Abstract
Diabetes is characterized, in part, by activation of toxic oxidative and glycoxidative pathways that are triggered by persistent hyperglycemia and contribute to diabetic complications. Inhibition of these pathways may benefit diabetic patients by delaying the onset of complications. One of such inhibitors, pyridoxamine (PM), had shown promise in clinical trials. However, the mechanism of PM action in vivo is not well understood. We have previously reported that hypohalous acids can cause disruption of structure and function of renal collagen IV in experimental diabetes (Brown et al., Diabetes, 2015). In the present study, we demonstrate that PM can protect protein functionality from hypochlorous and hypobromous acid-derived damage via a rapid direct reaction with and detoxification of these hypohalous acids. We further demonstrate that PM treatment can ameliorate specific hypohalous acid-derived structural and functional damage to renal collagen IV network in diabetic animal model. These findings suggest a new mechanism of PM action in diabetes, namely sequestration of hypohalous acids, which may contribute to known therapeutic effects of PM in human diabetic nephropathy.
Keywords: Diabetes, Nephropathy, Hypochlorous acid, Hypobromous acid, Protein halogenation, Post-translational modifications, Pyridoxamine
Introduction
Complications such as nephropathy are serious long-term consequences of diabetes pandemic. Among the proposed mechanisms of diabetic complications is the activation of oxidative pathways that cause damage to macromolecules thus promoting organ dysfunction [1]. Inhibition of these pathogenic pathways can protect macromolecules from damage and delay or prevent the development of diabetic complications.
Pyridoxamine (PM) is an investigational drug for treatment of diabetic nephropathy that is currently in Phase 3 clinical trial [2-3]. It has been shown to inhibit several oxidative pathways relevant to diabetes under in vitro conditions [4]. However, the information about PM therapeutic mechanism(s) in vivo is limited. The proposed mechanisms so far consider PM action upon lipid oxidation and metabolism [5-6].
Since more recent experimental evidence suggests that hypohalous acids may contribute to pathogenic mechanisms of diabetic complications [7-8], we set out to determine whether PM can alleviate structural and functional oxidative protein damage caused by hypohalous acids. We demonstrated that PM can rapidly react with hypohalous acids thus effectively inhibiting modification of protein amino acid side chains. This resulted in protection of protein functionalities, including enzymatic activity of RNase, binding of integrin α1β1 to extracellular matrix (ECM) and folding and assembly of NC1 domain of collagen IV, from damage by either hypochlorous (HOCl) or hypobromous (HOBr) acids. Furthermore, treatment of diabetic animals with PM significantly inhibited accumulation of HOCl-derived modifications in collagen IV compared to untreated diabetic animals. Our data suggest that PM treatment can target hypohalous acid-induced protein damage in diabetes. Since PM therapy has shown promise in early stages of diabetic nephropathy [3], these results also suggest a role for protein damage by hypohalous acids in pathogenesis of diabetic complications.
Experimental
Materials
Pancreatic ribonuclease A was purchased from Worthington Biochemical Corparation. L-Lysine, Nα-Acetyl-L-Lysine, NεAcetyl-L-Lysine, DLMethionine, NAcetyl-DL-Methionine, Histidine, NαAcetyl-DL-Histidine, Imidazole, NαAcetyl-L-Arginine, NαAcetyl-L-Glutamine, NαAcetyl-DL-Tryptophan, NαAcetyl-DLTyrosine, Ribonucleic acid, Chloramine T trihydrate, Potassium Bromide, Lanthanum nitrate hydrate, Sodium Iodide, 3,3′,5,5′-Tetramethylbenzidine (TMB), N,N-Dimethyl formamide (DMF), Pyridoxamine dihydrochloride, Guanidinium Hydrochloride, Sodium Hypochlorite solution, proteinase K and type IV collagen from Engelbreth-Holm-Swarm murine sarcoma basement membrane were purchased from Sigma-Aldrich. Human Integrin α1β1 was purchased from Millipore and β1 Integrin antibody was purchased from Santa Cruz Biotechnology Inc.
Measurements of RNase Activity
RNase activity was determined by measuring the formation of acid-soluble oligonucleotide as described by Kalnitsky et al[9], with some modifications previously described by Voziyan et al [10]. For the assay, 100 μL of 3 μg/mL RNase in 100 mM sodium acetate, pH 5.0, was mixed with 100 μL of 1% yeast RNA in the same buffer. After the incubation at 37°C for 5 min, the reaction was stopped by the addition of 100 μL of an ice-cold solution of 0.8% lanthanum nitrate in 18% perchloric acid. The incubations were kept on ice for 5 min to ensure complete precipitation of undigested RNA and then centrifuged at 12000g for 10 min. An aliquot of the supernatant (20 μL) was diluted to 1 mL with distilled water, the amount of digested (solubilized) RNA was determined by measuring the absorbance at 260 nm. The activity of RNase incubated either alone or with PM or an amino acid substitute at 37°C was monitored separately and used as a reference for each incubation time. This reference activity did not change significantly over the course of incubation.
Analysis of PM reactions with hypohalous acids by UPLC and ESI-MS
UPLC analyses were performed using Waters Acquity UPLC system equipped with Waters fluorescence and photodiode array detectors. The sample aliquot (10 μL) was loaded onto Phenomenex Synergy 4u Hydro-RP 80A (150 × 4.6 mm) column and eluted at a flow rate of 1 ml/min with the following gradient: 0-5 min, 100% buffer A; 5-15 min, linear gradient to 25% buffer B; 15-15.1 min, change to 100% buffer B; 15.1-20 min, 100% buffer B; 20-20.1 min, change to 100% buffer A; 20.1-25 min, 100% buffer A. Buffer A was 0.5% formic acid in water, buffer B was 0.5% formic acid in ACN. The UV detector was set at 295 nm. Electrospray ionization mass spectrometry (ESI-MS/MS, product ion sgans) was performed by infusion of column eluate into Thermo TSQ mass spectrometer in a positive ions mode with ranges from m/z 40-80 to m/z value of about 10 amu greater than that of each precursor ion. Tandem MS spectra were collected at CID voltage of 25V.
