, while they are selective against other kinases [78]. Developing strategies to utilize the Plk1-specific and -semispecific residues will be important for achieving a high level of Plk1 specificity.
Another possibility for improving specificity would be to utilize the structural plasticity of the catalytic domains and develop a new class of inhibitors that exploits a less-conserved allosteric pocket of a DFG-out conformation [99]. Intriguingly, recent reports suggest that Plk1 adopts a catalytically-inactive DFG-out conformation and SBE13 binds to this inactive form with outstanding Plk1 specificity [86, 118]. At present, the exact nature of SBE13 binding to Plk1 KD is not known. Since the I104, V172, and I192 residues are uniquely found around the conserved DFG motif of Plk1 (Figure 4D), investigating whether any of these residues interact with SBE13 and exploring how SBE13 can be further developed as a novel therapeutic agent will be worthwhile.
Targeting the PBD of Plk1
Although blocking PPIs affords an attractive strategy for specifically modulating biological processes, the development of PPI inhibitors can be made challenging by the extended, nondescript nature of most protein interfaces. These limitations can be overcome when significant contributions to overall protein-binding energies are made by confined “hot spot” regions that are amenable to targeting by small molecules [119]. Plk1 PBDs fall within this latter category because they engage in PPIs that can be disrupted with high affinity and specificity by compact motifs [47, 82, 120]. This suggests that the PBD may represent an alternate target to the KD for developing highly specific Plk1 inhibitors.
The molecular architecture of the PBDs
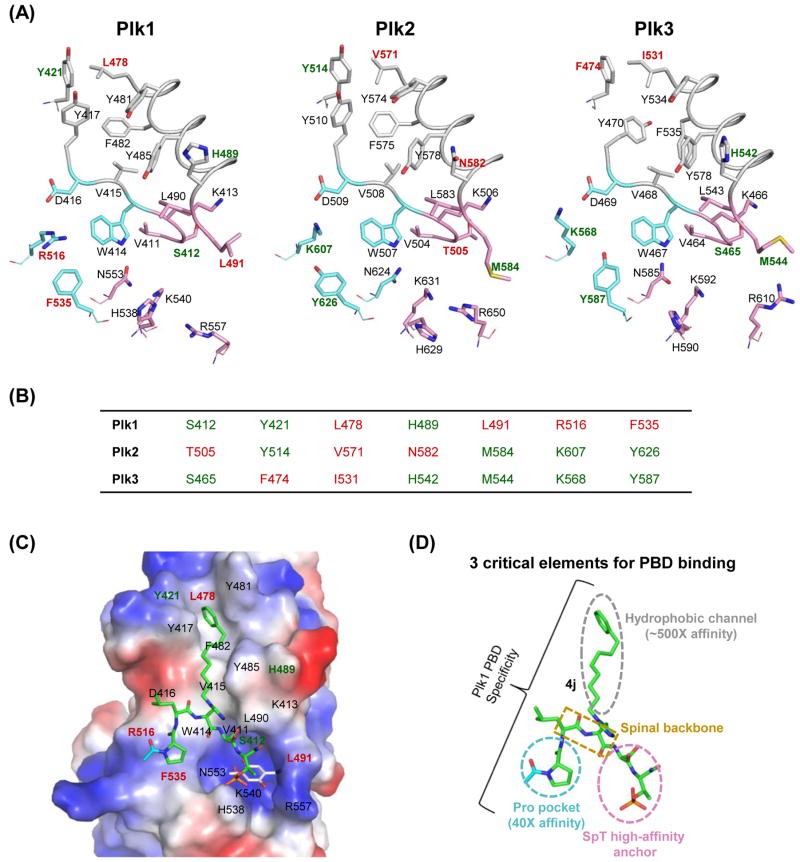
The PBDs of Plk1–3 function as phospho-recognition modules [47]. Although the overall protein sequences and domain folds are similar among the three PBDs, and only a small number of residues are specific to each (Figure 5A–B), the domains interact with their cognate pS/pT-binding targets in a highly specific manner [58, 59, 82]. In the case of the Plk1 PBD, four residues (L478, L491, R516, and F535) are uniquely found proximal to the phospho-binding pocket. This suggests that these residues may play important roles in conferring Plk1 PBD–binding specificity. Studies examining the interaction between the Plk1 PBD and the specific peptide ligand, PLHSpT, have revealed the key roles played by R516 and F535 [82]. Additionally, L478, which is located at the terminus of a hydrophobic channel, appears to be important for induced-fit interactions with an alkylphenyl group on the PLHSpT-derived peptide 4j [120] (Figure 5C). In the absence of co-crystal structures of the Plk2 and Plk3 PBDs bound with their cognate ligands, specificity-determining interactions of these domains remain unknown [121, 122].
Figure 5.
A structural comparison of Plk1–3 PBDs and the binding mode of Plk1 PBD with its ligands. (A) The apo structures of three Plk1–3 PBDs are represented in stick models: Plk1 PBD (PDB 1UMW), Plk2 PBD (PDB 4RS6), and Plk3 PBD (a model generated using the Robetta server [153]). The PBDs are arbitrarily divided into three regions based on the elements required for 4j binding to Plk1 PBD: a Pro pocket (cyan), a phospho-binding pocket (pink), and a hydrophobic channel (gray). The residues specific to each Plk are shown in red, and the overlapped residues in two of Plk1–3 are indicated in green. (B) Unique residues in each Plk are indicated in red, while residues overlapped in two of Plk1–3 are denoted in green. (C) 4j (green) bound to Plk1 PBD (PDB 3RQ7; [120]) is displayed in an electrostatic surface representation, with overlaid thymoquinone (gray) bound to Plk1 PBD (PDB 4H71; [130]). (D) A schematic diagram showing three elements that are critical for Plk1 PBD binding. The Pro pocket and the hydrophobic channel have been shown to increase the Plk1 PBD-binding affinity ~40-fold and ~500-fold, respectively [82, 120]. The critical role of the SpT anchor has been demonstrated previously [47, 82]. The spinal backbone of 4j is indicated in a dotted rectangular box.
