Abstract
Background
Cholesteryl ester transfer protein (CETP) plays a crucial role in lipid metabolism. Associations of common CETP variants with variation in plasma lipid levels, and/or CETP mass/activity have been extensively studied and well-documented; however, the effects of uncommon/rare CETP variants on plasma lipid profile remain undefined. Hence, resequencing of the gene in extreme phenotypes and follow-up rare-variant association analyses are essential to fill this gap.
Objective
To identify common and uncommon/rare variants in the CETP gene by resequencing the entire gene and test the effects of both common and uncommon/rare CETP variants on plasma lipid traits in two genetically distinct populations.
Methods and Results
The entire CETP gene plus flanking regions were resequenced in 190 individuals comprising 95 non-Hispanic Whites (NHWs) and 95 African blacks with extreme HDL-C levels. A total of 279 sequence variants were identified, of which 25 were novel. Selected variants were genotyped in the entire samples of 623 NHWs and 788 African blacks and 184 QC-passed variants were tested in relation to plasma lipid traits by using gene-based, single-site, haplotype and rare variant association analyses (SKAT-O). Two novel and independent associations of rs1968905 and rs289740 with HDL-C were identified in African blacks. Using SKAT-O analysis, we also identified rare variants with minor allele frequency <0.01 to be associated with HDL-C in both NHWs (P=0.024) and African blacks (P=0.009).
Conclusions
Our results point out that in addition to the common CETP variants, rare genetic variants in the CETP gene also contribute to the phenotypic variation of HDL-C in the general population.
Keywords: CETP, lipid metabolism, HDL-C, genetic association, rare variants, sequencing
1. INTRODUCTION
Human cholesteryl ester transfer protein (CETP protein; CETP gene) plays a crucial role in lipid metabolism by mediating the transfer of cholesteryl esters from high density lipoprotein (HDL) to apolipoprotein (apo) B rich lipoproteins in exchange for triglycerides (TG) [1]. High CETP activity contributes to unfavorable plasma lipoprotein profile by lowering HDL-cholesterol (HDL-C) and increasing low-density lipoprotein cholesterol (LDL-C) [1–3]. However, it has been shown that reduced CETP activity is associated with increased CAD risk despite resulting high levels of HDL-C (4–6). Even though the effects of CETP inhibition in the therapy of CAD are conflicting, inhibition of CETP activity has been used as a new approach to raise HDL-C levels (7–8). In order to understand the role of CETP in CAD and to develop therapeutic approaches, the elucidation of genetic variation in the CETP gene is important.
Several common variants in CETP have been reported to be associated with variation in plasma lipid levels, CAD risk and/or CETP mass/activity [9]. However, reported common CETP variants explain only 5–8% of the overall genetic contribution to lipid levels [6]. The so called 'missing heritability' likely lies in low-frequency and rare variants and they can only be uncovered by resequencing individuals with extreme lipid phenotypes [10–11]. This strategy has already been proven to be useful in identifying multiple rare variants in lipid genes affecting variation in plasma lipid levels (12–16). Toward this effort, Khovidhunkit et al. [16] sequenced the exons and exon-intron junctions of the CETP gene in 64 Thai individuals with hyperalphalipoproteinemia and identified rare CETP variants contributing to HDL-C. However, the contribution of rare CETP variants in regulating lipid levels in the general population requires further investigation.
The objective of this study was to resequence the entire CETP gene plus flanking regions in selected individuals having extreme HDL-C levels derived from two well-characterized population-based samples of U.S. non-Hispanic whites (NHWs) and African blacks in order to identify both common (minor allele frequency (MAF)≥5%), and rare and uncommon CETP variants (MAF <5%) followed by genotype-phenotype association analyses with lipid levels in the entire samples of 788 African blacks and 623 NHWs.
