Abstract
Production of β-lactamases of the four molecular classes (A, B, C, and D) is the major mechanism of bacterial resistance to β-lactams, the largest class of antibiotics that have saved countless lives since their inception 70 years ago. Although several hundred efficient class D enzymes have been identified in Gram-negative pathogens over the last four decades, they have not been reported in Gram-positive bacteria. Here we demonstrate that efficient class D β-lactamases capable of hydrolyzing a wide array of β-lactam substrates are widely disseminated in various species of environmental Gram-positive organisms. Class D enzymes of Gram-positive bacteria have a distinct structural architecture and employ a unique substrate binding mode quite different from that of all currently known class A, C, and D β-lactamases. They constitute a novel reservoir of antibiotic resistance enzymes.
The discovery of antibiotics and their introduction into clinical practice has revolutionized our ability to treat bacterial infections. Following their initial success, widespread and often uncontrolled use of antibiotics over more than seven decades has resulted in the selection of antibiotic-resistant pathogens. As a result, bacterial infections remain the number one killer in the world, claiming millions of human lives annually. It is estimated that by 2050 the global death rate will soar to 10 million at a cost of over $100 trillion if antibiotic-resistant pathogens remain unchecked (http://amr-review.org/). To address the ever expanding problem of antibiotic resistance in bacteria, more than a dozen structurally diverse classes of compounds have been introduced into clinical practice. Among them the β-lactams are represented by more than a hundred individual compounds that constitute over 60% of the world antibiotic market 1. The β-lactams kill bacteria by inactivating their penicillin-binding proteins (PBPs), essential enzymes involved in the assembly and morphogenesis of the bacterial cell wall2–4. To withstand the deleterious effects of β-lactam antibiotics, bacteria exploit several protection mechanisms. In Gram-negative bacteria production of the antibiotic-inactivating enzymes, β-lactamases, is by far the most prevalent mechanism of resistance. β-lactamases confer resistance to β-lactams via cleavage of the four-membered ring of the antibiotics and the subsequent release of an inactive product. More than 1300 individual enzymes belonging to the four molecular classes (A, B, C, and D) have been characterized 5. The β-lactamases of classes A, C, and D are active-site serine enzymes, and those of class B are zinc-dependent 6–8. The first β-lactamases identified were narrow-spectrum enzymes capable of producing resistance only to early penicillins and cephalosporins. Subsequent introduction into clinics of the next generations of β-lactams triggered selection of mutant enzymes capable of hydrolyzing these novel compounds. As an outcome of this successful evolution, substrate profiles of modern β-lactamases of Gram-negative bacteria widely range from narrow to expanded-spectrum, with many enzymes capable of producing resistance to virtually every available β-lactam antibiotic.
In Gram-positive pathogens, enzymes of only molecular classes A and B have currently been implicated in antibiotic resistance, with the vast majority of them belonging to class A9. The lack of reports regarding the existence of efficient class C and D β-lactamases in Gram-positive bacteria is puzzling in light of how widespread these enzyme classes are in Gram-negative pathogens10,11. Class D OXA-type enzymes of Gram-negative bacteria currently constitute the fastest growing and largest class of β-lactamases with almost 500 members being recognized (http://www.lahey.org/Studies/). Named for their ability to hydrolyze the penicillin antibiotic oxacillin, members of this family of enzymes have evolved to confer resistance to β-lactams of “last resort”, including the carbapenems12,13.
Here we describe the discovery of putative class D β-lactamases in genomes of the Bacillaceae, Clostridiaceae, and Eubacteriaceae families of Gram-positive bacteria. We demonstrate that these enzymes are most common in Bacillaceae, where they are present in at least 12 different Bacillus species. We show that Bacillus atrophaeus, B. pumilus, and B. subtilis encode active class D β-lactamases that produce high levels of resistance to β-lactam antibiotics when expressed in Escherichia coli. We also demonstrate that class D β-lactamases from Gram-positive bacteria exploit a unique substrate-binding mode due to the lack of the conserved arginine that is involved in anchoring of the carboxylate of β-lactam antibiotics in all previously known class A, C, and D β-lactamases.
RESULTS
Class D enzymes do exist in Gram-positive bacilli
Over the past decade thousands of bacterial genomes have been sequenced providing new opportunities for identification of antibiotic resistance genes. We screened the available genomic sequences of Gram-positive bacteria for the genes encoding putative class C and D β-lactamases. While no sequences for putative class C enzymes were found, several hundred open reading frames with conserved amino acid motifs characteristic for the class D β-lactamases have been identified in the Bacillaceae, Clostridiaceae, and Eubacteriaceae families of the phylum Firmicutes, and none have been found in the genomes of Gram-positive cocci, such as staphylococci, streptococci, and enterococci. Although the vast majority of the identified enzymes were from non-pathogenic soil bacteria, two were found in the chromosomes of Clostridium botulinum, a deadly Gram-positive pathogen. One also has to keep in mind that the number of enzymes identified in Gram-positive pathogenic organisms may significantly expand following progress in bacterial genome sequencing. Among currently available genomic sequences putative OXA-type β-lactamases were most common in Bacillaceae, where we have identified them in twelve different Bacillus species. Based on the extent of amino acid sequence identity, they can be subdivided into 9 groups (Supplementary Results, Supplementary Table 1), with sequences of enzymes within each group being ≥ 80% identical. The amino acid sequence identity between various groups of enzymes ranged from 45 to 79%. For two of these species (B. atrophaeus and B. pumilus), these putative OXA-type β-lactamases are native, as they are present in all strains whose genomes have been sequenced. All genomes of B. atrophaeus and some genomes of B. subtilis and B. endophyticus harbor two putative class D enzymes that share between 53% and 78% amino acid sequence identity.
