Abstract
Proteins comprise a majority of the dry weight of a cell, rendering them a major target for oxidative modification. Oxidation of proteins can result in significant alterations in protein molecular mass such as breakage of the polypeptide backbone, and/or polymerization of monomers into dimers, multimers and sometimes into insoluble aggregates. Protein oxidation can also result in structural changes to amino acid residue side chains, conversions which have only a modest effect on protein size but can have widespread consequences for protein function. There are a wide range of rate constants for amino acid reactivity, with cysteine, methionine, tyrosine, phenylalanine and tryptophan having the highest rate constants with commonly encountered biological oxidants. Free tryptophan and tryptophan protein residues react at a diffusion limited rate with hydroxyl radical, and also have high rate constants for reactions with singlet oxygen and ozone. Although oxidation of proteins in general and tryptophan residues specifically can have effects detrimental to the health of cells and organisms, some modifications are neutral while others contribute to the function of the protein in question or may act as a signal that damaged proteins need to be replaced. This review provides a brief overview of the chemical mechanisms by which tryptophan residues become oxidized, presents both the strengths and weaknesses of some of the techniques used to detect these oxidative interactions and discusses selected examples of the biological consequences of tryptophan oxidation in proteins from animals, plants and microbes.
1. Introduction
Half or more of the total dry mass of a cell comprises proteins, making them a major target for oxidative modification and damage [1, 2]. Protein oxidation can result in fragmentation due to cleavage of the polypeptide backbone, aggregation by cross-linking of amino acid residues, and modification of the molecular structure of amino acid residues [1, 2]. Depending on the specific protein and/or the specific site(s) of oxidation, these oxidative changes have variable effects on protein function, stability, and the capacity for non-functional proteins to be cleared from the cell. There is a strong bias in non-scientific reports and publications towards equating oxidation with detrimental health effects, and some of the same tendency to conflate oxidation with negative consequences is also prominent in the scientific literature. In actuality, however, the biological outcomes of protein oxidation and their amino acids components vary from necessary for normal physiological function, to neutral in effect, to highly deleterious. It is also possible that within a single protein, oxidative alteration of multiple amino acid residues can have dissimilar functional results for the protein.
Tryptophan (Trp) is one of the amino acids most susceptible to oxidation, reacting with hydroxyl radicals at a diffusion limited rate [2] and with both singlet oxygen (1O2) and ozone at significant rates under physiological conditions [1, 3]. Tryptophan, unlike other amino acids, reacts with 1O2 via both a physical and chemical route, with rate constants of ca. 2–7 × 107 M−1 sec−1 and 3 × 107 M−1 sec−1, respectively [1]. Reaction of Trp with ozone has been reported to occur at 7 × 106 M−1 sec−1 [4]. Most documented instances of Trp oxidation result in the production of Trp derivatives. Recent research with human SOD1 (hSOD1), however, has determined that Trp radicals can dimerize to form new intermolecular bonds between hSOD1 monomers [5].
A number of reviews covering protein oxidation [1, 2, 6, 7] examine the range of amino acid residue modifications resulting from interactions with oxygen-centered radicals and singlet oxygen. Other recent reviews cover Trp residue modifications engendered by reactive nitrogen species [8–10]. The classical review by Saito et al. [11] describes the chemistry that occurs during photooxidation of free Trp. The aim, however, of this review, is not to focus on the chemistry of free Trp and Trp residue oxidation. We intend, instead, to provide examples describing the biological consequences of Trp residue oxidation in order to illustrate the divergent effects, both documented and hypothesized, on the function of specific proteins that undergo Trp residue oxidation by reactive oxygen species. To that end we briefly examine how reactive oxygen species react with Trp to form specific products and also discuss the strengths and weaknesses of the various methods and technologies that are used to study proteins containing oxidized Trp residues. Lastly, we discuss Trp oxidation in the context of how residue oxidation affects the protein as a whole and its function in normal physiology (Table 1).
Table 1.
Biological consequence(s) of Trp oxidation in selected proteins
| Protein | Oxidant | Modification | Biological Consequence | Refs |
|---|---|---|---|---|
| Lysozyme | Ozone | Trps 108 & 111 to NFK | No change | [57] |
| Lysozyme | Ozone | Trp62 to NFK | Loss of activity | [59] |
| hSOD1 | Bicarbonate radical | Intermolecular Trp32 dimerization | Protein polymerization/aggregation, potential link to ALS | [5,46,63,64] |
| α-crystallin | Singlet oxygen (porphyrins, hypericin, Fenton chemistry), lens aging (unknown oxidant) | Trp to NFK, hydroxytryptophans, kynurenine | Darkening of lens, potential link to cataractogenesis | [53, 67–72] |
| Apolipoprotein B-100 | Hypochlorous acid | Trp to kynurenine | Modification of LDL into high-uptake form | [87] |
| Apolipoprotein A-1 | Hypochlorous acid | Trp to hydroxytryptophan and dihydroxytryptophan | Modification of HDL that decreases cholesterol efflux capacity | [88,89] |
| D1 | Singlet oxygen | Trp to NFK | Photoinhibition of photosynthesis | [91–93] |
| CP43 | Singlet oxygen | Trp to NFK | Photoinhibition of photosynthesis | [91–93] |
| MopE (Methylococcus capsulatus) | Unknown/Endogenous | Trp130 to kynurenine, | Gain of copper binding function | [94] |
| CorA (Methylomicrobium album BG8) | Unknown/Endogenous | Trp62 to kynurenine | Gain of copper binding function | [95] |
2. Tryptophan oxidation; mechanisms and products
Tryptophan and Trp residues interact with radicals, ozone and 1O2, and these oxidations can result in the formation of a mix of various intermediates and end-products both with and without intact indole rings (Fig. 1). The tryptophan radical can react with superoxide or molecular oxygen to form tryptophan hydroperoxide, which can then rearrange into N-formylkynurenine (NFK) and kynurenine [1, 12]. Direct reactions between Trp and 1O2 or ozone can also lead to the formation of the indole-cleaved products NFK and kynurenine [1].
Fig. 1.
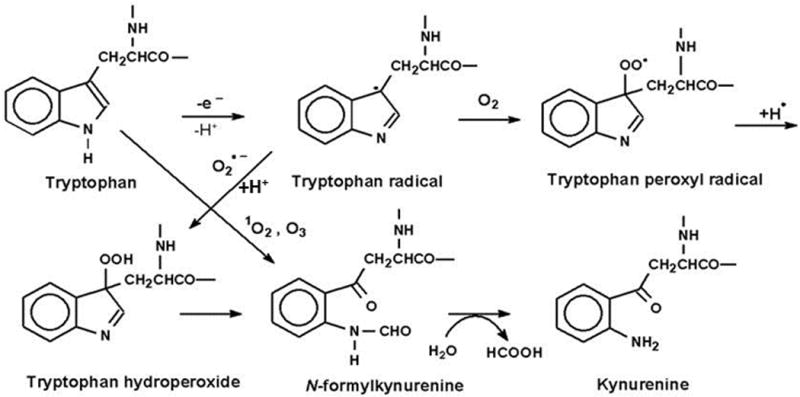
Oxidation of tryptophan to N-formylkynurenine (NFK) and kynurenine. Modified from GieβSauf et al. (59). Originally published in FRBM 46:1260–66,2009 (33).
Plowman et al. [13] synthesized a small Trp-containing peptide (LLWLR) with the goal of identifying molecular markers that could be used to distinguish 1O2-mediated Trp residue oxidation products from those of the hydroxyl radical. Fenton chemistry was used to generate the hydroxyl radical, while 1O2 was generated by visible light irradiation in the presence of the photosensitizer rose bengal. MS/MS (mass spectrometry) analysis comparing reaction products to those of untreated controls detected that hydroxyformylkynurenine and hydroxytryptophan were formed in the 1O2- and hydroxyl-radical-exposed peptides, respectively.