2. METHODS
2.1. Subjects
The study was carried out on two well-characterized and population-based epidemiological samples comprising unrelated 623 non-Hispanic whites (NHWs) from the US and 788 African blacks from Nigeria. NHWs were collected as a part of the San Luis Valley Diabetes Study, a population-based case-control study of type 2 diabetes in the San Luis Valley, Southern Colorado [17,18]. The 623 NHWs subjects used in the current study did not have diabetes and a detailed description of this sample set can be found elsewhere [19]. African black samples were drawn from a study on coronary heart disease (CHD)-related risk factors in Benin City, Nigeria [20,21]. The characteristics of African black samples have been described in detail in Harris et al. (1998). Biometric and quantitative data of 623 NHWs and 788 African Blacks are summarized in Table 1. For the resequencing discovery stage, we selected 95 NHWs and 95 African blacks falling in the upper (47 NHWs, 48 African blacks) and lower (48 NHWs, 47 African blacks) 10th percentile distribution of plasma HDL-C levels (Table 2). This study was approved by the University of Pittsburgh and University of Colorado Denver Institutional Review Boards. All study participants provided written informed consent.
Table 1.
Biometric and quantitative data (mean±SD) of the entire American white (NHWs) and African black samples
| Variable Mean±sd or percentage |
NHWs (n=623) | African Blacks (n=788) |
|---|---|---|
| Males/Females (%) | 47.35/52.64 | 62.81/37.18 |
| Age (Yrs) | 52.83 ± 11.41 | 40.95±8.39 |
| BMI (kg/m2) | 25.51 ± 4.06 | 22.87±4.04 |
| LDL-C (mg/dl) | 136.9 ± 40.8 | 109.25±34.40 |
| HDL-C (mg/dl) | 50.8 ± 14.4 | 47.88±12.87 |
| TG (mg/dl) | 142.72 ± 93.5 | 72.96±39.32 |
| TC (mg/dl) | 217.0 ± 43.5 | 172.01±38.47 |
TC: Total cholesterol; LDL-C: Low-density lipoprotein cholesterol; HDL-C; High-density lipoprotein cholesterol; TG: Triglycerides
Table 2.
Biometric and quantitative data (mean±SD) of resequencing samples of 95 NHWs, and 95 African blacks
| NHWs (n=95) | African Blacks (n=95) | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| High HDL-C (n=47) | Low HDL-C (n=48) | P-value* | High HDL-C (n=48) | Low HDL-C (n=47) | P-value* | |
| Sex (M/F) | 24/23 | 24/24 | 1.00 | 24/24 | 23/24 | 1.00 |
| Age (years) | 55.45 ± 9.80 | 53.03 ± 10.54 | 0.25 | 41.29 ± 8.72 | 40.87 ± 7.12 | 0.80 |
| BMI (kg/m2) | 23.17 ± 3.17 | 27.35 ± 3.90 | 1.2E-07 | 22.06 ± 4.70 | 23.91 ± 5.51 | 0.08 |
| TC (mg/dl) | 227.34 ± 51.76 | 208.81± 44.65 | 0.07 | 201±39.68 | 141.68 ± 31.03 | 2.4E-12 |
| LDL-C (mg/dl) | 126.84 ± 46.95 | 125.54 ± 54.97 | 0.90 | 112.55 ± 39.75 | 95.04 ± 28.28 | 0.02 |
| HDL-C (mg/dl) | 77.68 ± 13.32 | 31.81 ± 4.37 | 2.2E-16 | 76.05 ± 7.53 | 25.51 ± 5.66 | 2.2E-16 |
| TG (mg/dl) | 114.09 ± 60.88 | 240.21 ±153.22 | 1.7E-06 | 61.98 ± 19.85 | 95.79 ± 73.21 | 0.004 |
TC: Total cholesterol; LDL-C: Low-density lipoprotein cholesterol; HDL-C; High-density lipoprotein cholesterol; TG: Triglycerides;
P-values were calculated using t-test. No covariates were included.
2.2. Lipid measurements
Fasting total cholesterol (TC) was determined by esterase-oxidase method [22]. Serum HDL-C and TG concentrations were measured by enzymatic procedures [23]. LDL-C was calculated by using the Friedewald equation if TG levels were less than 400 mg/dl (4.5 mmol/l) [24].
