Abstract
The Endosomal Sorting Complexes Required for Transport (ESCRT) proteins mediate fundamental membrane remodeling events that require stabilizing negative membrane curvature. These include endosomal intralumenal vesicle formation, HIV budding, nuclear envelope closure and cytokinetic abscission. ESCRT-III subunits perform key roles in these processes by changing conformation and polymerizing into membrane-remodeling filaments. Here, we report the 4 Å resolution cryo-EM reconstruction of a one-start, double-stranded helical copolymer composed of two different human ESCRT-III subunits, CHMP1B and IST1. The inner strand comprises “open” CHMP1B subunits that interlock in an elaborate domain-swapped architecture, and is encircled by an outer strand of “closed” IST1 subunits. Unlike other ESCRT-III proteins, CHMP1B and IST1 polymers form external coats on positively-curved membranes in vitro and in vivo. Our analysis suggests how common ESCRT-III filament architectures could stabilize different degrees and directions of membrane curvature.
The ESCRT pathway is best known for facilitating membrane remodeling and fission for processes such as the endosomal intralumenal vesicle (ILV) formation, enveloped virus budding, nuclear envelope closure, and cytokinetic abscission (1–3). In these reactions, the ESCRT machinery assembles on the interior of a negatively-curved, cytoplasm-filled membrane neck, and pulls the membrane toward itself to the fission point. These fission reactions are topologically distinct from reactions in which cytoplasmic BAR domain-containing proteins and dynamin-family GTPases assemble around and constrict positively-curved membrane tubules.
ESCRT components are recruited to different membranes by site-specific adaptors that ultimately recruit ESCRT-III subunits and their binding partners, including VPS4-family ATPases. ESCRT-III assemblies promote membrane constriction and fission, possibly in concert with VPS4. Humans express 12 related ESCRT-III proteins, called CHarged Multivesicular body Proteins (CHMPs) 1A-7 and Increased Sodium Tolerance 1 (IST1) (1–3). Crystal structures of CHMP3 and IST1 show a common structure in which the first two helices form a long hairpin, the shorter helices 3 and 4 pack against the open end of the hairpin, and helix 5 folds back and packs against the closed end of the helical hairpin (4–6). This “closed” conformation appears to auto-inhibit ESCRT-III membrane binding and oligomerization (4, 7, 8). ESCRT-III subunits can also adopt a second, more extended “open” conformation that has been characterized biochemically, but not visualized in molecular detail. The open conformation appears to be the active, polymerization-competent state because mutations or solution conditions that favor this conformation typically promote polymerization (1–3). Many ESCRT-III subunits form spiraling homo- and heteromeric filaments, both in vitro (4, 9–15) and in cells (10, 16–19), but the structural basis for filament assembly is unclear.