Amino acids that react at a biologically relevant rate with 1O2 are methionine, cysteine, tyrosine, histidine and Trp, and these have reaction rate constants of approximately 1–7 × 107 M−1 sec−1 [1]. To more clearly comprehend how Trp reacts with 1O2 as an amino acid residue within a protein, Jensen et al. [14] measured rate constants for the reaction of Trp with 1O2 using 5 proteins, each with a single Trp residue. 1O2 was generated using visible light and an aqueous photosensitizer that did not bind to any of the proteins. The reaction rate constants of the proteins with solvent-exposed or partially exposed Trp residues did not vary much from that of free Trp. The constants for the two proteins with buried Trp residues, however, were not only much lower than that of free Trp but more than an order of magnitude different from each other. This suggests that while solvent exposure of Trp residues plays a major role in susceptibility to 1O2 oxidation, the local environment also contributes extensively.
Both NFK and kynurenine have been characterized as common degradation products of the reaction of Trp and Trp residue interaction with 1O2 [1]. The pathway through which NFK is produced is believed to be through either the formation of a dioxetane across the C2-C3 double bond or through a hydroperoxide at C3, which decomposes by cleavage of the C2-C3 bond to yield NFK. Alternatively, a multi-step process of ring closure followed by subsequent decomposition can also yield NFK [1, 6, 15]. Because tryptophan hydroperoxides are relatively unstable under physiological conditions [15], interactions between 1O2 and Trp lead to a mixture of end-products including hydrotryptophans, kynurenines, and NFK, with the relative amounts of each varying from protein to protein [2, 15, 16]. Ronsein et al. [17] characterized the photooxidation chemistry of free Trp using labeled hydroperoxides, HPLC/mass spectrometry and NMR. This work showed that the initial products of the reaction between 1O2 and Trp are a mixture of trans and cis hydroperoxides and that the two oxygen molecules of NFK derive from these hydroperoxides.
Because of the difficulties inherent in analyzing oxidation products of large heterogeneous molecules such as proteins, work done on amino acid chemical interactions tends to rely on information gleaned from reactions of free amino acids that is then applied to the corresponding amino acid residues within proteins. Furthermore, studies of Trp oxidation can be complicated by the presence of dual targets for hydroxyl radical attack. Both experimental [18] and computational data [19] demonstrate that the mixture of products that can potentially result from hydroxyl radical attack on Trp can be explained by interactions with both the aromatic and pyrrole rings. The hydroxyl radical attacks not only the aromatic center of the aromatic ring, but also the 2 and 3 positions of the pyrrole ring, and while attack upon the aromatic ring produces hydroxyl derivatives, attack upon the pyrrole tends to promote formation of oxindolyalamines, NFK and kynurenine [18, 19].
3. Detection methods
An optimal, completely informative study of biological tryptophan oxidation would include identification of the oxidant or modifying species, elucidation of the step by step mechanism by which the modification occurs, yield analysis of the end-products, and an understanding of the physical consequences to the structure of the protein and how transformation of Trp residues impacts protein biological function. While there are a number of techniques used to study Trp interactions with reactive oxygen species, there is no single technique that can be used to illuminate the entire story. Additionally, all techniques have inherent strengths and weaknesses, and all experimental questions can pose idiosyncratic challenges, and, if we are fortunate, provide unique points of view as well.
Direct, real-time detection of the Trp radical requires electron spin resonance (ESR) and/or ESR spin trapping. Direct ESR is the gold standard for detection of free radicals, and is most useful for studies of relatively stable radicals. Connor et al. [20], for example, published the first report of an L-tryptophan radical cation detected using fast-flow ESR and acidic Ce4+ as the oxidizing agent. Detection of tryptophan radical in proteins, however, is complicated by the ability of other amino acid residues to form radicals, and the possibility that radical transfer could occur between residues within a single molecule [21, 22] or between residues of two individual molecules [2]. In addition, the ESR spectrum of the tryptophan radical is very similar to that of the tyrosyl radical in proteins, and care must be taken to distinguish between them, which can be done by careful analyses of the dihedral angles for the methylene hydrogens attached to the carbon bound to the aromatic ring [20]. The dihedral angles can be determined from the magnitude of the hyperfine coupling constants of the methylene hydrogens. The hyperfine coupling constant is maximal when the methylene hydrogen is as far from the plane of the tryptophan ring as possible, and, conversely, approaches zero when the methylene hydrogen becomes coplanar to the tryptophan ring. If the Trp radical containing protein has a known crystal structure, the dihedral angles of the methylene hydrogens can identify which Trp residue was oxidized to a free radical. This approach was first used to assign the location of the tyrosyl radical in horse myoglobin [23]. To our knowledge, however, this approach has not been used to determine the location of Trp protein radicals, perhaps because the assignment of specific Trp radicals becomes increasingly difficult as the number of residues increases, unless the dihedral angles of the tryptophan radical are unique.
Other work using direct ESR detected a radical of unknown origin from the reaction of horse myoglobin with hydrogen peroxide [24]. This radical, which oxidized GSH, styrene, and arachidonic acid, was identified as a peroxyl radical from its g tensor and the 17O hyperfine coupling constants obtained by reacting molecular oxygen isotopically labelled with 17O. Additional work [25] using recombinant sperm whale myoglobin 13C-labeled at the C-3 indole ring, enabled the localization of the peroxyl radical to a specific tryptophan. Site-directed mutagenesis of Trp14 prevented peroxyl radical formation in the recombinant protein, whereas mutagenesis of Trp7 did not, implicating Trp14 as the site for peroxyl radical formation. Also, oxidation of tryptophan by myoglobin and hydrogen peroxide resulted in spin trap-inhibitable oxygen consumption, consistent with the formation of a tryptophan-derived peroxyl radical [26].
Spin trapping extends the sensitivity of ESR radical detection by using a double bond-containing molecule, the spin trap, which reacts with a reactive radical to produce a longer lived radical. A study of sperm whale metmyoglobin used spin trapping to identify a Trp radical resulting from hydrogen peroxide-mediated self-peroxidation [27]. In addition, this same work was also able to delineate the different hydrogen peroxide concentrations that favor either the Trp or the tyrosine radical in the self-peroxidation reaction. Low hydrogen peroxide to protein ratios favors Trp radical formation while higher ratios are more conducive to tyrosine radical formation. The Trp radical has been reported to be trapped by DMPO via ESR [28] and more recently using MS/MS analysis [29].
While direct ESR and ESR spin trapping have proven highly useful in detecting and identifying radicals in small molecules and in some purified proteins, these techniques have only very limited applicability for detection of free radicals in cells. Additional limitations are that not all biological oxidations of Trp are radical-mediated and that ESR requires milligram quantities of protein, making radical studies of scarce proteins prohibitively expensive. Finally, while ESR allows specific identification of radicals it provides no information about end-products.
In lieu of detection and/or identification of the chemical event that oxidizes Trp residues in proteins, many studies focus on the decrease in Trp concentration, the increase in product or both. Tryptophan has the highest fluorescent quantum yield amongst all the amino acids, both as a free amino acid and as a residue in proteins. This characteristic allows “before and after” spectra to be used as evidence for Trp residue modification through oxidation [30–33]. While fluorescence changes in free Trp are usually clearly detectable, fluorescence measurements of intact proteins can be complicated by factors such as neighboring amino acid residues and by protein tertiary structure. And while excitation of Trp and Trp-containing peptides and proteins at 290 nm produces a strong emission with a peak at 350 nm, the use of fluorescence to identify the products of Trp oxidation is more problematic because both kynurenine and NFK have lower fluorescence yields than Trp [34]. Finally, as mentioned above (see section 2), Trp residue oxidation generally results in a mixture of products, which makes the identification of specific fluorescence signatures more difficult. Although fluorescence assessments can be made with relative ease, the tendency of biological samples to have high background fluorescence coupled with the fact that the relative position (buried vs. exposed) of Trp residues affects light absorbance and emission confounds the usefulness of fluorescence as a quantitative tool. In most cases, however, tracking the disappearance of Trp fluorescence subsequent to 1O2 or free radical exposure can provide useful, if not definitive, evidence of alterations in Trp residue structure [30, 35].
