Abstract
Adipose tissue plays important roles in regulating carbohydrate and lipid homeostasis, though less is known about the regulation of amino acid metabolism in adipocytes. Here we applied isotope tracing to pre–adipocytes and differentiated adipocytes to quantify the contributions of different substrates to tricarboxylic acid metabolism and lipogenesis. In contrast to proliferating cells that use glucose and glutamine for acetyl–coenzyme A (AcCoA) generation, differentiated adipocytes increased branched chain amino acid (BCAA) catabolic flux such that leucine and isoleucine from media and/or protein catabolism accounted for as much as 30% of lipogenic AcCoA pools. Medium cobalamin deficiency caused methylmalonic acid accumulation and odd–chain fatty acid synthesis. B12 supplementation reduced these metabolites and altered the balance of substrates entering mitochondria. Finally, inhibition of BCAA catabolism compromised adipogenesis. These results quantitatively highlight the contribution of BCAAs to adipocyte metabolism and suggest that BCAA catabolism plays a functional role in adipocyte differentiation.
Introduction
Adipose tissue plays a major role in glucose and lipid homeostasis via storage of excess nutrients in lipid droplets and the release of bioenergetic substrates through lipolysis. Adipocytes, the major cellular constituent of adipose tissue, execute important regulatory functions through endocrine and paracrine signaling1. For example, the synthesis and release of lipids and adipokines influence fatty acid metabolism in the liver, appetite, inflammation, and insulin sensitivity2–4. Dysfunction in these pathways can contribute to insulin resistance5. Beyond these signaling functions, the increased adiposity associated with obesity and type 2 diabetes mellitus (T2DM) has highlighted the need to better understand metabolic regulation and activity in adipocytes.
Insulin stimulates glucose utilization and de novo lipogenesis (DNL) in the liver and adipose tissue, and glucose and fatty acids are considered the primary carbon sources fueling anaplerosis and acetyl–coenzyme A (AcCoA) generation in these sites6. Beyond carbohydrates and fat, both essential and non–essential amino acids also contribute significantly to AcCoA metabolism in cells. The branched chain amino acids (BCAAs) leucine, isoleucine, and valine are important ketogenic and/or anaplerotic substrates in a number of tissues7,8. In fact, clinical metabolomics studies have recently suggested that plasma levels of BCAAs, their downstream catabolites (e.g., acylcarnitines), and other essential amino acids become elevated in the context of insulin resistance9–11. However, the mechanisms leading to these changes and ultimate consequences in the context of metabolic syndrome are not fully understood.
Several past studies provide evidence that adipose tissue plays a role in BCAA homeostasis, though the quantitative contribution of these amino acids to TCA metabolism relative to other nutrients is not well–defined. Enzyme activity, substrate oxidation, and systems–based profiling of 3T3–L1 metabolism suggest that BCAA consumption increases precipitously during differentiation to adipocytes12–14. In addition, BCAA catabolic enzyme transcription increases significantly during 3T3–L1 differentiation15. While genetic modulation of Bcat2 in mice alters circulating BCAA levels, transplantation of wild–type adipose tissue to Bcat2–/– mice normalizes plasma BCAAs16–18. Finally, treatment of human subjects and animals with thiazolidinedione (TZDs) increases the transcription of BCAA catabolic genes in adipose tissue19,20, suggesting this metabolic activity may have beneficial effects in the context of T2DM.
To date, the extent that BCAA oxidation contributes to anaplerosis and DNL relative to other substrates has not been quantitatively determined in adipocytes. Here we employed [13C]–labeled isotope tracers, mass spectrometry, and isotopomer spectral analysis (ISA) to quantify BCAA utilization in proliferating pre–adipocytes and differentiated adipocytes. While proliferating pre–adipocytes did not appreciably catabolize these substrates, BCAAs accounted for as much as one–third of the mitochondrial AcCoA in terminally differentiated 3T3–L1 adipocytes as well as adipocytes isolated from human subcutaneous and visceral adipose tissues. Furthermore, inadequate cobalamin availability in 3T3–L1 cultures perturbed BCAA and fatty acid metabolism and led to the non–physiological accumulation of methylmalonate (MMA) and odd–chain fatty acids (OCFAs) synthesis. Finally, inhibition of BCAA catabolism negatively influenced 3T3–L1 adipogenesis. These results highlight the complex interplay between cellular differentiation and metabolic pathway flux in adipocyte biology.
Results
Adipogenesis reprograms amino acid metabolism
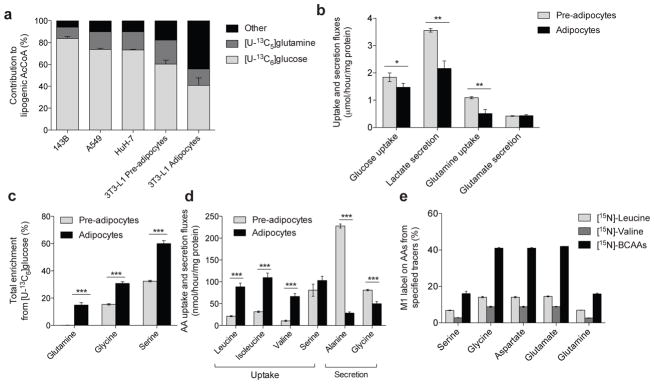
To understand how mitochondrial substrate utilization changes during adipogenesis we quantified the source of lipogenic AcCoA before and after differentiation. 3T3–L1 adipocytes displayed substantial lipid accumulation 7 days after induction of differentiation (Supplementary Results, Supplementary Fig. 1a). Palmitate (total from saponified non–polar extracts) in proliferating and differentiated 3T3–L1 cells cultured with [U–13C6]glucose (Supplementary Fig. 1b) had the expected pattern of labeling arising via pyruvate dehydrogenase (PDH)–mediated generation of M2 AcCoA (i.e., AcCoA containing two 13C isotopes). Isotopic enrichment from [U–13C5]glutamine (Supplementary Fig. 1c) was significantly lower compared to that observed with labeled glucose. ISA was employed to quantify the relative contributions of [U–13C6]glucose and [U–13C5]glutamine to de novo palmitate synthesis in 3T3–L1 pre–adipocytes versus adipocytes and compared to results in cancer cell lines of various tissue origins (Fig. 1a). Notably, glucose and glutamine accounted for ≥80% of the lipogenic AcCoA in all proliferating cells. In contrast to cells with an active cell cycle, a substantially greater fraction of lipid carbon in differentiated cells arose from substrates other than glucose and glutamine. As expected, absolute lipogenic flux from glucose and glutamine to palmitate was significantly increased after differentiation (Supplementary Fig. 1d).
Figure 1. Characterization of metabolic reprogramming during adipocyte differentiation.
(a) Contribution of [U–13C6]glucose and [U–13C5]glutamine to lipogenic AcCoA for palmitate synthesis in 143B, A549, HuH–7, and 3T3–L1 pre–adipocytes and adipocytes.
(b) Uptake and secretion fluxes in 3T3–L1 pre–adipocytes and adipocytes. Data shown are from 3 biological replicates; *represents p<0.05, **represents p<0.01 by Student’s 2–tailed t–test.