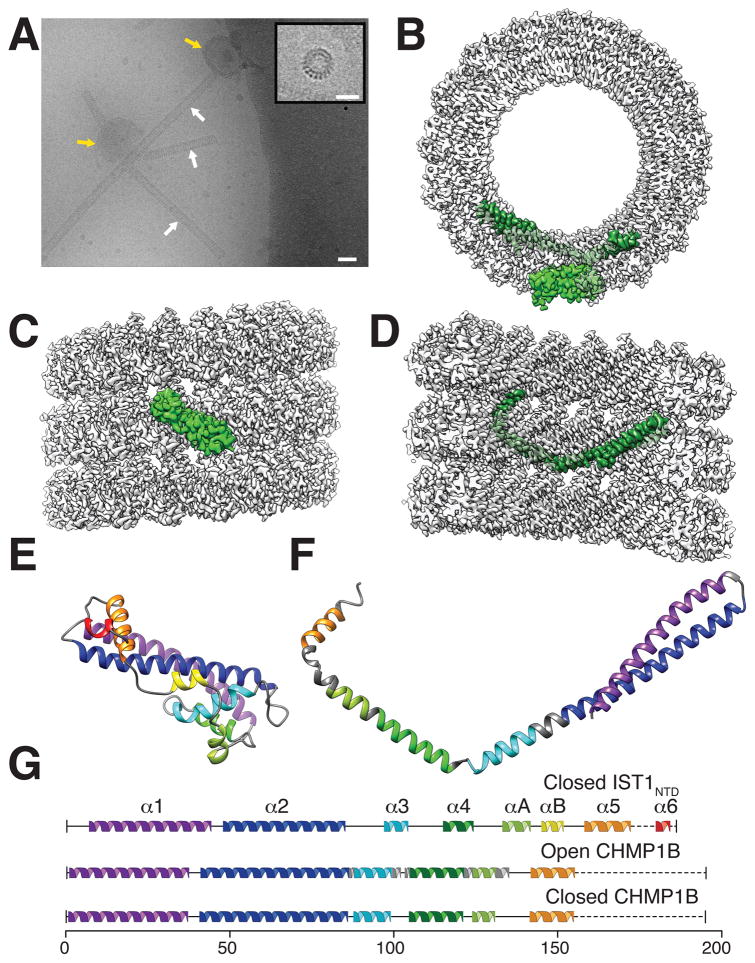
We used cryo-EM to determine the molecular structure of a helical copolymer comprising human IST1 and CHMP1B. Full-length IST1 and CHMP1B spontaneously co-assembled under low ionic strength conditions into well-ordered helical tubes. Helical order was further enhanced by using a truncated IST1 construct that spanned residues 1–189, hereafter termed IST1NTD, and by including small, acidic unilamellar vesicles (SUVs) to nucleate polymer formation. The resulting IST1NTD-CHMP1B tubes were long, straight, and 24 nm in diameter (Fig. 1A). The 3D structure of IST1NTD-CHMP1B assemblies was determined to a resolution of ~4 Å by real space helical reconstruction (Methods and figs. S1–4). Each tube comprised a right-handed one-start helical filament that packed with an inter-filament spacing of 5.1 nm/turn (Fig. 1B–D, Movie S1). Each filament was double-stranded, with distinct inner and outer strands (at 7.7 and 10.2 nm radii, respectively). Segmented densities from subunits in the outer strand corresponded well to the crystal structure of IST1NTD in its closed conformation (PDB: 3FRR) (5), with only minor refinements required to optimize the position of helix A (Fig. 1E, G, and Movie S2). By contrast, the CHMP1B subunits of the inner strand adopted a very different, open conformation. These subunits were almost entirely α-helical and side chain densities were clearly evident in the EM density (Fig. 1B, D, F, Movie S3). The open CHMP1B conformation resembled an arm, with helices 1–3 forming the upper arm and biceps, helix 4 and helix A forming the forearm, and helix 5 forming the hand. Joints between helices 3 and 4 and between helices A and 5 correspond to the elbow and wrist, respectively (Fig. 1F).
Fig. 1. IST1NTD and CHMP1B copolymerized into helical tubes comprising polar, double-stranded helical filaments.
(A) Electron cryomicrograph showing IST1NTD-CHMP1B tubes (white arrows) assembled by incubating equimolar IST1NTD and CHMP1B in the presence of polymer-nucleating small acidic unilamellar vesicles (SUVs, yellow arrows). Inset: end-on view of a short IST1NTD-CHMP1B tube. Bars: 40 nm (A), 20 nm (inset). (B) End-on view of the reconstructed IST1NTD-CHMP1B tube highlighting single subunits of IST1NTD (light green, outer strand) and CHMP1B (dark green, inner strand). (C) External view of the reconstructed helix with a highlighted IST1NTD subunit. (D) Internal cutaway view of the reconstructed helix with a highlighted CHMP1B subunit. (E) Ribbon diagram of the modeled IST1NTD subunit (closed conformation). (F) Ribbon diagram of the modeled CHMP1B subunit (open conformation). (G) Secondary structure diagrams for closed IST1NTD (top), open CHMP1B (middle), and closed CHMP1B (bottom).