Difficulties that protein structure can impose on chemical and physical analyses of specific amino acid residues such as Trp and its oxidation products can be overcome by digestion either chemical or enzymatic. The resulting peptide fragments or component amino acid residues can then be subjected to HPLC analysis. While chemical digestion is often more thorough, it can also produce artifacts through oxidation, a critical concern when studying Trp modification by radicals, 1O2 or other oxidative events. Because artifactual oxidation during chemical hydrolysis has been reported with Trp and methionine residues [36], care must be taken to eliminate oxygen at this step, a precautionary measure which can be somewhat problematic.
The mass spectrometry (MS) technique most commonly used for the identification of modified residues is electrospray ionization in combination with liquid chromatography mass spectrometry (ESI-LCMS). Typically LCMS-based proteomic workflows require the proteins to be converted into peptides for identification and quantification. Direct measurement of intact proteins is another analytical option. These approaches have considerably different pros and cons that must be weighed before deciding which is optimal for a specific course of study [37]. Because the low abundance of a modified protein in complex protein mixtures can be a fundamental challenge for MS characterization of protein-centered oxidation products the development of highly sensitive MS techniques is an active area of research.
Peptides with an oxidized Trp residue generally show an increase in mass corresponding to kynurenine, hydroxytryptophan, doubly oxidized tryptophan or a combination of these modifications [36]. Standard LCMS/MS approaches are generally successful in identifying sites of oxidation when proteins contain only a few modifications, however, even when other analytical approaches confirm that an oxidation event on a protein has occurred, LCMS analysis frequently fails to provide information about the sites of modification. One inherent reason is the low abundance of the modified residues. Thus, to improve the detection of low-abundance peptide adducts derived from proteolytic digests, selective enrichment methods must be employed prior to analysis [38–41]. Challenges affecting LCMS analyses in a bottom-up proteomic workflow stem from the wide range of peptide and protein physicochemical properties that give rise to large differences in MS responses. Sample handling, digestion efficiency, and separation can also impact results. As such, relative peptide intensities might not directly reflect the relative abundances of different proteins.
A major factor that influences LCMS-based analyses via electrospray ionization is ion suppression [42–44]. Possible ways to diminish ion suppression issues include changing sample modifiers, preparation techniques, and/or chromatographic conditions. Peptide intensity depends on the quantity of the peptide being ionized as well as on ionization efficiency and, under some conditions, on the properties of co-eluting peptides. The use of lower flow rates (e.g. 100 nL/min) or internal standards can help alleviate ion suppression. Other issues in LCMS-based analyses include the separation peak capacity, the reproducibility of the chromatography, and the mass measurement accuracy and resolving power of the mass spectrometer. Significant technological advances such as the development and commercialization of ultra-performance LC and high mass accuracy/resolution mass spectrometers have substantially overcome these issues, making LCMS-based analyses more reliable and accessible to biologists. Basically, there is no single recognized method that fulfills every analytical need, and available options for MS-based analyses can make it difficult for an investigator to choose the most appropriate approach to answer specific biological questions.
A mass spectrometric study [45] of free Trp exposed to hydroxyl radical via Fenton chemistry analyzed the products using ES-MS and ES-MS/MS. In addition to the expected mono- and dihydroxytryptophans and NFK, products were also identified that resulted from the reaction of Trp with oxidized Trp, and methyl indole derivatives. A dimer resulting from cross-linking of two Trp radicals was also detected. This same type of Trp dimer has recently been reported to occur due to the bicarbonate-dependent peroxidase activity of hSOD1, resulting in the production of a hSOD1 covalently linked dimeric protein [5, 46]. Domingues et al. [45] also used ES-MS to analyze products that accumulated in the presence of the spin trap DMPO. Adducts of Trp radicals were identified as monohydroxy-tryptophan and dihydroxy-tryptophan, and these results were confirmed with tandem mass spectrometry.
In contrast to the highly technological and instrument-dependent methods of mass spectrometry, the more available techniques of immunology have found utility in studies of free radical biology and medicine through the development and application of antibodies specific to the spin trap DMPO [47, 48] and to end-products of free radical chemistry and/or related oxidative processes such as nitrotyrosine [49] and nitrotryptophan [50]. Because NFK is a commonly encountered product of Trp oxidation, anti-serum specific to NFK was developed and validated by in vitro experiments with photooxidized myoglobin, milk proteins and cells, and with bicarbonate radical-oxidized hSOD1. This work showed that the anti-NFK antiserum was specific to NFK and showed little and no recognition of either kynurenine or Trp [51, 52 ].
The use of immunological techniques to detect an endogenous oxidation product has both strengths and weaknesses. One advantage is the ability to perform studies without the potentially interfering influence inherent in the addition of an exogenous reporter molecule. This advantage, however, also has a potential and concomitant disadvantage in that it can be difficult to distinguish detection of endogenous product that exists independently of the experiment from non-specific binding of the antibody. And, as with fluorescence, NFK detection does not provide information as to the chemistry responsible for the observed Trp oxidation. On the other hand, a distinctive advantage in the use of antibodies is their capacity to allow us a window into Trp oxidation in cells and tissues, thus providing snapshots of biological post-translational oxidative modifications. Accordingly, anti-NFK antiserum has provided the means to localize photosensitizer-mediated Trp oxidation of proteins in organelles [51], in specific proteins such as α-crystallin and actin [53], and in tissue of animals suffering late sepsis [54].
Another substantial advantage to the use of anti-NFK is the ability to perform relatively “low tech” experiments such as westerns before committing time and resources to the more complex modes of analyses such as HPLC and MS. Work with acetylcholinesterase (AChE), the enzyme that catabolizes the neurotransmitter acetylcholine, illustrates this point. Weiner et al. [33] determined that photooxidation of AChE with methylene blue led to loss of enzyme activity, and they conjectured that loss of activity was due to a 1O2-mediated conversion of an active site Trp to NFK. Western analysis of the 14 Trp residue-containing AChE did indeed show a correlation between NFK accumulation and photosensitized loss of activity. This observation empowered the decision to pursue a detailed MS analysis that detected Trp to NFK oxidation in two Trp residues in the active gorge [55, 56]. In the absence of the initial detection of NFK in the photosensitized AChE using western analysis, it would have been less tempting to initiate and complete a full scale MS investigation due to the potentially prohibitive presence of a total of 14 Trp residues in the intact protein.
4. Biological roles for Trp residues, effects of oxidation
Lysozyme
An early study looked at the role of Trp residues in the biological activity of egg white lysozyme [57]. While limited ozone exposure caused two lysozyme Trp residues (Trp108 and Trp111) to be converted to NFK, this oxidation occurred without loss of enzymatic activity. Continued ozonation, however, eventually resulted in complete inactivation of lysozyme with a corresponding increase in the conversion of the remaining four Trp residues to NFK. Subsequent work showed that bronchoalveolar lavage fluid recovered from ozone-exposed animals had significantly decreased lysozyme activity [58], while an in vitro study [59] determined that the ozone oxidation that converted a Trp residue in the polysaccharide binding domain to NFK also eliminated lysozyme activity.
hSOD1
Amyotrophic lateral sclerosis (ALS) (also known as Lou Gehrig’s disease) is a motor neuron disorder characterized by degeneration of neurons in the spinal cord and brain. Familial ALS (fALS) accounts for about 2% of all ALS cases and is linked to mutations in the gene encoding copper/zinc superoxide dismutase (hSOD1). There are more than 100 catalogued hSOD1 gene mutations that result in a gain of function phenotype leading to decreased protein solubility and increased protein aggregation [60]. Clinically, sporadic ALS and fALS are indistinguishable, with several lines of evidence suggesting that aberrant hSOD1 post-translational modifications are analogous to the genetically-encoded protein alterations found in fALS. Protein aggregates are a histopathological hallmark associated with all cases of ALS, and the protein aggregates found in sporadic ALS are immunoreactive with antibodies to hSOD1 [61], suggesting that aggregation of SOD-1 is basic to ALS etiology. Additionally, antibodies against misfolded hSOD1 provide some therapeutic protection, extending survival in transgenic mice expressing a mutant SOD1 (G93A) [62].