(c) Percentage of intracellular glutamine, serine, and glycine pools that were newly synthesized (labeled) from [U–13C6]glucose in 3T3–L1 pre–adipocytes and adipocytes. Data shown are 3 technical replicates representative of 3 biological replicates; ***represents p<0.001 by Student’s 2–tailed t–test.
(d) Net amino acid uptake and secretion in 3T3–L1 pre–adipocytes and adipocytes. Data shown are 3 technical replicates representative of 3 biological replicates; ***represents p<0.001 by Student’s 2–tailed t–test.
(e) Percentage of amino acids that contain an M1 label indicating transamination from indicated [15N]amino acid tracer.
Data presented in (a) represents model output ± 95% c.i and (b)–(e) represent mean ± s.d., Data shown in (b)–(d) are 3 technical replicates representative of 3 biological replicates; asterisks represent significant differences between groups by Student’s 2–tailed t–test where * represents p<0.05, ** represents p<0.01, and *** represents p<0.001.
When normalized to protein levels, glucose uptake, lactate secretion, and glutamine uptake were all significantly reduced in differentiated 3T3–L1 cells compared to those at the pre–adipocyte stage (Fig. 1b). Proliferating 3T3–L1 pre–adipocytes secreted more lactate per mole of glucose taken up – a hallmark of rapidly proliferating cells21. Importantly, the use of stable isotope tracers allowed us to measure the percentage of each amino acid pool that was newly synthesized in each cell type. Despite their lack of proliferation, several non–essential amino acids initially present at high levels in culture media were synthesized at strikingly high rates in differentiated 3T3–L1 adipocytes, including glutamine, glycine, and serine (Fig. 1c), while none of the proliferating cells tested synthesized glutamine de novo. After 24 hours of culture in medium containing 4 mM glutamine, approximately 20% of the pool was newly synthesized such that glutamine was secreted into the culture media at a significant rate (while still being consumed) (Supplementary Fig. 1e), in line with studies in adipose tissue22. Given that de novo synthesis of glutamine and other amino acids was occurring at significant rates in differentiated cells, we hypothesized that consumption of other amino acids increased upon differentiation to provide the necessary nitrogen for these reactions. Indeed, quantitation of amino acid fluxes indicated that BCAA consumption increased dramatically in differentiated 3T3–L1 adipocytes compared to pre–adipocytes (Fig. 1d), consistent with previous observations13. Furthermore, metabolic tracing using [15N]BCAAs indicated that nitrogen from these substrates contributed significantly to amino acid metabolism in differentiated adipocytes (Fig. 1e).
BCAAs fuel TCA metabolism and lipogenesis in adipocytes
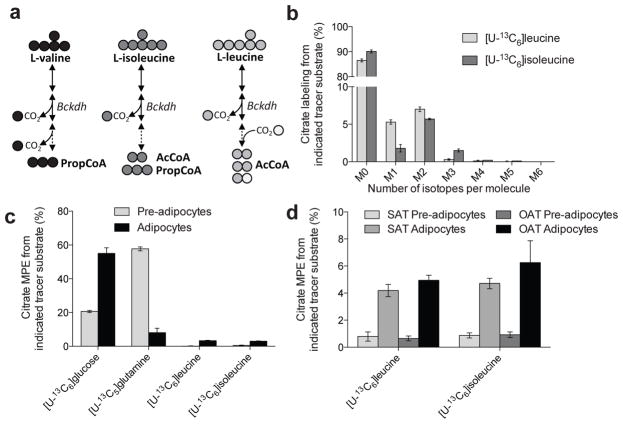
To better quantify the extent that leucine or isoleucine contributed to mitochondrial metabolism and lipogenesis in cultured adipocytes we applied [U–13C6]leucine or [U–13C6]isoleucine tracers to 3T3–L1 cultures. Figure 2a summarizes the carbon atom transitions associated with BCAA catabolism and resultant labeling in mitochondrial intermediates. For a more detailed map of BCAA catabolism, see Supplementary Figure 2a. Two M2 AcCoA and one M1 AcCoA molecules are produced from [U–13C6]leucine catabolism, and [U–13C6]isoleucine catabolism yields one M2 AcCoA and one M3 propionyl–CoA (PropCoA), which is subsequently converted to Succinyl–CoA. AcCoA molecules arising from leucine and isoleucine were readily incorporated into citrate pools in differentiated adipocytes (Fig. 2b). To compare changes before and after differentiation, 3T3–L1 pre–adipocytes and those differentiated for 6–7 days were cultured in the presence of [U–13C6]glucose, [U–13C5]glutamine, [U–13C6]leucine, or [U–13C6]isoleucine and the steady–state mole percent enrichment (MPE) in the citrate pool was measured (Fig. 2c and Supplementary Fig. 2b). While incorporation of carbon atoms from leucine and isoleucine into citrate was barely detectable in pre–adipocytes, their contribution increased significantly upon differentiation to 3.3% and 3.0%, respectively (Fig. 2c). Other TCA intermediates showed comparable levels of leucine and isoleucine enrichment (Supplementary Fig. 2c), beginning about two days into differentiation (Supplementary Fig. 2d).
Figure 2. BCAA catabolism is initiated upon adipocyte differentiation.
(a) Summary of BCAA catabolism and carbon atom transitions from each BCAA tracer. Abbreviations: Bckdh: branched-chain ketoacid dehydrogenase; AcCoA: acetyl-CoA; PropCoA: propionyl-CoA
(b) Citrate labeling in 3T3–L1 adipocytes from [U–13C6]leucine and [U–13C6]isoleucine.
(c) Mole percent enrichment (MPE) of citrate from each tracer substrate in 3T3–L1 pre–adipocytes and adipocytes.
(d) Mole percent enrichment (MPE) of citrate from [U–13C6]leucine and [U–13C6]isoleucine in primary human pre–adipocytes and adipocytes isolated from subcutaneous (SAT) or omental adipose tissue (OAT) depots.
Data presented in (b)–(d) represent mean ± s.d.
We also quantified the relative extent of BCAA catabolism in human pre–adipocytes isolated from subcutaneous or omental depots to observe whether induction of BCAA catabolism also occurs during human adipogenesis23,24. Trends comparing citrate MPE from both [U–13C6]leucine or [U–13C6]isoleucine in human pre–adipocytes versus differentiated adipocytes were similar to those observed in 3T3–L1 cells, suggesting that human adipocytes metabolize BCAAs at significant rates as well25 (Fig. 2d).
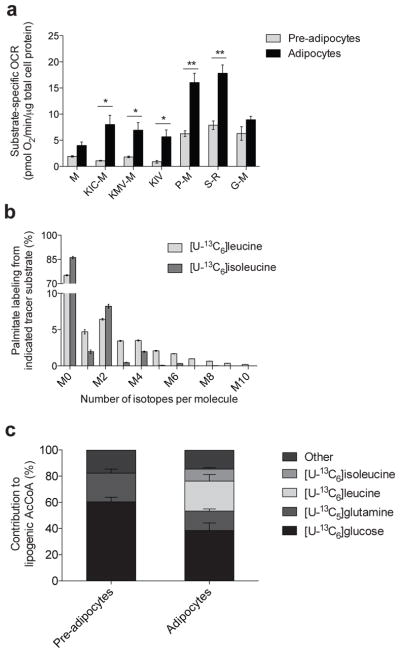
In addition to carbon atom enrichment, evidence of the oxidative capacity of these cells for each of the respective branched chain keto–acids (BCKAs) was obtained via respirometry analysis of permeabilized adipocytes26,27. When comparing uncoupler–stimulated respiration of adipocytes to pre–adipocytes we observed a marked increase in oxygen consumption from all substrates tested, but a disproportionate increase in the oxidation of the BCAT2 reaction products keto–isocaproate (KIC), keto–methylvalerate (KMV) and keto–isovalerate (KIV) was evident (Fig. 3a). The capacity to oxidize these BCKAs increased by 14–25 fold post–differentiation, a much greater increase than was observed for other substrates, suggesting that differentiation–induced mitochondrial biogenesis does not entirely explain the increased capacity to oxidize BCKAs (Supplementary Fig. 3a).