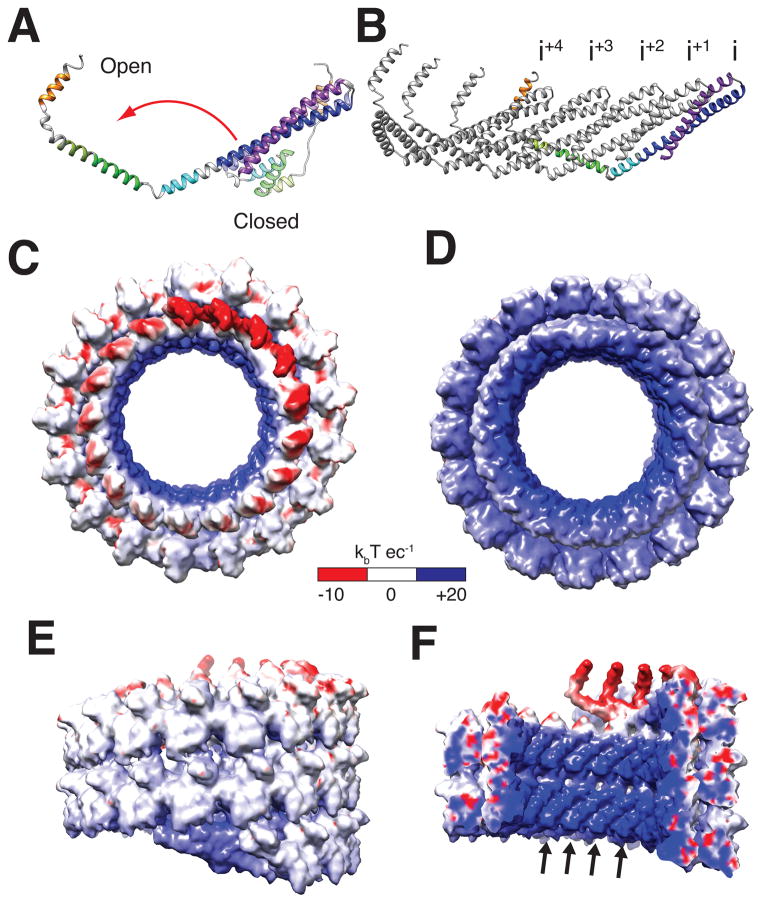
High ionic strength conditions (4, 8) typically favor the monomeric, closed ESCRT-III subunit conformation (4, 8, 20, 21), and CHMP1B also remained monomeric under high ionic strength conditions (fig. S5). Lowering the ionic strength triggered coassembly of IST1 and CHMP1B, implying that CHMP1B subunits are captured as they open. To visualize this conformational change, we generated a structure-based homology model for the CHMP1B closed state (see Methods). In comparison to the modeled closed state, the helix 5 hand is displaced by ~100 Å when CHMP1B opens. This global reorganization requires only three local rearrangements: the elbow angle between helices 3 and 4 must change, and the loops that connect helix 2/3 and helix 4/A must become helical to create the longer, continuous helices that extend the upper arm to the elbow and create the forearm in the open state (Figs. 1F–G, 2A and Movie S4).
Fig. 2. CHMP1B opening, strand structure, and electrostatic surface potentials of the IST1NTD-CHMP1B assembly.
(A) Superposition of the open and closed CHMP1B conformations. (B) Five interlocked CHMP1B molecules from the inner strand of the filament. (C) “Top-end”, electrostatic surface view of the IST1NTD-CHMP1B tube, highlighting the acidity of the CHMP1B inner strand (including Glu130, Asp131, Asp147, Glu152, Asp155, Glu156, Asp160) and the IST1NTD outer strand (including Asp49, Glu50, Glu57, Glu163, Glu168, Glu178, Asp180, Glu186). (D) “Bottom-end”, electrostatic surface view of the IST1NTD-CHMP1B tube, highlighting the strongly basic characters of the CHMP1B inner strand (including Lys3, Lys87, Lys94, Lys101, Lys107, and Lys114) and the IST1NTD outer strand (including Lys7, Arg10, Lys90, Arg109, Lys118, Lys127, Lys130, Lys134, Arg137). (E) Exterior, electrostatic surface view of the IST1NTD-CHMP1B tube, revealing the modestly basic character of the IST1NTD outer strand. (F) Internal cutaway electrostatic surface view of the IST1NTD-CHMP1B tube, revealing the strongly basic character of the lumenal surface, contributed primarily by basic residues in CHMP1B helix 1 (arrows), including Lys3, Lys13, Arg17, Lys20, Lys21, Lys24, Lys32, and Lys35.