N-Chloramine Assay
The presence of N-chloramines were determined by the method of Witko et al.[11] and Dypbukt et al.[12]. The former method is based on the colorimetric measurement of triiodide ions formed by the oxidation of potassium iodide (KI) in solution. Chloroamine-T, a commercially available source of N-chloramine, was used to calibrate the assay. 100 mM chloramine-T solution was made fresh in distilled H2O. The 100 mM chloramines-T was diluted to final concentrations ranging from 20 to 100 μM immediately before use. The direct oxidation of KI by NaOCl was also determined and these values were subtracted as background from the corresponding treated samples. In the alternative method the developing reagent was composed of 2 mM TMB in 400 mM acetate buffer, pH 5.4, containing 10% DMF and 100 μM sodium iodide. This solution was prepared by dissolving TMB in 100% DMF, diluting with acetate buffer to get the desired final concentration of TMB, and then adding sodium iodide. When the developing reagent was mixed with the solutions of chloramines, the final concentration of TMB was always at least 10 times that of the concentration of chloramines. This prevented further oxidation of the reaction product.
Cell Culture and Preparation of ECM
PFHR-9 cells were grown in 6 well culture treated plates and maintained at confluency for 7 days in the presence of 50 μM ascorbic acid. To remove cells from matrix, wells were briefly rinsed in 2 mL of hypotonic buffer (10 mM tri [pH 7.4], 0.1% CaCl2, 0.1% BSA) and subsequently incubated for 10 min in 1 mL of fresh hypotonic buffer. After this, wells underwent two 5 minute washes in 1 mL hypotonic buffer + 0.5% Trition X-100, and then two quick washes in 1 mL hypotonic buffer + 0.1% sodium deoxycholate. Sample material was returned to saline conditions by three brief washes of 2 mL each in PBS. Following this procedure, wells contained plate-bound matrix that is devoid of cells. After the removal of the buffer, plates were stored at −20°C until use.
Chlorination of Type IV Collagen and PFHR-9 Matrix Proteins
Type IV collagen from Engelbreth-Holm-Swarm (EHS) murine sarcoma basement membrane was immobilized on 96-well plates in 100 mM sodium phosphate buffer at 4°C overnight. Plates coated with EHS collagen or with ECM deposited by PFHR-9 cells were washed twice with 100 mM sodium phosphate, pH 7.5, and incubated in the same buffer with or without different concentrations of HOCl alone or HOCl and PM. Incubations were carried out for 1 hr in the dark at RT, washed twice with 100 mM sodium phosphate buffer and developed with N-Chloramine Assay for 2 h followed by detection at 340 nm.
Modification of Collagen IV and Integrin Binding Assay
20 μg/mL of type IV collagen in 20 mM sodium phosphate buffer, 7.5, was immobilized on 96 well plates at 4°C overnight. Nonspecific binding sites were blocked with 1% BSA in TBS for 2 hr at 30°C as previously described [13]. Wells were washed five times with either TBS or 100 mM sodium phosphate, pH 7.5, before being incubated in the same buffer in the presence of either HOCl or HOBr with or without PM. Incubations were carried out for 1 hr in the dark at RT and later washed five times with TBS before purified α1β1 integrin was overlaid in binding buffer (TBS, 0.1% BSA, 1 mM MgCl2, 0.2 mM MnCl2, 5 mM octylglucoside) and incubated for 2 hr at 30°C. In some incubations, EDTA (10 mM) was added to the binding buffer. The plates were washed with TBS, 1 mM MgCl2, 0.2 mM MnCl2, 0.01% Tween 20 and incubated with β1 integrin antibody (4B7R,1:500) for 1 hr. After extensive washing, the bound antibodies were detected using alkaline phosphatase-conjugated antimouse IgG antibodies. p-Nitrophenyl phosphate substrate (Sigma) was added to the wells, and aborbance was read at 410 nm. Readings from the samples containing EDTA were subtracted from all the data to obtain the baseline corrected magnesium dependent binding.
Animal experiments
The Principles of Laboratory Animal Care were followed according to institutional IACUC guidelines and approved protocol. Male Sprague-Dawley rats (Charles River, Wilmington, MA) were housed in a 12-hour light : dark cycle and temperature controlled animal facility. Standard food and water were available ad libitum. Animals were randomly assigned to control, diabetic or diabetic/PM groups. Diabetes was induced with intravenous administration of streptozotocin (Sigma, St. Louis, MO), 60 mg/kg of body weight, in a saline solution. Control rats were injected only with saline solution. Diabetes was confirmed if serum glucose concentration was at least 250 mg/dl on two consecutive days, with glucose testing beginning on the second day post STZ administration. Thereafter, in the diabetic animals, serum glucose was measured three times per week for 12 weeks using the glucose oxidase method (Glucose Analyzer II, Beckman Instruments, Palo Alto, CA). Diabetic rats in PM-treatment group were given 2 mg/mL PM in drinking water as previously described [14].
Isolation of collagen IV NC1 domains from renal tissues
Collagen IV NC1 domains were isolated from the kidneys of control rats, STZ-diabetic rats, and STZ-diabetic rats treated with PM as previously described [15], with minor modifications. Rat kidneys were homogenized in protease inhibitor cocktail with a homogenizer, and washed 3 times with inhibitors (precipitated by centrifugation at 8,000 × g for 10 min). The pellet was washed three times with the same solution and then suspended in 0.05% sodium azide and shaken for 1 h on ice and precipitated by centrifugation at 8,000 × g for 10 min. The pellet was washed with the same solution and then suspended in 1 M NaCl containing 200 Kunitz units/ml DNase, 2 mM phenylmethylsulfonyl fluoride, 0.1% protease inhibitors and gently shaken for 90 min at room temperature. The solution was precipitated by centrifugation at 8,000 × g for 10 min and the pellet re-suspended in 1% sodium deoxycholate and gently shaken for 1 h on ice. The resulting pellet was washed three times with distilled water. Detergent-prepared basement membranes from rat kidneys were solubilized by digestion with bacterial collagenase as previously described [16]. Collagenase digest was dialyzed against 50 mM Tris-HCl, pH 7.5, passed through DEAE column and a pass-through fraction was collected. This fraction was further purified using Superdex 200 10/300 GL gel-filtration column (GE Healthcare) as previously described [17].
For hexamer stability study, NC1 domains isolated from kidneys of control rats, diabetic rats and diabetic rats treated with PM were incubated in 50 mM Tris acetate buffer, pH 7.4 with 6M GdnCl at 80°C for 30 min followed by the injection onto Superdex 200 gel-filtration column equilibrated with 50 mM Tris acetate buffer, pH 7.4 and elution with the same buffer using AKTApurifier FPLC system (General Electric).