Current inhibitors targeting the PBD of Plk1
As an alternative approach to developing KD inhibitors, a great deal of effort has been made to develop inhibitors that target the PBD. Table 4 summarizes examples of small molecules and peptide mimetics that have been reported to inhibit the function of the Plk1 PBD, as measured through in vitro binding and cell-based assays. Notably, the phosphopeptide-binding interface of the PBD is shallow, and high-affinity binding relies heavily on interactions of a di-anionic phosphoryl moiety in the ligand with protein H538 and K540 residues. This places challenges on the development of small-molecule inhibitors.
Poloxin is a thymoquinone derivative that was identified through a screening campaign designed to isolate compounds capable of disrupting the Plk1 PBD–dependent interaction with its cognate peptide ligand in vitro. Poloxin is reported to inhibit Plk1 PBD binding with a moderate level of affinity (an IC50 value of 4.8 μM) and exhibit several-fold selectivity over the Plk2 and Plk3 PBDs [123]. The compound also shows anti-proliferative effects against a panel of cancer cells, and it suppresses tumor growth in xenograft mouse models [127]. Although Poloxin was suggested to bind to the Plk1 PBD irreversibly [128], whether its cellular effects stem from Plk1 inhibition remain to be clarified. As an iminoquinone, poloxin can potentially undergo Michael-type additions with biological thiols in cellular proteins [129]. Although this has been ruled out as a mechanism for the covalent PBD binding of poloxin [128], the ability to alkylate biological nucleophiles may cause an unacceptable level of cytotoxicity in vivo.
Poloxipan (“polo-box domain inhibitor, pan-specific”) (Table 4) is a benzylidene-thiazolotriazenedione that has been shown to inhibit the three PBDs of Plk1–3 with similar IC50 values of ~2–3 μM [124]. Although the manners in which poloxin and poloxipan bind to the Plk1 PBD are not known, the Plk1 PBD co-crystal complexes bound with the structurally similar thymoquinone and the oxime fragment of poloxin (called, poloxime), have been determined [130]. Interestingly, thymoquinone (gray; Figure 5C) binds within the phospho-recognition site, where it appears to interact with K540, which is critical for phosphoryl interactions. This suggests that thymoquinone could potentially serve as a template to substitute for the role of the SpT dipeptide. However, thymoquinone interacts poorly with other features that have been shown to be important for phospho-dependent PBD recognition.
The natural product benzotropolone derivative, purpurogallin (PPG; Table 4), inhibits Plk1 PBD–dependent interactions with an IC50 value of 0.3 μM in vitro, while efficiently inducing mitotic block and apoptosis in cultured cells [125]. PPG contains a hydroxyl group in the 2-position of its benzocycloheptenone ring system that appears to be required for PBD binding [131]. Although PPG shows greater PBD-binding affinity in vitro than thymoquinone-derived inhibitors, it exhibits pleiotropic effects against numerous targets that limit its therapeutic potential.
The ligand-binding surfaces of the Plk1 PBD have been explored extensively using PBD-binding peptides and peptide mimetics. To date, the highest affinity analogs have resulted from a development campaign starting with the PLHSpT sequence. These agents are characterized by their inclusion of alkylphenyl groups originating from various positions on the peptide chain [120, 132, 133]. Among this family of inhibitors, 4j (Table 4), which has an alkylphenyl group on the π-nitrogen of its His imidazole ring, exhibits a Plk1 PBD–binding affinity of Kd = 2 nM [120]. Structural analysis has shown that the alkylphenyl moiety of 4j (green) binds within a hydrophobic channel of the Plk1 PBD that is normally occluded in the unbound state (Figure 5C) [120]. Plk1 PBD co-crystal structures of other low-nanomolar-affinity peptide derivatives indicate that alkylphenyl groups originating from regions of the peptide more amino-distal than the His residue can access the cryptic pocket [132, 133]. The presence of this binding channel has been confirmed by co-crystal structures of longer PLHSpT-containing peptides and their derivatives [134–136]. It has also been found that replacing the His residue in PLHSpT with a variety of alkylphenyl-containing non-proteinogenic amino acids can enhance Plk1 PBD–binding affinity [137]. However, significant loss of binding affinity is incurred by shortening the PLHSpT sequence to HSpT, indicating that interactions provided by the amino-terminal region of the peptide are also important [82]. Optimization of the N-terminal interactions has been attempted to improve the affinity of the parent sequence by introducing bulky hydrophobic groups into the N-terminal Pro residue [138].
Although PLHSpT derivatives have provided a wealth of information for understanding binding surfaces of the Plk1 PBD (Figure 5D), the direct use of phosphopeptides for in vivo applications is limited by their poor bioavailability, which arises from reduced membrane transit of di-anionic phosphoryl species and the hydrolytic lability of phosphate esters in the presence of intracellular phosphatases. This presents a conundrum, because the phospho group plays a key role in overall binding affinity. In order to address this problem, peptide 4j was selected, due to its high Plk1 PBD–binding affinity and its selectivity, and the fact that its alkylphenyl group is located on the His residue, which is situated proximal to the minimal key “SpT” recognition motif [120]. It was found that, while the replacement of the pThr residue in 4j with non-phosphorus-containing acidic residues results in a dramatic loss of Plk1 PBD–binding affinity, use of the phosphonic acid–based pThr mimetic, Pmab [139], can be accomplished with complete retention of binding affinity [126, 140]. Whole-cell studies with the PEGylated Pmab-containing version of 4j (designated as 4j*) showed that effective mitotic block in cultured HeLa cells could be achieved, but only at high extracellular concentrations (IC50 = 320 μM), indicating poor cellular uptake [120, 126]. In further work, it was found that selective on-resin alkylation of the His N(τ) position of 4j provided bis-alkylated peptides that retained full Plk1 PBD–binding affinity (3c, IC50 = 1 nM; Table 4) [126, 141]. This is noteworthy since His bis-alkylation improves cellular efficacy in cultured HeLa cells (3c, IC50 = 85 μM for mitotic block), presumably by neutralizing (“charge masking”) one of the phosphoryl anionic charges arising from the cationic imidazolium moiety in the His residue [126]. Further improvement of cellular efficacy was gained by converting one phosphoryl hydroxyl of 3c to its neutral bio-reversible pivaloyloxymethyl ester (4c, IC50 = 55 μM for mitotic block in cultured HeLa cells; Table 4) [126].