In the filament, the open CHMP1B conformation is stabilized by extensive intersubunit interactions along the inner strand (Fig. 2B). Each CHMP1B molecule interacts with four other CHMP1B subunits that pack together and cross the forearm of the original subunit. In addition, the helix 5 hand grasps the shoulder of the hairpin four subunits away, making a domain-swapped contact that is analogous to the intrasubunit interaction between the hairpin and helix 5 in the closed ESCRT-III conformations (Fig. 2A–B). Opening and assembly reduces the total solvent accessible surface area of CHMP1B from ~10,720Å2 to ~6350Å2. The IST1NTD-CHMP1B assembly is further stabilized by three additional types of interactions, which differ completely from crystallized contacts for soluble IST1-CHMP1B heterodimers (Figs. 2C–F, figs. S6–7 (6)).
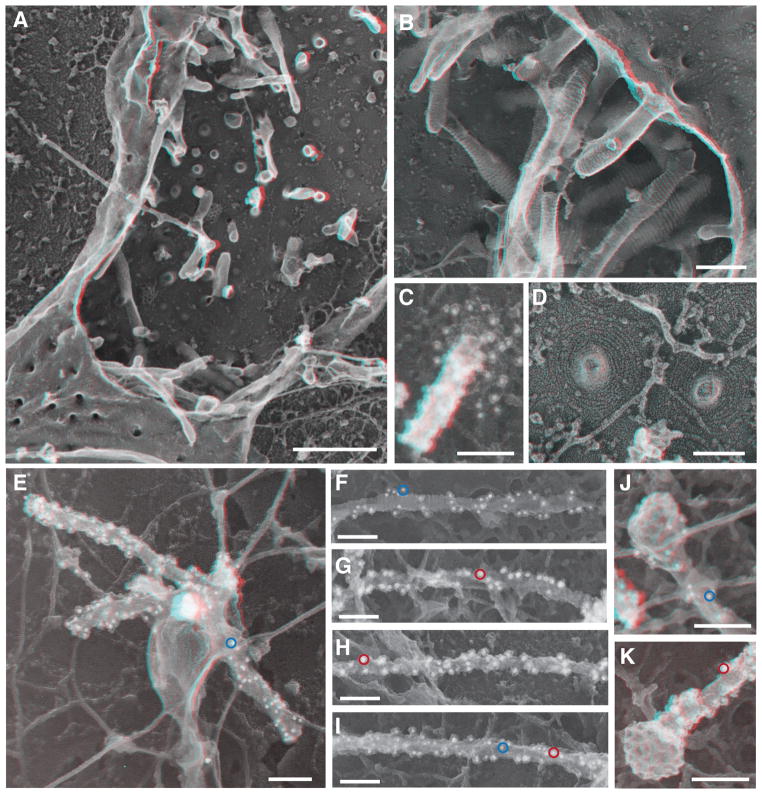
A final notable feature of the IST1-CHMP1B tube is the remarkably cationic interior. This surface is created by a series of conserved basic residues from helix 1 of CHMP1B (Fig. 2D, F), which forms the principal membrane-binding site in other ESCRT-III proteins (5, 22). Its position inside the IST1-CHMP1B copolymer was unexpected because well characterized ESCRT-mediated membrane remodeling events require that membranes interact with the exterior surface of coiled ESCRT-III filaments, e.g., as in the neck of a nascent ILV or viruses. The functional roles of IST1 and CHMP1B in such canonical ESCRT activities have been enigmatic, however, because neither protein is required for ILV biogenesis (23–26) or virus budding (27). Moreover, IST1 functions in the resolution of endosomal tubules that project into the cytoplasm and recycle cargoes back to the plasma membrane (28). Looking in cells, we found that moderately overexpressed CHMP1B polymerized into elongated cytoplasmic structures (fig. S8). Deep-etch EM of the plasma membranes revealed filaments that were similar to those of CHMP4A and other well studied ESCRT-III proteins (17), except that CHMP1B filaments coated tubules ~35–60 nm in diameter that extended into rather than away from the cytoplasm (Fig. 3A–D) (17, 18). Replicas of cells transfected and immunolabeled for CHMP1B alone, or with IST1, also revealed immunodecorated organelles and tubules that were not attached to the plasma membrane but again had recognizable striations (Fig. 3E–K). The resemblance of these organelles to early endosomes (29) including the occasional presence of clathrin-coated buds (Fig. 3J–K) indicates that CHMP1B and IST1 can coat and potentially remodel endosomal tubules.