Modification of NC1 hexamers in vitro and proteolytic digestion studies
Purified NC1 hexamer (1 mg/mL) was incubated in 50 mM TBS, pH 7.5 alone, with 1 mM HOCl or with 1 mM HOCl and 1 mM PM for 2 h at 37°C. Samples were washed 3 times with 50 mM Tris-HCl, pH 8.0 using centrifugal ultrafiltration (MWCO=10 KDa, Merck Millipore) and concentrated to 0.8 mg/mL NC1 hexamer. NC1 hexamer samples were digested at 50°C for 30 min with proteinase K (5:1, w/w) in 50 mM Tris-HCl, pH 8.0, 1 mM DTT. Digestions were stopped by addition of the denaturing loading buffer containing 100 mM DTT and heating at 90°C for 30 min. Samples (8 μg of NC1) were fractionated on 12% SDS-PAGE followed by Coomassie Brilliant Blue staining.
LC-MS/MS analysis of site-specific modifications in RNase and NC1 domain of collagen IV
The site-specific modifications in RNase or in purified collagen IV NC1 domains from rat kidneys were analyzed by liquid chromatography-tandem mass spectrometry. In preparation for LC-MS/MS analysis, RNase and NC1 domain samples were precipitated with 25% trichloroacetic acid on ice for 1 hour, and following centrifugation at 14,000×g, the pellets were washed with cold acetone, dried by speed-vac centrifugation, and reconstituted in 50mM Tris buffer, pH 8, containing 50% trifluoroethanol. Samples were then treated with TCEP to reduce disulfide bonds, cysteine residues were carbamidomethylated with iodoacetamide, and samples were diluted 5-fold with 100 mM Tris to obtain a final 10% TFE solution prior to proteolytic digestion. RNase samples were digested with either sequencing-grade trypsin or endoproteinase LysC, and the collagen IV NC1 domain samples were digested with trypsin. All digests were performed overnight at 37°C, after which, samples were acidified and diluted with 0.1% formic acid. The resulting peptides from each sample were then loaded onto a capillary reverse phase analytical column (360 μm O.D. × 100 μm I.D.) using an Eksigent NanoLC HPLC and autosampler. The analytical column was packed with 20cm of C18 reverse phase material (Jupiter, 3 μm beads, 300Å, Phenomenex), directly into a laser-pulled emitter tip. Peptides were gradient-eluted at a flow rate of 500nL/min, and the mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). A 90-minute gradient was performed, consisting of the following: 0-15 min, 2% B (loading phase); 15-55 min, 2-40% B; 55-65 min, 40-90% B; 65-68 min, 90% B, 68-70 min, 90-2% B, 70-90 min, 2% B. Upon gradient-elution, peptides were mass analyzed on a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific), equipped with a nanoelectrospry ionization source. The instrument was operated using a data-dependent method with dynamic exclusion enabled. For all analyses, full scan (m/z 300-2000) spectra were acquired with the Orbitrap as the mass analyzer (resolution 60,000). For the RNase samples, the twelve most abundant ions in each MS scan were selected for fragmentation via collision-induced dissociation (CID) in the LTQ, while for the NC1 domain samples, the data-dependent method included fragmentation of the sixteen most abundant ions in each MS scan. An isolation width of 2 m/z, activation time of 10 ms, and 35% normalized collision energy were used to generate MS2 spectra. Dynamic exclusion settings allowed for a repeat count of 1 within a repeat duration of 10 seconds, and the exclusion duration time was set to 15 seconds. All tandem mass spectra were converted into DTA files using Scansifter, and RNase data were searched against a bovine subset of the UniProtKB protein database (uniprot.org), while NC1 domain data were searched against a rat database. Protein databases were appended with reversed (decoy) protein sequences to facilitate false discovery rate estimates. Database searches were performed using SEQUEST [18], and results were assembled in Scaffold v 3.6.4 (Proteome Software) with minimum filtering criteria of 95% peptide probability and 99% protein probability. Searches were configured to use variable protein residue modifications, i.e. carbamidomethylation on cysteine (ΔM=57.0215); oxidation of methionine and histidine (ΔM=15.9949); mono- and double-chlorination of lysine, histidine, tyrosine, and tryptophan (ΔM=33.9611 and 67.9222); and mono- and double-bromination of lysine, histidine, tyrosine, and tryptophan (ΔM=77.9105 and 155.8210). For RNase, only modifications of functionally critical residues at positions K1, K7, H12, M13, K41, Y92, Y97, and H119 were considered. Sites of modification were validated by manual interpretation of the raw tandem mass spectra using QualBrowser software (Xcalibur 2.1.0, Thermo Scientific).
To facilitate quantitative analysis of modified RNase peptides, samples were spiked with solutions of three synthetic peptides to be used as internal reference standards during data analysis. The three peptides, bradykinin (Sigma B4184), angiotensin I (Sigma A9650), and angiotensin II (Sigma A8846) were reconstituted in 0.1% formic acid, and serial dilutions were made from 50 pmol/μL to 500 fmol/μL solutions. A combined peptide stock solution was then generated and spiked into each of the samples to arrive at approximately 25 fmol/μL of each peptide synthetic standard. Accurate mass measurements of the modified RNase and internal standard peptides, acquired in the Orbitrap, were used to generate extracted ion chromatograms (XICs). A window of 10 ppm around the theoretical monoisotopic m/z values of the observed precursor ions was utilized for making XICs of the standard peptides and the unmodified, Cl-modified, and Br-modified RNase peptide forms. Using QualBrowser (Xcalibur 2.1.0), the integrated area under the curve (AUC) for each XIC peak was determined. Peak areas calculated for RNase peptides were then normalized to the average of the peak areas of the three internal standard peptides. To determine the relative abundance of modified collagen IV peptides, XICs were similarly generated for the identified modified peptides as well as the corresponding unmodified collagen IV peptide forms. After calculating the AUCs for these peptides, the percent relative abundance of each modified peptide was calculated as a percentage of the summed AUC obtained for the modified and unmodified peptide pairs.
Statistical analyses
Data were expressed as means ± S.D., and statistical analysis was performed using Student's t test for unpaired samples or ANOVA followed by post-hoc Student-Newman-Keuls or Turkey tests. Differences were considered statistically significant if p values were less than 0.05.
