Abstract
The identification of RNA-binding proteins that physically associate with viral RNA molecules during infection can provide insight into the molecular mechanisms of RNA virus replication. Until recently, such RNA-protein interactions have been identified predominantly with the use of in vitro assays that may not accurately reflect associations that occur in the context of a living cell. Here we describe a method for the specific affinity purification of dengue virus RNA and associated proteins using in vivo cross-linking followed by antisense-mediated affinity purification. RNA-binding proteins that specifically co-purify with viral RNA using this method can be identified en masse by mass spectrometry. This strategy can potentially be adapted to the purification of any viral RNA species of interest.
Keywords: dengue virus, ribonucleoprotein complex, RNA affinity chromatography
1. Introduction
Viral RNAs must navigate a complex cellular environment and be successfully translated, replicated, and/or packaged into progeny virions during the course of a productive infection. Throughout various stages of the viral life cycle, the viral RNA is exposed to, and likely interacts directly with, a number of host cell and viral proteins. Some of these host RNA-binding proteins (RBPs) may bind to viral RNA and impede the establishment of productive infection [1–5]. Alternatively, the virus may co-opt the normal function of cellular RBPs to promote one or more phases of the viral life-cycle. For example, several well-characterized RBPs have been shown to interact with viral RNA and mediate the translation and/or synthesis of viral RNA genomes [6–12]. Therefore, the identification of novel interactions between viral RNA and host RBPs can provide insight into the molecular mechanisms of virus replication.
Until recently, the identification of interactions between viral RNA and host proteins has relied predominantly on in vitro systems. One commonly used strategy has been immobilization of in vitro transcribed subgenomic regions of viral RNA on solid state resins, followed by incubation with crude or fractionated cell lysates. Proteins that specifically bind to the RNA of interest can be enriched after washing away non-specific interactions and then identified using mass spectrometry [13]. More targeted approaches for the identification of interactions between protein and viral RNA, such as RNA-immunoprecipitation and electrophoretic mobility shift assay, have been used to test the ability of specific RBPs to bind discrete viral RNA regions [3, 4, 14]. These techniques can be powerful tools for identification and characterization of RNA-protein interactions. However, an important limitation of these strategies is that interactions occurring in vitro may not accurately reflect those that occur in the context of virus infection.
One strategy for the characterization of polyadenylated RNP complexes formed in vivo involves UV crosslinking followed by purification of RBPs on immobilized oligo(dT) [15–19] A recently described method couples this purification strategy with mass spectrometry for en masse identification of interactions between cellular mRNA and protein in living cells. This protocol utilizes an in vivo cross-linking approach in which growing cells are exposed to 254 nm UV light, forming covalent cross-links at sites of direct contact between protein and RNA. The cells are then lysed under denaturing conditions and ribonucleoprotein (RNP) complexes are purified from the lysate using immobilized oligo(dT) to specifically purify polyadenylated mRNA with its associated proteins [20, 21]. A modification of this strategy was recently used to identify proteins bound to the polyadenylated poliovirus RNA genome in infected cells [22]. Here we report an adaptation of this approach to specifically purify dengue virus (DENV) RNPs from infected cells. The identification of proteins associated with the non-polyadenylated viral genomes during infection, and characterization of their roles in virus replication, will lead to novel insights into the molecular mechanisms of DENV replication.
2. Method Overview
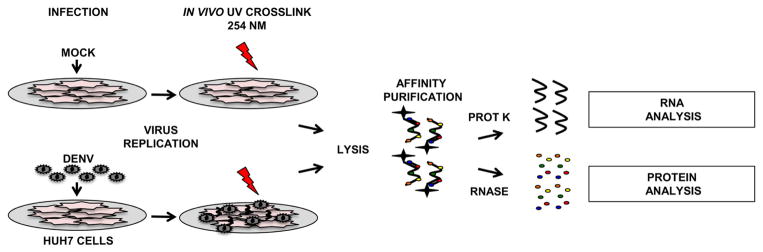
Figure 1 depicts a general overview of the affinity purification method. Infected cells are exposed to 254 nm UV light, inducing covalent cross-links between protein and RNA to which it is directly bound. Mock-infected cells serve as a control for background binding of cellular RNPs. Cells are lysed and viral RNPs are purified by hybridization of antisense biotinylated DNA oligonucleotides to the viral RNA, followed by capture of RNP complexes on streptavidin-coated magnetic beads. After a series of increasingly stringent washes, the bound material is eluted for analysis of RNA and/or protein. RNA recovery is assessed using RT-qPCR. Bound proteins can be liberated from the RNA for targeted or unbiased en masse identification.