Figure 3. BCAA catabolism fuels mitochondrial metabolism and lipogenesis in adipocytes.
(a) Substrate–specific oxygen consumption rate (OCR) in permeabilized 3T3–L1 pre–adipocytes and adipocytes. Substrate key: M: malate; KIC–M: keto–isocaproate and malate; KMV–M: keto–methylvalerate and malate; KIV: keto–isovalerate; P–M: pyruvate and malate; S–R: succinate and rotenone; G–M: glutamate and malate. 250 nM AdoCbl and biotin were supplemented to the respirometry medium. Data shown are from at least 5 biological replicates each with 4 technical replicates; *represents p<0.05, **represents p<0.01 by Student’s 2–tailed t–test.
(b) Palmitate labeling in 3T3–L1 adipocytes from [U–13C6]leucine and [U–13C6]isoleucine. Minimal label was detected in palmitate from [U–13C5]valine.
(c) Contribution of each tracer substrate to lipogenic AcCoA in 3T3–L1 pre–adipocytes and adipocytes after correction due to tracer dilution. BCAA contributions were adjusted to account for dilution of intracellular amino acids from protein turnover using the average BCAA labeling over the course of the experiment.
Data presented in (a) is mean ± s.e.m., (b) is mean ± s.d., and (c) is model output ± 95% c.i.
In contrast to cultured human adipocytes, 3T3–L1 cells exhibit high levels of DNL, which facilitates quantitation of isotope enrichment in the lipogenic AcCoA pool (Fig. 3b). To determine the relative contributions of [U–13C6]glucose, [U–13C5]glutamine, [U–13C6]leucine, and [U–13C6]isoleucine to this metabolic pool we modified our ISA model to account for partially labeled AcCoA arising from [U–13C6]leucine catabolism (Fig. 2a and Supplementary Results, Supplementary Table 1). The contributions of both glucose and glutamine to AcCoA decreased significantly upon differentiation (Fig. 3c). On the other hand, leucine and isoleucine contributions increased from undetectable levels in pre–adipocytes to 23% and 9%, respectively, in adipocytes. These results are consistent with the pronounced induction of BCAA oxidation we observed upon adipocyte differentiation. Finally, this metabolic reprogramming toward BCAA utilization dramatically increased 3 days post–induction and continued until 6 days post–induction (Supplementary Fig. 3b), coinciding with the accumulation of palmitate and other fatty acids (Supplementary Fig. 3c).
Protein catabolism supports BCAA metabolism
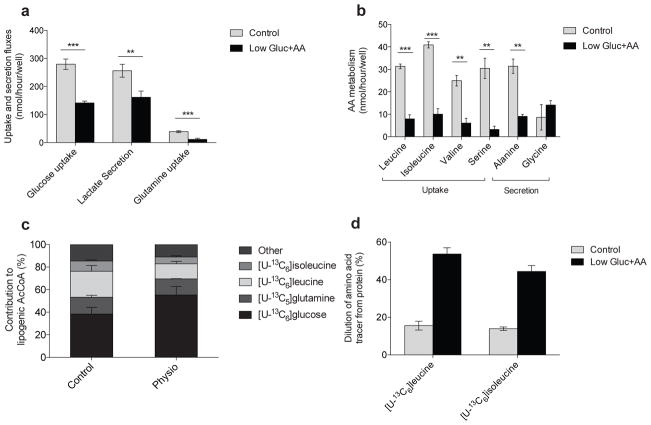
Elevated nutrient concentrations can influence metabolic activity through mass action or the regulation of cell signaling pathways, particularly in the case of glucose and BCAAs28. Indeed, BCAA levels in standard adipocyte media (i.e., DMEM) are over 4–fold higher than those measured in human and mouse plasma9. To test whether the high concentrations of amino acids and glucose in 3T3–L1 cultures influenced the extent of BCAA catabolism we cultured adipocytes in medium with glucose and the full complement of amino acids present at more physiological levels (termed “Low gluc+AA;” e.g., 6 mM glucose, 1 mM glutamine, and 200 μM for each BCAA; see Supplementary Results, Supplementary Table 2 for complete formulation). In comparing the metabolism of 3T3–L1 adipocytes in control versus Low gluc+AA medium we observed a significant reduction in glycolysis and amino acid uptake/secretion (Fig. 4a and 4b). On the other hand, total fatty acid pools and synthesis rates were unchanged (Supplementary Fig. 4a–b). ISA using specific tracers indicated that glucose contribution to AcCoA increased significantly, while the relative contribution of BCAAs to fatty acids decreased (Fig. 4c). Notably, the increased glucose contribution could be almost entirely largely accounted for by increased labeling of glutamine from glucose (Supplementary Fig. 4c). Perhaps more importantly, the decrease in BCAA–derived label in AcCoA pools was strongly impacted by dilution from unlabeled protein turnover (the only other source of essential amino acids present); within 24 hours approximately 50% of the intracellular BCAA pool became unlabeled (Fig. 4d). Notably, the final concentrations of each BCAA (25 μM leucine, 40 μM isoleucine, and 75 μM valine) were well below those observed in fasting human plasma29 (125 μM leucine, 75 μM isoleucine, and 200 μM valine), which would strongly induce protein catabolism at the end of culture. However, these data provide evidence that amino acids (BCAAs in particular) from both extracellular sources and protein catabolism are highly utilized by differentiated adipocytes. Though not surprising given the coordinate regulation of protein synthesis and catabolism30, the significant dilution of [13C]BCAAs observed here highlights the importance of amino acid recycling in metabolically active cells and tissues.
Figure 4. BCAA utilization is supported by protein catabolism.
(a) Uptake and secretion fluxes in 3T3–L1 adipocytes cultured in control and Low Gluc+AA media.
(b) Amino acid uptake and secretion in control and Low Gluc+AA media.
(c) Contribution of each tracer to lipogenic AcCoA in 3T3–L1 adipocytes cultured in control and Low Gluc+AA media. BCAA contributions were adjusted to account for dilution of intracellular amino acids from protein turnover using the average BCAA labeling over the course of the experiment. Glutamine dilution occurred primarily via glucose–derived synthesis.
(d) Percent of pool without label when cultured in indicated tracer substrate in control and Low Gluc+AA for 24 hours.
Data presented in (a)–(d) represent mean ± s.d., except (c) which represents model output ± 95% c.i. Data shown in (a)–(b) are 3 technical replicates representative of 3 biological replicates; **represents p<0.01 and *** represents p<0.001 by Student’s two–tailed t–test.
