Abstract
The function of RNA switches involved in the regulation of transcription and translation relies on their ability to adopt different, structurally well-defined states. A new class of ligand-responsive RNA switches, which we recently discovered in positive strand RNA viruses, are distinct from conventional riboswitches. The viral switches undergo large conformational changes in response to ligand binding while retaining the same secondary structure in their free and ligand-bound forms. Here, we describe FRET experiments to study folding and ligand binding of the viral RNA switches. In addition to reviewing previous approaches involving RNA model constructs which were directly conjugated with fluorescent dyes, we outline the design and application of new modular constructs for FRET experiments, in which dye labeling is achieved by hybridization of a core RNA switch module with universal DNA fluorescent probes. As an example, folding and ligand binding of the RNA switch from the internal ribosome entry site of hepatitis C virus is studied comparatively with conventional and modular FRET constructs.
Keywords: RNA switch, FRET, RNA virus, IRES, hepatitis C virus
1. Introduction
The genomes of several positive strand RNA viruses, including members of Flavi- and Picornaviridae, contain an internal ribosome entry site (IRES) element that controls a cap-independent mechanism for translation initiation by orchestrating ribosome assembly at the start codon without the need for many host initiation factors [1, 2]. IRES elements are located in the 5′ untranslated region (UTR) of genomes and adopt complex domain architectures which share similarity across viruses but are distinct in sequence and details of secondary structure [3]. The hepatitis C virus (HCV) contains a well characterized IRES element which is comprised of four independently folding domains [4]. Domain II is highly conserved in HCV clinical isolates [5] and a key participant in several steps of IRES-driven translation [6], including the stable entry of mRNA at the 40S decoding groove [7–9], initiation factor dissociation prior to subunit joining [10], and the transition from initiation to elongation stages [11]. We have previously discovered a ligand-captured conformational switch motif in the HCV domain II and functionally analogous switches in IRES elements of 11 other flavi- and picornaviruses [12–14]. The RNA switch consists of an internal loop motif (subdomain IIa) that can adopt two stable conformations, characterized by a bent fold in the ligand-free state and an extended structure captured by ligand binding in a deep pocket (Fig. 1). Three-dimensional structures for both conformational states of the HCV IRES switch have been determined by X-ray crystallography [15, 16], revealing the molecular basis of RNA fold stabilization in the absence and presence of a ligand. Recently, we obtained a crystal structure of a functionally homologous RNA switch from the IRES element of Seneca Valley virus (SVV), a picornavirus unrelated to HCV [13]. The IRES switch from SVV adopts a fold that is overall identical to the switch from HCV despite the two RNA motifs sharing little sequence or local secondary structure similarity. The SVV switch and analogous switches from other viral IRES elements interact with benzimidazole translation inhibitors as well as guanosine. Beyond the similarity of static structure and the ability to adopt two distinct conformational states, the viral IRES switch motifs are functionally homologous. Domain swap experiments of switches between different viruses demonstrate that biological function is completely conserved between these RNA building blocks [14].
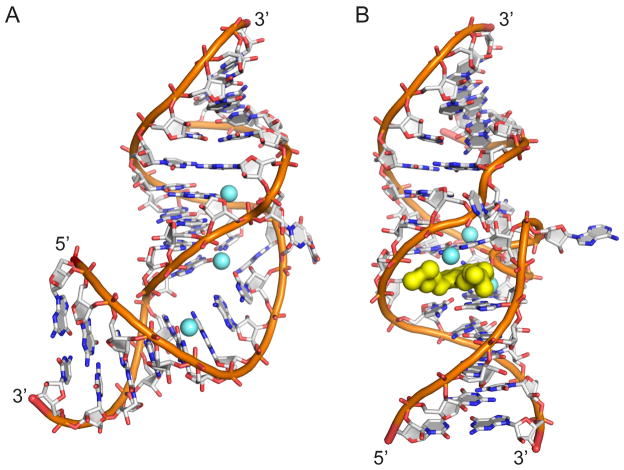
Fig. 1.