Results
Reaction of PM with hypohalous acids
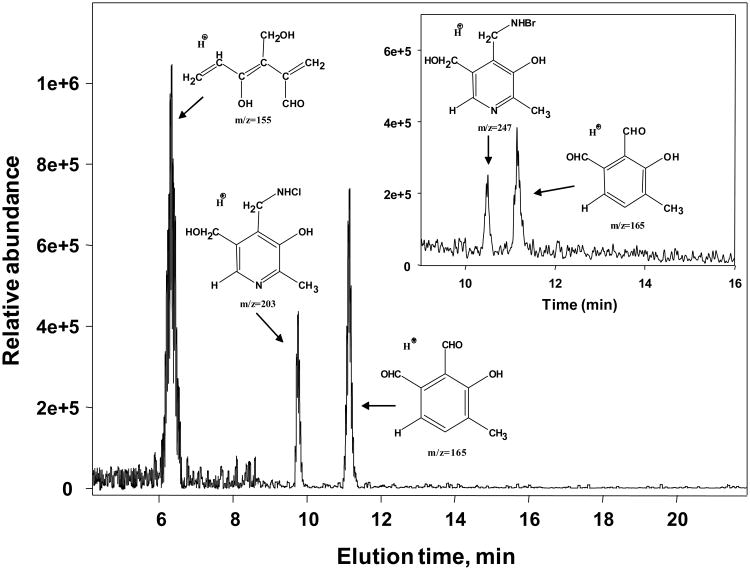
PM was consumed in the presence of either HOCl or HOBr. Analysis of the reaction products using RP-UPLC followed by tandem mass-spectrometry identified several halogenated and oxidized PM species (Fig. 1). The predicted structures of these species (Fig. 1) are based on m/z values and fragmentation patterns shown in Fig. S1. Thus, PM can scavenge hypohalous acids due, in part, to high reactivity of its 4-aminomethyl group [19].
Figure 1.
Predicted structures of major reaction products of PM and hypohalous acids. PM (0.5 mM) and equimolar concentrations of either HOCl (main panel) or HOBr (inset panel) were incubated in 100 mM sodium phosphate buffer in the dark at 37°C for 1 h. The LC-MS/MS analysis was performed using a ThermoScientific TSQ triple quadrupole mass-spectrometer as described under Experimental procedures.
Protection by PM of RNase structure and activity from damage by hypohalous acids
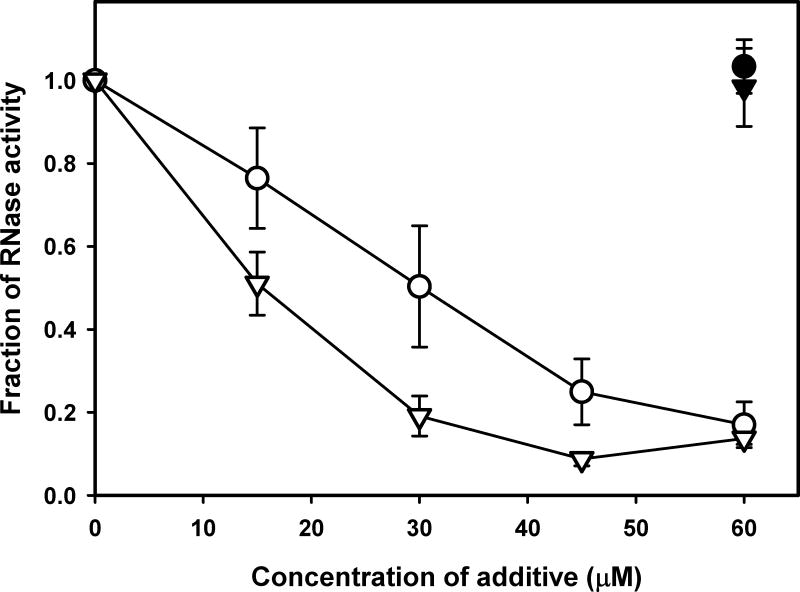
To investigate the impact of halogenation on protein functionality and whether it can be protected by PM, we determined mechanism of inhibition of enzymatic activity by hypohalous acids using RNase as a model. Both HOCl and HOBr inhibited RNase activity in concentration-dependent fashion while PM protected enzymatic activity from hypohalous acid-induced inhibition (Fig. 2). Competition experiments using protected NαAc derivatives of free amino acids were performed to determine relative reactivity of different amino acid side chains and PM towards hypohalous acids (Fig. S2). The results demonstrated that side chains of Met, His, Trp, and Tyr as well as N-terminal α-amino groups are the most likely protein sites to be modified by hypohalous acids (Fig. S2). The lack of protection by free NαAc-Tyr in the case of HOCl (Fig. S2A) is probably due to the fact that protein Tyr residues can be modified by HOCl indirectly via intramolecular reactions with chloramine intermediates on structurally juxtaposed lysine side chains [20]. On the other hand, the direct bromination of Tyr side chain by HOBr may be responsible for the high degree of protection (Fig. S2B). The observed higher reactivity of α-amino group compared to ε-amino group is due to enhancement of the reaction by further conversion of α-chloramine product to aldehyde [21]. PM demonstrated the highest IC50 values compared to amino acids (Fig. S2), indicating that it can effectively protect protein amino acid side chains from modification by hypohalous acids.
Figure 2.
Inhibition of RNase activity by HOCl or HOBr and protection by PM. RNase (15 μg/mL) was incubated alone or with different concentrations of either HOCl (open circles) or HOBr (open triangles) without PM or with 60 μM PM (closed symbols) at 37°C for 2 h. Enzymatic activity was determined as described under Experimental procedures.