2.3. DNA sequencing
DNA samples were extracted from blood clots and buffy coats from African blacks and NHWs, respectively, by using standard DNA extraction procedures. Polymerase chain reaction (PCR) was performed to amplify the entire CETP gene, including 16 exons and adjacent introns plus 1 kb from both 3' and 5' flanking regions of the gene (total ~26 kb genomic fragment). The Primer 3 software (http://frodo.wi.mit.edu/primer3/) was used for designing primers. The accession number of CETP reference sequence used in this study is: NC_000016 derived from Genbank in NCBI site (http://www.ncbi.nlm.nih.gov). A total of 47 overlapping resequencing amplicons were sequenced in both directions in selected 95 NHW and 95 African black individuals with extreme HDL-C levels. PCR conditions and primer sequences are available upon request. After PCR amplification, plates were sent to a commercial lab (Beckman Coulter Genomics, Danvers, MA) where automated Sanger sequencing was performed on ABI 3730x1DNA Analyzers. Sequencing data analysis was performed in our lab by using Variant Reporter (Applied Biosystems, Foster City, CA) and Sequencher (Gene Codes Corporation, Ann Arbor, MI) softwares. All amplicons were successfully analyzed except a repetitive region in intron 2 which did not pass the quality control (QC) and was not analyzed (positions between 6938 bp and 8664 bp).
2.4. Variant selection and Genotyping
Single-nucleotide variant (SNV) selection for genotyping in the total samples of 623 NHWs and 788 African blacks was based on our sequencing data plus information obtained from public databases, and literature including candidate and genome-wide association studies (GWAS) [25–31]. Sequencing-derived SNVs included tagSNPs and uncommon/rare variants that were selected based on their locations in the gene and their frequency distribution among individuals with extreme HDL-C included in resequencing. TagSNP selection was performed by tagger and LD analyses in Haploview by using the following parameters: r2≥0.9, MAF≥0.05. Forty-one common tagSNPs in NHWs and 83 tagSNPs in African blacks were identified which capture 81 and 126 common variants (MAF≥0.05) in NHWs and African blacks, respectively (see Supplementary Tables 1–2). We compared our sequencing-derived tagSNPs with the tagSNPs of HapMap data for CEU (Utah residents with ancestry from northern and western Europe) and YRI (Yoruba in Ibadan, Nigeria) populations and determined that all of the HapMap SNVs (MAF≥0.05) in the same region were captured by our tagSNPs. In addition to tagSNPs, 40 uncommon and low frequency (MAF< 0.05) variants in NHWs and 77 in African blacks were selected from our sequencing data. Detailed information for selection criteria of SNVs included in association analyses can be found in Supplementary Table 3 for NHWs and Supplementary Table 4 for African blacks. In addition, we selected 16 SNVs for genotyping (8 in NHWs and 8 in African blacks) that were not detected in our sequencing samples but they have been previously reported in dbSNP build 137 in European and African descent populations. We also genotyped 26 additional SNVs in NHWs and 7 in African blacks that were not identified tagSNPs in our sequencing cohort but previously they have been shown to be associated with lipid levels in candidate or GWAS. Altogether, 115 variants in NHWs and 175 variants in African blacks were selected for genotyping and 251 of them were successfully genotyped in at least one population (111 SNVs in NHWs and 140 SNVs in African blacks).
2.5. Genotyping
Selected variants were genotyped in the total sample of 623 NHWs and 788 African blacks by using either TaqMan (Applied Biosystems) or iPLEX Gold (Sequenom, San Diego, CA) genotyping methods following manufacturer’s protocols. The 384-well plates containing dried whole genome amplified DNAs were used in TaqMan and iPLEX Gold genotyping methods. The ABI Prism 7900HT Sequence Detection Systems was used for endpoint fluorescence reading of the 384-well plates after TaqMan protocol. The iPLEX Gold genotyping was performed in the Genomics and Proteomics Core laboratories of the University of Pittsburgh. Sequences of primers and probes used for custom TaqMan and iPLEX Gold genotyping are available upon request.