Medium B12 deficiency impacts BCAA and lipid metabolism
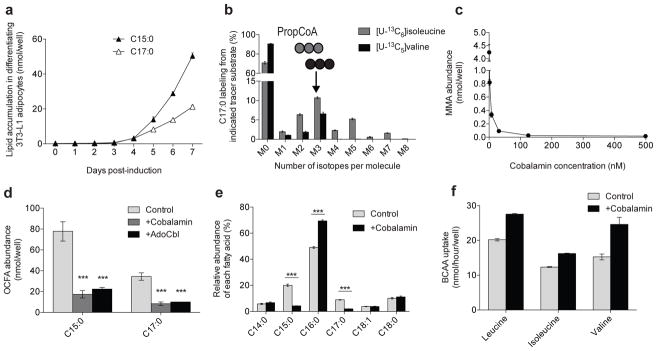
Intriguingly, we failed to observe prominent M3 labeling of citrate (Fig. 2b) or other TCA intermediates (not shown) when culturing differentiated 3T3–L1 cells with [U–13C6]isoleucine, suggesting that PropCoA and methylmalonyl CoA (MMA–CoA) derived from isoleucine did not enter the TCA cycle (Supplementary Fig. 2a). We also failed to detect 13C incorporation in any TCA metabolite from [U–13C5]valine (not shown). Notably, we measured a significant accumulation of methylmalonic acid (MMA) in differentiated 3T3–L1 cells, which was surprisingly present at similar intracellular levels to BCAAs (Supplementary Fig. 5a). Furthermore, high levels of odd–chain fatty acids (OCFAs) accumulated in 3T3–L1 adipocytes but not pre–adipocytes, including C15:0 and C17:0 species (Fig. 5a). These fatty acids have previously been observed in other reports using 3T3–L1 cells but their origin has remained unclear31.
Figure 5. BCAAs contribute to MMA, OCFAs, and BCFAs in differentiated 3T3–L1 adipocytes.
(a) Odd–chain fatty acid (OCFA) accumulation in 3T3–L1 adipocytes 0 to 7 days post–induction.
(b) C17:0 labeling from [U–13C6]isoleucine and [U–13C5]valine in 3T3–L1 adipocytes.
(c) MMA abundance in cells cultured in DMEM +10% FBS and 0, 2, 8, 32, 125, or 500 nM cobalamin beginning on Day 0 of differentiation.
(d) Odd–chain fatty acid (OCFA) levels in 3T3–L1 adipocytes after culture with 500 nM cobalamin or 100 nM AdoCbl. **represents p<0.01 and ***represents p<0.001 compared to control condition by two–way ANOVA and Holm–Sidak’s multiple comparison test.
(e) Relative abundance of the most abundant fatty acids in control and +cobalamin conditions. ***represents p<0.001 compared to control condition by Student’s two–tailed t–test.
(f) Uptake of BCAAs in control and +cobalamin condition.
Data presented in (a)–(f) represent mean ± s.d. Data shown in (d)–(e) are from 3 technical replicates representative of 3 biological replicates.
Distinct M3 labeling was observed in MMA after 24 hours of culture with [U–13C6]isoleucine or [U–13C5]valine (Supplementary Fig. 5b), as expected based on known pathway architecture. Similarly, the C15:0 and C17:0 OCFAs also contained a prominent M3 peak after culture with these BCAA tracers, (Fig. 5b and Supplementary Fig. 5c), providing evidence that these fatty acids arose from BCAA–derived DNL rather than via α–oxidation. These data suggest that MMA and PropCoA accumulate in differentiated 3T3–L1 adipocytes, presumably due to defects in BCAA catabolism.
MMA accumulates in patients with the genetic disorder methylmalonic aciduria. This phenotype may be caused by mutations in the gene encoding MMA–CoA mutase (MUT) or genes involved in cobalamin metabolism, since conversion of MMA–CoA to Succinyl–CoA requires the coenzyme 5′–deoxyadenosylcobalamin (AdoCbl)32. Notably, the human adipocytes analyzed in this study were cultured in DMEM/F12, which contains 500 nM cobalamin, and both OCFAs and MMA were undetectable in these cultures. Since typical 3T3–L1 culture conditions employ DMEM lacking cobalamin and supplementation with 10% dialyzed or untreated serum might not be sufficient to support this metabolism, we hypothesized that deficiencies in cobalamin availability led to the accumulation of MMA and OCFAs. Indeed, addition of cobalamin to media at levels ranging from 2 nM (ca. human plasma) to 500 nM (DMEM/F12) prevented the accumulation of these metabolites (Fig. 5c and Supplementary Fig. 5d). We obtained similar results when adding cobalamin or AdoCbl to 3T3–L1 culture medium (Supplementary Fig. 5e and Fig. 5d).
Supplementation of cobalamin impacted the relative abundance of fatty acid species as well as the contribution of BCAAs to TCA metabolism. For example, addition of cobalamin from day 0 to day 7 of differentiation reduced OCFA levels as a percentage of the total intracellular fatty acids, with a concomitant increase in the relative amount of palmitate (Fig. 5e). At the same time, cobalamin supplementation promoted anaplerosis of valine, as evidenced by increased enrichment of fumarate, malate, and other TCA intermediates from [U–13C5]valine (Supplementary Fig. 5f–h). Furthermore, 3T3–L1 adipocytes took up more BCAAs, including leucine, when cultured in cobalamin–supplemented media (Fig. 5f), suggesting cobalamin deficiency and/or MMA/OCFA accumulation results in broader regulation of the pathway. On the other hand, enrichment from [U–13C6]glucose decreased across all TCA intermediates in 3T3–L1 adipocytes cultured with 500 nM cobalamin (Supplementary Fig. 5i). These data indicate that accumulation of MMA, OCFAs, and presumably their precursor, PropCoA due to inadequate cobalamin availability perturbs BCAA, fatty acid, and mitochondrial metabolism in cultured adipocytes.