Two conformational states of the subdomain IIa RNA switch from the IRES element of HCV as visualized by X-ray crystallography. Both structures contain three magnesium ions (light blue spheres). (A) The RNA switch adopts a bent fold in the ligand-free state. (B) An extended architecture of the RNA switch is captured by benzimidazole viral translation inhibitors and guanine which bind at a deeply encapsulating ligand binding site (yellow surface).
The viral IRES switches differ from metabolite-sensing riboswitches with regard to their small size as well as intrinsic stability and structural definition of the constitutive conformational states. While benzimidazole viral translation inhibitors were initially discovered as fortuitous ligands of the subdomain IIa switch in HCV [12, 17], it was later recognized that the RNA switch contains a binding site for guanosine which likely serves as the cognate biological ligand [5, 13]. The benzimidazole inhibitors bind and lock the RNA switch in an extended conformation which permits association of the IRES with the ribosomal 40S subunit but ultimately prevents translation initiation. The cognate guanosine ligand, however, likely as part of a trigger sequence within the viral genome, has a weaker affinity and captures the extended state of the RNA switch transiently while allowing the assembled 80S ribosome to undock from the IRES and initiate translation [13].
To study conformational changes in viral RNA switches, we have conceived a FRET method that monitors the distance between base-paired stems flanking the switch motif [12]. In this article, we will describe the development and application of dye-labeled oligonucleotide model constructs for FRET experiments that include the observation of folding and ligand binding of viral RNA switches.
2. Materials and methods
2.1. Design of RNA FRET constructs
RNA conventional model FRET constructs were designed as described previously [12] guided by crystal structures to identify sites for 5′ terminal Cy3 or Cy5 modification near to their Förster radius. RNA modular FRET constructs were designed from optimized conventional model FRET constructs [12] with several modifications. The lower stem of this construct was lengthened from seven to eight base pairs to accommodate the Cy3-conjugated DNA oligonucleotide of five bases while maintaining three Watson-Crick base pairs on the lower side of the internal loop for stability of the central RNA core. The upper stem was shortened from thirteen to twelve base pairs to partially offset the change in distance between dyes due to the lengthening of the lower stem. Additionally, the U○U closing pair of the RNA core’s upper stem was changed to a Watson-Crick G–C pair for added construct stability. We chose a length of five bases for the dye-modified DNA oligonucleotides as we thought this might be the minimum length required for stable hybridization to the RNA core.
2.2. Preparation of RNA constructs for FRET experiments
Cyanine dye labeled and unlabeled RNA and DNA oligonucleotides were obtained by chemical synthesis (Integrated DNA Technologies, Coralville, IA). Unlabeled RNA oligonucleotides were purified by standard desalting while dye labeled RNA and DNA oligonucleotides were purified by HPLC to remove any excess free dye. Lyophilized single stranded oligonucleotides were dissolved in 10 mM sodium cacodylate buffer, pH 6.5, to prepare stock solutions.
Working solutions for conventional model FRET constructs were prepared at 1μM concentration from terminally Cy3/Cy5-labeled single stranded RNA oligonucleotides diluted with 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer, pH 7.0.
Working solutions for modular FRET constructs were prepared at 1 μM concentration from single stranded RNA oligonucleotides and 5′ terminally Cy3/Cy5-labeled single stranded DNA oligonucleotides diluted with 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer, pH 7.0.
2.3. FRET titration experiments monitoring folding and ligand binding
FRET folding experiments at increasing concentration of Mg2+ were prepared in PCR tubes with conventional or modular FRET construct at 100 nM concentration in 10 mM HEPES buffer, pH 7.0, in a total volume of 110 μL. RNA concentration and volume may be increased to achieve larger absolute fluorescence signals and minimize pipetting error. FRET constructs were annealed from oligonucleotides and folded in the presence of Mg2+ by heating PCR tube samples at 65°C for 5 minutes followed by snap cooling and incubating on ice for 15 minutes. 100 μL of each sample was then transferred to a 96-well plate and incubated in the dark at 25°C for 10 minutes. FRET measurements were performed on a Spectra Max Gemini XS monochromator plate reader (Molecular Devices, Sunnyvale, CA) at 25°C by exciting the Cy3 label at 520 nm and reading the transferred fluorescence as Cy5 emission at 670 nm. Emission filters were set at 550 and 665 nm. Mg2+ dependent FRET folding was calculated and analyzed as described previously [18].