2.6. Statistical Analysis
Variants identified by sequencing were analyzed by using Haploview [32] (www.broadinstitute.org/haploview) to test the concordance of the genotype distribution with Hardy-Weinberg equilibrium, to determine allele frequencies and their distributions among high and low HDL groups and their linkage disequilibrium (LD) patterns. For those SNVs that were genotyped in the entire sample, the additive linear regression model was used to test for the effects of genotypes on the means of plasma HDL-C, TG, LDL-C, and TC levels. While all plasma lipid levels were transformed to natural logarithms using Box-Cox transformation to improve normality in African blacks, only HDL-C and TG were transformed in NHWS. The significant covariates were identified using stepwise regression in both directions. In NHWs, the covariates included in the analyses were gender, age, BMI, smoking and, in African blacks, they were gender, age, BMI, smoking, waist (waist measurement (cm) at the narrowest point), staff level (junior/senior) and exercise (minutes walking or bicycling to work each day (min)). The R statistical software package (http://www.r-project.org) was used to perform all computations. False discovery rate (FDR) method [33] was used to control for multiple testing. A P <0.05 was considered as suggestive evidence of association and FDR value (q-value) of <0.05 as statistically significant. The versatile gene-based associations (VEGAS) were performed to assess the relationship between traits and CETP [34]. For haplotype association analysis, the generalized linear model (GLM) was used [35]. Since including too many haplotypes can make GLM inefficient and impractical, we used sliding windows consisting of 4 SNVs per window in order to reduce the number of haplotypes considered in association analysis. A global P-value for testing overall effect of haplotypes with frequency >0.01 was used to assess the association between the trait and haplotypes in each window. Sliding-window haplotype analysis was performed using the haplo.glm function in the Haplo.Stats R package (version 1.5.0). We analyzed the cumulative effects of uncommon/rare variants by using burden tests and the SKAT-O method [36]. SKAT-O method has been proposed to be the optimal test for rare variant analysis over the SKAT and burden tests. Three different minor allele frequency bin thresholds (<1%, <2% and <5%) were used to perform the analyses. The SKAT-O method was implemented using the “SKAT” R package.
2.7. Functional annotation of significant SNVs
We used information from RegulomeDB [37] online database (http://regulome.stanford.edu/) version 1.0 to retrieve the regulatory annotations for the identified significant SNVs.
3. RESULTS
3.1. DNA sequencing
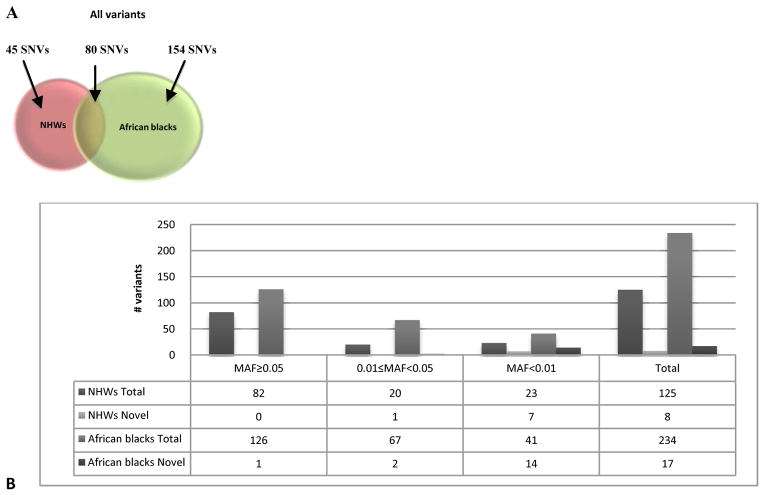
We identified a total of 279 variants by resequencing the CETP gene in 190 NHWs and African blacks, including 263 diallelic (181 transitions vs. 82 transversions), 2 triallelic and 14 indels. Of 279 variants, 254 are known and 25 are novel (submitted to dbSNP database: http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH) (Supplementary Tables 5 and 6). Eighty of these variants were present in both population groups, 45 were found only in NHWs and 154 variants were found only in African blacks. The locations of the identified 279 variants were as follows: 224 in introns, 39 in the flanking regions, 1 in the 5'UTR of exon 1, 1 in the 3'UTR of exon 16, and 14 in coding exons. Of the identified 14 coding variants, 7 were non-synonymous and 7 were synonymous. The seven non-synonymous coding variants were: aspartic acid to valine (D6V) and alanine to glycine (A15G) in exon 1; arginine to tryptophan (R154W) in exon 5; alanine to proline (A390P), and valine to methionine (V385M) in exon 12; isoleucine to valine (I405V) in exon 14; and arginine to glutamine (R451Q) in exon 15. All observed 14 indels were located in introns, including two insertions and 12 deletions, ranging in size from one to twelve bases. The characteristics of the identified variants are given in Fig. 1.