BCAA metabolism contributes to adipogenesis
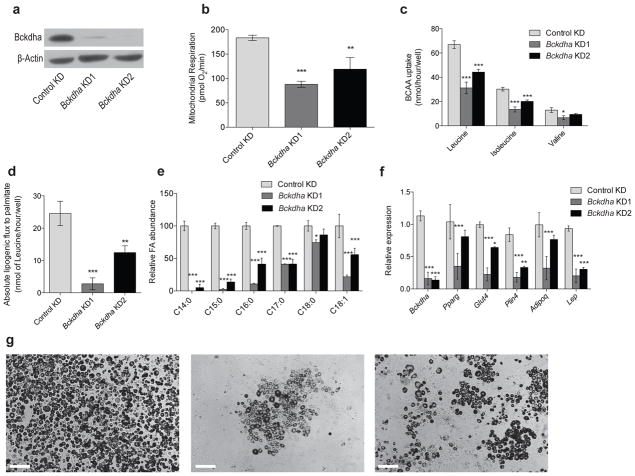
Given the high catabolic flux of BCAAs observed in differentiated adipocytes we next determined whether this pathway plays a functional role in adipogenesis. To address this question we generated polyclonal populations of 3T3–L1 cells that stably express shRNAs targeting Bckdha or non–specific sequences (Control). At confluence these cells were subjected to differentiation in the absence of cobalamin and rosiglitazone and characterized each with respect to their metabolism, morphology, and differentiation markers. After differentiation, knockdown (KD) cells exhibited decreased Bckdha protein levels compared to Control (Fig. 6a). Respiration and BCAA uptake were also significantly decreased in KD cells compared to controls (Figs. 6b and 6c). Consistent with this observation, lipogenic flux of [U–13C6]leucine to fatty acids was markedly lower in Bckdha KD cells (Fig. 6d). Furthermore, the abundance of numerous fatty acids was decreased in differentiated KD cells, including fatty acids synthesized de novo as well as those taken up from the medium (Fig. 6e). Finally, the transcription of selected adipogenic genes (Pparg, Glut4, Plin4, Adipoq, and Lep) was significantly lower in the Bckdha KD conditions compared to Control (Fig. 6f), further suggesting that BCAA catabolism contributes to adipogenesis. Representative images of differentiated cultures also suggested that Bckdha KD negatively impacted adipogenic differentiation, as evidenced by decreased lipid droplet formation (Fig. 6g). Supplementation of cobalamin did not observably impact adipogenic differentiation (not shown), suggesting that some metabolite or pathway flux upstream of MMA–CoA mutase contributes to adipogenesis.
Figure 6. Inhibition of BCAA catabolism impairs adipocyte differentiation.
(a) Western blot of Bckdha and β–Actin in differentiated 3T3–L1 adipocytes.
(b) Basal respiration in Control and Bckdha KD adipocytes normalized to nuclear quantitation.
(c) BCAA uptake in Control KD and Bckdha KD adipocytes.
(d) Absolute lipogenic flux of leucine to palmitate synthesis.
(e) Relative abundance of the most abundant fatty acids.
(f) Quantitative PCR analysis of adipocyte–specific gene expression. Data represents 2 biological replicates analyzed via 2–way ANOVA with Holm–Sidak’s multiple comparisons test.
(g) Representative images of adipocyte differentiation (scale bar: 200 μm) From left: Control KD, Bckdha KD1, Bckdha KD2.
Data presented in (b)–(e) represent mean ± s.d. (f) is presented as mean ± s.e.m. Asterisks in (b)–(e) represent significance compared to Control KD where *represents p<0.05, **represents p<0.01, and ***represents p<0.001. Unless otherwise specified, data is 3 technical replicates representative of 3 biological replicates analyzed via two–way ANOVA with Holm–Sidak’s multiple comparison test.
Discussion
Here we have explored how specific nutrients differentially contribute to mitochondrial metabolism as a function of adipocyte differentiation. Human and murine cell lines from diverse tissue origins, including human visceral and subcutaneous pre–adipocytes, predominantly obtained AcCoA for DNL from glucose and glutamine when proliferating. Upon differentiation the relative conversion of glucose and glutamine carbon to fatty acids was reduced while significant de novo glutamine synthesis was initiated. At the same time, differentiated adipocytes catabolized BCAAs such that leucine and isoleucine accounted for approximately one–third of lipogenic AcCoA. Even in the presence of more physiological concentrations of BCAAs this catabolism was sustained via protein catabolism, as significant dilution arising from protein turnover was observed in differentiated adipocytes. These data indicate that proteinogenic amino acids serve as a significant carbon source for mitochondrial metabolism (and DNL) in differentiated adipocytes, similar to results obtained with pancreatic cancer cells cultured in low glutamine medium33.
The suppression of BCAA catabolism during proliferation is consistent with the dedicated utilization of these nutrients for protein biosynthesis. Mammalian target of rapamycin (mTOR) activity and associated protein translation required for proliferation is sensitive to leucine availability in particular28,34. The differentiation–dependent induction of BCAA catabolism in 3T3–L1 cells and human pre–adipocytes demonstrated here may therefore serve as a model system for identifying key activators or suppressors of BCAA metabolism. PPARγ activity increases significantly during early adipogenesis and has recently been associated with BCAA catabolic enzyme expression in human tissues, animals, and cell models13,15,19,20. However, additional transcription factors and co–activators driving this process may be identified through more systematic analyses.
Our results also demonstrate the critical role of vitamins as enzyme cofactors in mediating important metabolic processes and highlight potential deficiencies that could affect various model systems. In our hands and other published studies, differentiated 3T3–L1 cells accumulated significant levels of OCFAs31. We confirmed these metabolites arose from the accumulation of PropCoA and extension by fatty acid synthase (FASN) rather than α–oxidation35. Supplementation of cobalamin suppressed OCFA and MMA abundances in culture, suggesting that 3T3–L1 cells have the appropriate mitochondrial machinery for converting cobalamin to the activated AdoCbl cofactor necessary for MUT activity. Given the severe pathologies associated with acute, untreated methylmalonic aciduria36, our results highlight the importance of investigating mitochondrial function as it correlates with cobalamin availability in other cell types that catabolize BCAAs. These findings also demonstrate that standard culture conditions for differentiated 3T3–L1 cells are inadequate to support complete BCAA oxidation. Indeed, the heart, muscle, and kidney are known to metabolize BCAAs and may be particularly affected by deficiencies in this cofactor7,8,37. Numerous biological models that are routinely used to represent these tissues (e.g., rat cardiomyocytes, primary murine myocytes and adipocytes) employ media that may lack sufficient cobalamin for BCAA catabolism, as has been observed in glial cells38. MMA and OCFA accumulation may affect results obtained from such systems given the demonstrated pathology associated with these metabolites39.
Elevated plasma BCAAs, methionine, OCFAs, as well as C3– and C5–acylcarnitine species have all been associated with obesity and insulin resistance40,41. While biotin availability is hypothesized to be a potential unifying cause of perturbations in these pathways10, cobalamin–deficiency might also compromise BCAA catabolism in a similar manner to biotin. Indeed, a recent flux model of adipocyte metabolism that incorporated clinical proteomic and transcriptional data identifies MMA–CoA conversion to Succinyl–CoA as a downregulated pathway in obese compared to lean patients42. We observed that cobalamin deficiency induces the accumulation of downstream intermediates arising from BCAA catabolism (carnitine is also notably limited in culture systems employing dialyzed serum). Notably, several studies indicate that patients with T2DM are commonly deficient in cobalamin due to malabsorption caused by metformin or other causes43,44. Furthermore, the expression of cobalamin transporters and cobalamin availability itself can influence the composition of the gut microbiome45, highlighting the complexity in which this cofactor can impact human health.
While BCAA catabolism is clearly induced during adipogenesis, functional knockdown of the pathway in pre–adipocytes impaired lipid accumulation and differentiation. These data provide the first direct evidence that BCAA oxidation is important for adipogenic differentiation. Notably, the presence or absence of cobalamin did not elicit similar effects on differentiation to Bckdha KD (data not shown). Presumably, some metabolite(s) along the BCAA catabolic pathway that are upstream of PropCoA and AcCoA could mediate the observed effects on adipogenesis, though additional studies are required to identify such a regulatory effect. Our demonstration of a link between BCAA catabolism and adipogenesis is consistent with previous observations in Bcat2–/– mice, which exhibit decreased adiposity amongst other metabolic changes16–18. These results may also provide insights into potential adipose–mediated pathogenesis in patients with maple syrup urine disease (MSUD). However, cell autonomous BCAA catabolism is likely not a prerequisite for adipogenic differentiation, as demonstrated by the moderate effects noted here and the (relative) health of MSUD patients. Collectively, our quantitative findings highlight critical roles for BCAA catabolism and vitamin availability in adipocyte differentiation, lipogenesis, and bioenergetics.