Fig. 3. CHMP1B and IST1 tubulated cellular membranes.
(A) Survey view of the cytoplasmic surface of the plasma membrane in an unroofed COS-7 cell expressing FLAG-CHMP1B. Tubular invaginations extending into the cell interior are apparent along the exposed plasma membrane and as stabilized openings at the edges of the cell. Use view glasses for 3D viewing of anaglyphs (left eye = red). (B) Higher magnification view of tubular invaginations induced and coated by FLAG-CHMP1B filaments. (C) Immunodecoration confirmed the presence of CHMP1B around and along a tubule in a cell expressing untagged CHMP1B. Antibody detected with 12 nm gold is white in these contrast reversed EM images. (D) Higher magnification view of FLAG-CHMP1B filament spirals on exposed plasma membrane. (E) CHMP1B-immunoreactive organelle in an unroofed COS-7 cell expressing untagged CHMP1B. Antibody detected with 12 nm gold is white in these contrast reversed EM images; a representative gold particle is circled in blue. (F to I) Representative internal tubules from cells co-expressing untagged CHMP1B (12 nm gold, example circled in blue) and IST1-myc (18 nm gold); examples circled in red. (J and K) Clathrin coated bud capping the end of CHMP1B (J) and IST1-myc (K) immuno-labeled tubules from co-transfected cells. Importantly, measurements of filament diameter (and interstrand spacing) showed that when apparently unitary filaments were resolvable, their diameter varied from 5–10 nm including platinum. These measurements are generally consistent with the dimensions of IST1-CHMP1B and CHMP1B filaments formed in vitro. Scale bars 500nm (A), 100nm (B) to (K).
To determine whether distinct membrane remodeling topologies are intrinsic properties of different ESCRT-III filaments, we compared the structures induced by spirals of CHMP1B to those induced by the prototypical ESCRT-III protein CHMP4A. In earlier studies, CHMP4 proteins only deformed the membrane when bound to ATPase-deficient VPS4B (17) or an activated CHMP2A mutant (18). Here we found that deleting C-terminal sequences yielded a mutant CHMP4A that formed tight, membrane-deforming spirals (Fig. 4A). The deformations induced by CHMP4A spirals were directed away from the cytoplasm, as expected (17, 18). Comparable views of cells transfected with full-length and C-terminal truncations of CHMP1B confirmed that CHMP4A and CHMP1B induced cellular membranes to tubulate in opposite directions (Fig. 4B, fig. S9).
Fig. 4. Topology of ESCRT-III membrane deformation in cells and in vitro.