Fig. 1. Properties of the CETP variants identified in American whites (NHWs) and African blacks.
a. Number of all SNVs identified in each population are shown by venn diagrams. b. Chart showing the distribution of the minor allele frequencies (MAF) of all variants
Of the 125 variants found in NHWs, 82 were common (MAF≥0.05), 20 were uncommon (0.01≤MAF<0.05) and 23 were rare (MAF<0.01) (see Supplementary Table 5). Of the 234 variants identified in African blacks, two were triallelic [rs289712 (intron 8) and rs7192120 (exon 12; synonymous)]. Among the 234 bi-allelic variants, 126 were common (MAF≥0.05), 67 were uncommon (0.01≤MAF<0.05) and 41 were rare (MAF<0.01) (see Supplementary Table 6).
3.2. Distribution of identified CETP variants in extreme HDL-C groups
Minor allele frequency distributions of the 125 variants in NHWs and 234 variants in African blacks between the two extreme HDL-C groups are shown in Supplementary Table 5 and Supplementary Table 6, respectively. In NHWs, 14 variants were present only in the low HDL-C group vs 11 only in the high HDL-C group; the remaining were present in both groups. In African blacks, 27 variants were present only in the low HDL-C group vs 29 only in the high HDL-C group; the remaining were present in both groups.
3.3. Genotyping of CETP variants
Two hundred and fifty one CETP variants were successfully genotyped in at least one population group (111 in NHWs and 140 in African blacks) and 51 of them were present in both groups. The genotype call rates, selection criteria and other features of the genotyped SNVs in NHWs (39 tagSNPs; 72 others) and African blacks (73 tagSNPs; 67 others) are shown in Supplementary Tables 3–4. The LD bins (r2≥0.80) of the final QC passed SNVs are listed in Supplementary Tables 7–8. Of the 140 SNVs genotyped in African blacks, two triallelic SNVs (rs289712C/T/A and rs7192120C/G/T) were excluded from analysis. In NHWs, two SNVs (rs247615, rs12720918), which did not meet Hardy-Weinberg equilibrium after Bonferroni correction (P<10E-04), were excluded along with five monomorphic SNVs [rs4784741, rs9924087, rs1968905, rs189866004, rs5887]. In African blacks 7 SNVs were excluded from the analyses; rs158477 had low call rate (<85 %), and five SNVs (rs71383212, rs11860407, rs12708985, rs12720939, rs17231611) deviated from Hardy-Weinberg equilibrium (P<10E-04) and rs9936680 was non-polymorphic. Thus, a total of 104 SNVs (69 common and 35 uncommon/rare based on their MAF in the entire sample) in NHWs and 131 variants (86 common and 45 uncommon/rare) in African blacks were included in subsequent association analyses. The discrepancy rate was determined to be 0–0.5% for the genotyped variants based on the random repeats of ~10% of the samples.
3.4. Gene-based association analysis
Gene-based association test was performed to assess the joint effects of all successfully genotyped CETP variants on plasma lipid traits (HDL-C, TG, LDL-C, TG). Significant association was observed only with HDL-C in both NHWs (P=8.87E-04) and African blacks (P=9.00E-06) (Table 3). In view of the strong association of CETP with HDL-C, subsequent single-site, haplotype, and rare variant analyses were conducted with HDL-C only.
Table 3.