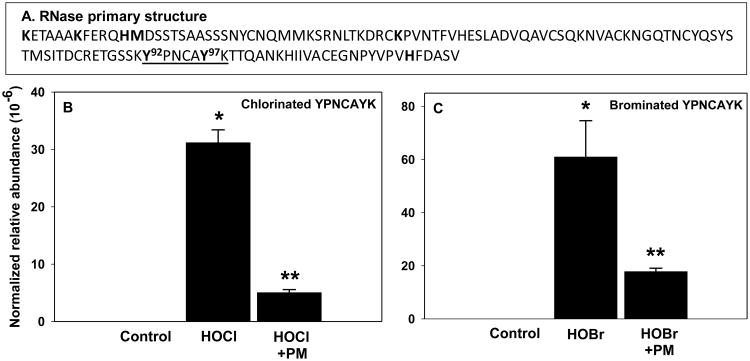
We then determined specific hypohalous acid-derived modifications of critical functional sites of RNase that may be responsible for inhibition of enzymatic activity. Using LC-MSMS, we investigated potential modifications at the following functional sites [22-24]: halogenation of Lys1, Lys7, Lys41, His12, His119, Tyr92 and Tyr97 as well as oxidation of His12, His119, and Met13 (Fig. 3A). We found high levels of tryptic RNase peptides containing singly and doubly halogenated Tyr92 and Tyr97 residues in HOCl- or HOBr-treated samples but not in the untreated controls (Fig. 3B and C, Fig. S3 and Table S1). Oxidation of Met13 was relatively high in controls and was not significantly increased upon RNase treatment with hypohalous acids (data not shown), consistent with a known artifact of methionine residue oxidation during sample preparation [25]. We did not find modifications of any other functionally important RNase residues. The absence of chloramine modifications of lysine and histidine side chains is consistent with the transient nature of these chloramines [20]. Thus, stable chlorine and bromine adducts of Tyr92 and Tyr97 were major modifications detected in RNase functional sites upon treatment with HOCl or HOBr. Halogenation of Tyr92 and Tyr97 was significantly inhibited in the presence of PM (Fig. 3B and C).
Figure 3.
Halogenation of functionally important RNase residues Y92 and Y97 and protection by PM. Location of key functional sites (bold font) and tryptic YPNCAYK peptide (underlined) within RNase sequence (A). RNase (50 μg/mL) was modified with either 60 μM HOCl (B) or 60 μM HOBr (C) with or without 60 μM PM and analyzed using LC-MSMS as described under Experimental procedures. Normalized relative abundance = sum of all modified forms of YPNCAYK peptide/average of three internal standard peptides. *P<0.05, control vs. hypohalous acid (n=3); **P<0.05, hypohalous acid vs. hypohalous acid + PM (n=3).
PM inhibits modification of collagen IV by hypohalous acids and protects collagen IV-integrin binding
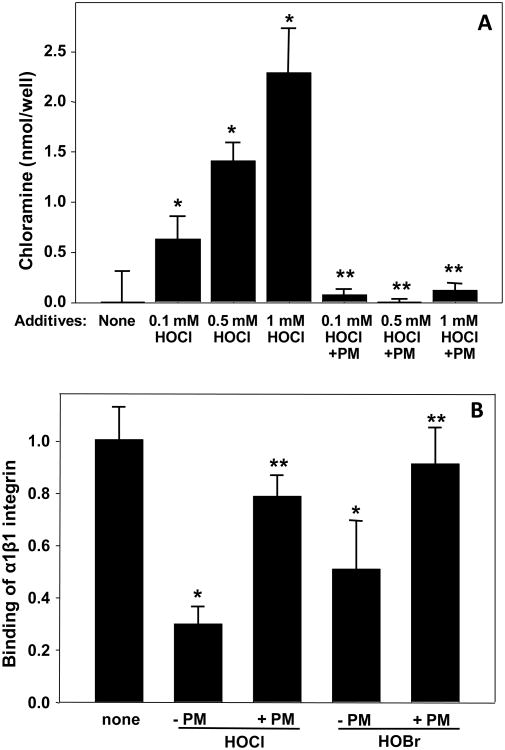
Long-lived ECM proteins are particularly susceptible to modification and functional damage due to non-enzymatic PTMs, including halogenation [8]. Therefore, we wanted to determine whether PM can protect collagen IV, an ECM protein, from hypohalous acid-derived modification and functional damage. Incubation of EHS collagen IV with HOCl caused concentration-dependent increase in protein chloramines; this chloramine accumulation was inhibited in the presence of PM (Fig. 4A). Moreover, PM also protected binding of collagen IV to integrin α1β1, a principal collagen IV receptor, from inhibition by hypohalous acids (Fig. 4B).
Figure 4.
(A) Chlorination of collagen IV and protection by PM. EHS collagen IV coated onto 96-well plate was incubated with indicated concentrations of HOCl with or without the equimolar concentrations of PM for 1 h at 37°C. Protein chloramine content was determined as described under Experimental procedures. *P<0.05, control vs. HOCl; **P<0.05, HOCl vs. HOCl+PM (n=4). (B) Inhibition of integrin binding to HOCl- or HOBr-modified collagen IV and protection by PM. EHS collagen IV coated onto 96-well plate was incubated with 0.1 mM HOCl or 0.1 mM HOBr either with or without 0.1 mM PM for 1 h at 37°C. Binding of α1β1 integrin was determined after 2 h at 30°C using solid phase binding assay as described under Experimental procedures. The bars represent background-subtracted Mn2+-dependent binding±SD (n=4). *P<0.05, control vs. HOCl or HOBr; **P<0.05, HOCl or HOBr vs. HOCl+PM or HOBr+PM.