New strategies
Ample data suggests that PBD-directed agents constitute a fascinating and strategically new approach to Plk1 inhibitors. Although substantial progress has been made in this area, to date, low–molecular weight compounds exhibit suboptimal levels of PBD-binding affinity [131]. By contrast, peptide and peptide mimetic–based inhibitors, including 4j, exhibit high Plk1 PBD–binding affinities in assays in vitro [120, 132, 133], yet many of these inhibitors suffer from poor membrane permeability and low bioavailability in cell-based assays [120, 126]. Nevertheless, studies with a variety of 4j-based inhibitors have identified three structural elements that are critical for high affinity and specificity (Figure 5D). These elements include 1) an “SpT” high-affinity anchor pocket; 2) a region accessed by the N-terminal Pro residue that facilitates Plk1 PBD–binding affinity and specificity; and 3) a hydrophobic channel that is capable of enhancing the binding affinity ~500–1000 fold with undiminished specificity (Figure 5D). The future challenge will be to develop inhibitors that combine good cellular uptake and intracellular stability with high Plk1 PBD–binding affinity and selectivity. Because the canonical phosphopeptide-binding cleft is shallow, it may be difficult to achieve adequate affinity and selectivity with non-covalent inhibitors whose interactions are confined to the pThr-binding pocket. It will probably be necessary to access more extensive regions of the protein that take advantage of Plk1-specific residues (L478, L491, R516, and F535; Figure 5A) and maximize affinity-inducing interactions (Figure 5D). Ideally, this should be accomplished while minimizing overall molecular size and charge. It is easy to imagine that initial leads for such inhibitors may arise from high-throughput screens or from minimal phosphopeptide mimetics. Plk1 PBD co-crystal structures of these low–molecular weight compounds could then be used as the basis for further optimization of biological properties. To date, high-affinity peptides and peptide mimetics have included either a phosphoryl- or phosphonic acid–containing component, and such a requirement may carry over into viable low–molecular weight inhibitors. Should this be the case, minimizing anionic charge through intramolecular charge masking or bio-reversible prodrug protection (as exemplified by 3c and 4c; Table 4) may be needed to yield membrane-permeable agents that retain high intracellular Plk1 PBD–binding affinities. Macrocyclization offers a potential means of advancing peptide leads. While head-to-tail cyclized analogs based on PLHSpT and its N-alkyl-His derivative 4j have yielded Plk1-specific peptomer ligands with low micromolar IC50 values [142], more recent mono-anionic macrocyclic peptide mimetics based on peptides such as 3c (Table 4) have been reported, which utilize a cationic bis-alkylated His residue as the ring-closing junction. Certain of these analogs retain low-nanomolar Plk1 PBD-binding affinities [143]. An alternative approach may be found by tethering small-molecule PBD-binding compounds with agents that interact within the KD. If successful, the resulting bivalent constructs could exhibit substantial increases in overall affinity and selectivity relative to either monomeric component [144].
Treatment strategies for Plk1 inhibitors
Developing a proper dosing and administration regimen is very important for achieving the maximum level of efficacy for each therapeutic agent. Plk1 inhibitors have been administered either as monotherapy or in combination with other already-approved anti-cancer agents. Although monotherapy can provide multiple advantages, such as simple drug regimens, absence of drug–drug interactions, and fewer adverse events, combination therapy can improve drug efficacy even at lower doses of each agent, thus maintaining a low toxicity profile. Volasertib has been extensively tested either as monotherapy or in combination with other therapeutics to treat hematologic malignancies, advanced solid tumors, and multiple other types of cancers [145–147]. Since combination therapy generally targets more than one pathological process, it can be more effective and can help reduce drug resistance. To identify effective regimens for combination therapy against Plk1-addicted cancers, more systematic investigation has to be carried out with various chemotherapeutic agents. In the event that both Plk1 catalytic and PBD inhibitors are available, they can be used either alone or together to reduce their dose levels.
While employing an effective drug treatment strategy is important, selecting a cohort of patients suitable for the anti-Plk1 regimen will be equally critical to achieving the maximum possible benefit. Cancer cells with an inactivated TP53 or activated KRAS mutation appear to be more sensitive to Plk1 inhibition [67–72], suggesting that anti-Plk1 therapy may be more effective in patients with one or both of these mutations. It has been suggested that p53 inactivation confers a selective advantage for clonal outgrowth during AML progression [148–150]. Since the effect of Plk1 inhibition is synergized by the loss of TP53 in a mouse tumor model [70] and Plk1 overexpression is closely associated with TP53 mutations in primary human breast cancer [71], anti-Plk1 therapy appears to be a particularly promising method of treating AML with TP53 mutations. Whether an activated RAS mutation also synergizes the effect of Plk1 inhibition has not been closely examined in human cancers [8]. In any case, since TP53 and RAS mutations are highly frequent in a broad spectrum of human cancers, patient stratification by cancer gene profiling will be important for providing treatment guidelines.