CETP gene-based association test in NHWs and African blacks
| NHWs | African blacks | |||||||
|---|---|---|---|---|---|---|---|---|
| Trait | Number of SNVs* | Test Statistics | P | b Smallest P | Number of SNVs* | Test Statistics | P | b Smallest P |
| HDL-C | 100 | 322.1438 | 8.87E-04 | 7.41E-05 (rs247617) | 131 | 470.565 | 9.00E-06 | 7.88E-10 (rs34065661) |
| LDL-C | 100 | 81.92025 | 0.639 | 6.864E-03 (CTP13868) | 131 | 150.8933 | 0.277 | 0.019 (rs80296794) |
| TG | 100 | 52.73295 | 0.944 | 0.01562 (rs56208677) | 131 | 104.4884 | 0.733 | 3.097E-03 (rs114908369) |
| TC | 100 | 91.41172 | 0.538 | 2.05E-04(CTP13868) | 131 | 181.023 | 0.117 | 3.057E-03 (rs114203109) |
P: gene-based,
Smallest P detected for a single SNV
Variants with missing phenotype data were not included
3.5. Association of common variants (MAF≥0.05) with HDL-C levels
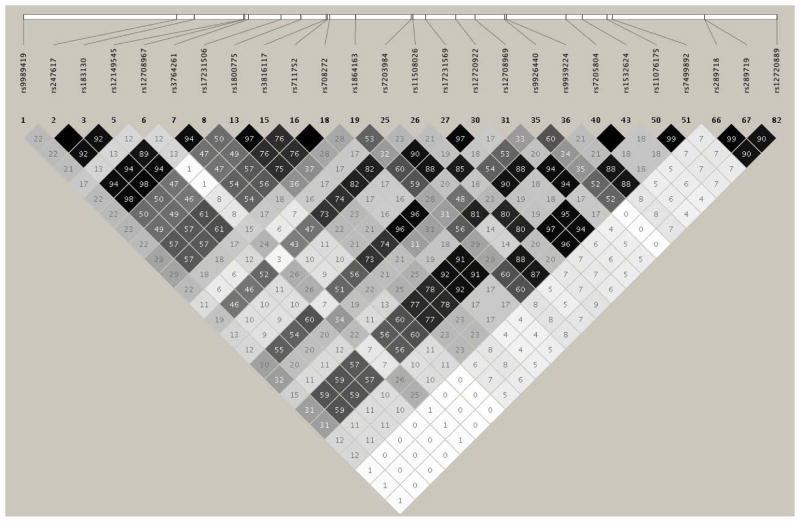
In NHWs, we identified 26 common SNVs exhibiting associations with HDL-C levels (see Table 4). The most significant variant was rs247617 (β=0.043; P=7.41E-05; located ~3kb upstream of the CETP gene) and this was correlated (r2>0.46) with twelve other HDL-C-associated SNVs (see Fig. 2) in NHWs. The remaining thirteen SNVs showed independent associations with HDL-C of the top SNV, rs247617. Among African blacks, 31 SNVs were associated with HDL-C; the most significant SNV was non-synonymous, rs34065661 (β=2.07, P=7.88E-10) and this was strongly correlated (r2=0.99) with a promoter SNV rs17231520 (β=1.99, P=1.86E-09) (see Table 4 and Fig. 3). We also observed two novel independent associations of rs1968905 (β=0.63, P=0.002) and rs289740 (β=0.87, P=0.003) with HDL-C. Detailed results of the association of the CETP variants with HDL-C in NHWs and African blacks can be found in Supplementary Table 9 and Supplementary Table 10, respectively.
Table 4.
Association results (P<0.05) of common CETP variants with HDL-C levels in NHWs (n=623) and African blacks (n=788)
| NHWs | African blacks | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| † RefSNP ID | †† Chr16 Position | Locations | MAF | Beta | P | FDR | MAF | Beta | P | FDR | ‡ RegulomeDB Scores |
| rs9989419 | 56951227 | 5' flanking | 0.4 | −0.026 | 0.014 | 0.067 | - | - | - | - | 1f |
| rs247617 | 56956804 | 5' flanking | 0.347 | 0.043 | 7.41E-05 | 0.004 | - | - | - | - | 5 |
| rs183130 | 56957451 | 5' flanking | 0.347 | 0.04 | 1.91E-04 | 0.004 | 0.244 | 1.03 | 2.25E-06 | 7.37E-05 | No data |
| rs12149545 | 56959249 | 5' flanking | 0.33 | 0.042 | 1.66E-04 | 0.004 | - | - | - | - | No data |
| rs12708967 | 56959299 | 5' flanking | 0.205 | -0.034 | 1.14E-02 | 0.065 | - | - | - | - | No data |
| rs3764261 | 56959412 | 5' flanking | 0.328 | 0.043 | 1.63E-04 | 0.004 | 0.323 | 0.62 | 0.002 | 0.02 | No data |