PM protects NC1 hexamer folding/assembly and proteolytic stability from HOCl-induced damage
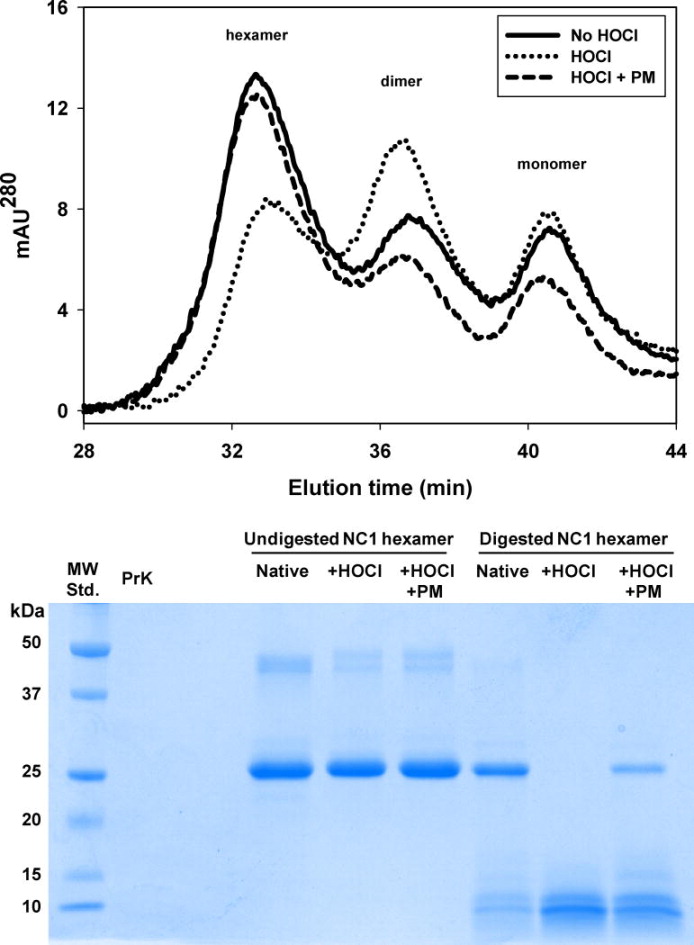
Chlorination and oxidation of NC1 hexamer of collagen IV with HOCl can affect hexamer folding and assembly and its susceptibility to proteolytic degradation [8]. When NC1 hexamers isolated from rat kidney were denatured with 6 M GdnCl and injected onto a gel-filtration column to allow refolding and reassembly, the hexameric form was prevalent with some dimers and monomers also present (Fig. 5A). However, specimens pre-treated with HOCl prior to denaturation had lower hexamer and higher dimer content compared to those without HOCl pre-treatment (Fig. 5A). This partial loss of hexamer folding and assembly competence did not occur when NC1 hexamers were pre-treated with both HOCl and PM (Fig. 5A). Similarly, NC1 hexamer pre-treated with HOCl was more susceptible to proteolysis by proteinase K and this increase in proteolytic susceptibility was partially ameliorated by PM (Fig. 5B). These results are consistent with a perturbed, less compact structure of the HOCl-modified NC1 hexamer.
Figure 5.
Effect of HOCl treatment on folding and structure of NC1 hexamer of collagen IV isolated from rat kidneys. (Upper panel) Purified NC1 hexamers were incubated in 50 mM TBS, pH 7.4 (solid line) or in the same buffer supplemented either with 50 μM HOCl (dotted line) or with 50 μM HOCl and 100 μM PM (dashed line) for 2 h at 37°C. Final protein concentration was 0.2 mg/mL. Samples were then supplemented with 6 M GdnCl and denatured at 80°C for 30 min. Samples were injected onto gel-filtration FPLC column equilibrated with 50 mM TBS to allow for NC1 domain refolding and hexamer reassembly and analyzed as described under Experimental procedures. (Lower panel) Purified NC1 hexamers (1 mg/mL) were incubated in 50 mM TBS, pH 7.5 alone, with 1 mM HOCl or with 1 mM HOCl and 1 mM PM for 2 h at 37°C. Samples were washed and concentrated to 0.8 mg/mL NC1 hexamer. Samples were then subjected to a limited proteolysis with proteinase K (PrK) as described under Experimental procedures. Samples (8 μg of NC1) were fractionated on 12% SDS-PAGE followed by Coomassie Brilliant Blue staining.
PM treatment protects renal collagen IV from hypohalous acid-derived damage in experimental diabetes
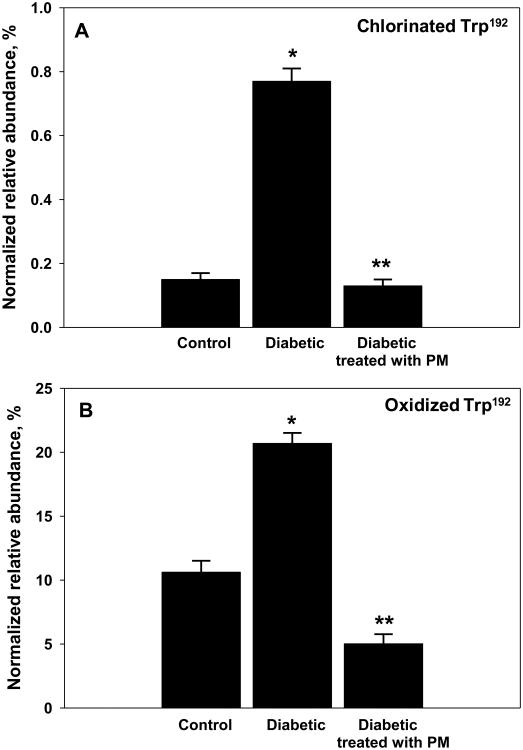
In order to determine whether PM can inhibit ECM damage by hypohalous acids in vivo, NC1 domains of collagen IV were isolated from kidneys of control rats, diabetic rats, and diabetic rats treated with PM. Preparations were analyzed using LC-MS/MS for the levels of chlorination and oxidation at a specific residues, Trp192 and Trp28 of NC1 domains of collagen IV [8]. There was a significant increase in the level of chlorination and oxidation of Trp192 of α1NC1 domain in diabetic animals compared to controls (Fig. 6 and Table S2). However, in diabetic animals treated with PM, the modification levels at this site did not increase and were comparable to those in non-diabetic control animals (Fig. 6). Similar inhibition of chlorination and oxidation modifications by PM treatment was detected at Trp28 in α2NC1 domain (Tables S3 and S4).
Figure 6.
Chlorination (A) and oxidation (B) levels of Trp192 within α1NC1 domain of collagen IV in experimental diabetes and protection by PM. Collagen IV NC1 domains were isolated from kidneys of control rats, STZ-diabetic rats and STZ-diabetic rats treated with PM and analyzed using LC-MS/MS as described under Experimental procedures. Tryptic peptide containing Trp192 was identified and tryptophan residue chlorination and oxidation levels quantified. Results are expressed as mean+/-SD. *P<0.05, diabetic vs. control; **P<0.05, diabetic + PM vs. diabetic (n=3). Normalized relative abundance = (modified peptide/all modified forms of peptide + unmodified peptide) × 100%.
Discussion
In this study, we demonstrated that PM can protect protein functionality from damage by hypohalous acids. Most importantly, PM treatment ameliorated HOCl-induced damage to renal collagen IV in the animal model of diabetes. This PM treatment protected NC1 domain of collagen IV from disruption of local structure, decrease in hexamer assembly competence and increase in proteolytic degradation. Therefore, it is likely that PM treatment may protect the integrity of renal collagen IV networks in diabetes.