Figure 1. Schematic of affinity purification technique.
Cells are infected with dengue virus at an MOI of 1. Mock infected cells serve as a negative control. RNA-protein cross-links are induced 30 hours post-infection by exposing the cells to 254 nm UV. Cells are lysed under denaturing conditions and incubated with biotinylated antisense DNA oligonucleotides. RNA-protein complexes are captured on streptavidin-coated magnetic beads. Protein can be digested with proteinase K for subsequent RNA analysis by RT-qPCR. Alternatively, proteins can be liberated from the RNA by RNase treatment and analyzed by Western blot or en masse by mass spectrometry.
3. Detailed Methods
3.1. Rational design of antisense oligonucleotides by RNase H mapping
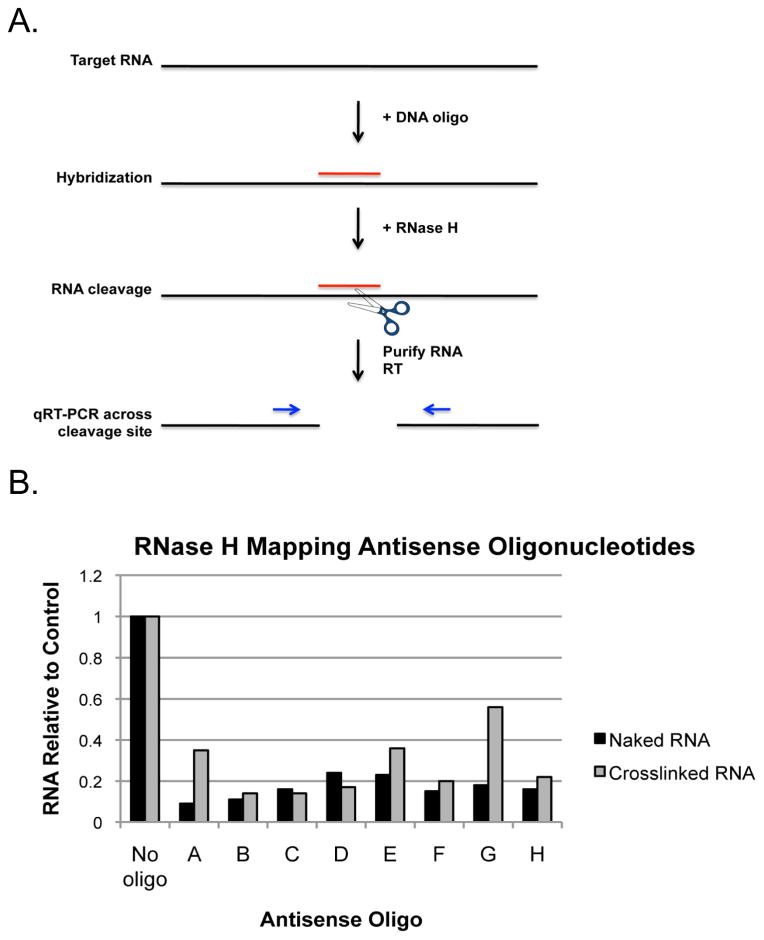
RNA molecules can exhibit significant secondary structure and in living cells are associated with RNA-binding proteins. As a result, certain regions of an RNA molecule after UV cross-linking to partner proteins may be unavailable for hybridization to an antisense oligonucleotide. The likelihood that a given antisense oligonucleotide sequence will be able to bind its target RNA and thus be effective in mediating affinity purification may be predicted using theoretical or empirical structural data where available. However, these data are often derived from naked RNA in the absence of protein. One way to determine the accessibility of a given RNA sequence in the context of bound protein is by RNase H mapping of the target RNA (Figure 2A). This is accomplished by incubation of candidate antisense DNA oligonucleotides with cross-linked cell lysate, followed by treatment with RNase H, which will specifically cleave the RNA in an RNA:DNA hybrid. Cleavage efficiency of the RNA can then be assessed by RT-qPCR using PCR primers that span the predicted cleavage site.
Figure 2. RNase H mapping antisense oligonucleotide positions.