FRET ligand binding experiments were performed as above, except a 2 mM Mg2+ concentration was held constant while increasing the concentration of compound 1. Ligand binding occurred in the annealing and folding steps as described above. Following FRET measurements, Cy3 fluorescence was recorded by exciting the Cy3 label at 520 nm and reading the emission at 570 nm with an emission filter set at 550 nm. Cy5 fluorescence was recorded by exciting the Cy5 label at 620 nm and reading the emission at 670 nm with an emission filter set at 665 nm. Ligand induced FRET changes were calculated and analyzed as described previously [18].
3. Results and discussion
3.1. FRET approach to monitor folding of viral RNA switches
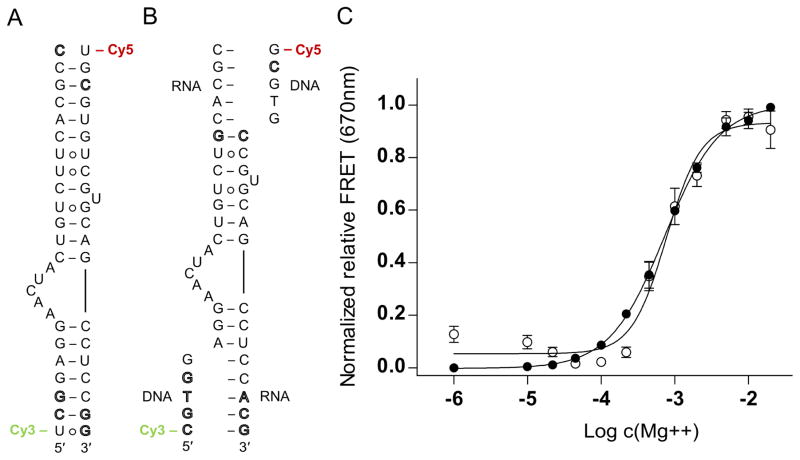
The viral IRES switches adopt a unique bent fold in the absence of a ligand, as revealed by the crystal structures of the RNA motifs from HCV (Fig. 1A) and SVV [13, 15]. In the HCV IRES, which has been visualized by cryo-EM of ribosomal complexes [19–21], the sharp turn introduced by the switch motif directs the IRES domain II at the E site. Since the curved topology of the domain II would prevent progression of the ribosome from initiation to elongation [19, 20], conformational dynamics of the subdomain IIa switch has been invoked as a mechanism facilitating ribosome release from the IRES [13]. Model oligonucleotides representing viral IRES switches enable the investigation of conformational changes in isolation from the background of the complex IRES-ribosome interaction. In previous studies, we used the crystal structure of the HCV IRES subdomain IIa switch to design RNA constructs carrying 5′ terminal cyanine dye labels for FRET experiments (Fig. 2A). The length of the stems flanking the internal loop was chosen such that the distance of the cyanine-modified 5′ termini in the bent state of the switch is close to the Förster radius of the dye pair. Therefore, maximum FRET signal is achieved for the folded switch in the ligand-free state.
Fig. 2.
Secondary structure and folding of viral RNA switches monitored by FRET experiments with cyanine dye-labeled oligonucleotides. (A) Secondary structure of previously developed, conventional model construct of the HCV IRES subdomain IIa switch 5′ terminally conjugated with Cy3 and Cy5 dyes. (B) Modular FRET construct consisting of unmodified oligonucleotides that contain the HCV RNA switch and carry overhanging single strands which hybridize with cyanine dye-conjugated DNA oligonucleotides. In both panels A and B, outlined letters represent nucleotides deviating from HCV genotype 1b sequence. (C) Normalized relative FRET signal for the Mg2+ titration of the HCV conventional (●) and modular (○) FRET constructs. Dose-response fitting curves gave EC50 values for Mg2+ dependent folding of 730 ± 40 μM (conventional) and 770 ± 80 μM (modular), respectively, for the two different constructs. Error bars represent ±1 s.d. calculated from triplicate experiments.