Enzymology of the Gram-positive class D β-lactamases
To evaluate whether putative OXA-type enzymes of Gram-positive bacteria possess β-lactamase activity and are capable of producing resistance to β-lactam antibiotics, we cloned the genes for five of them. Two of these genes from B. atrophaeus (we named the encoded enzymes BAT-1 and BAT-2) and one from B. pumilus (the encoded enzyme BPU-1) were chosen because they are native for these species. Two other genes from B. subtilis (the encoded enzymes BSU-1 and BSU-2) were chosen as their host is well studied. When expressed in an Escherichia coli background, three of the encoded enzymes, BPU-1, BAT-1, and BSU-1, significantly (up to 2048-fold) elevated the minimal inhibitory concentrations (MICs) of the β-lactam antibiotics ampicillin, ticarcillin, and oxacillin. The two other putative β-lactamases, BAT-2 and BSU-2, did not produce antibiotic resistance in E. coli. Among the three active β-lactamases, BPU-1 produces the highest levels of resistance to penicillins, and was chosen to study in more detail. This enzyme elevates the MICs of various penicillins up to 2048-fold and the MICs of the expanded-spectrum cephalosporins ceftazidime and cefepime and the monobactam aztreonam by 128-fold (Table 1). The MICs of the narrow-spectrum cephalosporins cephalothin and cephalexin, the expanded-spectrum cephalosporins cefotaxime and ceftriaxone, and carbapenems, imipenem, meropenem, and doripenem, are elevated by only 1.5- to 4-fold in the presence of the enzyme. BPU-1 is resistant to inhibition by classical class A β-lactamase inhibitors sulbactam and tazobactam, whereas a significant decrease in resistance to ampicillin is observed in the presence of another inhibitor, clavulanic acid. Based on the ability of the enzyme to produce resistance to expanded-spectrum cephalosporins, BPU-1 is classified as an extended-spectrum β-lactamase14. To evaluate whether BPU-1 is capable of producing resistance in a Gram-positive host, we expressed its gene from an E. coli/Bacillus shuttle vector in the β-lactam sensitive B. pumilus strain. Prior to performing MIC evaluation, we grew the strain and tested whether growth medium or sonicated pellet could turn over the chromogenic substrate nitrocefin or the β-lactam antibiotic ampicillin (both are good substrates for BPU-1). No activity was observed against these substrates which demonstrates that this strain does not express BPU-1 or any other β-lactamase. When expressed in the B. pumilus host, BPU-1 increased the MICs of ampicillin, ticarcillin, and piperacillin 4-, 2-, and 2-fold with bacterium inoculum of 5×105 CFU/ml and 64-, 32-, and 32-fold with bacterium inoculum of 5×106 CFU/ml, respectively, which is well below the increase in MICs observed in E. coli (Table 1). The difference in the MIC values in these two hosts could result from a number of factors that include enzyme expression levels and sensitivity of penicillin-binding proteins (targets of β-lactams) to antibiotics. Moreover, Gram-negative bacteria have an outer membrane that could significantly impede the penetration of antibiotics into the cell. In addition, presence of the outer membrane creates a periplasmic space in Gram-negative bacteria which is absent in Gram-positive organisms. The periplasmic space allows for a very high concentration of β-lactamases which would efficiently hydrolyze incoming antibiotic thus producing high levels of resistance. Although the levels of resistance produced by BPU-1 in Gram-positive hosts are relatively low when compared to those in E. coli, this could be advantageous for bacterial survival in environmental niches where a gradient of drug concentrations exist as a result of biosynthesis of antibiotics by β-lactam-producing organisms. An important question yet to be answered is whether various enzymes from this new family are expressed in their hosts and, if so, what levels of resistance to β-lactams they produce. This would require the extensive analysis of dozens of bacterial strains harboring multiple variants of these enzymes. Such analysis is further complicated by the potential impact of other β-lactamases and/or alternative mechanisms on the observed phenotype and levels of resistance. Given that it has taken more than four decades to generate similar data for class D β-lactamases of Gram-negative bacteria, this would be a daunting task.
Table 1.
Antimicrobial susceptibility profile of E. coli JM83 expressing the BPU-1 β-lactamase
| Antimicrobial | MIC (µg/ml) | ||
|---|---|---|---|
| BPU-1 | Control1 | Fold change | |
| Ampicillin | 1,024 | 2 | 512 |
| Ampicillin-Clavulanic acid2 | 32 | 2 | 323 |
| Ampicillin-Sulbactam2 | 1,024 | 2 | 03 |
| Ampicillin-Tazobactam2 | 1,024 | 2 | 03 |
| Piperacillin | 256 | 2 | 128 |
| Ticarcillin | 8,192 | 4 | 2048 |
| Carbenicillin | 8,192 | 4 | 2048 |
| Oxacillin | 8,192 | 256 | 32 |
| Cephalothin | 8 | 4 | 2 |
| Cephalexin | 8 | 4 | 2 |
| Cefotaxime | 0.125 | 0.031 | 4 |
| Ceftriaxone | 0.125 | 0.031 | 4 |
| Ceftazidime | 32 | 0.25 | 128 |
| Cefoxitin | 8 | 4 | 2 |
| Cefepime | 4 | 0.03 | 128 |
| Aztreonam | 8 | 0.06 | 128 |
| Imipenem | 0.25 | 0.125 | 2 |
| Meropenem | 0.030 | 0.015 | 2 |
| Doripenem | 0.045 | 0.030 | 1.5 |
Control strain E. coli JM83 with the pHF016 vector.
Clavulanic acid, sulbactam and tazobactam were used at a constant concentration of 4 µg/mL.
Fold change in comparison to the MIC value for ampicillin.
We next evaluated the catalytic efficiency of BPU-1, the most active of the five studied enzymes, by determining its steady-state kinetic parameters for the turnover of a wide range of β-lactam substrates. In agreement with the high MIC values for penicillins, the enzyme has high catalytic efficiency (kcat/Km values 1.1 – 4.7 × 106 M−1s−1) against these antibiotics (Table 2). Moreover, the catalytic efficiency of BPU-1 against penicillins matches or exceeds those of the clinically important OXA-23, OXA-48, and OXA-58 class D β-lactamases of Gram-negative pathogens15–17. Among the other structural groups of β-lactam antibiotics studied, the fourth generation cephalosporin, cefepime, is the best substrate for BPU-1 (kcat/Km = 1.2 × 105 M−1s−1), while the catalytic efficiency of the enzyme against other cephalosporins, the monobactam aztreonam, and carbapenems is 3- to 36-fold lower. The substrate profile and catalytic activity of BPU-1 is characteristic of the extended spectrum β-lactamases.
Table 2.