A) Schematic of the RNase H mapping assay. Target RNA is incubated with complementary antisense DNA oligonucleotides and RNase H. If hybridization occurs, RNase H will cleave the RNA involved in the RNA:DNA hybrid. Cleavage is measured using RT-qPCR. B) Infected cells were cross-linked, lysed, and incubated with candidate antisense DNA oligonucleotides and RNase H. Purified RNA from mock cross-linked cells was used as a positive control for RNase H cleavage. RNA was purified from RNase H reactions and cleavage assessed by RT-qPCR using qPCR primers that amplify across the oligonucleotide hybridization site. Data is represented as the amount of RNA remaining relative to the no oligo control.
3.2. Materials
cross-linked lysate from DENV-infected Huh7 cells (see Note 1), ~2.5 × 104 cell equivalents per μl. This corresponds to an approximate protein concentration of 15 mg/ml.
purified RNA from mock-cross-linked cells, heat denatured, 100 ng per reaction.
RNase H (NEB).
antisense DNA oligonucleotide (100 μM stock).
RQ1 DNase (Promega).
60 mM CaCl2.
60 mM MgCl2.
20 mM EGTA, pH 8.0.
RNA purification reagent, e.g. Trizol (Life Technologies).
cDNA synthesis and PCR reagents.
3.3. Procedure
assemble 10 μL reactions containing 8.5 μL cell lysate or naked RNA, 1 μL antisense oligonucleotide (100 pmol), and 0.5 μL RNase H (2.5 U).
incubate reactions at 37°C for 30 minutes.
add 1 μL RQ1 DNase, 2.5 μL 60 mM CaCl2, and 2.5 μL 60 mM MgCl2.
incubate reactions at 37°C for 30 minutes to hydrolyze oligonucleotides.
add 1.5 μL 20 mM EGTA pH 8.0 to stop the DNase reaction.
incubate at 65°C for 10 minutes to inactivate DNase.
purify RNA and perform cDNA synthesis and PCR using pre-validated qPCR primers that span the antisense hybridization site.
3.4. Notes
3.5. Results
Figure 2B shows the results of RNase H mapping using selected antisense oligonucleotides. Oligonucleotides A-H were designed to hybridize to regions distributed across the length of the 10.6 kb DENV type-2 strain New Guinea C (DENV2 NGC) viral genome at approximately 1 kb intervals. The no oligo condition serves as a negative control for background RNA cleavage. Denatured RNA purified from mock-cross-linked cells serves as a positive control. The relative levels of RNase H cleavage under each condition is measured using qPCR primers designed to amplify across the predicted site of cleavage. The amount of RNA remaining is expressed relative to the amount of uncleaved RNA in the no oligo control.
We find that different regions of the viral genome exhibit variable levels of accessibility for hybridization based on the observed differences in cleavage efficiency between naked and cross-linked RNA. RNA cleavage after incubation of the cross-linked viral RNA with oligonucleotides A, E, and G was inhibited compared to the naked RNA control, suggesting that these regions of the viral RNA are either structured or occluded by the presence of bound proteins. Therefore, these candidate oligonucleotides were not selected for use in the affinity purification procedure. For the selection of oligonucleotides to be used in the affinity purification of DENV RNP, we chose oligonucleotides which resulted in cleavage of at least 80% of the RNA relative to the no oligonucleotide control and which did not exhibit a difference in cleavage levels in the presence of crosslinked protein.
For the purification of DENV RNP, we chose to use an equimolar mixture of 10 unique DNA oligonucleotide sequences. The oligonucleotides were biotinylated at the 5′ end and contained a triethyleneglycol (TEG) spacer between the biotin moiety and the first DNA base. Unmodified DNA oligonucleotides were used for several reasons. First, they are much less costly to synthesize than oligonucleotides containing stabilizing base modifications or RNA. Second, RNase H was not active in the buffer composition used for the affinity purification, and therefore cleavage of the RNA due to endogenous RNase H was not a concern. Finally, we found that the use of oligonucleotides containing modifications such as 2′-O-methyl RNA bases or phosphorothioate bonds resulted in an increase in the amount of background RNAs in the affinity purified material (data not shown).
4. Cross-linking and Lysis
Exposure of growing cell cultures to 254 nm UV light prior to cell lysis induces formation of covalent bonds between proteins and nucleic acids that are in direct physical association [24]. In the context of our protocol, subsequent recovery of the RNP of interest during affinity purification depends in part on solubility of the RNP after cell lysis. Whole cell lysates may be prepared simply by using Affinity Purification Buffer containing 0.5% LiDS (see section 5). Alternatively, milder lysis conditions may be used to facilitate removal of nuclei prior to performing the affinity purification procedure. For purification of DENV RNP, we perform an initial lysis in buffer containing 2% n-dodecyl-beta-D-maltoside, which efficiently solubilizes ER membranes [C. Nicchitta (Duke University), personal communication]. Intact nuclei are then removed from the lysate by centrifugation prior to supplementation of the lysate with Affinity Purification Buffer.