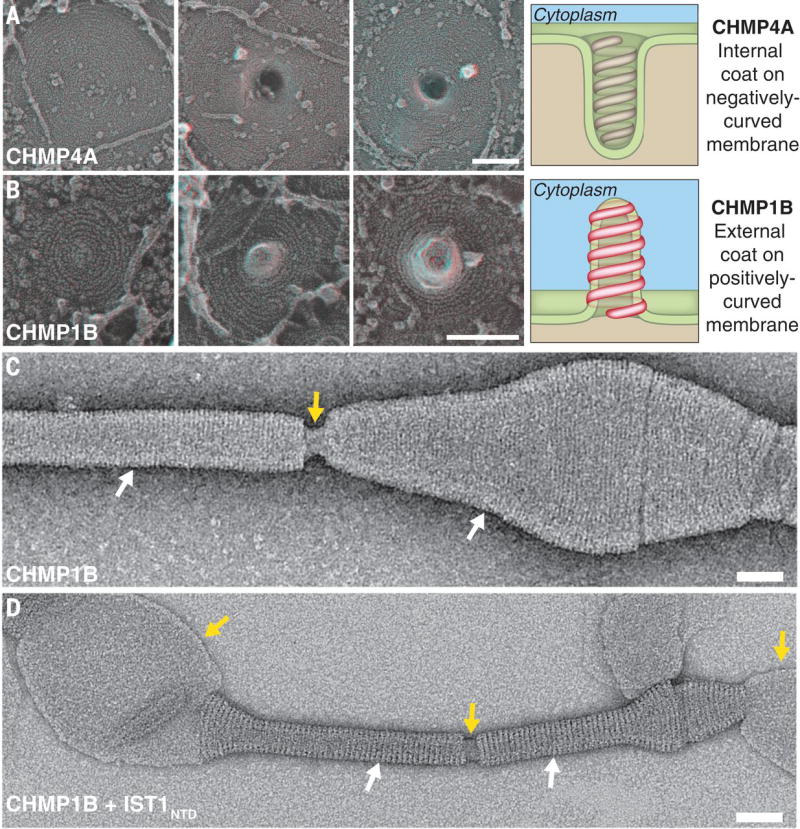
(A) Series of filament spirals on the plasma membrane of COS-7 cells expressing CHMP4A1–164 show development of the outwardly directed protrusions previously associated with ESCRT-III filaments (15, 16). Drawing highlights relationship between CHMP4A filament spiral and a negatively-curved plasma membrane tubule. (B) Series of filament spirals on the plasma membrane of COS-7 cells expressing FLAG-CHMP1B show development of invaginations directed into the cell. Drawing highlights relationship between CHMP1B filament spiral and a positively-curved plasma membrane tubule. (C) Negative stain electron micrograph showing that CHMP1B tubulates liposomes and forms a filamentous coat on the outside of the tubule. White arrows highlight regions coated by the CHMP1B helices, and the yellow arrow highlights a break in the coat where the internal lipid is visible. (D) Negative stain electron micrograph showing that the IST1NTD-CHMP1B copolymer forms on the outside of membrane tubules. White arrows highlight regions coated by the IST1NTD-CHMP1B copolymer, and the yellow arrows highlight breaks in the helical coat or uncoated regions of the liposome where the internal membrane is visible. Scale bars 100nm (A) and (B), 50 nm (C) and (D).
We also tested how CHMP1B polymers bind and remodel liposomal membranes in vitro. Under physiological solution conditions, CHMP1B formed single and double-stranded one-start helices and spirals around membrane tubules, with interstrand spacing of 4.7 ± 0.1 nm (Fig. 4C, figs. S10–S11). Adding IST1 to CHMP1B-induced membrane tubules generated copolymeric helices that were structural analogs of the membrane-free IST1-CHMP1B assemblies described above, as judged by their interstrand spacing (5.1 ± 0.1 nm vs. 5.2 ± 0.1 nm) and by the similarities of 2D class averages of the two assemblies (Fig. 4D, figs. S10–S11). The positively-curved membrane tubules within the protein coats could be visualized with negative staining of the CHMP1B and IST1-CHMP1B assemblies (Fig. 4C–D, fig. S10), in cryo-EM images, and in 2D class averages of the molecules along the tangential surface of the bilayer (fig. S11). Thus, CHMP1B and IST1 form external coats on positively-curved membranes in vitro and in cells.