Both oxidative modifications in general and specific Trp alterations have been linked to increases in protein misfolding in numerous proteins, including hSOD1. The ability of the sole Trp residue of hSOD1 to potentiate protein aggregation in the presence of bicarbonate was first studied by Zhang et al. [46, 63]. These reports showed that the bicarbonate/hydrogen peroxide-dependent peroxidase activity of hSOD1 leads to formation of protein polymers, that the presence of the Trp residue is essential for this aggregation to occur, and that the process is probably radical-mediated. This latter observation was confirmed by Medinas et al. [5], who used MS to determine that an hSOD1 ditryptophan crosslink formed via a radical-radical reaction.
In vivo evidence for a link between hSOD1 aggregation and ALS comes from analysis of hSOD1 isolated from human and transgenic mouse blood [64] which identified Trp32 modifications in both. These modifications were found independently of whether the samples were prepared under aerobic or anaerobic conditions, signifying that the alterations occurred in vivo, and not as a result of sample preparation. Experiments also compared WT hSOD1 with G93A hSOD1 (a fALS disease-causing mutant protein) expressed in motor neurons [64]. The G93A-expressing motor neurons died much sooner than those expressing WT hSOD1. It is also significant that substituting a less oxidation-prone amino acid (phenylalanine) for Trp32 in the G93A protein reduced the cell death rate to almost WT levels, suggesting that interfering with the Trp-mediated aggregation of the protein also decreases protein toxicity.
The aging lens, cataracts and crystallin
The human eye is comprised of a succession of transparent tissues that allow transmission of light onto the retina, enabling vision. The cornea, at the ocular surface, contributes two thirds of the focusing capacity and filters out all wavelengths below 295 nm while long wavelength UVB (295–314 nm) and UVA (315–400 nm) radiation are absorbed by the adult human lens. More than 90% of the total dry mass of the lens is made up of the crystallin proteins, with α-crystallin representing about 35% of this total [65]. In contrast with other proteins in other organs, the proteins of the mature human lens do not turn over, and while new cells and new proteins are added to the lens during its lifetime, the process is one of protein accumulation rather than replacement. The modifications and damage that occur in the lens, therefore, also accumulate. Cataracts develop because the cumulative damage to the lens proteins causes them to become less soluble leading to lens clouding. Despite advances in cataract surgery, cataracts are still the leading cause of blindness in the world [66].
Studies on post-translational modifications of α-crystallin and their potential contribution to cataract formation have chronicled a number of alterations to light-absorbing aromatic amino acids such as tryptophan. In vitro studies with bovine α-crystallin have described Trp oxidation products in α-crystallin photosensitized with porphyrins [67, 68] and hypericin [53, 69], and in α-crystallin subjected to Fenton chemistry [70]. A proteomic comparison [71] of young, aged and early stage cataract-containing lenses found several post-translational modifications correlated with age, cataracts or both. Semi-quantitative analysis determined that an increase in the amount of oxidized Trp residues correlated with lens aging and cataract development, with specific Trp modifications found only in cataractous lenses. Lens protein mass spectrometric studies have also shown that accumulation of Trp oxidation products in human lens α-crystallin correlates with both aging and cataract [71, 72]. Analysis of post-translational modifications in lens protein from a four-year-old congenital cataract [72] also detected oxidized Trp residues.
A further consideration in Trp oxidation to kynurenine and NFK is that these two amino acid derivatives are both more efficient photosensitizers than their parent molecule [1]. This becomes biologically significant due to the unique nature of the human lens and the proteins that comprise it. The role of α-crystallin is to intercept and focus light, and because the protein itself, damaged or otherwise, is not turned over, increasing α-crystallin kynurenine and NFK content permanently increases the potential for photosensitized lens damage.
Although all proteins are constructed from L-amino acids, D-aspartyl residues have been detected in proteins from metabolically inactive or inert tissue, including the lens. An investigation into the mechanism by which this racemization occurs revealed that is accelerated by exposure to UVB radiation [73]. While aspartyl residues do not themselves absorb UV radiation, a study using synthetic peptides showed that racemization of aspartyl residues was contingent upon their proximity to and position relative to photooxidizable Trp residues [74]. This suggests the likelihood that oxidation of Trp residues can not only result in alteration of the Trp residue itself, but may also potentiate permanent transformations in protein content and structure at other residues less prone to oxidation.
Apolipoproteins
Each particle of low density lipoprotein (LDL) contains a single molecule of apolipoprotein B-100 (apoB-100). This large (ca. 550 kDa.) protein contains a highly hydrophilic domain at the N-terminus in addition to several other hydrophobic segments in the remainder of the molecule. ApoB-100 plays a key role in LDL recognition and binding to the LDL receptor found at the surface of the majority of human cells. Binding of LDL to the receptor is followed by degradation of the lipoprotein and release of cholesterol into the cell. Alterations to apoB-100, such as those produced by oxidation, can result in a complete loss of or a decrease in receptor affinity, and oxidatively modified LDL is considered a risk factor in atherosclerosis [75].
Early research showed that incubation of LDL with Cu2+ resulted in oxidized LDL with properties similar to that of LDL from cells [76]. Several subsequent studies using Cu2+ oxidation as a model have detected oxidation of Trp residues to various derivatives [68–70]. Gieβauf et al. [77] used HPLC to analyze Cu2+-oxidized LDL following delipidation and pronase digestion, and detected a loss of Trp-specific fluorescence with a corresponding increase in fluorescence consistent with NFK. In another study that used LCMS, Cu2+ oxidation of LDL was seen to convert Trp residues to kynurenine [78]. Work by Batthyány et al. [79] using both LDL and apoB-100 purified from LDL showed that the purified protein itself was capable of copper reduction and comprises one of the elements of LDL responsible for this reduction. This study also used EPR spin trapping to show that a spectrum consistent with a tryptophanyl radical could be detected from the aqueous, protein-containing portion of LDL incubated with Cu2+, and that this spectrum matched that of purified, lipid-independent apoB-100 incubated with Cu2+ [79]. Interestingly, Borén et al. [80] reported that a mutation in the apoB-100 gene resulting in a Trp4369 to Tyr4369 substitution results in defective receptor binding of LDL, suggesting that oxidation of apoB-100 Trp residues could have far-reaching effects on the biology of LDL, and subsequently on the pathogenesis of atherosclerosis.
Myeloperoxidase (MPO) is a component of circulating immune system cells such as polymorphic neutrophil granulocytes and monocytes, and MPO-deficient humans and animal show increased susceptibility to yeast and fungal infections [81]. MPO has also been observed in human atherosclerotic tissue [82] with products of MPO catalyzed reactions being observed in all stages of atherosclerosis [83]. Because oxidation of LDLs is believed to be an early event in development of atherosclerotic lesions, a number of studies [84–87] have focused on the effects of myeloperoxidase-derived (MPO) oxidants on modifications of apoB-100.