4.1 Materials
(10) 500 cm2 dishes (~1e9) confluent Huh7 cells infected with DENV2 NGC at an MOI of 1 for 30 hours (see Notes 2 and 3).
(10) 500 cm2 dishes of confluent Huh7 cells, mock-infected.
Ice cold PBS.
UV Stratalinker 2400 (Stratagene) equipped with standard 254 nm bulbs (see Note 4).
Lysis Buffer: 200 mM KCl, 20 mM HEPES pH 7.2, 2% n-dodecyl-beta-D-maltoside (Sigma), 1% NP-40, 40 U/ml murine RNase inhibitor (NEB), 1X protease inhibitor cocktail (Roche).
4.2 Procedure
Wash cells twice with 30 ml cold PBS making sure to remove all traces of growth medium. Leave ~ 5 ml PBS remaining after the last wash or just enough to cover the cell monolayer.
Remove lid and place dish on ice in the cross-linker, approximately 10 cm from UV light source.
Irradiate the cells with 0.15 J/cm2.
Immediately add 15 ml cold PBS to dish upon completion of UV exposure.
Scrape to collect cells and transfer to a 50 ml conical tube on ice, pooling two dishes per tube (~1e8 cells).
Pellet cells by centrifugation at 4°C for 5 minutes at 1500 × g.
Remove PBS and lyse the cell pellet in 2 ml cold Lysis Buffer (see Note 5).
Snap freeze lysates in liquid nitrogen and store at −80°C until the affinity purification procedure is performed.
4.3. Notes
4.4. Results
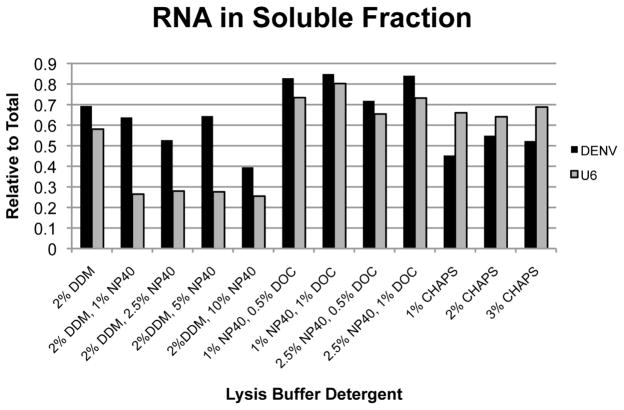
For the purpose of purification of DENV RNP from infected cells, we found optimization of the cell lysis conditions to be an important parameter in enhancing the efficiency and specificity of the affinity purification procedure. Because DENV replicates exclusively in the cytoplasm, we sought to identify conditions under which nuclei would remain intact and could be removed from the lysate, minimizing interference from nuclear nucleic acids. In addition, DENV RNAs are tightly associated with ER membranes, which are not efficiently solubilized by many detergents that will leave nuclear membranes largely intact. To identify appropriate lysis conditions, we tested a number of detergent combinations and measured the relative amount of viral RNA released into the soluble fraction under each condition using RT-qPCR. In parallel, we measured the levels of the cellular U6 RNA as a measure of nuclear integrity (Figure 3).
Figure 3. Optimization of cell lysis conditions.
Infected Huh7 cells were lysed in buffer containing 200 mM KCl, 20 mM HEPES pH 7.2, and the indicated detergents. Insoluble material was cleared from the lysate by centrifugation and total RNA was purified from both soluble and pellet fractions. DENV and nuclear U6 RNAs were quantitated by RT-qPCR. DDM = n-dodecyl β-D-maltoside, NP40 = igepal CA-360, DOC = deoxycholic acid
We found that while buffers containing the detergents deoxycholic acid (DOC) and CHAPS efficiently released the viral RNA into the soluble fraction, they also solubilized nuclear membranes as indicated by the presence of U6 RNA. In contrast, combinations of n-dodecyl-beta-D-maltoside (DDM) and NP-40 resulted in approximately 50% recovery of viral RNA in the soluble fraction, with minimal (~20%) recovery of U6 RNA. Based on these data, a combination of 2% DDM and 1% NP-40 was chosen for lysis of cell pellets.