To validate the FRET approach, folding of the RNA switch from HCV induced by metal ion binding was investigated. The crystal structure of the HCV IRES subdomain IIa revealed three tightly bound Mg2+ ions which are integral to the RNA switch architecture (Fig. 1A). We anticipated that the internal loop of the subdomain IIa RNA may not be stably folded in the absence of Mg2+, leaving the flanking helices arranged in an overall extended conformation and separating the terminally attached dyes beyond the Förster radius. Folding of the switch RNA was induced by addition of an increasing amount of Mg2+ and monitored by FRET measurement of the Cy5 acceptor dye emission while exciting the Cy3 donor. A dose-dependent increase of the FRET signal was observed which indicated localization of the dye labels within the Förster radius due to formation of the bent RNA switch architecture (Fig. 2C). The apparent EC50 value of the Mg2+ dose response for folding of the HCV subdomain IIa RNA was determined at 730 ± 40 μM, comparable to previously measured values with similar constructs [12, 13]. Corresponding dye-labeled model oligonucleotides for RNA switches from four other viruses, including classic swine fever virus (CSFV), bovine viral diarrhea virus (BVDV), avian encephalomyelitis virus (AEV) and Seneca Valley virus (SVV) were designed with guidance from secondary structure comparisons with HCV subdomain IIa RNA as no high-resolution structural data has been determined for BVDV and AEV. CSFV and SVV RNA FRET construct design was also supported by NMR [10] and X-ray crystal structures [15]. These RNA constructs were tested in analogous FRET experiments, resulting in EC50 values for folding and Mg2+ binding of 130–220 ± 20 μM (CSFV, BVDV, AEV) and 580 ± 90 μM (SVV) [13].
While these previous investigations required the preparation of covalently dye-modified oligonucleotides for each viral IRES switch, we have now developed a modular FRET approach in which unmodified model RNA constructs are hybridized with dye-labeled DNA oligonucleotides (Fig. 2B). The modular design for the HCV RNA switch was tested in the salt-induced folding experiment. Titration with Mg2+ resulted in a dose-dependent increase of the FRET signal with an EC50 value of 770 ± 80 μM which was in excellent agreement with the affinity determined with the conventional construct (Fig. 2C).
The modular constructs will be advantageous to investigate conformational dynamics of other candidate RNA switches, especially those identified in subdomain IIa of various IRES-containing viruses [14], since the same pair of universal Cy3- and Cy5-conjugated DNA sequences can be used to prepare various dye-labeled FRET constructs. The cyanine dye-conjugated DNA strands are only 5 nucleotides in length and therefore readily prepared by robust chemical synthesis from which they are obtained in high purity by HPLC and significantly more cost effective than longer modified RNA oligonucleotides. Labeling of RNA by hybridization with dye-modified DNA oligonucleotides has been used extensively in fluorescence studies of complex RNA architectures, such as the ribosome, for which site-specific covalent modification of large nucleic acid components is difficult to achieve [22, 23]. A prime concern associated with the fluorescent probe hybridization approach is the integrity of the target RNA structure which ideally is only minimally disturbed by binding of the dye-conjugated DNA oligonucleotide. In the modular FRET constructs of viral RNA switches described here, structure and conformational flexibility of the switch motif are not affected by the DNA probes which hybridize distantly from the internal loop (Fig. 2B), as attested by the consistent EC50 values for Mg2+-induced folding of the conventional and modular RNAs (Fig. 2C). The implementation of this method for studying other RNA switches may require core RNA length modification to achieve optimal FRET distances as the required number of base pairs flanking an internal loop of interest for stability may vary. Changes in stem length may also place a dye closer or further from its FRET pair based on its helical position. Alternatively, a suspected RNA switch may not be amenable to this method of study if it does not adopt well-defined stable conformations but remains flexible.
3.2. Monitoring ligand binding at viral RNA switches
While a bent conformation is preferred by RNA switches from viral IRES elements in the ligand-free state, binding of benzimidazole translation inhibitors or guanosine captures an extended structure containing a deep pocket that encapsulates the ligand (Fig. 1). The transition between conformational states can be monitored by FRET measurements similar to the folding experiments outlined in the previous section. The same model constructs have been used to investigate conformational changes during ligand binding, including the conventional dye-conjugated RNA and a modular oligonucleotide system (Fig. 2A, B). Previously, cyanine dye-labeled RNA, similar to the construct shown in Fig. 2A, was instrumental in establishing target binding and elucidating the molecular mechanism of action of benzimidazole inhibitors of HCV IRES-driven translation [5, 12]. A related RNA construct was used to develop a high-throughput screen for translation inhibitor ligands targeting the HCV subdomain IIa RNA switch [18].