The hypochlorous acid (HOCl) produced by MPO reacts with LDL in a concentration dependent manner [88], and oxidizes apoB-100 Trp amino acid residues, which can be detected by decreases in Trp residue fluorescence [84, 87, 88]. Exposure of LDL to HOCl converts it into a high-up take form that results in the accumulation of lipids within cells [87], suggesting that MPO-mediated apoB-100 Trp residue oxidation plays an important role in the development of atherosclerotic lesions [85]. Additionally, MPO polymorphisms resulting in MPO deficiency or loss of function have been associated with decreased coronary disease [81].
Apolipoprotein A-1 (apoA-1), the major protein component of high density lipoprotein (HDL), is also a target for MPO-catalyzed oxidation [86, 89, 90], and this oxidation is also believed to contribute to development of atherosclerotic lesions [85]. Fluorescence analysis indicated that HOCl oxidizes HDL more rapidly than LDL while a mass spectroscopy study [90] showed that HOCl oxidized all four apoA-1 Trp residues to hydroxytryptophan and dihydroxytryptophan, although to different extents. Hadfield et al. [89] exposed lipid-free apoA-1 and apoA-1-containing reconstituted high-density lipoprotein (rHDLs) to HOCL and to hypothiocyanous acid (HOSCN), another major MPO-derived oxidant, and both oxidized apoA-1 Trp residues. And, while oxidation with either HOCl or HOSCN decreased the cholesterol efflux capacity of apoA-1, only HOCl increased the inflammatory properties of rHDLs containing apoA-1.
Photosynthesis
The plant photosynthetic apparatus is a membrane-bound, multicomponent assembly of molecules composed of two photosystems (PSI and PSII), each of which contains multiple polypeptides. The photosystems also contain chlorophylls as well as ancillary light-harvesting pigments that enable the plant to convert light into chemical energy [91]. Intensive research over many years has focused on the goal of increasing photosynthetic efficiency. The combined structural and functional complexity of the photosynthetic apparatus, however, has proved a hindrance to a more thorough understanding of how the components interact. While numerous post-translational modifications to structural, enzymatic and regulatory protein subunits have been described, the role/result of these modifications is not always well understood.
In the gathering of light energy, the plant photosynthetic machinery is continuously faced with conditions that generate both radical and non-radical oxidative species [92]. Leakage from the electron transport chains in both photosystems I and II, especially under high light conditions, can result in the production of 1O2, H2O2 and hydroxyl radical. While the potential for damage to its biomolecular components is intrinsic to the process of photosynthesis, an equally intrinsic system for both sensing damage and initiating repair is also present. One aspect of the sensing and repair mechanism seems to rely on the oxidation of Trp residues to NFK in core proteins of PSII, a reaction occurring readily in the presence of 1O2. In PSII, increasing oxidation of Trp365 to NFK in the CP43 subunit correlates with increasing light stress and increasing photoinhibition in thylakoid membranes [91, 93]. While a small increase in ionic strength results in a decrease in NFK365 and leads instead to a Trp to NFK conversion in a different intrinsic polypeptide (D1), both the CP43 and the D1 Trp oxidations to NFK appear to be linked to D1 degradation and subsequent replacement of the damaged D1 proteins. [91]. Because D1 protein degradation and replacement is a key step in photosynthetic recovery from high light stress Kasson et al. [91] have proposed that Trp to NFK oxidation is a key signal for photosynthetic damage repair through protein turnover.
MopE
Studies on Trp residue oxidation in proteins focus almost exclusively on those modifications resulting in loss or diminution of protein function. In contrast, a unique gain of biological function requiring Trp residue oxidation has been described in the methane-oxidizing bacterium Methylococcus capsulatus [94]. Methane-oxidizing bacteria, such as M. capsulatus, use a methane monooxygenase to catalyze the oxidation of methane to methanol as their first step in energy production, and use copper for both catalytic activity and regulation of this enzyme.
During copper-limited growth, M. capsulatus secretes large amounts of the copper-binding protein MopE. When the crystal structure of this protein was determined it was found to contain a single, partially buried copper ion located in a pocket surrounded by two histidine residues and what was, in the primary sequence, predicted to be a Trp residue. The involvement of the Trp residue in copper binding was unexpected because the indole side chain had not previously been reported to coordinate metal ions in enzymes. Electron density maps and other data, however, suggested that the Trp residue had been oxidized to kynurenine, a supposition subsequently confirmed by mass spectrometry. Furthermore, expression of MopE in Escherichia coli produces a protein which cannot bind copper because the Trp remains unoxidized. Subsequent work [95] with another methanotrophic bacterium, Methylomicrobium album, described a protein with a copper binding site homologous to that of MopE, and with a comparable Trp to kynurenine oxidation.
5. Concluding remarks
Free amino acids, as well as protein and peptide amino acid residues can be oxidized by a number of reactive species such as ozone, 1O2 and free radicals. These modifications alter the chemical nature of the amino acid and have the potential to also alter the structure of the protein in which they are contained. The amino acids and amino acid residues most susceptible to oxidation include methionine, histidine, cysteine, tyrosine and Trp. While most reactions with Trp involve chemical alteration to the amino acid itself that lead to deleterious alterations in protein function, there are also reports of Trp dimerization (hSOD1), Trp residue photosensitization in α-crystallin leading to racemization of a non-UV absorbing amino acid, and Trp oxidation leading to a gain of function (MopE). There is also work that strongly suggests that Trp oxidation in plants serves as a signal that protein turnover is needed to ensure proper function of the photosynthetic machinery. As the number of detection methods increases and they become more sensitive, it is likely there will be further reports of how Trp residue oxidations impact biology and human health.
Acknowledgments
We thank B. Jean Corbett, Dr. Ann Motten and Mary J. Mason for valuable help in the preparation of the manuscript. This work was supported by the Intramural Research Program of the NIH, NIEHS.