5. Affinity Purification
Prior to affinity purification, cell lysates are centrifuged to remove insoluble material and intact nuclei. The affinity purification procedure is then performed under denaturing conditions of high salt and anionic detergent that prevent indirect or non-specific interactions from being maintained or forming post-lysis. Therefore, the only proteins that specifically co-purify with the RNA of interest are those that covalently cross-link to RNA in intact living cells. Due to the scale of material required for affinity purification of dengue viral RNP, we do not routinely perform a no cross-link control. However, others have demonstrated that these buffer conditions for affinity purification are effective in preventing RNA-protein binding in the absence of UV cross-linking [20].
5.1. Materials
frozen lysates (~1e8 cells per tube).
2X Affinity Purification Buffer: 40 mM Tris HCl pH 7.5, 1 M LiCl, 1% LiDS, 2 mM EDTA, 10 mM DTT, 40 U/ml murine RNase Inhibitor (NEB), 1X protease inhibitor cocktail (Roche).
10 μM stock biotinylated antisense DNA oligonucleotides.
Dynabeads MyOne Streptavidin C1 beads (Life Technologies).
Magnetic tube rack.
Wash Buffer 1: 20 mM Tris HCl pH 7.5, 500 mM LiCl, 0.5% LiDS, 1 mM EDTA, 5 mM DTT.
Wash Buffer 2: 20 mM Tris HCl pH 7.5, 500 mM LiCl, 0.1% LiDS, 1 mM EDTA, 5 mM DTT.
Wash Buffer 3: 20 mM Tris HCl pH 7.5, 500 mM LiCl, 1 mM EDTA, 5 mM DTT.
Wash Buffer 4: 20 mM Tris HCl pH 7.5, 200 mM LiCl, 1 mM EDTA, 5 mM DTT.
5.2. Procedure
Thaw lysates rapidly to ensure complete cell lysis and place immediately on ice. All steps subsequent to thawing are performed on ice or in cold room.
Mix lysates and incubate on ice for 30 minutes.
Centrifuge lysates at 4°C for 10 minutes at 1500 × g to pellet nuclei, transfer supernatant to a fresh tube.
-
Supplement lysates with an equal volume of 2X Affinity Purification Buffer. The final buffer composition for annealing and capture is:
100 mM KCl
10 mM HEPES pH 7.2
1% n-dodecyl-beta-D-maltoside
0.5% NP-40
20 mM Tris HCl pH 7.5
500 mM LiCl
0.5% LiDS
1 mM EDTA
5 mM DTT
40 U/ml murine RNase inhibitor (NEB)
1X protease inhibitor cocktail (Roche)
Transfer aliquots (~0.05% of total) of each sample to a fresh tube and store at −80°C for use as input when performing RNA and protein analysis.
Add 250 pmol biotinylated oligonucleotide (see Note 6) to lysate and anneal by rotating samples at 70°C for 10 minutes followed by slow cooling to 40°C in a hybridization oven (see Note 7).
Place samples on ice once lowest annealing temperature is achieved to prevent further non-specific hybridization of oligonucleotides.
Capture complexes on 2.5 mg Dynabeads MyOne Streptavidin C1 beads by rotation at 4°C for 2 hours to overnight.
Capture magnetic beads by placing tubes in an appropriate magnetic rack, e.g. Dynamag (Life Technologies).
Remove the supernatant containing unbound material, saving an aliquot for RNA analysis if desired (see Note 8).
Wash the beads once with Wash Buffer 1, twice with Wash Buffer 2, twice with Wash Buffer 3, and once with Wash Buffer 4 by resuspending beads in a volume equal to that used for hybridization, rotating at 4°C for 5 minutes, and capturing beads with magnetic rack.
Remove all traces of the final wash and proceed to sample analysis.
5.3. Notes
5.4. Sample Analysis
Beads can be resuspended in buffer containing 10 mM Tris HCl pH 7.5 and 50 mM NaCl and aliquoted for evaluation of both RNA and protein in the same sample. If only RNA or protein is to be analyzed, the beads can be resuspended directly in either Proteinase K or RNase reaction solutions.
6. RNA Analysis to evaluate affinity purification efficiency and specificity
6.1. Materials
5X Proteinase K Buffer: 50 mM Tris HCl pH 7.5, 750 mM NaCl, 1% SDS, 50 mM EDTA pH 8.0, 2.5 mM DTT, and 25 mM CaCl2
Proteinase K 20 mg/ml (Life Technologies)
Trizol Reagent (Life Technologies)
GlycoBlue Coprecipitant (Life Technologies)
cDNA synthesis and qPCR amplification reagents
6.2. Procedure
Make a reaction master mix containing 1X Proteinase K Buffer and 1 μg Proteinase K per 20 μL.