Substrate profile of the BPU-1 β-lactamase
| Antimicrobial | kcat (s−1) | Km (µM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| Ampicillin | 56 ± 2 | 27 ± 3 | (2.1 ± 0.3) × 106 |
| Piperacillin | 60 ± 2 | 15 ± 2.5 | (3.9 ± 0.6) × 106 |
| Ticarcillin | 39 ± 2 | 34 ± 5 | (1.1 ± 0.2) × 106 |
| Oxacillin | 19 ± 1 | 4 ± 1 | (4.7 ± 1.4) × 106 |
| Cephalosporin C | 0.093 ± 0.003 | 200 ± 16 | (4.7 ± 0.4) × 102 |
| Cephalothin | 0.70 ± 0.03 | 60 ± 6 | (1.2 ± 0.1) × 104 |
| Ceftazidime1 | >0.9 | >150 | (8.1 ± 0.6) × 103 |
| Cefotaxime1 | >0.7 | >150 | (7.2 ± 1.0) × 103 |
| Cefepime1 | >13 | >180 | (1.2 ± 0.3) × 105 |
| Aztreonam | 0.51 ± 0.02 | 35 ± 5 | (1.5 ± 0.2) × 104 |
| Imipenem | 0.26 ± 0.02 | 6.6 ± 0.8 | (3.9 ± 0.6) × 104 |
| Meropenem | 0.014 ± 0.001 | 4.2 ± 0.4 | (3.3 ± 0.4) × 103 |
| Doripenem | 0.014 ± 0.02 | 4.1 ± 1.1 | (3.4 ± 0.9) × 103 |
| Nitrocefin | 830 ± 30 | 82 ± 8 | (1.0 ± 0.1) × 107 |
Only the lower limit for kcat and Km could be measured
We also analyzed whether BAT-2 and BSU-2, enzymes that do not confer resistance, possess any β-lactamase activity. Our kinetic studies show that these two enzymes are active but exhibit low catalytic efficiencies (kcat/Km) against ampicillin of 7.9 × 102 and 1.8 × 103 M−1s−1, respectively. These values are three orders of magnitude below that for BPU-1, which explains why these enzymes do not produce resistance in E. coli. BSU-2 and BAT-2 are only 61 to 63% identical to BSU-1 and BAT-1 and even less to BPU-1 (52% and 54%, respectively; Supplementary Table 1). At this point it is unclear which of these differences in the amino acid sequences of the enzymes are responsible for differences in their catalytic activities. Although BAT-2 and BSU-2 are very poor β-lactamases, their catalytic efficiency is significantly higher than that reported for the class D-like protein encoded by the ybxI gene of B. subtilis 16818. YbxI was postulated to be a class D β-lactamase, but subsequent kinetics studies demonstrated that the enzyme has extremely low catalytic efficiency against very few β-lactams. Thus it was concluded that YbxI is most likely a PBP with very low β-lactamase activity18.
Structural characterization of BPU1
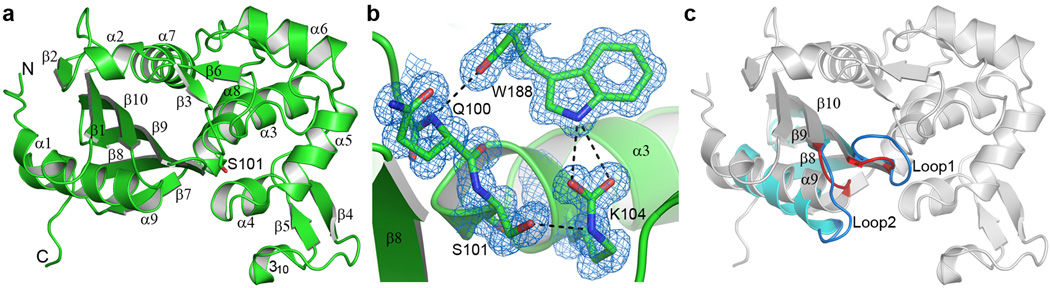
The crystal structure of BPU-1 determined at 1.0 Å resolution (Fig. 1a and Supplementary Table 2) shows that the overall fold of the enzyme is topologically similar to that of known Gram-negative class D OXA enzymes (Supplementary Fig. 1)16,19–27. Despite having relatively low sequence identities (ranging from 26% to 34%) the structures superimpose well, with root-mean-squares differences (rmsds) between 1.2 and 1.4 Å for 85–90% of the Cα atoms (Supplementary Table 3). Residues and sequence motifs identified in the Gram-negative class D enzymes as being critical for activity are structurally conserved, including the catalytic serine residue (Ser101 in BPU-1 numbering) in the SxxK motif at the N-terminus of helix α3 (Fig. 1a and Supplementary Fig. 2), the SxV motif (containing the universally conserved Ser149) on the short loop between helices α4 and α5, the YGN motif at the end of helix α6, the KTG motif (containing the universally-conserved Lys239) on strand β8, and the WxxG motif on strand β9 (Supplementary Fig. 2). Moreover, it is important to note that the conserved lysine (Lys104) three residues downstream of Ser101 is fully carboxylated in the BPU-1 structure (Fig. 1b). This post-translational modification is observed only in the Gram-negative OXA β-lactamases, and is critical for activity of the enzymes28. These structural and functional similarities establish that the Gram-positive enzymes we have identified and characterized are bona fide class D β-lactamases.
Figure 1. The BPU-1 structure.
(a) Ribbon representation of the BPU-1 crystal structure. The secondary structure assignment is indicated. The sidechain of the catalytic serine residue, Ser101, indicates the location of the enzyme active site. Single letter amino acid abbreviations are used throughout the Figures for clarity. (b) Final 2Fo-Fc electron density near the BPU-1 active site, contoured at 1.2σ. Electron density for the carboxylate moiety covalently attached to the side chain of Lys104 is evident. The carboxylated lysine is anchored by two hydrogen bonding interactions to a highly-conserved tryptophan residue (Trp188) from a loop adjacent to the active site. This loop is in turn anchored to the N-terminus of helix α3 by an additional hydrogen bonding interaction. (c) Superposition of BPU-1 (gray ribbons) and OXA-23 (cyan ribbons). Only strands equivalent to β8, β9, and β10, along with helix α9, are shown for OXA-23 for clarity. Two loops (Loop1 and Loop2) which show the largest structural deviation between BPU-1 and the OXA enzymes are shown in red for BPU-1 and blue for OXA-23.
Structural deviations are observed, however, between BPU-1 and the OXA enzymes, primarily in the loops connecting strands β8 and β9 (designated Loop 1) and strand β10 with the terminal helix α9 (Loop 2) (Fig. 1c and Fig. 2). Both of these loops are near the opening of the enzyme’s active site. In BPU-1 Loops 1 and 2 are very short, comprising only two residues in both cases, in distinct contrast to the OXA enzymes where the equivalent loops are significantly longer (from six to nine residues). It is noted in several of the OXA enzymes that a partially-conserved methionine at the beginning of Loop 1 is involved in the formation of a “hydrophobic bridge” across the opening of the active site cleft 24,25 that has been implicated in extending the activity of these enzymes to include carbapenems 29. In BPU-1 methionine is not present at the equivalent position, and the tight type II β-turn between strands β8 and β9 completely changes the direction of the polypeptide chain relative to the equivalent loops in the OXA enzymes.