Abbreviations
- Trp
Tryptophan
- NFK
N-formylkynurenine
- ESR
electron spin resonance
- AChE
acetylcholinesterase
- LDL
low density lipoprotein
- apoB-100
apolipoprotein B-100
- apoA-1
apolipoprotein A-1
- HDL
high density lipoproteins
- 1O2
singlet oxygen
- hSOD1
human SOD1
- ALS
amyotrophic lateral sclerosis
- MS
mass spectrometry
- MPO
myeloperoxidase
- HOCl
hypochlorous acid
- HOSCN
hypothiocyanous acid
- PSII
photosystem II
References
- 1.Davies MJ. Singlet oxygen-mediated damage to proteins and its consequences. Biochem Biophys Res Commun. 2003;305:761–770. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 2.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Mudd JB, Leavitt R, Ongun A, McManus TT. Reaction of ozone with amino acids and proteins. Atmos Environ. 1969;3:669–682. doi: 10.1016/0004-6981(69)90024-9. [DOI] [PubMed] [Google Scholar]
- 4.Pryor WA, Giamalva DH, Church DF. Kinetics of ozonation. 2. Amino acids and model compounds in water and comparisons to rates in nonpolar solvents. J Am Chem Soc. 1984;106:7094–7100. [Google Scholar]
- 5.Medinas DB, Gozzo FC, Santos LFA, Iglesias AH, Augusto O. A ditryptophan cross-link is responsible for the covalent dimerization of human superoxide dismutase 1 during its bicarbonate-dependent peroxidase activity. Free Radic Biol Med. 2010;49:1046–1053. doi: 10.1016/j.freeradbiomed.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Davies MJ. Reactive species formed on proteins exposed to singlet oxygen. Photochem Photobiol Sci. 2004;3:17–25. doi: 10.1039/b307576c. [DOI] [PubMed] [Google Scholar]
- 7.Pattison DI, Rahmanto AS, Davies MJ. Photo-oxidation of proteins. Photochem Photobiol Sci. 2012;11:38–53. doi: 10.1039/c1pp05164d. [DOI] [PubMed] [Google Scholar]
- 8.Nuriel T, Hansler A, Gross SS. Protein nitrotryptophan: Formation, significance and identification. J Proteomics. 2011;74:2300–2312. doi: 10.1016/j.jprot.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebrin I, Bregere C, Gallaher TK, Sohal RS. Detection and characterization of peroxynitrite-induced modifications of tyrosine, tryptophan, and methionine residues by tandem mass spectrometry. Methods Enzymol. 2008;441:283–294. doi: 10.1016/S0076-6879(08)01215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 11.Saito I, Matsuura T, Nakagawa M, Hino T. Peroxidic intermediates in photosensitized oxygenation of tryptophan derivatives. Acc Chem Res. 1977;10:346–352. [Google Scholar]
- 12.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 13.Plowman JE, Deb-Choudhury S, Grosvenor AJ, Dyer JM. Protein oxidation: identification and utilisation of molecular markers to differentiate single oxygen and hydroxyl radical-mediated oxidative pathways. Photochem Photobiol Sci. 2013;12:1960–1967. doi: 10.1039/c3pp50182e. [DOI] [PubMed] [Google Scholar]
- 14.Jensen RL, Arnbjerg J, Ogilby PR. Reaction of singlet oxygen with tryptophan in proteins: A pronounced effect of the local envrionment on the reaction rate. J Am Chem Soc. 2012;134:9820–9826. doi: 10.1021/ja303710m. [DOI] [PubMed] [Google Scholar]
- 15.Gracanin M, Hawkins CL, Pattison DI, Davies MJ. Singlet-oxygen-mediated amino acid and protein oxidation: Formation of tryptophan peroxides and decomposition products. Free Radic Biol Med. 2009;47:92–102. doi: 10.1016/j.freeradbiomed.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Ronsein GE, de Oliveira MCB, de Medeiros MHG, Di Mascio P. Characterization of O2 (1Δ g)-derived oxidation products of tryptophan: a combination of tandem mass spectrometry analyses and isotopic labeling studies. J Am Soc Mass Spectrom. 2009;20:188–197. doi: 10.1016/j.jasms.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Ronsein GE, Oliveira MCB, Miyamoto S, Medeiros MHG, Di Mascio P. Tryptophan oxidation by singlet molecular oxygen [O2 (1Δg)]: Mechanistic studies using 18O-labeled hydroperoxides, mass spectrometry, and light emission measurements. Chem Res Toxicol. 2008;21:1271–1283. doi: 10.1021/tx800026g. [DOI] [PubMed] [Google Scholar]
- 18.Maskos Z, Rush JD, Koppenol WH. The hydroxylation of tryptophan. Arch Biochem Biophys. 1992;296:514–520. doi: 10.1016/0003-9861(92)90605-v. [DOI] [PubMed] [Google Scholar]
- 19.Mujika JI, Uranga J, Matxain JM. Computational study on the attack of OH radicals on aromatic amino acids. Chemistry-A European Journal. 2013;19:6862–6873. doi: 10.1002/chem.201203862. [DOI] [PubMed] [Google Scholar]
- 20.Connor HD, Sturgeon BE, Mottley C, Sipe HJ, Jr, Mason RP. L-Tryptophan radical cation electron spin resonance studies: connecting solution-derived hyperfine coupling constants with protein spectral interpretations. J Am Chem Soc. 2008;130:6381–6387. doi: 10.1021/ja0780277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese B, Graber M, Cordes M. Electron transfer in peptides and proteins. Curr Opin Chem Biol. 2008;12:755–759. doi: 10.1016/j.cbpa.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 22.Moser CC, Anderson JLR, Dutton PL. Guidelines for tunneling in enzymes. Biochem Biophys Acta. 2010;1797:1573–1586. doi: 10.1016/j.bbabio.2010.04.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunther MR, Sturgeon BE, Mason RP. A long-lived tyrosyl radical from the reaction between horse metmyoglobin and hydrogen peroxide. Free Radic Biol Med. 2000;28:709–719. doi: 10.1016/s0891-5849(00)00164-7. [DOI] [PubMed] [Google Scholar]
- 24.Kelman DJ, DeGray JA, Mason RP. Reaction of myoglobin with hydrogen peroxide forms a peroxyl radical which oxidizes substrates. J Biol Chem. 1994;269:7458–7463. [PubMed] [Google Scholar]
- 25.DeGray JA, Gunther MR, Tschirret-Guth R, Ortiz de Montellano PR, Mason RP. Peroxidation of a specific tryptophan of metmyoglobin by hydrogen peroxide. J Biol Chem. 1997;272:2359–2362. doi: 10.1074/jbc.272.4.2359. [DOI] [PubMed] [Google Scholar]
- 26.Gunther MR, Kelman DJ, Corbett JT, Mason RP. Self-peroxidation of metmyoglobin results in formation of an oxygen-reactive tryptophan-centered radical. J Biol Chem. 1995;270:16075–16081. doi: 10.1074/jbc.270.27.16075. [DOI] [PubMed] [Google Scholar]
- 27.Gunther MR, Tschirret-Guth RA, Lardinois OM, Ortiz de Montellano PR. Tryptophan-14 is the preferred site of DBNBS spin trapping in the self-peroxidation reaction of sperm whale metmyoglobin with a single equivalent of hydrogen peroxide. Chem Res Toxicol. 2003;16:652–660. doi: 10.1021/tx0256580. [DOI] [PubMed] [Google Scholar]
- 28.McArthur KM, Davies MJ. Detection and reactions of the globin radical in haemoglobin. Biochem Biophys Acta-Prot Struct Molec Enzymol. 1993;1202:173–181. doi: 10.1016/0167-4838(93)90002-9. [DOI] [PubMed] [Google Scholar]
- 29.Vallelian F, Garcia-Rubio I, Puglia M, Kahraman A, Deuel JW, Engelsberger WR, Mason RP, Buehler PW. Spin trapping combined with quantitative mass spectrometry defines free radical redistribution within the oxidized hemoglobin:haptoglobin complex. Free Radic Biol Med. 2015;85:259–286. doi: 10.1016/j.freeradbiomed.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 30.Dalsgaard TK, Otzen D, Nielsen JH, Larsen LB. Changes in structures of milk proteins upon photo-oxidation. J Agric Food Chem. 2007;55:10968–10976. doi: 10.1021/jf071948g. [DOI] [PubMed] [Google Scholar]
- 31.Sancataldo G, Vetri V, Fodera V, Di Cara G, Militello V, Leone M. Oxidation enhances human serum albumin thermal stability and changes the routes of amyloid fibril formation. Plos One. 2014;9:e84552. doi: 10.1371/journal.pone.0084552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utrera M, Rodriguez-Carpena JG, Morcuende D, Estévez M. Formation of lysine-derived oxidation products and loss of tryptophan during processing of porcine patties with added avocado byproducts. J Agric Food Chem. 2012;60:3917–3926. doi: 10.1021/jf3001313. [DOI] [PubMed] [Google Scholar]