Add 20 μL master mix to 5 μL of the recovered beads.
Incubate at 55°C for 60 minutes with occasional mixing to resuspend beads.
Add 500 μL Trizol reagent and purify RNA following the manufacturers protocol, adding 1 μL GlycoBlue Coprecipitant prior to precipitation with isopropanol and resuspending purified RNA in 20 μL RNase-free water.
Use 10 μL of purified RNA for cDNA synthesis (see Note 9).
Use cDNA as template for qPCR amplification using primers specific for dengue RNA as well as selected cellular RNAs to assess the specificity of the affinity purification procedure (see Note 10). Calculate the efficiency of the affinity purification by comparison of target RNA in the eluates vs. input samples.
6.3. Notes
6.4. Results
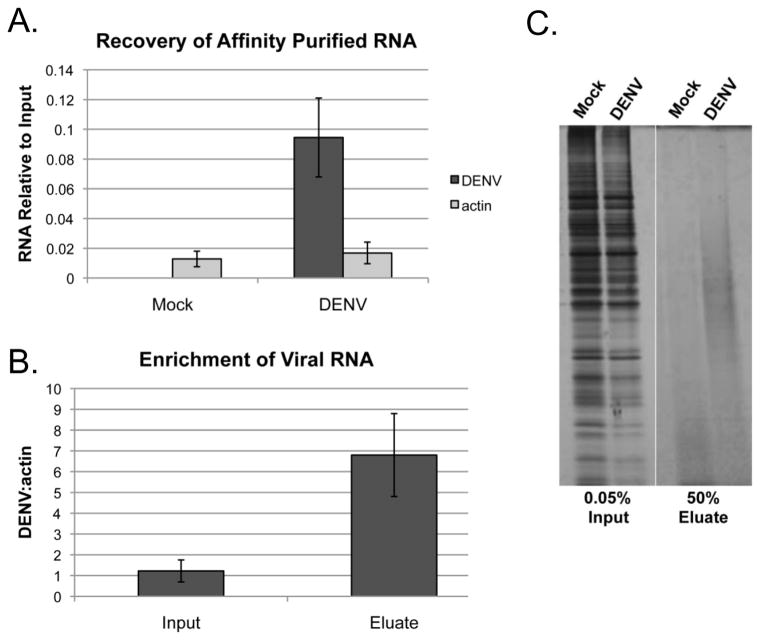
Figure 4A shows the results of RNA analysis from affinity-purified material across several independent experiments (N=14). An average recovery of ~9% of the viral RNA relative to input is obtained after affinity purification with DENV-specific antisense oligonucleotides. In contrast, the recovery of actin, which serves as a control for specificity is <2% in both mock and infected conditions. While relatively inefficient for unknown reasons, the recovery of the viral RNA observed is consistent with or exceeds the yields reported for other RNAs using similar approaches [21, 23, 26].
Figure 4. Validation of RNA affinity purification procedure.
A) RNA was purified from affinity purified eluates (N=14) and the yield of DENV and actin RNA was determined by RT-qPCR. B) The enrichment of viral RNA per actin after affinity purification was determined by RT-qPCR. C) Affinity purified protein prepared from Mock and DENV infected samples was separated by SDS-PAGE and visualized by silver stain.
The percentage of the RNA recovered after affinity purification is perhaps less informative than whether there is a specific enrichment of the target RNA relative to a non-specific RNA. Figure 4B shows that the relative amount of viral RNA present in affinity purified samples is enriched by ~7-fold compared to input in relation to actin RNA. This demonstrates that the affinity purification procedure successfully enriches for viral RNA.
7. Protein Analysis to evaluate affinity purification efficiency and specificity
7.1. Materials
RNase A and T1 cocktail diluted 1:2.5 (Life Technologies).
RNase buffer: 20 mM Tris HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.5 mM DTT.
SDS-PAGE sample buffer.
Silver Stain Plus (Bio-Rad).
7.2. Procedure
Make an RNase reaction cocktail containing 1X RNase buffer and 1 μL diluted RNase cocktail (8 U RNase T1, 0.2 U RNase A) per material from 1e8 cells.
Resuspend magnetic beads in 50 μL RNase reaction cocktail and incubate at 37°C for 60 minutes.