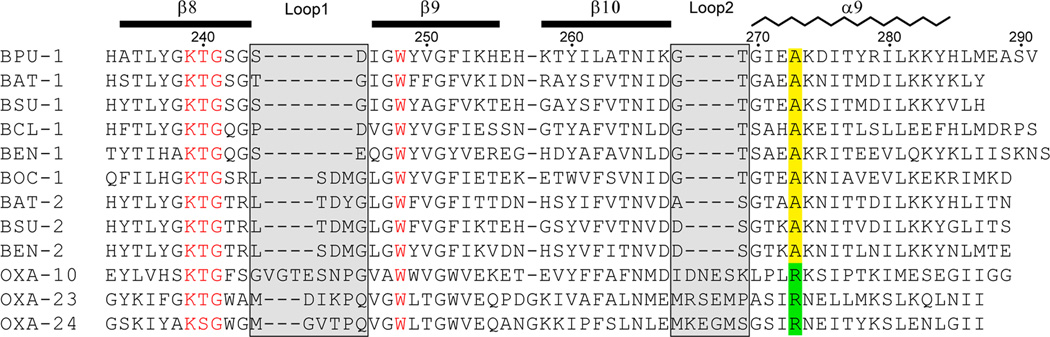
Figure 2. Structure-based sequence alignment of Gram-positive class D β-lactamases.
A segment of the sequence alignment of BPU-1 and other class D β-lactamase enzymes identified in various Bacillus species is shown, covering the two fingerprint loops (Loop1 and Loop2, highlighted in light gray) which show the largest structural variation compared to the Gram-negative OXA enzymes. OXA-10, OXA-23 and OXA-24 are shown as the representative Gram-negative β-lactamases. The secondary structure assignment for BPU-1 is indicated at the top. The conserved arginine residue in the Gram-negative enzymes is highlighted green, and the equivalent alanine residue is highlighted yellow. Residues and motifs which are highly conserved in the Gram-negative and Gram-positive enzymes are colored red.
An important arginine residue is missing
The shortening of Loop 2 in BPU-1 results in significant structural rearrangements in the active site of the enzyme. The short BPU-1 Loop 2 causes a rotation of helix α9 of approximately 25° in comparison to the OXA enzymes, and pulls the N-terminus of the helix approximately 5 Å closer to the β8-β10 sheet (Fig. 3a). This movement of the helix effectively blocks an internal cavity formed by the concave face of the β-sheet. In the class D OXA-type β-lactamases of Gram-negative bacteria this cavity houses the side chain of a highly conserved arginine residue. In BPU-1 and the other Gram-positive class D-like enzymes, this arginine is missing, substituted in all cases by alanine (Ala273 in BPU-1, Fig. 2). The rotation of helix α9 into this internal arginine pocket positions the helix significantly closer to the active site, such that the N-terminus of this helix occupies the same spatial location as the guanidinium group of the arginine side chain in the Gram-negative OXA enzymes (Fig. 3a). This arginine residue in the OXA enzymes plays a significant role in the stabilization of the acyl-enzyme intermediate by formation of a salt bridge with the invariant carboxylate moiety of β-lactam antibiotics (Fig. 3b). The arginine (Arg259 using OXA23 numbering 25) is on the first turn of the final helix, with its guanidinum group anchored in the arginine pocket by hydrogen bonds to the carbonyl oxygen atoms of Gly218 and Ala 256 (Fig. 3a). Not surprisingly, given its role in substrate binding, an arginine pocket at this location is conserved also in all class A and class C β-lactamases. In the class C enzymes the arginine (Arg349, AmpC numbering 30) is structurally and sequentially equivalent to the arginine of the class D β-lactamases, and in the class A enzymes three different arginine residues perform the same role, Arg244 in TEM-, SHV- and GES-like enzymes (using the Ambler numbering system 31), Arg220 in SME- and KPC-like enzymes, and Arg276 in the CTX-M enzymes and TOHO-1 32. Although the arginine residues in these class A β-lactamases come from different parts of the structure, with the exception of CTX-M and TOHO-1 the arginine side chains project into a cavity highly reminiscent of that observed in the OXA enzymes, with the guanidinum groups of Arg244 and Arg220 overlapping almost exactly with the equivalent groups from the class D Arg259 (Supplementary Fig. 3), and giving rise to similar electrostatic interactions with the carboxylate group of the substrates. In contrast to all other studied class A β-lactamases, the Arg276 residue in CTX-M and TOHO-1 is slightly further from the conserved guanidinium cavity and does not appear to interact with the substrate.
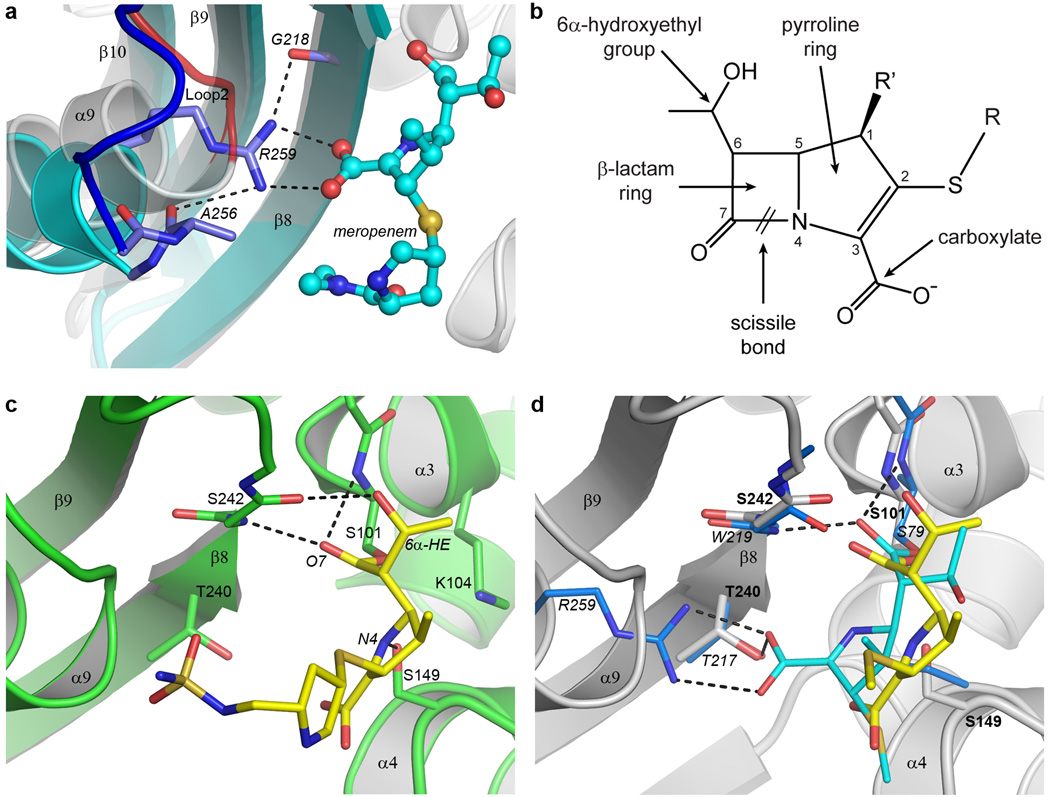
Figure 3. BPU-1 substrate binding.