Concluding remarks
Anti-mitotic drugs have been the most successful anticancer drugs. However, widely used microtubule-targeting chemotherapeutic agents, such as taxanes and vinca alkaloids, also affect non-dividing cells and tissues. Plk1 is highly expressed in actively dividing cells and tissues, and its overexpression promotes tumorigenesis. More importantly, cancer cells are addicted to a high level of Plk1, and the reversal of Plk1 addiction is sufficient for inducing cancer cell–selective mitotic block and apoptotic cell death. Therefore, the addiction of mitotically active cancer cells to a high level of Plk1 makes Plk1 a discriminating target for anticancer therapy. In line with this notion, the attractiveness of targeting Plk1 has been amply indicated by numerous target validation trials, which have been described above. However, there are still many open questions (Outstanding Questions) that need to be addressed to maximize the effect of anti-Plk1 therapy. For instance, although an impressive number of inhibitors have been tested during preclinical and clinical development, most Plk1 inhibitors developed to date exhibit dose-limiting toxicity, which narrows therapeutic windows for treatment. To overcome this obstacle and to reduce the high attrition rate for anti-Plk1 drug discovery, it seems imperative to increase the specificity against Plk1. To this end, future efforts should be directed towards improving the specificity of currently available ATP-competitive inhibitors and developing clinically applicable PBD inhibitors by exploring Plk1-specific residues surrounding PBD-binding pockets. Unlike the KD inhibitors aimed at annihilating Plk1 catalytic activity in both cancer cells and normal cells equally, PBD inhibitors can be developed in such a way that they inhibit a subset of PBD-dependent interactions enriched in biochemically rewired cancer cells, but not normal cells, thereby imposing an additional layer of cancer cell–selective killing effect. Gaining anti-Plk1 specificity and cancer cell–selective killing are the two important goals that need to be accomplished in order to generate next-generation anti-Plk1 therapeutic agents. A better understanding of the molecular basis of how Plk1 interacts with its binding targets and substrates in normal vs. cancer cells will be fundamentally important for providing new insights into the development of novel strategies that can be explored for future drug discovery against Plk1.
Outstanding Questions.
Can Type II (allosteric) inhibitors be developed against Plk1 by trapping it in a DFG-out, inactive state?
Can PBD inhibitors be developed to specifically interfere with a subset of PBD-dependent interactions vital for cancer cell viability, and therefore exhibit superior selectivity over KD inhibitors in killing cancer cells but not normal cells?
Can one achieve a synergistic drug effect by using Plk1 inhibitors in combination or in sequence with other anti-cancer agents?
How can we determine whether an inhibitor functions effectively against Plk1 in vivo? There is a pressing need to identify biomarkers that could serve as surrogate endpoints to evaluate anti-Plk1 therapeutic activity.
What are the underlying mechanism(s) that sensitizes Plk1 inhibition in cancer cells bearing activated RAS or inactivated TP53 mutations?
Can anti-Plk1 regimen be effective in cancer cells with a genetic background other than RAS or TP53 mutation? Profiling genetic alterations that exhibit synthetic lethality with Plk1 interrogation will be important to provide patient selection guidelines for anti-Plk1 therapy.
TRENDS BOX.
A large body of evidence suggests that Plk1 is an attractive anticancer drug target. Plk1 inhibition appears to be particularly vulnerable in cancer cells bearing an inactivating TP53 or activating RAS mutations because of their addictions to a high level of Plk1.
Plk1 offers two biochemically distinct drug targets—the N-terminal KD and the C-terminal protein-protein interaction domain, called PBD.
A remarkable progress has been made in generating KD inhibitors, yielding several ATP-competitive analogs currently in clinical trials. However, these inhibitors commonly exhibit dose-limiting toxicities by inhibiting other structurally similar kinases.
The PBD has emerged as an alternative target for developing a new class of Plk1 inhibitors. Although PBD inhibitors could confer superb binding affinity and specificity, currently available inhibitors are still at an early stage of development.
Acknowledgments
We would like to thank Klaus Strebhardt for critical reading of the manuscript and many colleagues for generously sharing their views and insights. We apologize to all authors whose work could not be cited due to space limitations. This research was supported in part by the Intramural Research Program of the NIH, NCI (K.S.L. and T.R.B), the Korea Basic Science Institute grant T35418 (J.K.B.), and a Korean Biomedical Scientist Fellowship from the KRIBB Research Initiative Program, Korea Research Institute of Bioscience and Biotechnology, Republic of Korea.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanks SK, et al. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 2.Lahiry P, et al. Kinase mutations in human disease: interpreting genotype–phenotype relationships. Nat Rev Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P, Alessi DR. Kinase drug discovery--what’s next in the field? ACS Chem Biol. 2013;8:96–104. doi: 10.1021/cb300610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melnikova I, Golden J. Targeting protein kinases. Nat Rev Drug Discov. 2004;3:993–994. doi: 10.1038/nrd1600. [DOI] [PubMed] [Google Scholar]
- 5.Garuti L, et al. Polo-like kinases inhibitors. Curr Med Chem. 2012;19:3937–3948. doi: 10.2174/092986712802002455. [DOI] [PubMed] [Google Scholar]
- 6.Yim H. Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs. 2013;24:999–1006. doi: 10.1097/CAD.0000000000000007. [DOI] [PubMed] [Google Scholar]
- 7.Strebhardt K, et al. Thoughts on the current assessment of Polo-like kinase inhibitor drug discovery. Expert Opin Drug Discov. 2015;10:1–8. doi: 10.1517/17460441.2015.962510. [DOI] [PubMed] [Google Scholar]