Our analyses provide insight into how ESCRT-III helices can assemble structures of decreasing diameter, as might be required to draw membranes together to the fission point. In addition to uniform 24 nm helices, we frequently observed conical helical assemblies of membrane-associated CHMP1B and IST1NTD-CHMP1B (Fig. 4C–D, figs. S10–11), as well as conical IST1-CHMP1B filaments assembled in the absence of nucleating vesicles (fig. S12). One class of membrane-free IST1NTD-CHMP1B cones was sufficiently common to be reconstructed at low resolution, which confirmed that the cones are composed of the same double-stranded filaments as those seen in the IST1NTD-CHMP1B helices (Fig. 4D, figs. S1, S11–12, Movie S5–6). Within the conical spiral, both the degree of filament curvature and the lateral interactions between adjacent filaments varied continuously. Small changes in filament curvature are likely accommodated by altering the angles of the “elbow” and “wrist” joints. Larger changes could, in principle, be accommodated (or even driven) by ratcheting the buttressing intersubunit interactions made by the “forearm” and “hand”. For example, changing the connectivity from i+4 to i+5 would tend to straighten the CHMP1B filament. The required continuum of differing lateral interactions in the IST1NTD-CHMP1B spirals is apparently accommodated by small shifts in ionic inter-strand interactions, which is plausible because the basic charges are distributed almost uniformly along one edge of the double strand (Fig. 2C–D). Wider IST1-CHMP1B spirals will tend to propagate toward their preferred 24 nm diameter. At this diameter, the narrow lumen (~10 nm) would force internal opposing lipid bilayers (each ~4.7 nm wide) toward hemi-fission (30, 31), potentially providing a driving force for membrane constriction and fission. Finally, we note that other ESCRT-III subunits, such as CHMP4A, tubulate membranes in the opposite direction to that seen with CHMP1B. Such stabilization of negative, rather than positive, membrane curvature could be achieved simply by altering the intrinsic degree and direction of filament curvature, while retaining an analogous membrane-binding surface and subunit connectivity.
Supplementary Material
Acknowledgments
Electron microscopy was performed at UCSF, the University of Utah and the OHSU Multiscale Microscopy Core with technical support from the FEI Living Lab and the OHSU Center for Spatial Systems Biomedicine (OCSSB). We thank B. Carragher, C. Potter and the National Resource for Automated Molecular Microscopy—which is supported by the National Institutes of Health (NIH) through the National Center for Research Resources (NCRR) P41 program (RR17573)—where some initial imaging studies were performed. We also thank B. Bammes and Direct Electron, LP (San Diego, CA), W. Chiu and J. Jakana at the National Center for Macromolecular Imaging at Baylor College of Medicine (Houston, TX) where other images were collected. Computing support was provided by the Center for High Performance Computing at the University of Utah and the NSF XSEDE consortium. We also thank C. Hill, F. Whitby, H. Schubert, B. Barad and J. Fraser for guidance regarding model refinement and validation, D. Eckert for performing analytical ultracentrifugation experiments, J. Iwasa for artwork, M. LaLonde, C. Rodesch, G. Mercenne, and M. Gudheti for help with supporting experiments, and T. Goddard for Chimera 2 developments. We thank R. Roth for deep-etch EM replicas; J. Heuser for sharing deep-etch EM with us and advice; E. Clipperton for CHMP4A mutants; M. Pretz for IST1-myc constructs; and M. Hartstein for artwork. We thank K. Blumer, M. Babst, and other members of our laboratories and colleagues for helpful discussions. This research was supported by the Searle Scholars Program (A.F.), the Jane Coffin Childs Foundation (M.G.S.), the Phenix Industrial Consortium (P.A.), an American Cancer Society grant PF-11-279-01-CCG (L.A.C.), a US Department of Energy contract DE-AC02-05CH11231 (P.A.), NIH 2P50GM082545-06 (A.F.), 1DP2GM110772-01 (A.F.), R01GM112080 (W.I.S. and A.F.), and R01AI051174 (W.I.S.), 1P01 GM063210 (P.A.), R01GM076686 (P.I.H.), R01NS050717 (P.I.H.) and a NSF fellowship DGE-1143954 (A.K.C.). All of the expression constructs used in our study have been or will be deposited in the public DNASU plasmid repository (http://dnasu.org/DNASU/Home.jsp). The density map has been deposited in the EMDB (EMD-6461) and the fitted models deposited in the PDB (3JC1).