Our data also suggest that PM treatment ameliorates hypohalous acid-induced disruption of interactions between ECM and integrins, key cellular receptors that regulate cell adhesion, migration and proliferation as well as ECM synthesis [26]. Interestingly, PM has been previously shown to protect integrin-ECM interactions from inhibition by glyoxal and methylglyoxal, hyperglycemia-derived reactive dicarbonyl compounds, which can modify specific integrin binding sites in ECM proteins [27-28]. Thus, PM appears to protect cell-ECM interactions via at least two mechanisms, i.e. sequestration of reactive carbonyl species and hypohalous acids.
Hypohalous acids may play an important role in diabetic complications. Overproduction of HOCl and/or HOBr by a family of peroxidase enzymes such as MPO and peroxidasin/VPO-1 [29-30], may cause protein modification with pathogenic consequences. Indeed, diabetes-induced activation of MPO and overproduction of HOCl has been previously reported [7]. Our previous work shows that one of the potential pathogenic consequences of peroxidase activation in diabetes may be damage by hypochlorous acid of critical functional sites in ECM proteins [8]. In the in vitro competition studies (Fig. S2), reactivity of tryptophan side chain with HOCl was among the highest, suggesting that this residue is one of the preferred targets for modification within protein microenvironment. In experimental diabetes, PM therapy effectively prevented HOCl-derived modification of key tryptophan residues located within subunit assembly interface of NC1 domains of collagen IV.
Conclusion
Mechanism of PM action towards hypohalous acids utilizes the very high reactivity of 4-aminomethyl substituent of pyridinium ring. This reaction mechanism allows PM to protect protein structure and function from hypohalous acid-induced damage including that in experimental diabetes. This is in agreement with the fact that PM therapy is effective at the early stages of diabetic nephropathy [3], since oxidative post-translational modifications are likely to accumulate early in the course of diabetes. Thus, PM ability to scavenge hypohalous acids may contribute to its therapeutic effects in diabetic nephropathy demonstrated in clinical trials [2-3].
Supplementary Material
Highlights.
- Pyridoxamine can efficiently compete with amino acid side chains in the reaction with hypochlorous and hypobromous acids.
- Pyridoxamine protects protein structure and function from damage by hypohalous acids.
- Pyridoxamine therapy inhibits chlorination of renal ECM in experimental diabetes.
Acknowledgments
Authors would like to thank Ms. Parvin Todd and Ms. Salisha Hill for expert technical help and Dr. Vadim Pedchenko for helpful discussions. This work was supported by the grant DK65138 from the National Institutes of Health. Mr. Hartman Madu was supported by the summer student research grant DK65123 from the National Institutes of Health. Dr. Carl Darris was supported by the research fellowship award T32DK007569-24S from the National Institutes of Health. Mr. Josh Avance was supported by Vanderbilt Aspirnaut program and 1R25DK096999 and NIDDKD Step-up grants from the National Institutes of Health.
Abbreviations
- AGE
advanced glycation end products
- CAM
carbamidomethylation
- CID
collision-induced dissociation
- ECM
extracellular matrix
- GdnCl
guanidinium hydrochloride
- HDL
high density lipoprotein
- HOCl
hypochlorous acid
- HOBr
hypobromous acid
- HPLC
high performance liquid chromatography
- LC-MS/MS
liquid-chromatography coupled with tandem mass spectrometry
- PM
pyridoxamine
- ROS
reactive oxygen species
- RCS
reactive carbonyl species
- TFE
trifluoroethanol
Footnotes
Appendix A. Supporting information: Supplementary data associated with this article can be found in the online version at http://….
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baynes JW, Thorpe SR. Role of oxidative stress in diabetic complications: a new perspective on an old paradigm. Diabetes. 1999;48:1–9. doi: 10.2337/diabetes.48.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Williams ME, Bolton WK, Khalifah RG, Degenhardt TP, Schotzinger RJ, McGill JB. Effects of Pyridoxamine in Combined Phase 2 Studies of Patients with Type 1 and Type 2 Diabetes and Overt Nephropathy. Am J Nephrol. 2007;27:605–614. doi: 10.1159/000108104. [DOI] [PubMed] [Google Scholar]
- 3.Lewis EJ, Greene T, Spitalewiz S, Blumenthal S, Berl T, Hunsicker LG, Pohl MA, Rohde RD, Raz I, Yerushalmy Y, Yagil Y, Herskovits T, Atkins RC, Reutens AT, Packham DK, Lewis JB. Pyridorin in type 2 diabetic nephropathy. J Am Soc Nephrol. 2012;23:131–136. doi: 10.1681/ASN.2011030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voziyan PA, Hudson BG. Pyridoxamine as a multifunctional pharmaceutical: targeting pathogenic glycation and oxidative damage. Cell Mol Life Sci. 2005;62:1671–1681. doi: 10.1007/s00018-005-5082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metz TO, Alderson NL, Chachich ME, Thorpe SR, Baynes JW. Pyridoxamine traps intermediates in lipid peroxidation reactions in vivo: evidence on the role of lipids in chemical modification of protein and development of diabetic complications. J Biol Chem. 2003;278:42012–42019. doi: 10.1074/jbc.M304292200. [DOI] [PubMed] [Google Scholar]
- 6.Grove KJ, Voziyan PA, Spraggins JM, Wang S, Paueksakon P, Harris RC, Hudson BG, Caprioli RM. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J Lipid Res. 2014;55:1375–1385. doi: 10.1194/jlr.M049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Yang J, Jennings LK. Leukocyte-derived myeloperoxidase amplifies high-glucose--induced endothelial dysfunction through interaction with high-glucose--stimulated, vascular non--leukocyte-derived reactive oxygen species. Diabetes. 2004;53:2950–2959. doi: 10.2337/diabetes.53.11.2950. [DOI] [PubMed] [Google Scholar]