- 8.Yim H, Erikson RL. Plk1-targeted therapies in TP53- or RAS-mutated cancer. Mutat Res Rev Mutat Res. 2014 doi: 10.1016/j.mrrev.2014.02.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 10.Barr FA, et al. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 11.Petronczki M, et al. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Zitouni S, et al. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 13.Elia AE, et al. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 14.Hudson JW, et al. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol. 2001;11:441–446. doi: 10.1016/s0960-9822(01)00117-8. [DOI] [PubMed] [Google Scholar]
- 15.Seong YS, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–32293. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- 16.de Carcer G, et al. Plk5, a polo box domain-only protein with specific roles in neuron differentiation and glioblastoma suppression. Molecular and cellular biology. 2011;31:1225–1239. doi: 10.1128/MCB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung GC, et al. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 18.Kishi K, et al. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnaud L, et al. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- 20.Golsteyn RM, et al. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KS, et al. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns TF, et al. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthew EM, et al. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007;6:2571–2578. doi: 10.4161/cc.6.20.5079. [DOI] [PubMed] [Google Scholar]
- 24.Bahassi eM, et al. Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene. 2002;21:6633–6640. doi: 10.1038/sj.onc.1205850. [DOI] [PubMed] [Google Scholar]
- 25.Xie S, et al. Genotoxic stress-induced activation of Plk3 is partly mediated by Chk2. Cell Cycle. 2002;1:424–429. doi: 10.4161/cc.1.6.271. [DOI] [PubMed] [Google Scholar]
- 26.Xie S, et al. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem. 2001;276:43305–43312. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 27.Xie S, et al. Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. J Biol Chem. 2001;276:36194–36199. doi: 10.1074/jbc.M104157200. [DOI] [PubMed] [Google Scholar]
- 28.Xie S, et al. Regulation of cell cycle checkpoints by polo-like kinases. Oncogene. 2005;24:277–286. doi: 10.1038/sj.onc.1208218. [DOI] [PubMed] [Google Scholar]
- 29.Helmke C, et al. The role of Plk3 in oncogenesis. Oncogene. 2015 doi: 10.1038/onc.2015.105. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Seeburg DP, et al. Polo-like kinases in the nervous system. Oncogene. 2005;24:292–298. doi: 10.1038/sj.onc.1208277. [DOI] [PubMed] [Google Scholar]
- 31.Seeburg DP, Sheng M. Activity-induced Polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci. 2008;28:6583–6591. doi: 10.1523/JNEUROSCI.1853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauselmann G, et al. The polo-like protein kinases Fnk and Snk associate with a Ca(2+)- and integrin-binding protein and are regulated dynamically with synaptic plasticity. EMBO J. 1999;18:5528–5539. doi: 10.1093/emboj/18.20.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habedanck R, et al. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 34.Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Lu LY, et al. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol Cell Biol. 2008;28:6870–6876. doi: 10.1128/MCB.00392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma S, et al. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol Cell Biol. 2003;23:6936–6943. doi: 10.1128/MCB.23.19.6936-6943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, et al. Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1 alpha under hypoxic conditions. Cancer Res. 2008;68:4077–4085. doi: 10.1158/0008-5472.CAN-07-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland AJ, et al. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012;26:2684–2689. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schvartzman JM, et al. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrino R, et al. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- 42.Dai W, et al. PRK, a cell cycle gene localized to 8p21, is downregulated in head and neck cancer. Genes, chromosomes & cancer. 2000;27:332–336. doi: 10.1002/(sici)1098-2264(200003)27:3<332::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 43.Li B, et al. prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J Biol Chem. 1996;271:19402–19408. doi: 10.1074/jbc.271.32.19402. [DOI] [PubMed] [Google Scholar]
- 44.Ko MA, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 45.Macmillan JC, et al. Comparative expression of the mitotic regulators SAK and PLK in colorectal cancer. Annals of surgical oncology. 2001;8:729–740. doi: 10.1007/s10434-001-0729-6. [DOI] [PubMed] [Google Scholar]
- 46.Marina M, Saavedra HI. Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Frontiers in bioscience. 2014;19:352–365. doi: 10.2741/4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elia AE, et al. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 48.Lowery DM, et al. Structure and function of polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 49.Lowery DM, et al. The Polo-box domain: a molecular integrator of mitotic kinase cascades and Polo-like kinase function. Cell Cycle. 2004;3:128–131. [PubMed] [Google Scholar]
- 50.Park JE, et al. Feed-forward mechanism of converting biochemical cooperativity to mitotic processes at the kinetochore plate. Proc Natl Acad Sci USA. 2011;108:8200–8205. doi: 10.1073/pnas.1102020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JE, et al. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67:1957–1970. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng KY, et al. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kothe M, et al. Selectivity-determining residues in Plk1. Chem Biol Drug Des. 2007;70:540–546. doi: 10.1111/j.1747-0285.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, et al. Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol. 2013;20:1047–1053. doi: 10.1038/nsmb.2623. [DOI] [PubMed] [Google Scholar]
- 55.Archambault V, et al. Understanding the Polo Kinase machine. Oncogene. 2015 Jan 26; doi: 10.1038/onc.2014.451. Epub ahead of print Review. [DOI] [PubMed] [Google Scholar]