Footnotes
www.sciencemag.org/cgi/content/full/science.aad8305/DC1
Materials and Methods
REFERENCES AND NOTES
- 1.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 3.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 8.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 10.Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL, Bell SD, Jensen GJ. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–2327. doi: 10.1091/mbc.E12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, Schoehn G, Weissenhorn W. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell Microbiol. 2013;15:213–226. doi: 10.1111/cmi.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, Schoehn G, Forterre P, Weissenhorn W, Renesto P. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLOS ONE. 2011;6:e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen QT, Schuh AL, Zheng Y, Quinney K, Wang L, Hanna M, Mitchell JC, Otegui MS, Ahlquist P, Cui Q, Audhya A. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, Schoehn G, Weissenhorn W, Sadoul R. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cashikar AG, Shim S, Roth R, Maldazys MR, Heuser JE, Hanson PI. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife. 2014;3:e02184. doi: 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 20.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Göttlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 22.Buchkovich NJ, Henne WM, Tang S, Emr SD. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev Cell. 2013;27:201–214. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol Biol Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita E. Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. FEBS J. 2012;279:1399–1406. doi: 10.1111/j.1742-4658.2012.08534.x. [DOI] [PubMed] [Google Scholar]
- 28.Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, Seaman MN, Hazan J, Reid E. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J Cell Biol. 2013;202:527–543. doi: 10.1083/jcb.201211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoorvogel W, Oorschot V, Geuze HJ. A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlovsky Y, Kozlov MM. Membrane fission: Model for intermediate structures. Biophys J. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Please refer to supplemental material online for 12 additional figures, 6 movies, materials and methods, and additional citations.
- 32.von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kräusslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/S0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 33.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laue TM, Shah BD, Ridgeway TM, Pelletier . Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 37.Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth DS, Avila-Sakar A, Cheng Y. J Vis Exp. 2011;58:1–8. doi: 10.3791/3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrigley NG. The lattice spacing of crystalline catalase as an internal standard of length in electron microscopy. J Ultrastruct Res. 1968;24:454–464. doi: 10.1016/S0022-5320(68)80048-6. [DOI] [PubMed] [Google Scholar]
- 40.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 41.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/S0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 44.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Pintilie G, Chiu W. Comparison of Segger and other methods for segmentation and rigid-body docking of molecular components in cryo-EM density maps. Biopolymers. 2012;97:742–760. doi: 10.1002/bip.22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD, Baker D. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine PV, et al. Comput Crystallogr Newsletter. 2013:43–44. [Google Scholar]
- 58.Grosse-Kunstleve RW, et al. Int Union Crystallogr Comput Comm Newsletter. 2004;4:19–36. [Google Scholar]
- 59.Echols AN, et al. Comput Crystallogr Newsletter. 2010;1:4–11. [Google Scholar]
- 60.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. Web Server. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522. doi: 10.1093/nar/gkm276. Web Server. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244. doi: 10.1093/nar/gki408. Web Server. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(S9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pei J, Kim BH, Grishin NV. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 68.Gilbert P. Iterative methods for the three-dimensional reconstruction of an object from projections. J Theor Biol. 1972;36:105–117. doi: 10.1016/0022-5193(72)90180-4. [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heuser JE. Membrane traffic in anaglyph stereo. Traffic. 2000;1:35–37. doi: 10.1034/j.1600-0854.2000.010106.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.