- 8.Brown KL, Darris C, Rose KL, Sanchez OA, Madu H, Avance J, Brooks N, Zhang MZ, Fogo A, Harris R, Hudson BG, Voziyan P. Hypohalous Acids Contribute to Renal Extracellular Matrix Damage in Experimental Diabetes. Diabetes. 2015 doi: 10.2337/db14-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalnitsky G, Resnick H. The effect of an altered secondary structure on ribonuclease activity. J Biol Chem. 1959;234:1714–1717. [PubMed] [Google Scholar]
- 10.Voziyan PA, Metz TO, Baynes JW, Hudson BG. A post-Amadori inhibitor pyridoxamine also inhibits chemical modification of proteins by scavenging carbonyl intermediates of carbohydrate and lipid degradation. J Biol Chem. 2002;277:3397–3403. doi: 10.1074/jbc.M109935200. [DOI] [PubMed] [Google Scholar]
- 11.Witko V, Nguyen AT, Descamps-Latscha B. Microtiter plate assay for phagocyte-derived taurine-chloramines. J Clin Lab Anal. 1992;6:47–53. doi: 10.1002/jcla.1860060110. [DOI] [PubMed] [Google Scholar]
- 12.Dypbukt JM, Bishop C, Brooks WM, Thong B, Eriksson H, Kettle AJ. A sensitive and selective assay for chloramine production by myeloperoxidase. Free Radic Biol Med. 2005;39:1468–1477. doi: 10.1016/j.freeradbiomed.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Pedchenko V, Zent R, Hudson BG. Alpha(v)beta3 and alpha(v)beta5 integrins bind both the proximal RGD site and non-RGD motifs within noncollagenous (NC1) domain of the alpha3 chain of type IV collagen: implication for the mechanism of endothelia cell adhesion. J Biol Chem. 2004;279:2772–2780. doi: 10.1074/jbc.M311901200. [DOI] [PubMed] [Google Scholar]
- 14.Degenhardt TP, Alderson NL, Arrington DD, Beattie RJ, Basgen JM, Steffes MW, Thorpe SR, Baynes JW. Pyridoxamine inhibits early renal disease and dyslipidemia in the streptozotocin-diabetic rat. Kidney Int. 2002;61:939–950. doi: 10.1046/j.1523-1755.2002.00207.x. [DOI] [PubMed] [Google Scholar]
- 15.Fox JW, Butkowski RJ, Hudson BG. Detergent-prepared glomerular basement membrane is composed of a heterogeneous group of polypeptides. J Biol Chem. 1981;256:9313–9315. [PubMed] [Google Scholar]
- 16.Wieslander J, Kataja M, Hudson BG. Characterization of the human Goodpasture antigen. Clin Exp Immunol. 1987;69:332–340. [PMC free article] [PubMed] [Google Scholar]
- 17.Kahsai TZ, Enders GC, Gunwar S, Brunmark C, Wieslander J, Kalluri R, Zhou J, Noelken ME, Hudson BG. Seminiferous tubule basement membrane. Composition and organization of type IV collagen chains, and the linkage of alpha3(IV) and alpha5(IV) chains. J Biol Chem. 1997;272:17023–17032. doi: 10.1074/jbc.272.27.17023. [DOI] [PubMed] [Google Scholar]
- 18.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. J Am Soc Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Caldes C, Vilanova B, Adrover M, Munoz F, Donoso J. Phenol group in pyridoxamine acts as a stabilizing element for its carbinolamines and Schiff bases. Chem Biodivers. 2011;8:1318–1332. doi: 10.1002/cbdv.201000296. [DOI] [PubMed] [Google Scholar]
- 20.Bergt C, Fu X, Huq NP, Kao J, Heinecke JW. Lysine residues direct the chlorination of tyrosines in YXXK motifs of apolipoprotein A-I when hypochlorous acid oxidizes high density lipoprotein. J Biol Chem. 2004;279:7856–7866. doi: 10.1074/jbc.M309046200. [DOI] [PubMed] [Google Scholar]
- 21.Hazen SL, d'Avignon A, Anderson MM, Hsu FF, Heinecke JW. Human neutrophils employ the myeloperoxidase-hydrogen peroxide-chloride system to oxidize alpha-amino acids to a family of reactive aldehydes. Mechanistic studies identifying labile intermediates along the reaction pathway. J Biol Chem. 1998;273:4997–5005. doi: 10.1074/jbc.273.9.4997. [DOI] [PubMed] [Google Scholar]
- 22.Moussaoui M, Guasch A, Boix E, Cuchillo C, Nogues M. The role of non-catalytic binding subsites in the endonuclease activity of bovine pancreatic ribonuclease A. J Biol Chem. 1996;271:4687–4692. doi: 10.1074/jbc.271.9.4687. [DOI] [PubMed] [Google Scholar]
- 23.Eberhardt ES, Wittmayer PK, Templer BM, Raines RT. Contribution of a tyrosine side chain to ribonuclease A catalysis and stability. Protein Sci. 1996;5:1697–1703. doi: 10.1002/pro.5560050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackburn P, Gavilanes JG. The role of lysine-41 of ribonuclease A in the interaction with RNase inhibitor from human placenta. J Biol Chem. 1980;255:10959–10965. [PubMed] [Google Scholar]
- 25.Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS. Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J Biol Chem. 2003;278:19587–19590. doi: 10.1074/jbc.C300135200. [DOI] [PubMed] [Google Scholar]
- 26.Pozzi A, Voziyan PA, Hudson BG, Zent R. Regulation of matrix synthesis, remodeling and accumulation in glomerulosclerosis. Curr Pharm Des. 2009;15:1318–1333. doi: 10.2174/138161209787846748. [DOI] [PubMed] [Google Scholar]
- 27.Pedchenko VK, Chetyrkin SV, Chuang P, Ham AJ, Saleem MA, Mathieson PW, Hudson BG, Voziyan PA. Mechanism of perturbation of integrin-mediated cell-matrix interactions by reactive carbonyl compounds and its implication for pathogenesis of diabetic nephropathy. Diabetes. 2005;54:2952–2960. doi: 10.2337/diabetes.54.10.2952. [DOI] [PubMed] [Google Scholar]
- 28.Chetyrkin S, Mathis M, Pedchenko V, Sanchez OA, McDonald WH, Hachey DL, Madu H, Stec D, Hudson B, Voziyan P. Glucose Autoxidation Induces Functional Damage to Proteins via Modification of Critical Arginine Residues. Biochemistry. 2011 doi: 10.1021/bi200757d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malle E, Buch T, Grone HJ. Myeloperoxidase in kidney disease. Kidney Int. 2003;64:1956–1967. doi: 10.1046/j.1523-1755.2003.00336.x. [DOI] [PubMed] [Google Scholar]
- 30.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol. 2012;8:784–790. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.