- 56.Johnson EF, et al. Pharmacological and functional comparison of the polo-like kinase family: insight into inhibitor and substrate specificity. Biochemistry. 2007;46:9551–9563. doi: 10.1021/bi7008745. [DOI] [PubMed] [Google Scholar]
- 57.Park J-E, et al. Direct quantification of polo-like kinase 1 activity in cells and tissues using a highly sensitive and specific ELISA assay. Proc Natl Acad Sci USA. 2009;106:1725–1730. doi: 10.1073/pnas.0812135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reindl W, et al. Development of high-throughput assays based on fluorescence polarization for inhibitors of the polo-box domains of polo-like kinases 2 and 3. Anal Biochem. 2009;395:189–194. doi: 10.1016/j.ab.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 59.Reindl W, et al. A high-throughput assay based on fluorescence polarization for inhibitors of the polo-box domain of polo-like kinase 1. Anal Biochem. 2008;383:205–209. doi: 10.1016/j.ab.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 60.Lee KS, et al. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 62.van Vugt MA, et al. Polo-like kinase 1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 63.Raab M, et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat Commun. 2011;2:395–405. doi: 10.1038/ncomms1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 65.Luo J, et al. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurray HR, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, et al. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sur S, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degenhardt Y, et al. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther. 2010;9:2079–2089. doi: 10.1158/1535-7163.MCT-10-0095. [DOI] [PubMed] [Google Scholar]
- 70.Guan R, et al. Small interfering RNA-mediated Polo-like kinase 1 depletion preferentially reduces the survival of p53-defective, oncogenic transformed cells and inhibits tumor growth in animals. Cancer research. 2005;65:2698–2704. doi: 10.1158/0008-5472.CAN-04-2131. [DOI] [PubMed] [Google Scholar]
- 71.King SI, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast cancer research: BCR. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knecht R, et al. PLK (polo-like kinase), a new prognostic marker for oropharyngeal carcinomas. Int J Cancer. 2000;89:535–536. [PubMed] [Google Scholar]
- 74.Knecht R, et al. Prognostic significance of polo-like kinase (PLK) expression in squamous cell carcinomas of the head and neck. Cancer Res. 1999;59:2794–2797. [PubMed] [Google Scholar]
- 75.Strebhardt K, et al. Prognostic value of polo-like kinase expression in melanomas. JAMA. 2000;283:479–480. doi: 10.1001/jama.283.4.479. [DOI] [PubMed] [Google Scholar]
- 76.Kneisel L, et al. Expression of polo-like kinase (PLK1) in thin melanomas: a novel marker of metastatic disease. J Cutan Pathol. 2002;29:354–358. doi: 10.1034/j.1600-0560.2002.290605.x. [DOI] [PubMed] [Google Scholar]
- 77.Elez R, et al. Tumor regression by combination antisense therapy against Plk1 and Bcl-2. Oncogene. 2003;22:69–80. doi: 10.1038/sj.onc.1206038. [DOI] [PubMed] [Google Scholar]
- 78.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 79.Spankuch-Schmitt B, et al. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–1877. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 80.Nogawa M, et al. Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J Clin Invest. 2005;115:978–985. doi: 10.1172/JCI23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith P, et al. Epigenetic inactivation implies a tumor suppressor function in hematologic malignancies for Polo-like kinase 2 but not Polo-like kinase 3. Cell Cycle. 2006;5:1262–1264. doi: 10.4161/cc.5.12.2813. [DOI] [PubMed] [Google Scholar]
- 82.Yun SM, et al. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat Struct Mol Biol. 2009;16:876–882. doi: 10.1038/nsmb.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duffey MO, et al. Discovery of a potent and orally bioavailable benzolactam-derived inhibitor of Polo-like kinase 1 (MLN0905) Journal of medicinal chemistry. 2012;55:197–208. doi: 10.1021/jm2011172. [DOI] [PubMed] [Google Scholar]
- 84.Chen S, et al. Identification of novel, potent and selective inhibitors of Polo-like kinase 1. Bioorg Med Chem Lett. 2012;22:1247–1250. doi: 10.1016/j.bmcl.2011.11.052. [DOI] [PubMed] [Google Scholar]
- 85.Santamaria A, et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol Biol Cell. 2007;18:4024–4036. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keppner S, et al. Identification and validation of a potent type II inhibitor of inactive polo-like kinase 1. ChemMedChem. 2009;4:1806–1809. doi: 10.1002/cmdc.200900338. [DOI] [PubMed] [Google Scholar]
- 87.McInnes C, et al. Inhibitors of Polo-like kinase reveal roles in spindle-pole maintenance. Nature chemical biology. 2006;2:608–617. doi: 10.1038/nchembio825. [DOI] [PubMed] [Google Scholar]
- 88.Peters U, et al. Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat Chem Biol. 2006;2:618–626. doi: 10.1038/nchembio826. [DOI] [PubMed] [Google Scholar]
- 89.Sato Y, et al. Imidazopyridine derivatives as potent and selective Polo-like kinase (PLK) inhibitors. Bioorganic & medicinal chemistry letters. 2009;19:4673–4678. doi: 10.1016/j.bmcl.2009.06.084. [DOI] [PubMed] [Google Scholar]
- 90.Gilmartin AG, et al. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 2009;69:6969–6977. doi: 10.1158/0008-5472.CAN-09-0945. [DOI] [PubMed] [Google Scholar]
- 91.Beria I, et al. NMS-P937, a 4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline derivative as potent and selective Polo-like kinase 1 inhibitor. Bioorganic & medicinal chemistry letters. 2011;21:2969–2974. doi: 10.1016/j.bmcl.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 92.Valsasina B, et al. NMS-P937, an orally available, specific small-molecule polo-like kinase 1 inhibitor with antitumor activity in solid and hematologic malignancies. Molecular cancer therapeutics. 2012;11:1006–1016. doi: 10.1158/1535-7163.MCT-11-0765. [DOI] [PubMed] [Google Scholar]