Online Methods
Imaging
A 0.35% (w/v) Oil Red O stock was prepared in isopropanol and filtered through a 0.22 μm filter. Cells were washed with PBS, fixed with 4% paraformaldehyde for 30 minutes at room temperature, washed 2X in milli-Q water, washed once with 60% isopropanol, and finally stained with 3:2 (stock:water) solution for 30 minutes. Stain solution was removed from cells and cells were washed 4X with milli-Q water.
Pre-adipocyte isolation
All procedures for AT explant culture were carried out using sterile techniques. Fresh SAT was minced and placed into a 4% BSA/HEPES salts buffer containing 333 units collagenase (Worthington Biochemical Corp.) per mL buffer (~1000 units per gram of AT) and incubated for 1 hour in a 37°C shaking water bath. Following digestion, the suspension was filtered through a 450 μm nylon mesh (Component Supply Company), centrifuged at 50xg for 10 min at 25°C. The pellet and infernatant containing the SVF was aspirated and centrifuged through 250 μm nylon mesh, then centrifuged at 800xg for 10 min at 20°C. Further pre-adipocyte isolation and culture was performed as previously described23.
Respirometry
Respirometry was conducted using a Seahorse XF96 Analyzer. For respiration in intact cells, cells were plated at 5×103 cells/well and maintained and differentiated as described above. 10 days after differentiation was initiated, growth medium was replaced with unbuffered DMEM (Sigma #5030) supplemented with 8 mM glucose, 3 mM glutamine, 1 mM pyruvate, and 0.5 mM carnitine. Mitochondrial respiration is calculated as the oxygen consumption rate sensitive to 1 μM rotenone and 2 μM antimycin A. Immediately after the experiment, rates were normalized for cell number using CyQuant (Life Technologies) and scaled appropriately.
For respiration in permeabilized cells, cells were seeded onto XF96 plates at 1×104 cells/well and were maintained and differentiated as described above. Measurements of differentiated cells were conducted 10–14 days after differentiation was initiated. Respiration was initially measured in MAS medium supplemented with 0.2% (w/v) BSA, 4 mM ADP, and 3 nM recombinant Perfringolysin O (purchased as XF Plasma Membrane Permeabilizer, Seahorse Bioscience)26 and sequentially offered 2 μg/mL oligomycin, successive additions of 2 μM FCCP, and 2 μM antimycin A. Permeabilized adipocytes were offered 5 mM branched chain keto acids with 0.5 mM malate (except KIV, for which only FADH2-linked respiration was measured) and concentrations for commonly used respiratory substrates are as described previously27. Uncoupler-stimulated respiration was calculated as the difference between the maximum respiratory rate in response to FCCP that was sensitive to antimycin A. All data were normalized to total cell protein as measured by the bicinchoninic acid assay (Pierce) with matched plates of whole cells.
Extracellular Flux measurements
A Yellow Springs Instruments (YSI) 2950 was used to obtain extracellular flux measurements. Briefly, spent media was centrifuged at 300xg for 5 minutes to remove cell debris. Culture medium supernatants were centrifuged at 15,000 RPM for 15 minutes to remove impurities and analyzed for glucose, lactate, glutamine, and glutamate measurements. Amino acid uptake and secretion was calculated using GC-MS derivatization of fresh and spent media with internal labeled standards for quantification.
Isotopomer spectral analysis
Mass isotopomer distributions were determined by integrating metabolite ion fragments4 summarized in Table S3 and corrected for natural abundance using in-house algorithms. ISA analysis was conducted using INCA49 and a more complete model for fatty acid synthesis that accounted for production of myristate, palmitate, oleate, and stearate (Table S1).
Cell culture and reagents
All reagents were purchased form Sigma–Aldrich unless otherwise noted. All media and sera were purchased from Life Technologies unless otherwise stated. Murine 3T3–L1 pre–adipocytes were purchased from the American Type Culture Collection and cultured in high glucose Dulbecco’s modified Eagle medium (DMEM) (Life Technologies) supplemented with 10% bovine calf serum (BCS) below 70% confluence. Cells were regularly screened for mycoplasma contamination. For differentiation, 10,000 cells/well were seeded into 12–well plates (ThermoFisher) and allowed to reach confluence (termed Day –2). On Day 0 differentiation was induced with 0.5 mM 3–isobutyl–1–methylxanthine (IBMX), 0.25 μM dexamethasone, 1 μg/ml insulin, and 100 nM rosiglitazone in DMEM containing 10% fetal bovine serum (FBS). Media was changed on Day 2 to DMEM + 10% FBS with 1 μg/ml insulin. Day 4, and thereafter DMEM + 10% FBS was used. Cobalamin (500 nM) or 5′–adenosylcobalamin (AdoCbl; 100 nM) were supplemented to cultures when noted.
Human subjects
Weight stable obese subjects undergoing elective laparoscopic gastric bypass via Roux–en–Y for the treatment of obesity were recruited for the study to provide material for pre–adipocyte isolation. The Institutional Review Boards of Scripps Memorial Hospital and the University of California, San Diego approved the studies. All subjects gave informed consent.
Adipose tissue (AT) biopsy
Subcutaneous (S) AT biopsies were obtained from the superficial abdominal SAT depot and visceral (V) AT was obtained from the greater omentum25. Biopsy tissue was placed in a sterile HEPES salts solution as previously described46, and transported to the lab for immediate processing. Pre–adipocyte isolation is described in Supplementary Methods.
Human adipocyte differentiation
Human pre–adipocytes were maintained in DMEM/F12 + 10% FBS. At Day 0 (2 days after confluence was reached), cells were differentiated in DMEM/F12 + 3% FBS, 0.5 mM IBMX, 100 nM insulin, 100 nM dexamethasone, 2 nM T3, 10 μg/mL transferrin, 1 μM rosiglitazone, 33 μM biotin, and 17 μM pantothenic acid47 induction cocktail for 7 days with media changes every 2 days. After induction, cells were maintained in DMEM/F12 with 10 nM insulin and 10 nM dexamethasone until metabolic tracing experiments were conducted on Day 14.
Tracing experiments
All [13C] glucose and amino acid tracers were purchased from Cambridge Isotopes Inc. Stable isotope labeling of intracellular metabolites in differentiated 3T3–L1 cells was accomplished by culturing cells 7 days post–induction in tracer medium for 24 hours unless otherwise stated. Proliferating 3T3–L1, A549, HuH–7, and 143B cells were seeded at 10,000 cells/cm2 and cultured in tracer medium for 48 hours. Custom DMEM or DMEM/F12 (Hyclone Laboratories, Inc.) was formulated with the specified tracer, unlabeled versions of other chemical components, and 10% dialyzed FBS such that all nutrients were available to cells in all experiments (with one or more labeled). Complete formulation of DMEM and Low gluc+AA media can be found in Supplementary Table 3. Mole percent enrichment (MPE) of isotopes was calculated as the percent of all atoms within the metabolite pool that are labeled:
where n is the number of carbon atoms in the metabolite and Mi is the relative abundance of the ith mass isotopomer.