- 33.Weiner L, Roth E, Silman I. Targeted oxidation of Torpedo californica acetylcholinesterase by singlet oxygen. Photochem Photobiol. 2011;87:308–316. doi: 10.1111/j.1751-1097.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 34.Fukunaga Y, Katsuragi Y, Izumi T, Sakiyama F. Fluorescence characteristics of kynurenine and N′-formylkynurenine. Their use as reporters of the environment of tryptophan 62 in hen egg-white lysozyme. J Biochem. 1982;92:129–141. doi: 10.1093/oxfordjournals.jbchem.a133909. [DOI] [PubMed] [Google Scholar]
- 35.Paz A, Roth E, Ashani Y, Xu Y, Shnyrov VL, Sussman JL, Silman I, Weiner L. Structural and functional characterization of the interaction of the photosensitizing probe methylene blue with Torpedo californica acetylcholinesterase. Protein Sci. 2012;21:1138–1152. doi: 10.1002/pro.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perdivara I, Deterding LJ, Przybylski M, Tomer KB. Mass spectrometric identification of oxidative modifications of tryptophan residues in proteins: Chemical artifact or post-translational modification? J Am Soc Mass Spectrom. 2010;21:1114–1117. doi: 10.1016/j.jasms.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messana I, Cabras T, Iavarone F, Vincenzoni F, Urbani A, Castagnola M. Unraveling the different proteomic platforms. J Sep Sci. 2013;36:128–139. doi: 10.1002/jssc.201200830. [DOI] [PubMed] [Google Scholar]
- 38.Dunn JD, Reid GE, Bruening ML. Techniques for phosphopeptide enrichment prior to analysis by mass spectrometry. Mass Spectrom Rev. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 39.Gundry RL, White MY, Murray CI, Kane LA, Fu Q, Stanley BA, Van Eyk JE. Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Curr Protoc Mol Biol. 2009:10.25.11–10.25.23. doi: 10.1002/0471142727.mb1025s88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 41.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 42.Annesley TM. Ion suppression in mass spectrometry. Clin Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 43.Bonfiglio R, King RC, Olah TV, Merkle K. The effects of sample preparation methods on the variability of the electrospray ionization response for model drug compounds. Rapid Commun Mass Spectrom. 1999;13:1175–1185. doi: 10.1002/(SICI)1097-0231(19990630)13:12<1175::AID-RCM639>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 44.Sterner JL, Johnson MV, Nicol GR, Ridge DP. Signal suppression in electrospray ionization Fourier transform mass spectrometry of multi-component samples. J Mass Spectrom. 2000;35:385–391. doi: 10.1002/(SICI)1096-9888(200003)35:3<385::AID-JMS947>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 45.Domingues MRM, Domingues P, Reis A, Fonseca C, Amado FML, Ferrer-Correia AJV. Identification of oxidation products and free radicals of tryptophan by mass spectrometry. J Am Soc Mass Spectrom. 2003;14:406–416. doi: 10.1016/S1044-0305(03)00127-2. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Joseph J, Crow J, Kalyanaraman B. Mass spectral evidence for carbonate-anion-radical-induced posttranslational modification of tryptophan to kynurenine in human Cu, Zn superoxide dismutase. Free Radic Biol Med. 2004;37:2018–2026. doi: 10.1016/j.freeradbiomed.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 47.Gomez-Mejiba SE, Zhai Z, Della-Vedova MC, Munoz MD, Chatterjee S, Towner RA, Hensley K, Floyd RA, Mason RP, Ramirez DC. Immuno-spin trapping from biochemistry to medicine: Advances, challenges, and pitfalls. Focus on protein-centered radicals. Biochim Biophys Acta. 2014;1840:722–729. doi: 10.1016/j.bbagen.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic Biol Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 49.Ye YZ, Strong M, Huang ZQ, Beckman JS. Antibodies that recognize nitrotyrosine. Methods Enzymol. 1996;269:201–209. doi: 10.1016/s0076-6879(96)69022-3. [DOI] [PubMed] [Google Scholar]
- 50.Bregere C, Rebrin I, Sohal RS. Detection and characterization of in vivo nitration and oxidation of tryptophan residues in proteins. Methods Enzymol. 2008;441:339–349. doi: 10.1016/S0076-6879(08)01219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrenshaft M, Bonini MG, Feng L, Chignell CF, Mason RP. Partial colocalization of oxidized, N-formylkynurenine-containing proteins in mitochondria and golgi of keratinocytes. Photochem Photobiol. 2010;86:752–756. doi: 10.1111/j.1751-1097.2010.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrenshaft M, de Oliveira Silva S, Perdivara I, Bilski P, Sik RH, Chignell CF, Tomer KB, Mason RP. Immunological detection of N-formylkynurenine in oxidized proteins. Free Radic Biol Med. 2009;46:1260–1266. doi: 10.1016/j.freeradbiomed.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehrenshaft M, Roberts JE, Mason RP. Hypericin-mediated photooxidative damage of α-crystallin in human lens epithelial cells. Free Radic Biol Med. 2013;60:347–354. doi: 10.1016/j.freeradbiomed.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatterjee S, Lardinois O, Bhattacharjee S, Tucker J, Corbett J, Deterding L, Ehrenshaft M, Bonini MG, Mason RP. Oxidative stress induces protein and DNA radical formation in follicular dendritic cells of the germinal center and modulates its cell death patterns in late sepsis. Free Radic Biol Med. 2011;50:988–999. doi: 10.1016/j.freeradbiomed.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Triquigneaux MM, Ehrenshaft M, Roth E, Silman I, Ashani Y, Mason RP, Weiner L, Deterding LJ. Targeted oxidation of Torpedo californica acetylcholinesterase by singlet oxygen: identification of N-formylkynurenine tryptophan derivatives within the active-site gorge of its complex with the photosensitizer methylene blue. Biochem J. 2012;448:83–91. doi: 10.1042/BJ20120992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silman I, Roth E, Paz A, Triquigneaux MM, Ehrenshaft M, Xu Y, Shnyrov VL, Sussman JL, Deterding LJ, Ashani Y, Mason RP, Weiner L. The specific interaction of the photosensitizer methylene blue with acetylcholinesterase provides a model system for studying the molecular consequences of photodynamic therapy. Chem-Biol Interact. 2013;203:63–66. doi: 10.1016/j.cbi.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Previero A, Coletti-Previero MA, Jolles P. Localization of non-essential tryptophan residues for the biological activity of lysozyme. J Mol Biol. 1967;24:261–268. doi: 10.1016/0022-2836(67)90331-2. [DOI] [PubMed] [Google Scholar]
- 58.Holzman RS, Gardner DE, Coffin DL. In vivo inactivation of lysozyme by ozone. J Bact. 1968;96:1562–1566. doi: 10.1128/jb.96.5.1562-1566.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakiyama F, Natsuki R. Identification of tryptophan 62 as an ozonization-sensitive residue in hen egg-white lysozyme. J Biochem. 1976;79:225–228. doi: 10.1093/oxfordjournals.jbchem.a131051. [DOI] [PubMed] [Google Scholar]
- 60.Anderson PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci Reports. 2006;6:37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- 61.Sábado J, Casanovas A, Hernández S, Piedrafita L, Hereu M, Esquerda JE. Immunodetection of disease-associated conformers of mutant Cu/Zn superoxide dismutase 1 selectively expressed in degenerating neurons in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2013;72:646–661. doi: 10.1097/NEN.0b013e318297fd10. [DOI] [PubMed] [Google Scholar]
- 62.Gros-Louis F, Soucy G, Lariviere R, Julien JP. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J Neurochem. 2010;113:1188–1199. doi: 10.1111/j.1471-4159.2010.06683.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Andrekopoulos C, Joseph J, Chandran K, Karoui H, Crow JP, Kalyanaraman B. Bicarbonate-dependent peroxidase activity of human Cu, Zn-superoxide dismutase induces covalent aggregation of protein - Intermediacy of tryptophan-derived oxidation products. J Biol Chem. 2003;278:24078–24089. doi: 10.1074/jbc.M302051200. [DOI] [PubMed] [Google Scholar]