- 93.Hikichi Y, et al. TAK-960, a novel, orally available, selective inhibitor of polo-like kinase 1, shows broad-spectrum preclinical antitumor activity in multiple dosing regimens. Molecular cancer therapeutics. 2012;11:700–709. doi: 10.1158/1535-7163.MCT-11-0762. [DOI] [PubMed] [Google Scholar]
- 94.Rudolph D, et al. BI 6727, A Polo-like Kinase Inhibitor with Improved Pharmacokinetic Profile and Broad Antitumor Activity. Clin Cancer Res. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 95.Gumireddy K, et al. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer cell. 2005;7:275–286. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 96.Kothe M, et al. Structure of the catalytic domain of human polo-like kinase 1. Biochemistry. 2007;46:5960–5971. doi: 10.1021/bi602474j. [DOI] [PubMed] [Google Scholar]
- 97.Cohen MS, et al. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 100.Pargellis C, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 101.Yang W, et al. Design, synthesis and biological evaluation of bis-aryl ureas and amides based on 2-amino-3-purinylpyridine scaffold as DFG-out B-Raf kinase inhibitors. Eur J Med Chem. 2015;89:581–596. doi: 10.1016/j.ejmech.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 102.Dietrich J, et al. Application of a novel [3+2] cycloaddition reaction to prepare substituted imidazoles and their use in the design of potent DFG-out allosteric B-Raf inhibitors. Bioorg Med Chem Lett. 2010;18:292–304. doi: 10.1016/j.bmc.2009.10.055. [DOI] [PubMed] [Google Scholar]
- 103.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 104.Raab M, et al. Quantitative chemical proteomics reveals a Plk1 inhibitor-compromised cell death pathway in human cells. Cell research. 2014;24:1141–1145. doi: 10.1038/cr.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ciceri P, et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nature chemical biology. 2014;10:305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maire V, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73:813–823. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- 107.Lund-Andersen C, et al. PLK1-inhibition can cause radiosensitization or radioresistance dependent on the treatment schedule. Radiother Oncol. 2014;110:355–361. doi: 10.1016/j.radonc.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 108.Rudolph D, et al. Efficacy and mechanism of action of volasertib, a potent and selective inhibitor of Polo-like kinases, in preclinical models of acute myeloid leukemia. J Pharmacol Exp Ther. 2015;352:579–589. doi: 10.1124/jpet.114.221150. [DOI] [PubMed] [Google Scholar]
- 109.Emmitte KA, et al. Discovery of thiophene inhibitors of polo-like kinase. Bioorg Med Chem Lett. 2009;19:1018–1021. doi: 10.1016/j.bmcl.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 110.Russo MA, et al. The PLK1 inhibitor GSK461364A is effective in poorly differentiated and anaplastic thyroid carcinoma cells, independent of the nature of their driver mutations. Thyroid. 2013;23:1284–1293. doi: 10.1089/thy.2013.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olmos D, et al. Phase I study of GSK461364, a specific and competitive Polo-like kinase 1 inhibitor, in patients with advanced solid malignancies. Clin Cancer Res. 2011;17:3420–3430. doi: 10.1158/1078-0432.CCR-10-2946. [DOI] [PubMed] [Google Scholar]
- 112.Wang NN, et al. Molecular targeting of the oncoprotein PLK1 in pediatric acute myeloid leukemia: RO3280, a novel PLK1 inhibitor, induces apoptosis in leukemia cells. Int J Mol Sci. 2015;16:1266–1292. doi: 10.3390/ijms16011266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bowles DW, et al. Phase I study of oral rigosertib (ON 01910.Na), a dual inhibitor of the PI3K and Plk1 pathways, in adult patients with advanced solid malignancies. Clin Cancer Res. 2014;20:1656–1665. doi: 10.1158/1078-0432.CCR-13-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reddy MV, et al. Discovery of a clinical stage multi-kinase inhibitor sodium (E)-2-{2-methoxy-5-[(2′,4′,6′-trimethoxystyrylsulfonyl)methyl]phenylamino}acetate (ON 01910.Na): synthesis, structure-activity relationship, and biological activity. J Med Chem. 2011;54:6254–6276. doi: 10.1021/jm200570p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garland LL, et al. A phase I pharmacokinetic study of HMN-214, a novel oral stilbene derivative with polo-like kinase-1-interacting properties, in patients with advanced solid tumors. Clin Cancer Res. 2006;12:5182–5189. doi: 10.1158/1078-0432.CCR-06-0214. [DOI] [PubMed] [Google Scholar]
- 116.Takagi M, et al. In vivo antitumor activity of a novel sulfonamide, HMN-214, against human tumor xenografts in mice and the spectrum of cytotoxicity of its active metabolite, HMN-176. Invest New Drugs. 2003;21:387–399. doi: 10.1023/a:1026282716250. [DOI] [PubMed] [Google Scholar]
- 117.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 118.Keppner S, et al. Biological impact of freezing Plk1 in its inactive conformation in cancer cells. Cell Cycle. 2010;9:761–773. doi: 10.4161/cc.9.4.10644. [DOI] [PubMed] [Google Scholar]
- 119.Arkin MR, et al. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chemistry & biology. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu F, et al. Serendipitous alkylation of a Plk1 ligand uncovers a new binding channel. Nature chemical biology. 2011;7:595–601. doi: 10.1038/nchembio.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shan HM, et al. Crystal structure of the polo-box domain of polo-like kinase 2. Biochemical and biophysical research communications. 2015;456:780–784. doi: 10.1016/j.bbrc.2014.11.125. [DOI] [PubMed] [Google Scholar]
- 122.Kim JH, et al. Structural analysis of the polo-box domain of human Polo-like kinase 2. Proteins. 2015 doi: 10.1002/prot.24804. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reindl W, et al. Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem & Biol. 2008;15:459–466. doi: 10.1016/j.chembiol.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 124.Reindl W, et al. A pan-specific inhibitor of the polo-box domains of polo-like kinases arrests cancer cells in mitosis. Chembiochem: a European journal of chemical biology. 2009;10:1145–1148. doi: 10.1002/cbic.200900059. [DOI] [PubMed] [Google Scholar]