Gas Chromatography/Mass Spectrometry (GC/MS) analysis
Polar metabolites and fatty acids were extracted using methanol/water/chloroform and analyzed as previously described48. Proliferating 3T3–L1 cells and primary human adipocytes were cultured in 6–well plates, while differentiated 3T3–L1 cells were cultured in a 12–well plate and the volumes of extraction buffers were adjusted accordingly. Polar metabolites were derivatized in 20 μl of 2% (w/v) methoxyamine hydrochloride (Thermo Scientific) in pyridine and incubated at 37°C for 60–90 minutes. Samples were then silylated with 30 μl of N–tertbutyldimethylsilyl–N–methyltrifluoroacetamide (MTBSTFA) with 1% tert–butyldimethylchlorosilane (tBDMS) (Regis Technologies) at 37°C for 30–45 minutes. Samples were centrifuged at 15,000 RPM for 5 minutes and supernatant was transferred to GC sample vials for analysis. Extracted nonpolar metabolites were evaporated, saponified, and esterified to form fatty acid methyl esters (FAMEs) through addition of 500 μl 2% (w/v) H2SO4 in methanol and incubation at 50°C for 90–120 minutes. FAMEs were extracted after addition of 100 μl saturated NaCl solution with two 500 μl hexane washes and evaporated to dryness before redissolving in 50–100 μl of hexane and transfer to glass GC vials for analysis. Derivatized samples were analyzed by GC–MS using a DB–35MS column (30m × 0.25 mm i.d. × 0.25 μm, Agilent J&W Scientific) installed in an Agilent 7890A gas chromatograph (GC) interfaced with an Agilent 5975C mass spectrometer (MS).
For quantitation of amino acids in plasma samples an isotope–labeled internal standard mixture was prepared and added during extraction.
Lentiviral production and shRNA KD of Bckdha
Glycerol stocks of TRC2–pLKO.1–puro shRNA targeting mouse Bckdha (KD1: NM_007533.2–421s1c1: CCGGTCCTTCTACATGACCAACTATCTCGAGATAGTTGGTCATGTAGAAGG ATTTTTG; KD2: NM_007533.2–1188s1c1: CCGGGCAGTCACGAAAGAAGGTCATCTCGA GATGACCTTCTTTCGTGACTGCTTTTTG), and a non–targeting control construct were purchased from Sigma Aldrich, packaged in HEK293T cells using the transfection agent Fugene 6 and required packaging plasmids VSV–G, gag/pol, and rev. HEK293T medium containing lentiviral constructs was collected two days later and filtered (0.45 μm). Polybrene was added to a final concentration of 6 μg/mL. 3T3–L1 pre–adipocytes were infected with 0.5 mL of virus–containing medium in a 6–well plate for 4 hours before addition of 2 mL of virus–free medium. After 24 hours of recovery, transduced cells were selected with 2 μg/mL puromycin. Cells were then plated to 12–well plates for differentiation as described above but without rosiglitazone. Puromycin was removed from the medium beginning on Day 0.
RNA isolation and quantitative RT–PCR
Total RNA was purified from cultured cells using Trizol Reagent (Life Technologies) per manufacturer’s instructions. First–strand cDNA was synthesized from 1 μg of total RNA using iScript Reverse Transcription Supermix for RT–PCR (BioRad Laboratories) according to the manufacturer’s instructions. Individual 20 μL SYBR Green real–time PCR reactions consisted of 2 μL of diluted cDNA, 10 μL of SYBR Green Supermix (BioRad), and 1 μL of each 5μM forward and reverse primers. For standardization of quantification, 18S was amplified simultaneously. The PCR was carried out on 96 well plates on a CFX Connect Real time System (Bio–Rad), using a three–stage program provided by the manufacturer: 95°C for 3 min, 40 cycles of 95°C for 10 sec and 60°C for 30 sec. Gene–specific primers used are listed in Supplementary Table 4.
Western Blots
3T3–L1 adipocytes with Control KD or Bckdha KD were lysed in ice-cold RIPA buffer with 1x protease inhibitor (Sigma-Aldrich. 30 μg of total protein was separated on 12% SDS-PAGE gel. The proteins were transferred to a nitrocellulose membrane and immunoblotted with rabbit anti-Bckdha (Novus Biologicals NBP1-79616) (1:1000 dilution) and mouse anti-Beta-Actin (Cell Signaling 8H10D10) (1:5,000). Specific signal was detected with horseradish peroxidase-conjugated secondary antibody goat anti-rabbit (1:2,500) or rabbit anti-mouse (1:10,000) using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and developed using Blue Devil Autoradiography film (Genesee Scientific).
Statistical analysis
All results shown as averages of multiple independent experiments are presented as mean ± SEM; results shown as averages of technical replicates representative of biological replicates are presented as mean ± SD. P values were calculated using Student’s two–tailed t test or ANOVA, as required; *, P value between 0.01 and 0.05; **, P value between 0.001 and 0.01; ***, P value <0.001. Errors associated with MFA and ISA of lipogenesis are 95% confidence intervals determined via sensitivity analysis.
Supplementary Material
Acknowledgments
We thank Dr. Robert R. Henry for human adipocyte material and Dr. Todd Coleman for use of equipment. This work was supported, in part, by NIH grant R01CA188652 (C.M.M.), California Institute of Regenerative Medicine (CIRM) Award RB5–07356 (C.M.M.), DOD grant W81XWH–13–1–0105 (C.M.M.), and a Searle Scholar Award (C.M.M.) as well as grants from the American Diabetes Association 7–05–DCS–04 and the Medical Research Service 1 I010X00635–01A1, Department of Veterans Affairs, VA San Diego Healthcare System (R.R.H.), NIH grant 1R01NS087611 (A.N.S.), and Seahorse Bioscience.
Footnotes
Author Contributions
S.A.P. and T.P.C. obtained biopsies and isolated human pre–adipocytes from adipose tissue; A.S.D. and A.N.M. performed oxygen consumption experiments; C.R.G and M.W. performed all other experiments. C.R.G., M.W., A.S.D., A.N.M., T.P.C., and C.M.M. designed research; C.R.G., M.W., and C.M.M. wrote the paper with help from all authors.
No potential conflicts of interest relevant to this article were reported.