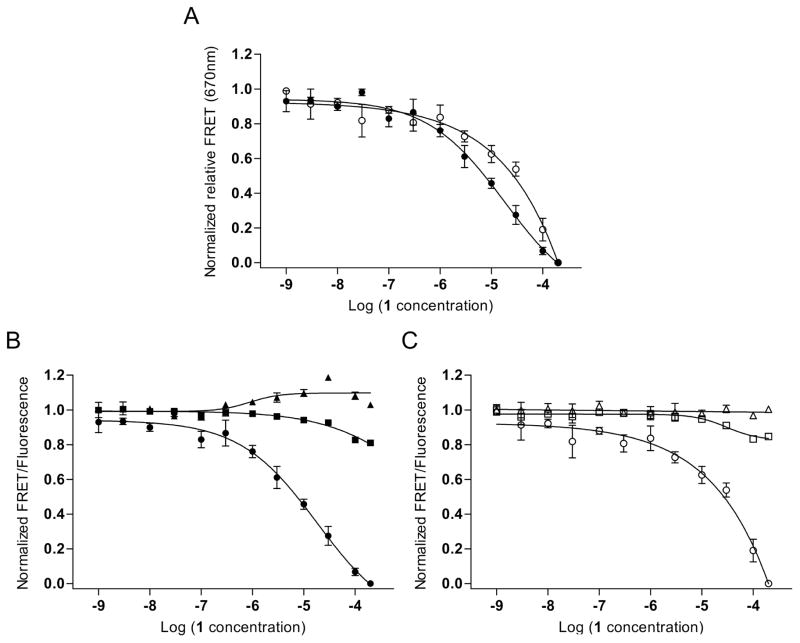
To monitor compound binding to viral RNA switches, dye-labeled oligonucleotide constructs were incubated in the presence of Mg2+ at physiological concentration (2 mM) to obtain the stably folded bent conformation, followed by addition of ligand and incubation. Dose-dependent decrease of the FRET signal coinciding with increased Cy3 donor emission is indicative of ligand capture of the extended switch conformation (Fig. 3). When the dyes move beyond the Förster radius, reduced efficiency of energy transfer by FRET causes growing Cy3 donor emission which, at the same time, provides a selectivity control for compounds that stabilize the extended switch state rather than nonspecifically quenching fluorescence. Since ligands capture extended states from an equilibrium of switch conformations, and bent structures are preferred by the unbound switch, complex formation is a relatively slow event and requires incubation of the RNA target with compound for 10–30 minutes to achieve maximum signal change [18]. Binding of a benzimidazole translation inhibitor (compound 1 [13, 24]), which was previously used to determine the structure of the RNA target complex [16], captured the extended state of the HCV switch at an EC50 value of 22 ± 12 μM (Fig. 3A, B).
Fig. 3.
Binding of a translation inhibitor ligand 1 [13, 24] to the HCV RNA switch as monitored by FRET experiments with cyanine dye-labeled oligonucleotides. (A) Normalized relative FRET signal for the titration of 1 to a conventional model construct of the HCV IRES subdomain IIa switch 5′ terminally conjugated with Cy3 and Cy5 dyes (●) and a modular FRET construct consisting of unmodified oligonucleotides that contain the HCV RNA switch and carry overhanging single strands which hybridize with cyanine dye-conjugated DNA oligonucleotides (○). Dose-response fitting curves gave an EC50 value for capture of the extended switch state of 22 ± 12 μM for the conventional construct and 37 ± 21 μM for the modular construct. As the FRET signal for the modular system did not reach saturation, affinity of compound 1 was calculated by dose-response fitting to the Cy5 emission signal. (B) Individual dye and FRET signals for the conventional model construct. (●)=FRET; (■)=Cy5; (▲)=Cy3. (C) Individual dye and FRET signals for the modular construct. (○)=FRET; (□)=Cy5; (△)=Cy3. Error bars represent ±1 s.d. calculated from triplicate experiments.