(a) Close up view of the arginine pocket in OXA-23 (cyan ribbons, blue loop and lighter blue sticks), superimposed on the equivalent region near Loop 2 in BPU-1 (gray ribbons and red loop). The conserved Gram-negative class D arginine residue (represented by Arg259 from OXA-23) responsible for anchoring the carboxylate group of meropenem (cyan ball-and-stick) in OXA-23 is indicated. (b) The general structure of the carbapenem antibiotics. Differences in the structure of the tail group (R) distinguish the various clinically-available antibiotics including imipenem, meropenem, doripenem, and ertapenem. The R’ position is occupied either by hydrogen in imipenem) or by a methyl group in meropenem, doripenem, and ertapenem. (c) The substrate binding site in the doripenem-BPU-1 complex. The BPU-1 structure is represented by green ribbons and sticks and the covalently-linked doripenem by yellow sticks. The hydrogen bonding interactions between the doripenem and the protein are shown as dashed black lines. (d) The difference in coordination of doripenem (yellow sticks) in BPU-1 (grey ribbons, grey sticks) and meropenem (cyan sticks) in OXA-23. Only the residues which interact with the meropenem are shown for OXA-23 (light blue sticks) for clarity, and only the hydrogen bonding in the meropenem-OXA-23 complex is shown. The OXA-23 residues are labeled in italics and the BPU-1 residues in plain bold text. The tail of the doripenem in BPU-1 is truncated at the sulfur atom for clarity.
Since arginine at this location is conserved in all currently known active-site serine β-lactamases, we have evaluated how the loss of the arginine pocket might affect substrate binding by BPU-1. Binding experiments were attempted by soaking pre-formed apo-BPU-1 crystals in solutions containing a range of different β-lactam antibiotics and inhibitors, and residual electron density in the BPU-1 active site was observed with several β-lactam compounds, indicative of the formation of covalently-linked acyl-enzyme intermediate species. The complex formed between BPU1 and the carbapenem doripenem was the most stable (Supplementary Fig. 4). There was no evidence of carboxylation of Lys104 in this complex. The carbonyl oxygen atom (O7, Fig. 3b) of the hydrolyzed β-lactam ring of doripenem sits in a small pocket generally referred to as the oxyanion hole33,34, between the N-terminus of helix α3 and the side of strand β8. In BPU-1 the O7 atom accepts hydrogen bonds from the amide nitrogen atoms of Ser101 and Ser242 (Fig. 3c). This mode of carbonyl oxygen anchoring in productive acyl-enzyme intermediates and enzyme-inhibitor complexes is conserved not only in class D β-lactamases (Fig. 3d) but also in class A and C enzymes35–39. Two additional hydrogen bonds, one between the 6α-hydroxyethyl (6α-HE) group of the doripenem and the carbonyl oxygen of Ser242 (Fig. 3c), and another between the N4 nitrogen on the doripenem pyrroline ring and the side chain of the conserved Ser149 (Fig. 3c and Supplementary Fig. 4), also serve to anchor doripenem in the active site of BPU-1. Hydrophobic residues lining one side of the active site cleft may also assist in substrate recognition and binding (Supplementary Fig. 4). In contrast to the acyl-enzyme intermediates observed in the Gram-negative class D β-lactamases25,29, the loss of the arginine anchor in BPU-1 results in a substantial movement of the five-membered pyrroline ring away from the arginine pocket. This displaces the carboxylate group approximately 3.5 Å from the equivalent position in the Gram-negative OXA-type enzymes (Fig. 3d) and swings the 6α-HE group approximately 3 Å towards strand β8 where the new hydrogen bonding interaction with the Ser242 carbonyl oxygen is formed (Fig. 3c). A potential hydrogen bonding interaction with the side chain of Thr240 on strand β8 is also lost due to the rotation of the pyrroline ring in BPU-1. This residue belongs to the Class D β-lactamase fingerprint motif KTG (Supplementary Fig. 2), and in the Gram-negative enzymes this threonine also sits in the arginine pocket where it interacts with the carboxylate (Fig. 3d). Overall, when compared to the Gram-negative enzymes OXA-23 and OXA-2425,40, there is a net loss of hydrogen bonding interactions between the protein and the carbapenem substrate, which is only partially compensated for by other residues within the active site, at least in the interaction of BPU-1 with doripenem. Such loss of hydrogen bonding interactions may result in diminished binding affinity and activity against carbapenem antibiotics and this is what we observe for these substrates15. Given that BPU-1 has a high catalytic efficiency against the penicillins that matches or exceeds those of OXA enzymes, there may be other areas within the BPU-1 active site which can specifically interact with these substrates and compensate for the loss of hydrogen bonding interactions with their carboxylate moiety and its mutation diminishes the catalytic activity of class A enzymes 32,41.
Comparison of the sequences of Gram-positive class D enzymes with those of OXA β-lactamases of Gram-negative bacteria shows that the two main regions of sequence variation coincide with Loops 1 and 2 (Supplementary Fig. 2). These two short loops thus serve as a fingerprint motif for the Gram-positive class D β-lactamases (Fig. 2). In all of the Gram-positive enzymes there is invariably an alanine residue (Ala273 in BPU-1) at the same sequence location as the arginine (Arg259 in OXA24) in the OXA enzymes of Gram-negative organisms (Fig. 2). This sequence difference is critical as the smaller alanine allows helix α9 to closely approach the β8-β10 sheet. This, coupled with the short Loop 2, strongly suggests that the arginine pocket would be absent in all of the Bacillus enzymes. Moreover, β-lactam binding in these enzymes would be reminiscent of the mode of binding that we observe in BPU-1.