- 125.Watanabe N, et al. Deficiency in chromosome congression by the inhibition of Plk1 polo box domain-dependent recognition. J Biol Chem. 2009;284:2344–2353. doi: 10.1074/jbc.M805308200. [DOI] [PubMed] [Google Scholar]
- 126.Qian WJ, et al. Mono-anionic phosphopeptides produced by unexpected histidine alkylation exhibit high plk1 polo-box domain-binding affinities and enhanced antiproliferative effects in hela cells. Biopolymers. 2014;102:444–455. doi: 10.1002/bip.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuan J, et al. Polo-box domain inhibitor poloxin activates the spindle assembly checkpoint and inhibits tumor growth in vivo. The American journal of pathology. 2011;179:2091–2099. doi: 10.1016/j.ajpath.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou Y, et al. Thymoquinone and Poloxin are slow-irreversible inhibitors to human Polo-like kinase 1 Polo-box domain. J Med Coll PLA. 2010;25:136–142. [Google Scholar]
- 129.Lee KS, Idle JR. Pinning down the polo-box domain. Chemistry & biology. 2008;15:415–416. doi: 10.1016/j.chembiol.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 130.Yin Z, et al. Thymoquinone blocks pSer/pThr recognition by Plk1 Polo-box domain as a phosphate mimic. ACS chemical biology. 2013;8:303–308. doi: 10.1021/cb3004379. [DOI] [PubMed] [Google Scholar]
- 131.Liao C, et al. Exploring Potential Binding Modes of Small Drug-like Molecules to the Polo-Box Domain of Human Polo-like Kinase 1. ACS Med Chem Lett. 2010;1:110–114. doi: 10.1021/ml100020e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu F, et al. Identification of high affinity polo-like kinase 1 (Plk1) polo-box domain binding peptides using oxime-based diversification. ACS Chem Biol. 2012;7:805–810. doi: 10.1021/cb200469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu F, et al. Peptoid-Peptide hybrid ligands targeting the polo box domain of polo-like kinase 1. Chembiochem. 2012;13:1291–1296. doi: 10.1002/cbic.201200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sledz P, et al. High-throughput interrogation of ligand binding mode using a fluorescence-based assay. Angewandte Chemie. 2012;51:7680–7683. doi: 10.1002/anie.201202660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.ŒledŸ P, et al. From Crystal Packing to Molecular Recognition: Prediction and Discovery of a Binding Site on the Surface of Polo-Like Kinase 1. Angew Chem Int Ed Engl. 2011;50:4003–4006. doi: 10.1002/anie.201008019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tan YS, et al. Using ligand-mapping simulations to design a ligand selectively targeting a cryptic surface pocket of polo-like kinase 1. Angewandte Chemie. 2012;51:10078–10081. doi: 10.1002/anie.201205676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qian WJ, et al. Non-proteinogenic amino acids in the pThr-2 position of a pentamer peptide that confer high binding affinity for the polo box domain (PBD) of polo-like kinase 1 (Plk1) Bioorganic & medicinal chemistry letters. 2012;22:7306–7308. doi: 10.1016/j.bmcl.2012.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Murugan RN, et al. Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1. PLoS One. 2013;8:e80043. doi: 10.1371/journal.pone.0080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu F, et al. Preparation of orthogonally protected (2S, 3R)-2-amino-3-methyl-4-phosphonobutyric acid (Pmab) as a phosphatase-stable phosphothreonine mimetic and its use in the synthesis of polo-box domain-binding peptides. Tetrahydron. 2009;65:9673–9679. doi: 10.1016/j.tet.2009.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qian W, et al. Effects on polo-like kinase 1 polo-box domain binding affinities of peptides incurred by structural variation at the phosphoamino acid position. Bioorg Med Chem. 2013;21:3996–4003. doi: 10.1016/j.bmc.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qian WJ, Burke TR., Jr Mitsunobu mischief: neighbor-directed histidine N(tau)-alkylation provides access to peptides containing selectively functionalized imidazolium heterocycles. Organic & biomolecular chemistry. 2015;13:4221–4225. doi: 10.1039/c5ob00171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Murugan RN, et al. Development of cyclic peptomer inhibitors targeting the polo-box domain of polo-like kinase 1. Bioorg Med Chem. 2013;21:2623–2634. doi: 10.1016/j.bmc.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qian W-J, et al. Neighbor-directed histidine N(τ)–alkylation: A route to imidazolium-containing phosphopeptide macrocycles. Biopolymers Pept Sci. 2015 doi: 10.1002/bip.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cox KJ, et al. Tinkering outside the kinase ATP box: allosteric (type IV) and bivalent (type V) inhibitors of protein kinases. Future medicinal chemistry. 2011;3:29–43. doi: 10.4155/fmc.10.272. [DOI] [PubMed] [Google Scholar]
- 145.Janning M, Fiedler W. Volasertib for the treatment of acute myeloid leukemia: a review of preclinical and clinical development. Future Oncol. 2014;10:1157–1165. doi: 10.2217/fon.14.53. [DOI] [PubMed] [Google Scholar]
- 146.Stadler WM, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer. 2014;120:976–982. doi: 10.1002/cncr.28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Döhner H, et al. Randomized, phase 2 trial of low-dose cytarabine with or without volasertib in AML patients not suitable for induction therapy. Blood. 2014;124:1426–1433. doi: 10.1182/blood-2014-03-560557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhao Z, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes & Dev. 2010;24:1389–1402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu G, et al. P53 gene mutations in acute myelogenous leukaemia. Br J Haematol. 1992;81:489–494. doi: 10.1111/j.1365-2141.1992.tb02979.x. [DOI] [PubMed] [Google Scholar]
- 150.Slingerland JM, et al. Mutation of the p53 gene in human acute myelogenous leukemia. Blood. 1991;77:1500–1507. [PubMed] [Google Scholar]
- 151.Kang YH, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 152.Aubele DL, et al. Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain. ChemMedChem. 2013;8:1295–1313. doi: 10.1002/cmdc.201300166. [DOI] [PubMed] [Google Scholar]
- 153.Kim DE, et al. Protein structure prediction and analysis using the Robetta server. Nucleic acids research. 2004;32:W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]