Similar to the folding studies outlined in section 3.1., we have extended the scope of the FRET assay for monitoring ligand binding to viral RNA switches by using modular dye-labeled constructs. For the HCV switch, the modular oligonucleotide system (Fig. 2B) reported an EC50 value for capture of the extended RNA by compound 1 at 37 ± 21 μM which was in good agreement with the affinity determined with the conventional non-modular construct (Fig. 3A, C). As was observed in the folding experiments, hybridization of the RNA switch with dye-labeled DNA probes did not interfere with the FRET signal change induced by ligand binding.
4. Concluding remarks
Ligand-responsive RNA switches, which are distinct from bacterial riboswitches, have recently been discovered in the IRES elements of positive strand RNA viruses [14]. These viral switches constitute a new class of conformationally flexible RNA modules which, unlike conventional metabolite-sensing riboswitches, adopt distinct three-dimensional folds in the free and ligand-bound state while retaining the same secondary structure. X-ray crystallographic studies have been instrumental to characterize the architecture of the viral RNA switches and to elucidate molecular recognition by ligands that capture the extended state. Investigation of conformational changes in real time, however, requires dynamic techniques applied in solution. In this article, we have outlined how FRET experiments have been used to monitor folding and ligand-captured switching action of the viral RNA modules. We have also described a new approach that uses fluorescently labeled modular oligonucleotide model constructs representing the switch from HCV which are readily adapted for FRET studies of various other RNA switches.
Highlights.
Viral RNA switches adopt distinct conformational states to regulate translation
RNA switch folding and ligand binding monitored by FRET
Modular FRET uses universal fluorescent DNA probes to study multiple RNA switches
Acknowledgments
M. A. Boerneke is the recipient of a GAANN fellowship from the US Department of Education. Research was supported by the UCSD Academic Senate, grant No. RM069B. Instrumentation at the Biomolecule Crystallography Facility was acquired with funding from the National Institutes of Health, grant OD011957.
Abbreviations
- AEV
avian encephalomyelitis virus
- BVDV
bovine vial diarrhea virus
- CSFV
classical swine fever virus
- FRET
Förster resonance energy transfer
- HCV
hepatitis C virus
- IRES
internal ribosome entry site
- SVV
Seneca Valley virus
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pisarev AV, Shirokikh NE, Hellen CU. Compt Rend Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Hellen CU, de Breyne S. J Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plank TD, Kieft JS. Wiley interdisciplinary reviews. RNA. 2012;3:195–212. doi: 10.1002/wrna.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 5.Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, Hermann T. J Med Chem. 2014;57:1694–1707. doi: 10.1021/jm401312n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto GA, Puglisi JD. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 7.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolupaeva VG, Pestova TV, Hellen CU. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filbin ME, Kieft JS. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locker N, Easton LE, Lukavsky PJ. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Nature Chem Biol. 2009;5:823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. Proc Nat Acad Sci USA. 2014;111:15952–15957. doi: 10.1073/pnas.1414678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boerneke MA, Hermann T. RNA Biol. 2015;12:780–786. doi: 10.1080/15476286.2015.1054592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Angew Chem Int Ed. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 16.Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. Proc Nat Acad Sci USA. 2012;109:5223–5228. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, Swayze EE, Griffey RH. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 18.Zhou S, Rynearson KD, Ding K, Brunn ND, Hermann T. Bioorg Med Chem. 2013;21:6139–6144. doi: 10.1016/j.bmc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 20.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Burger J, Mielke T, Spahn CM. Nature Struct Mol Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 22.Blanchard SC. Curr Opin Struct Biol. 2009;19:103–109. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrov A, Puglisi JD. Nucleic Acids Res. 2010;38:e143. doi: 10.1093/nar/gkq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker MA, Satkiewicz E, Hermann T, Bergdahl BM. Molecules. 2011;16:281–290. doi: 10.3390/molecules16010281. [DOI] [PMC free article] [PubMed] [Google Scholar]