DISCUSSION
Our in silico analysis of bacterial genomes demonstrates that the genes for the putative class D β-lactamases are widely disseminated in the Bacillaceae family of the phylum Firmicutes, and the open reading frames characteristic for class D enzymes are common also in the Clostridiaceae and Eubacteriaceae families of Gram-positive environmental bacteria. The discovery of efficient class D β-lactamases in Gram-positive bacteria expands our understanding of reservoirs, structure, and evolution of antibiotic resistance enzymes. Moreover, our microbiological and kinetic data show that chromosomes of bacilli encode both poor and efficient extended-spectrum class D β-lactamases capable of hydrolyzing a wide variety of β-lactam antibiotics. The atomic resolution structure of the enzyme from B. pumilus demonstrates that although the overall three-dimensional structure of this Gram-positive class D β-lactamase is highly reminiscent of the Gram-negative OXA enzymes, the Gram-positive enzymes employ a unique substrate binding mode due to lack of an arginine residue that is conserved in all currently known serine β-lactamases, and which plays a critical role in anchoring the carboxylate of β-lactam antibiotics. The binding mode observed in BPU-1 is unique for the class D enzymes from Gram-positive bacteria and distinguish them not only from OXA enzymes of Gram-negative organism but also from all currently know class A and C β-lactamases. Since we have demonstrated that the Gram-positive class D enzymes have a high catalytic efficiency against β-lactam substrates and produce high levels of resistance to various antibiotics when expressed in E. coli, the implication is that a productive acyl-enzyme intermediate forms in these enzymes, albeit with the carboxylate moiety remaining unanchored in the active site. These differences in the substrate binding modes strongly suggest that class D enzymes of Gram-positive organisms and all other known serine β-lactamases took different evolutionary pathways to engineer their substrate binding machinery.
ONLINE METHODS
Identification of class D β-lactamases in Gram-positive bacteria
We analyzed thousands of genomes of Gram-positive bacteria deposited in the PATRIC database42 for the presence of genes for putative class D β-lactamases. First, several keywords (lactamase, penicillin, transpeptidase) were used for the initial screening. Putative enzymes that were found were analyzed manually for the presence of the conserved motifs characteristic of class D β-lactamases of Gram-negative organisms (Supplementary Figure 2) to exclude other classes of β-lactamases. Only open reading frames up to 400 amino acids containing all of the conserved motifs were analyzed to exclude high molecular weight PBPs. After several hundred putative class D β-lactamases were identified, their amino acid sequence alignment was performed. Enzymes with less than 80% of amino acid sequence identity were used for a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to expand our search for additional enzymes. Those that were found were analyzed manually as described above. A search for class C β-lactamases was performed similarly using conserved motifs characteristic for these enzymes.
Cloning of the bpu-1, bat-1, bat-2, bsu-1, and bsu-2 genes
The genes for a putative class D β-lactamases from Bacillus pumilus (bpu-1; GenBank accession number ABV63006), Bacillus atropheus (bat-1; GenBank accession number CP002207.1 and bat-2; GenBank accession number YP003975776.1), and Bacillus subtilis (bsu-1; GenBank accession number ELS62907.1 and bsu-2; GenBank accession number NP388091.1) were optimized for expression in Escherichia coli and custom synthetized (GenScript). The predicted leader peptides for the five genes were replaced with the signal sequence of the outer-membrane protein A (OmpA) to ensure efficient transport of the mature enzymes into the periplasm. Unique NdeI and HindIII restriction sites were introduced at the 5’- and 3’-ends of the constructs, respectively, which were subsequently cloned into the pHF01643 vector between NdeI and HindIII sites. The resulting constructs pHF016:BPU-1, pHF016:BAT-1, pHF016:BAT-2, pHF016:BSU-1, and pHF016:BSU-2 were transformed into E. coli JM83 for antibiotic susceptibility testing. For expression of the BPU-1 enzyme, the OmpA leader was removed and the NdeI restriction site encoding the methionine start codon was introduced at the 5’-end of the bpu-1 gene by PCR. The gene was subsequently cloned into the pET24a(+) expression vector between the NdeI and HindIII sites and the resulting construct (pET24a:BPU-1) was transformed into E. coli BL21(DE3) cells. The nucleotide sequences of all constructs were verified by DNA sequencing.
Antibiotic susceptibility testing
Minimum inhibitory concentrations (MIC) of various β-lactam antibiotics were determined in Mueller-Hinton II broth by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute44. MICs of β-lactam antibiotics were determined for E. coli JM83 harboring the pHF016:BPU-1, pHF016:BAT-1, pHF016:BAT-2, pHF016:BSU-1, and pHF016:BSU-2 constructs while E. coli JM83 carrying the pHF016 vector43 was used as a control. To select a proper strain for MIC testing in Gram-positive background, we checked whether the B. pumilus strain that harbors the gene for BPU-1 expresses this or any other β-lactamases. We grew this strain in 100 ml of Mueller-Hinton II broth overnight and pelleted the bacteria by centrifugation. The supernatant was reserved and the pellet was resuspended in 10 ml of phosphate buffer, pH 7.0 and subsequently sonicated. Both the reserved supernatant and the sonicated pellet were tested spectrophotometrically for the ability to hydrolyze the chromogenic substrate nitrocefin and the β-lactam antibiotic ampicillin. MICs produced in the β-lactam sensitive B. pumilus were determined following cloning the gene for BPU-1 into the commercially available shuttle vector pHY300PLK (Takara Bio Company) in which the ampicillin resistance marker was substituted with that for kanamycin resistance from the pHF016 vector. The optical density of the cultures was monitored spectrophotometrically at 625nm and diluted accordingly to obtain the 5×105 colony-forming units/ml in the test well. MICs were determined in triplicate in 96-well plates after 16–20 hours incubation at 37 °C.
Protein expression and purification
The E. coli BL21(DE3) strain harboring the pET24a:BPU-1 plasmid were grown in shaker/incubator at 37 °C in LB broth supplemented with 50 µg/mL kanamycin A until the optical density reached 0.6 at 600 nm. Expression of the BPU-1 enzyme was initiated by the addition of 0.4 mM isopropyl-β-D-thiogalactopyranoside and the culture was incubated at 24°C for an additional 20 hours. Cells were collected by centrifugation (4,500 g/10 minutes, 4°C) then resuspended and sonicated in 25mM Hepes, pH 8.0, 1 mM EDTA, and 0.2 mM DTT. The lysate was centrifuged (20,000g/20minutes, 4°C), and the soluble proteins were separated by CM cation-exchange chromatography (Bio-Rad). The 50 ml column was washed with 5 column-volume of 25mM Hepes buffer, pH 8.0 and a 0–600 mM NaCl gradient was used for protein elution. Fractions were analyzed by 12% SDS-PAGE. BPU-1 was eluted with 220–260 mM NaCl, collected, and dialyzed against 25mM Hepes, pH 7.5. The protein concentration was measured spectrophotometrically using the predicted extinction coefficient (Δε280 = 61880 cm−1 M−1) and BCA kit (Pierce).