Capture beads using magnetic rack and transfer the protein-containing supernatant to a fresh tube, store at −80°C or proceed directly to analysis.
Add SDS-PAGE sample buffer and separate proteins using standard SDS-PAGE.
Analyze by silver stain using the manufacturer’s protocol (see Note 11).
7.3. Notes
7.4. Results
If the ultimate goal is to identify RBPs associated with an RNA of interest, it is important to validate that there is an enrichment of protein in affinity purified samples prepared from infected cells relative to control. Figure 4C shows silver stain analysis of total protein present in affinity purified samples prepared from ~5E7 mock or DENV infected cells. While the intensity of bands in the input samples demonstrates that the same amount of total protein was processed for each condition, we observe an apparent increase in the amount of protein present in eluate samples from infected cells relative to the mock control. These results demonstrate that while the amount of material recovered is relatively small, there is a concomitant enrichment of protein upon specific affinity purification of viral RNA.
8. Summary
Here we describe a method for the specific purification of viral RNPs directly from infected cells. The advantage of this technique over traditional in vitro strategies is the ability to confidently identify proteins that are bound to the viral RNA in vivo in the context of productive infection. We have successfully applied this methodology in conjunction with mass spectrometry to identify a candidate list of cellular proteins found to be specifically associated with dengue virus RNA in infected cells (Phillips, unpublished data). However, the general approach described here could theoretically be applied to any RNA of interest, provided it is abundant enough that the signal to noise ratio is sufficiently high for reliable protein identification. A technical limitation of this approach is the large amount of starting material necessary to recover a sufficient amount of protein for identification. This issue may be addressed in part by scaling up the amount of material processed and/or optimizing the efficiency of recovery through the use of additional antisense oligonucleotides [26]. Another important limitation of this strategy is that the efficiency with which covalent crosslinks will form between a given protein and RNA depends on the proximity of their association as well as the amino acid composition of the protein [27]. Therefore this in vivo crosslinking approach may fail to identify RBPs that bind transiently to the RNA of interest or that do not contain sufficient amino acid residues amenable to forming crosslinks. As with any approach for en masse identification of interacting molecules, the interactions identified using this strategy should be validated using an independent methodology, for example RNA-immunoprecipitation [28]. Validated interactions can then be further investigated for their potential importance in the viral life cycle, for example, by siRNA-mediated silencing of candidate RBPs followed by infection.
Highlights.
A method for purification of endogenous dengue virus ribonucleoprotein complexes is described.
The method incorporates cross-linking of RNA and protein in living cells using ultraviolet light prior to cell lysis.
Dengue virus RNA and cross-linked proteins are purified using complementary biotinylated oligonucleotides.
This method can be used in principle to study interactions between RNA-binding proteins and any viral RNA.
Acknowledgments
The authors would like to acknowledge Drs. Elizabeth Dethoff and Kevin Weeks (University of North Carolina Chapel Hill) for helpful discussion regarding DENV2 RNA secondary structure. This work was funded by NIH R01-AI101431 to MGB and NIH F32-AI109834 to SLP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
RNase H is not compatible with many common detergents used in cell lysis buffers, such as SDS and NP-40 {[23] and data not shown}. Lysis conditions under which RNase H maintains its activity, e.g. hypotonic swelling followed by mechanical lysis, may need to be identified prior to RNase H mapping.
The number and type of cells used will depend on the RNA of interest and its copy number per cell. Initial optimization experiments should be performed to determine the scale at which the affinity purification should be performed to ensure reliable identification of proteins.
DENV2 infections and subsequent processing of cells is performed according to biosafety level 2 standards.
Crosslinking can also be performed using 365 nm ultraviolet after incorporation of the photoreactive ribonucleoside analog 4-thiouridine (4SU) [25]. We found that for DENV RNP, the efficiency of crosslinking did not appear to be enhanced by 4SU incorporation based on the efficiency of RNA recovery after phenol:chloroform extraction (data not shown). It is also not clear from previous reports that 4SU incorporation will lead to the identification of a great number of RBPs [20]. The strategy employed may be best determined on a case-by-case basis.
The composition of the lysis buffer will depend on the subcellular localization of the RNA of interest, i.e. soluble cytoplasmic, nuclear, or membrane associated. Different detergent compositions may need to be tested to ensure the optimal lysis conditions are used.
The concentration and sequence complexity of the antisense oligonucleotide mixture will depend on the individual target RNA and needs to be determined empirically to optimize recovery of the RNA of interest while keeping the recovery of non-specific RNA to a minimum.