- 64.Taylor DM, Gibbs BF, Kabashi E, Minotti S, Durham HD, Agar JN. Tryptophan 32 potentiates aggregation and cytotoxicity of a copper/zinc superoxide dismutase mutant associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282:16329–16335. doi: 10.1074/jbc.M610119200. [DOI] [PubMed] [Google Scholar]
- 65.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76:145–153. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 66.Rao GN, Khanna R, Payal A. The global burden of cataract. Curr Opin Ophthamol. 2011;22:4–9. doi: 10.1097/ICU.0b013e3283414fc8. [DOI] [PubMed] [Google Scholar]
- 67.Ehrenshaft M, Zhao B, Andley UP, Mason RP, Roberts JE. Immunological detection of N-formylkynurenine in porphyrin-mediated photooxided lens α-crystallin. Photochem Photobiol. 2011;87:1321–1329. doi: 10.1111/j.1751-1097.2011.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McDermott M, Chiesa R, Roberts JE, Dillon J. Photooxidation of specific residues in α-crystallin polypeptides. Biochemistry. 1991;30:8653–8660. doi: 10.1021/bi00099a023. [DOI] [PubMed] [Google Scholar]
- 69.Schey KL, Patat S, Chignell CF, Datillo M, Wang RH, Roberts JE. Photooxidation of lens α-crystallin by hypericin (active ingredient in St. John’s Wort) Photochem Photobiol. 2000;72:200–203. doi: 10.1562/0031-8655(2000)072<0200:polcbh>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 70.Finley EL, Dillon J, Crouch RK, Schey KL. Identification of tryptophan oxidation products in bovine α-crystallin. Protein Sci. 1998;7:2391–2397. doi: 10.1002/pro.5560071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hains PG, Truscott RJW. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J Proteome Res. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 72.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR., III Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujii N, Momose Y, Ishibashi Y, Uemura T, Takita M, Takehana M. Specific racemization and isomerization of the aspartyl residue of αA-crystallin due to UV-B irradiation. Exp Eye Res. 1997;65:99–104. doi: 10.1006/exer.1997.0315. [DOI] [PubMed] [Google Scholar]
- 74.Cai S, Fuji N, Takeshi S, Fujii N. Simultaneous ultraviolet B-induced photo-oxidation of tryptophan/tyrosine and racemization of neighboring aspartyl residues in peptides. Free Radic Biol Med. 2013;65:1037–1046. doi: 10.1016/j.freeradbiomed.2013.08.171. [DOI] [PubMed] [Google Scholar]
- 75.Steinberg D, Parthasarathy S, Carew TE, Khoo JC, Witztum JL. Modifications of low-density lipoprotein that increase its atherogenicity. N Engl J Med. 1989;320:915–924. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- 76.Steinbrecher UP, Parthasarathy S, Leake DS, Witztum JL, Steinberg D. Modification of low density lipoprotein by endothelial cells involves lipid peroxidation and degradation of low density lipoprotein phospholipids. Proc Natl Acad Sci USA. 1984;81:3883–3887. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gieβauf A, van Wickern B, Simat T, Steinhart H, Esterbauer H. Formation of N-formylkynurenine suggests the involvement of apolipoprotein B-100 centered tryptophan radicals in the initiation of LDL lipid peroxidation. FEBS letters. 1996;389:136–140. doi: 10.1016/0014-5793(96)00546-7. [DOI] [PubMed] [Google Scholar]
- 78.Yang C, Gu ZW, Yang M, Lin SN, Siuzdak G, Smith CV. Identification of modified tryptophan residues in apolipoprotein B-100 derived from copper ion-oxidized low-density lipoprotein. Biochemistry. 1999;38:15903–15908. doi: 10.1021/bi991464g. [DOI] [PubMed] [Google Scholar]
- 79.Batthyány C, Santos CXC, Botti H, Cerveñansky C, Radi R, Augusto O, Rubbo H. Direct evidence for apo B-100-mediated copper reduction: studies with purified apo B-100 and detection of tryptophanyl radicals. Arch Biochem Biophys. 2000;384:335–340. doi: 10.1006/abbi.2000.2102. [DOI] [PubMed] [Google Scholar]
- 80.Borén J, Ekström U, Ågren B, Nilsson-Ehle P, Innerarity TL. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J Biol Chem. 2001;276:9214–9218. doi: 10.1074/jbc.M008890200. [DOI] [PubMed] [Google Scholar]
- 81.Kutter D, Devaquet P, Vanderstocken G, Paulus JM, Marchal V, Gothot A. Consequences of total and subtotal myeloperoxidase deficiency: risk or benefit? Acta Haematol. 2000;104:10–15. doi: 10.1159/000041062. [DOI] [PubMed] [Google Scholar]
- 82.Daugherty A, Dunn JL, Rateri DL, Heinecke JW. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J Clin Invest. 1994;94:437–444. doi: 10.1172/JCI117342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heinecke JW. Mechanisms of oxidative damage of low density lipoprotein in human atherosclerosis. Curr Opin Lipidol. 1997;8:268–274. doi: 10.1097/00041433-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 84.Carr AC, Decker EA, Park YJ, Frei B. Comparison of low-density lipoprotein modification by myeloperoxidase-derived hypochlorous and hypobromous acids. Free Radic Biol Med. 2001;31:62–72. doi: 10.1016/s0891-5849(01)00552-4. [DOI] [PubMed] [Google Scholar]
- 85.Nicholls SJ, Hazen SL. Myeloperoxidase, modified lipoproteins, and atherogenesis. J Lipid Res. 2009;50:S346–S351. doi: 10.1194/jlr.R800086-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jerlich A, Hammel M, Nigon F, Chapman MJ, Schaur RJ. Kinetics of tryptophan oxidation in plasma lipoproteins by myeloperoxidase-generated HOCl. Eur J Biochem. 2000;267:4137–4143. doi: 10.1046/j.1432-1327.2000.01449.x. [DOI] [PubMed] [Google Scholar]
- 87.Hazell LJ, Stocker R. Oxidation of low-density lipoprotein with hypochlorite causes transformation of the lipoprotein into a high-uptake form for macrophages. Biochem J. 1993;290:165–172. doi: 10.1042/bj2900165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jerlich A, Fabjan JS, Tschabuschnig S, Smirnova AV, Horakova L, Hayn M, Auer H, Guttenberger H, Leris HJ, Tatzber F, Waeg G, Schaur RJ. Human low density lipoprotein as a target of hypochlorite generated by myeloperoxidase. Free Radic Biol Med. 1998;24:1139–1148. doi: 10.1016/s0891-5849(97)00439-5. [DOI] [PubMed] [Google Scholar]
- 89.Hadfield KA, Pattison DI, Brown BE, Hou L, Rye KA, Davies MJ, Hawkins CL. Myeloperoxidase-derived oxidants modify apolipoprotein A-I and generate dysfunctional high-density lipoproteins: comparison of hypothiocyanous acid (HOSCN) with hypochlorous acid (HOCI) Biochem J. 2013;449:531–542. doi: 10.1042/BJ20121210. [DOI] [PubMed] [Google Scholar]
- 90.Shao B, Heinecke JW. Using tandem mass spectrometry to quantify site-specific chlorination and nitration of proteins: Model system studies with high-density lipoprotein oxidized by myeloperoxidase. In: Enrique C, Lester P, editors. Methods in Enzymology. Academic Press; 2008. pp. 33–63. [DOI] [PubMed] [Google Scholar]
- 91.Kasson TMD, Barry BA. Reactive oxygen and oxidative stress: N-formyl kynurenine in photosystem II and non-photosynthetic proteins. Photosynth Res. 2012;114:97–110. doi: 10.1007/s11120-012-9784-z. [DOI] [PubMed] [Google Scholar]
- 92.Kasson TMD, Rexroth S, Barry BA. Light-induced oxidative stress, N-formylkynurenine, and oxygenic photosynthesis. Plos One. 2012;7:e42220. doi: 10.1371/journal.pone.0042220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dreaden TM, Chen J, Rexroth S, Barry BA. N-Formylkynurenine as a marker of high light stress in photosynthesis. J Biol Chem. 2011;286:22632–22641. doi: 10.1074/jbc.M110.212928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J Biol Chem. 2008;283:13897–13904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- 95.Johnson KA, Ve T, Larsen O, Pedersen RB, Lillehaug JR, Jensen HB, Helland R, Karlsen OA. CorA is a copper repressible surface-associated copper(I)-binding protein produced in Methylomicrobium album BG8. Plos One. 2014;9:e87750. doi: 10.1371/journal.pone.0087750. [DOI] [PMC free article] [PubMed] [Google Scholar]