Enzyme kinetics
All kinetic data were collected with a Cary 50 spectrophotometer (Varian) at room temperature. The reaction mixtures contained 100 mM sodium phosphate buffer, pH 7.0, 50 mM sodium bicarbonate, various concentrations of β-lactam substrate, and 0.5 nM - 1.7 µM BPU-1 enzyme. The reaction was initiated by addition of the enzyme, and the reaction progress curve was followed spectrophotometrically. The hydrolysis of various β-lactams by the BPU-1 β-lactamase was measured using the following wavelengths (nm) and extinction coefficients (M−1cm−1): ampicillin 240/−538; piperacillin 235/−1070; ticarcillin 235/−660; oxacillin 260/+440; cephalothin 262/−7,960; cephalosporin C 280/−2390; cefotaxime 265/−6,260; ceftazidime 260/−10,800; cefepime 264/−8240; aztreonam 318/−640; imipenem 297/−10,930; meropenem 298/−7200; doripenem 299/−11540; nitrocefin 500/+15900. Steady-state velocities were calculated from the slopes of progress curves and plotted as a function of substrate concentrations. This allowed the determination of Km and kcat by nonlinear regression with Prism 5 software (GraphPad Software, Inc.) using the Michaelis-Menten equation: v = Vmax × S/(Km + S), where v is the initial velocity, S is the β-lactam concentration, Km is the Michaelis constant and Vmax= kcat × E where E is the enzyme concentration. In the cases when measurements were limited by a range of substrate concentrations which were much less than the Km, the ratio of kcat/Km was obtained by fitting the reaction time course to the following equation: At = A∞ + (A0 −A∞) e−kt where At is the absorbance at time t, Ao and A∞ is the initial and final absorbance and k = (kcat/Km) × E.
Crystallization, data collection, structure solution and refinement
Initial crystallization trials with the BPU-1 β-lactamase were set using the sitting drop method in Intelliplates (Art Robbins), using PEG/Ion screens I and II, and Crystal Screens I and II (Hampton Research). Crystals were observed in three conditions from PEG-Ion screen II; #16, 8% Tacsimate (pH 7.0), 20% PEG 3,350; #22, 0.2 M ammonium citrate tribasic (pH 7.0), 20% PEG 3,350; and #32, 2% Tacsimate (pH 5.0), 0.1 M sodium citrate pH 5.6, 16% PEG 3,350. Crystals were transferred to a cryoprotectant solution containing 30% glycerol in each crystallization condition, flash-cooled in liquid nitrogen, and screened for diffraction quality on beamline BL7-1 at the Stanford Synchrotron Radiation Lightsource (SSRL). Crystals from condition #32 were found to diffract close to atomic resolution so these crystals were used for subsequent data collection experiments. The crystals belonged to a primitive monoclinic space group with cell dimensions a = 47.66 Å, b = 79.87 Å, c = 65.01 Å, β = 92.3°. The Matthews coefficient45 assuming one molecule in the asymmetric unit was 1.9 Å3/Da (34.5% solvent content). Complete datasets were collected from two flash-cooled crystals on the SSRL beamline BL12-2, using X-rays at 12658 eV (0.9795 Å) and a PILATUS 6M PAD detector running in shutterless mode. The data sets, comprising 800 images and 760 images respectively, were processed with XDS46 and scaled and merged using XSCALE46 to give a final data set comprising 261147 unique reflections extending to 1.0 Å resolution. The program POINTLESS47 indicated that the space group was P21. Final data collection statistics are given in Supplementary Table 2.
The doripenem-BPU-1 complex was prepared by soaking pre-formed apo-BPU-1 crystals in a solution containing 50 mM doripenem in 2% Tacsimate, 0.1 M sodium citrate pH 5.6, 16% PEG 3,350 for varying lengths of time ranging from a few seconds up to 5 minutes. After soaking in the doripenem solution the crystals were transferred to a cryoprotectant solution containing the buffer augmented with 30% glycerol, and flash-cooled in liquid nitrogen. Data extending to 1.35 Å resolution were collected from a single crystal on SSRL beamline BL12-2, processed using XDS46 and scaled with AIMLESS48. Final statistics are also given in Supplementary Table 2.
Structure Solution and Refinement
The BPU-1 sequence was aligned with several other class D β-lactamases from Gram-negative bacteria whose structures had been determined. The pairwise sequence identities ranged from 28.4% (OXA-45, PDB code 4gn2) to 36.8% (OXA-48, PDB code 3g4p) (Supplementary Table 3). Since there was no significantly preferred alignment, molecular replacement (MR) models were generated from all eleven OXA structures using the CCP4 program CHAINSAW to convert the structures into a pseudo-BPU-1 model, where identical residues in the matched sequences were retained and those which differed were truncated at the Cβ atom. All models gave strong MR solutions, indicating two independent molecules in the asymmetric unit. These models were subsequently refined for 15 cycles using REFMAC 49 and 2Fo-Fc and Fo-Fc electron density maps calculated. Refinement statistics indicated that the model derived from OXA-48 was significantly better than any of the others, and inspection of the BPU-1-OXA-48 sequence alignment showed the least number of insertions or deletions between these two sequences. Refinement of the structure was completed with PHENIX.REFINE 50 and manual building of the model using the molecular graphics program COOT 51. Water molecules were added in structurally and chemically relevant positions, and the atomic displacement parameters (B-factors) for the protein atoms were refined anisotropically. In the final refinement cycle, hydrogen atoms were added in calculated positions for all protein atoms. Final refinement statistics are given in Supplementary Table 2, and the structure is shown in Fig. 1. Ramachandran statistics indicate that 99% of the residues lie in the allowed regions, 1% in the generously allowed regions and no residues fall in disallowed regions, as calculated using PROCHECK 52.
The doripenem-BPU-1 structure was solved by MR using the refined apo-BPU-1 structure as the starting model with all water molecules and alternate side chain conformations removed, and the carboxylate moiety removed from Lys104. Doripenem was added to residual electron density observed in the active site (Supplementary Fig. 4) of both independent BPU-1 molecules in the asymmetric unit. The structure was refined using PHENIX.REFINE with anisotropic B-factors, and final refinement statistics are given in Supplementary Table 2. Ramachandran statistics indicate that 99.1% of the residues lie in the allowed regions, 0.7% in the generously allowed regions and one residue falls in a disallowed region, as calculated using PROCHECK52.
Statistical analysis of data
Statistical analysis of the crystallographic data was automatically performed in all of the programs used (noted above) and is presented here without modification. All MICs were measured in triplicate. Other statistical analyses were performed using the Prism 5 software.