The final temperature of the annealing reaction will depend on the melting temperatures of the antisense oligonucleotides used.
Additional material can be captured by successive rounds of affinity purification using the unbound material and fresh beads.
Using an equal volume of purified RNA in each cDNA synthesis reaction eliminates skewing of RNA yield calculations due to differences in the affinity purification of RNA across conditions.
The amount of cDNA template used for qPCR should be determined empirically to ensure that detection occurs within the linear range of the qPCR assay. For our assay, we routinely use 5 μL of cDNA template that has been diluted 1:20 in water.
Due to the limiting amount of material recovered after affinity purification it can be technically challenging to acquire enough protein to detect, even with a method as sensitive as silver stain. An alternative approach to validating the recovery of protein is to perform Western blot analysis, however, this is only useful if there is prior knowledge of a protein that can serve as a positive control for binding.
References
- 1.Deo S, et al. Activation of 2′ 5′-oligoadenylate synthetase by stem loops at the 5′-end of the West Nile virus genome. PLoS One. 2014;9(3):e92545. doi: 10.1371/journal.pone.0092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deo S, et al. Characterization of the termini of the West Nile virus genome and their interactions with the small isoform of the 2′ 5′-oligoadenylate synthetase family. J Struct Biol. 2015;190(2):236–49. doi: 10.1016/j.jsb.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282(42):30497–508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 4.Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10(7):e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin RJ, et al. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41(5):3314–26. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt SL, Jackson RJ. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5(3):344–59. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y, et al. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11(12):1809–24. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L, et al. Polypyrimidine tract-binding protein influences negative strand RNA synthesis of dengue virus. Biochem Biophys Res Commun. 2009;385(2):187–92. doi: 10.1016/j.bbrc.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis WG, et al. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol. 2007;81(18):10172–87. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polacek C, Friebe P, Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol. 2009;90(Pt 3):687–92. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 11.Sokoloski KJ, et al. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe. 2010;8(2):196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei Y, et al. Functional interaction between cellular p100 and the dengue virus 3′ UTR. J Gen Virol. 2011;92(Pt 4):796–806. doi: 10.1099/vir.0.028597-0. [DOI] [PubMed] [Google Scholar]
- 13.Ward AM, et al. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8(6):1173–86. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anwar A, et al. The polypyrimidine tract-binding protein is required for efficient dengue virus propagation and associates with the viral replication machinery. J Biol Chem. 2009;284(25):17021–9. doi: 10.1074/jbc.M109.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreyfuss G, Choi YD, Adam SA. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4(6):1104–14. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Venrooij WJ, et al. In vivo cross-linking of proteins to mRNA in human cells. Mol Biol Rep. 1981;7(1–3):93–9. doi: 10.1007/BF00778738. [DOI] [PubMed] [Google Scholar]
- 17.Mayrand S, Pederson T. Nuclear ribonucleoprotein particles probed in living cells. Proc Natl Acad Sci U S A. 1981;78(4):2208–12. doi: 10.1073/pnas.78.4.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayrand S, et al. Structure of nuclear ribonucleoprotein: identification of proteins in contact with poly(A)+ heterogeneous nuclear RNA in living HeLa cells. J Cell Biol. 1981;90(2):380–4. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Eekelen CA, et al. Adenoviral heterogeneous nuclear RNA is associated with host cell proteins. Eur J Biochem. 1981;119(3):461–7. doi: 10.1111/j.1432-1033.1981.tb05630.x. [DOI] [PubMed] [Google Scholar]
- 20.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8(3):491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 22.Lenarcic EM, et al. Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J Virol. 2013;87(15):8697–712. doi: 10.1128/JVI.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108(51):20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markovitz A. Ultraviolet light-induced stable complexes of DNA and DNA polymerase. Biochim Biophys Acta. 1972;281(4):522–34. doi: 10.1016/0005-2787(72)90153-0. [DOI] [PubMed] [Google Scholar]
- 25.Favre A, et al. 4-Thiouridine photosensitized RNA-protein crosslinking in mammalian cells. Biochem Biophys Res Commun. 1986;141(2):847–54. doi: 10.1016/s0006-291x(86)80250-9. [DOI] [PubMed] [Google Scholar]
- 26.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith KC. Photochemical addition of amino acids to 14C-uracil. Biochem Biophys Res Commun. 1969;34(3):354–7. doi: 10.1016/0006-291x(69)90840-7. [DOI] [PubMed] [Google Scholar]
- 28.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1(1):302–7. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]