References
- 1.Rosen E, Spiegelman B. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steppan C, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 3.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell metabolism. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turer AT, Scherer PE. Adiponectin: mechanistic insights and clinical implications. Diabetologia. 2012;55:2319–2326. doi: 10.1007/s00125-012-2598-x. [DOI] [PubMed] [Google Scholar]
- 5.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahn BB, Flier JS. Obesity and insulin resistance. Journal of Clinical Investigation. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buse MG, Biggers JF, Friderici KH, Buse JF. Oxidation of branched chain amino acids by isolated hearts and diaphragms of the rat. The effect of fatty acids, glucose, and pyruvate respiration. The Journal of biological chemistry. 1972;247:8085–8096. [PubMed] [Google Scholar]
- 8.Rosenthal J, Angel A, Farkas J. Metabolic fate of leucine: a significant sterol precursor in adipose tissue and muscle. The American journal of physiology. 1974;226:411–418. doi: 10.1152/ajplegacy.1974.226.2.411. [DOI] [PubMed] [Google Scholar]
- 9.Newgard C, et al. A branched–chain amino acid–related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiehn O, et al. Plasma metabolomic profiles reflective of glucose homeostasis in non–diabetic and type 2 diabetic obese African–American women. PloS one. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nature medicine. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedishvili N, Popov K, Jaskiewicz J, Harris R. Coordinated expression of valine catabolic enzymes during adipogenesis: analysis of activity, mRNA, protein levels, and metabolic consequences. Archives of biochemistry and biophysics. 1994;315:317–322. doi: 10.1006/abbi.1994.1506. [DOI] [PubMed] [Google Scholar]
- 13.Si Y, Yoon J, Lee K. Flux profile and modularity analysis of time–dependent metabolic changes of de novo adipocyte formation. American journal of physiology Endocrinology and metabolism. 2007;292:E1637–1646. doi: 10.1152/ajpendo.00670.2006. [DOI] [PubMed] [Google Scholar]
- 14.Chuang DT, Hu CW, Patel MS. Induction of the branched–chain 2–oxo acid dehydrogenase complex in 3T3–L1 adipocytes during differentiation. The Biochemical journal. 1983;214:177–181. doi: 10.1042/bj2140177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackey D, et al. Regulation of adipose branched–chain amino acid catabolism enzyme expression and cross–adipose amino acid flux in human obesity. American journal of physiology Endocrinology and metabolism. 2013;304:E1175–E1187. doi: 10.1152/ajpendo.00630.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.She P, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell metabolism. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman M, She P, Peroni O, Lynch C, Kahn B. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. The Journal of biological chemistry. 2010;285:11348–11356. doi: 10.1074/jbc.M109.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman HA, Olson KC, Chen G, Lynch CJ. Adipose transplant for inborn errors of branched chain amino acid metabolism in mice. Molecular genetics and metabolism. 2013;109:345–353. doi: 10.1016/j.ymgme.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sears D, et al. Mechanisms of human insulin resistance and thiazolidinedione–mediated insulin sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18745–18750. doi: 10.1073/pnas.0903032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao G, et al. Multi–tissue, selective PPARy modulation of insulin sensitivity and metabolic pathways in obese rats. American Journal of Physiology Endocrinology and Metabolism. 2010;300:E164–174. doi: 10.1152/ajpendo.00219.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalski TJ, Watford M. Production of glutamine and utilization of glutamate by rat subcutaneous adipose tissue in vivo. The American journal of physiology. 1993;266:E151–154. doi: 10.1152/ajpendo.1994.266.1.E151. [DOI] [PubMed] [Google Scholar]
- 23.Tchkonia T, et al. Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. American journal of physiology. Endocrinology and metabolism. 2004;288:E267–277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- 24.Lee M-JJ, Wu Y, Fried SK. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Molecular aspects of medicine. 2013;34:1–11. doi: 10.1016/j.mam.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips SA, Ciaraldi TP, Oh DK, Savu MK, Henry RR. Adiponectin secretion and response to pioglitazone is depot dependent in cultured human adipose tissue. American journal of physiology. Endocrinology and metabolism. 2008;295:E842–850. doi: 10.1152/ajpendo.90359.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Divakaruni AS, Rogers GW, Murphy AN. Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark–Type Oxygen Electrode. Current protocols in toxicology. 2013;60:1–16. doi: 10.1002/0471140856.tx2502s60. [DOI] [PubMed] [Google Scholar]
- 27.Divakaruni AS, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5422–5427. doi: 10.1073/pnas.1303360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cynober L. Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition. 2002;18:761–766. doi: 10.1016/s0899-9007(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts L, Virtue S, Vidal–Puig A, Nicholls A, Griffin J. Metabolic phenotyping of a model of adipocyte differentiation. Physiological genomics. 2009;39:109–119. doi: 10.1152/physiolgenomics.90365.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapadia CR. Vitamin B12 in health and disease: part I––inherited disorders of function, absorption, and transport. Gastroenterologist. 1995;3:329–344. [PubMed] [Google Scholar]
- 33.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras–transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch CJ, et al. Potential role of leucine metabolism in the leucine–signaling pathway involving mTOR. American journal of physiology. Endocrinology and metabolism. 2003;285:854–863. doi: 10.1152/ajpendo.00153.2003. [DOI] [PubMed] [Google Scholar]
- 35.Su X, et al. Sequential Ordered Fatty Acid α Oxidation and Δ9 Desaturation Are Major Determinants of Lipid Storage and Utilization in Differentiating Adipocytes†. Biochemistry. 2004;43:5033–5044. doi: 10.1021/bi035867z. [DOI] [PubMed] [Google Scholar]
- 36.Haarmann A, et al. Renal involvement in a patient with cobalamin A type (cblA) methylmalonic aciduria: a 42–year follow–up. Molecular genetics and metabolism. 2013;110:472–476. doi: 10.1016/j.ymgme.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 37.Birn H. The kidney in vitamin B12 and folate homeostasis: characterization of receptors for tubular uptake of vitamins and carrier proteins. American Journal of Physiology – Renal Physiology. 2006;291:22–36. doi: 10.1152/ajprenal.00385.2005. [DOI] [PubMed] [Google Scholar]
- 38.Barley FW, Sato GH, Abeles RH. An effect of vitamin B12 deficiency in tissue culture. Journal of Biological Chemistry. 1972;247:4270–4276. [PubMed] [Google Scholar]
- 39.Kishimoto Y, Williams M, Moser HW, Hignite C, Biemann K. Branched–chain and odd–numbered fatty acids and aldehydes in the nervous system of a patient with deranged vitamin B12 metabolism. Journal of Lipid Research. 1973;14:69–77. [PubMed] [Google Scholar]
- 40.Newgard C. Interplay between lipids and branched–chain amino acids in development of insulin resistance. Cell metabolism. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin–resistant state. Advances in nutrition (Bethesda, Md ) 2011;2:445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mardinoglu A, et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Molecular Systems Biology. 2013:9. doi: 10.1038/msb.2013.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinstatler L, Qi YP, Williamson RS, Garn JV, Oakley GP., Jr Association of biochemical B12 deficiency with metformin therapy and vitamin B12 supplements: the National Health and Nutrition Examination Survey, 1999–2006. Diabetes care. 2012;35:327–333. doi: 10.2337/dc11-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang D, et al. Higher Prevalence of Metformin–Induced Vitamin B12 Deficiency in Sulfonylurea Combination Compared with Insulin Combination in Patients with Type 2 Diabetes: A Cross–Sectional Study. PLoS ONE. 2014;9:e109878. doi: 10.1371/journal.pone.0109878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell host & microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips SA, et al. Selective regulation of cellular and secreted multimeric adiponectin by antidiabetic therapies in humans. American journal of physiology. Endocrinology and metabolism. 2009;297:767–773. doi: 10.1152/ajpendo.00378.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee M-JJ, Wu Y, Fried SK. A modified protocol to maximize differentiation of human preadipocytes and improve metabolic phenotypes. Obesity. 2012;20:2334–2340. doi: 10.1038/oby.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metallo C, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.