Supplementary Material
Acknowledgments
The Stanford Synchrotron Radiation Lightsource (SSRL) is a national user facility operated by Stanford University and supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Author Contributions
S.B.V. and N.T.A. performed the genomic sequence screening and sequence alignments; M.T., N.T.A., N.K.S. and H.F designed and performed the microbiological and kinetic experiments, and analyzed that data; M.T., N.T.A. and C.A.S. designed and performed the crystallography experiments; C.A.S. collected and analyzed the crystallographic data; S.B.V. and C.A.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
Additional Information
Methods, along with any additional Supplementary Information display items, are available in the online version of the paper.
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5CTM and 5CTN for the apo and doripenem-bound forms of BPU-1.
REFERENCES
- 1.Poole K. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 2004;61:2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waxman DJ, Strominger JL. Penicillin-binding proteins and the mechanism of action of β-lactam antibiotics. Annu. Rev. Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 3.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 4.Cho H, Uehara T, Bernhardt TG. β-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K. Proliferation and significance of clinically relevant β-lactamases. Ann. NY Acad. Sci. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 6.Ambler RP. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. (Biol.) 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 7.Jaurin B, Grundstrom T. AmpC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc. Natl Acad. Sci. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouellette M, Bissonnette L, Roy PH. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: Nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kernodle DS. In: Gram-positive pathogens. Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Washington, D.C.: ASM Press; 2000. pp. 609–620. [Google Scholar]
- 10.Hedges RW, Datta N, Kontomichalou P, Smith JT. Molecular specificities of R factor-determined β-lactamases: Correlation with plasmid compatibility. J. Bacteriol. 1974;117:56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sykes RB, Matthew M. The β-lactamases of gram-negative bacteria and their role in resistance to β-lactam antibiotics. J. Antimicrob. Chemother. 1976;2:115–157. doi: 10.1093/jac/2.2.115. [DOI] [PubMed] [Google Scholar]
- 12.Leonard DA, Bonomo RA, Powers RA. Class D β-lactamases: A reappraisal after five decades. Acc. Chem. Res. 2012;46:2407–2425. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antunes NT, et al. Class D β-lactamases: Are they all carbapenemases? Antimicrob. Agents Chemother. 2014;58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Docquier JD, et al. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Verma V, et al. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D β-lactamase from Acinetobacter baumannii . J. Biol. Chem. 2011;286:37292–37303. doi: 10.1074/jbc.M111.280115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo M-L, et al. The ybxI gene of Bacillus subtilis 168 encodes a class D β-lactamase of low activity. Antimicrob. Agents Chemother. 2004;48:484–490. doi: 10.1128/AAC.48.2.484-490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Docquier JD, et al. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob. Agents Chemother. 2010;54:2167–2174. doi: 10.1128/AAC.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golemi D, et al. The first structural and mechanistic insights for class D β-lactamases: Evidence for a novel catalytic process for turnover of β-lactam antibiotics. J. Am. Chem. Soc. 2000;122:6132–6133. [Google Scholar]
- 21.Kaitany KC, et al. Structures of the class D carbapenemases OXA-23 and OXA-146: Mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins and aztreonam. Antimicrob. Agents Chemother. 2013;58:4848–4855. doi: 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paetzel M, et al. Crystal structure of the class D β-lactamase OXA-10. Nature Struct. Biol. 2000;7:918–924. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 23.Pernot L, et al. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J. Mol. Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 24.Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. 2007;104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CA, et al. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii . Chem. Biol. 2013;20:1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith CA, Antunes NT, Toth M, Vakulenko SB. Crystal structure of carbapenemase OXA-58 from Acinetobacter baumannii . Antimicrob. Agents Chemother. 2014;58:2135–2143. doi: 10.1128/AAC.01983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider KD, et al. Structures of the class D carbapenase OXA-24 from Acinetobacter baumanii in complex with doripenem. J. Mol. Biol. 2011;406:583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beadle BM, Shoichet BK. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob. Agents Chemother. 2002;46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambler RP, et al. A Standard numbering scheme for the class-A β-lactamases. Biochemical J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marciano DC, Brown NG, Palzkill T. Analysis of the plasticity of location of the Arg244 positive charge within the active site of the TEM-1 β-lactamase. Protein Science. 2009;18:2080–2089. doi: 10.1002/pro.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannouli M, et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 2010;48:1223–1230. doi: 10.1128/JCM.02263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curley K, Pratt RF. The oxyanion hole in serine β-Lactamase catalysis: Interactions of thiono substrates with the active site. Bioorg. Chem. 2000;28:338–356. doi: 10.1006/bioo.2000.1184. [DOI] [PubMed] [Google Scholar]
- 35.Maveyraud L, et al. Crystal structure of 6α-(hydroxymethyl)penicillanate complexed to the TEM-1 β-lactamase from Escherichia coli: Evidence on the mechanism of action of a novel inhibitor designed by a computer-aided process. J. Am. Chem. Soc. 1996;118:7435–7440. [Google Scholar]
- 36.Mourey L, et al. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative, and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J. Am. Chem. Soc. 1998;120:9382–9383. [Google Scholar]
- 37.Patera A, Blaszczak LC, Shoichet BK. Crystal structures of substrate and inhibitor complexes with AmpC β-lactamase: Possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 2000;122:10504–10512. [Google Scholar]
- 38.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry. 2001;40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 39.Stewart NK, Smith CA, Frase H, Black DJ, Vakulenko SB. Kinetic and structural requirements for carbapenemase activity in GES-type β-lactamases. Biochemistry. 2015;54:588–597. doi: 10.1021/bi501052t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider KD, Karpen ME, Bonomo RA, Leonard DA, Powers RA. The 1.4 Å crystal structure of the class D β-lactamase OXA-1 complexed with doripenem. Biochemistry. 2009;48:11840–11847. doi: 10.1021/bi901690r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson JM, Distler AM, Prati F, Bonomo RA. Probing active site chemistry in SHV β-lactamase variants at Ambler position 244, Understanding unique properties of inhibitor resistance. J. Biol. Chem. 2006;281:26734–26744. doi: 10.1074/jbc.M603222200. [DOI] [PubMed] [Google Scholar]
- 42.Wattam AR, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucl. Acids Res. 42. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toth M, Smith C, Frase H, Mobashery S, Vakulenko S. An antibiotic-resistance enzyme from a deep-sea bacterium. J. Am. Chem. Soc. 2010;132:816–823. doi: 10.1021/ja908850p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 45.Matthews BW. Solvent contents of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 47.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. 2011;D67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. 1999;D55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.