| rs17231506 | 56960616 | 5' flanking | 0.343 | 0.042 | 1.25E-04 | 0.004 | - | - | - | - | 6 |
| rs4783961 | 56960982 | 5' flanking | - | - | - | - | 0.454 | 0.64 | 0.001 | 0.011 | 4 |
| rs1800775 | 56961324 | 5' flanking | 0.489 | -0.027 | 1.25E-02 | 0.004 | 0.395 | −0.53 | 0.006 | 0.041 | 3a |
| rs17231520 | 56961915 | 5' flanking | - | - | - | - | 0.09 | 1.99 | 1.86E-09 | 1.22E-07 | 5 |
| rs34065661 | 56962023 | Exon 1 (A15G) | - | - | - | - | 0.088 | 2.07 | 7.88E-10 | 1.03E-07 | 5 |
| rs3816117 | 56962246 | Intron 1 | 0.488 | −0.025 | 1.84E-02 | 0.065 | 0.394 | −0.54 | 0.006 | 0.041 | 5 |
| rs711752 | 56962299 | Intron 1 | 0.443 | 0.033 | 2.19E-03 | 0.083 | 0.232 | 0.91 | 3.70E-05 | 0.001 | 5 |
| rs708272 | 56962376 | Intron 1 | 0.442 | 0.033 | 2.42E-03 | 0.031 | 0.236 | 0.88 | 6.33E-05 | 0.001 | 5 |
| rs1864163 | 56963321 | Intron 2 | 0.265 | −0.035 | 3.28E-03 | 0.031 | 0.278 | −0.7 | 0.001 | 0.013 | 5 |
| rs7203984 | 56965346 | Intron 2 | 0.206 | −0.032 | 0.011 | 0.034 | - | - | - | - | 5 |
| rs11508026 | 56965416 | Intron 2 | 0.472 | 0.025 | 0.012 | 0.065 | - | - | - | - | 5 |
| rs17231569 | 56965867 | Intron 2 | 0.19 | -0.031 | 0.019 | 0.065 | - | - | - | - | 5 |
| rs142058276 | 56966072 | Intron 2 | - | - | - | - | 0.082 | 1.66 | 2.06E-06 | 7.37E-05 | No data |
| rs12720922 | 56966973 | Intron 2 | 0.192 | −0.03 | 0.022 | 0.091 | - | - | - | - | 5 |
| rs12708969 | 56967672 | Intron 2 | 0.439 | 0.028 | 0.009 | 0.065 | - | - | - | - | No data |
| rs9924087 | 56968330 | Intron 2 | - | - | - | - | 0.207 | 0.57 | 0.011 | 0.061 | |
| rs9926440 | 56968751 | Intron 2 | 0.302 | −0.027 | 0.019 | 0.083 | - | - | - | - | 4 |
| rs9939224 | 56968820 | Intron 2 | 0.205 | −0.034 | 0.008 | 0.065 | - | - | - | - | 4 |
| rs11076174 | 56969234 | Intron 2 | - | - | - | - | 0.132 | −0.71 | 0.013 | 0.068 | 5 |
| rs891141 | 56969811 | Intron 4 | - | - | - | - | 0.13 | 0.79 | 0.004 | 0.031 | 4 |
| rs891143 | 56970068 | Intron 5 | - | - | - | - | 0.09 | 1.29 | 1.13E-04 | 0.002 | No data |
| rs7205804 | 56970977 | Intron 5 | 0.453 | 0.031 | 0.004 | 0.039 | 0.097 | −0.76 | 0.015 | 0.071 | 5 |
| rs1532624 | 56971567 | Intron 7 | 0.452 | 0.032 | 0.003 | 0.031 | 0.097 | −0.77 | 0.014 | 0.071 | 5 |
| rs11076175 | 56972466 | Intron 8 | 0.184 | −0.034 | 0.01 | 0.065 | 0.243 | −0.51 | 0.02 | 0.086 | No data |
| rs7499892 | 56972678 | Intron 8 | 0.185 | −0.033 | 0.011 | 0.065 | - | - | - | - | No data |
| rs9930761 | 56973280 | Intron 8 | - | - | - | - | 0.148 | 0.72 | 0.01 | 0.059 | 4 |
| rs5883 | 56973441 | Exon 9 (F287F) | - | - | - | - | 0.155 | 0.65 | 0.017 | 0.075 | 4 |
| rs11076176 | 56973534 | Intron 9 | - | - | - | - | 0.403 | −0.43 | 0.027 | 0.106 | 5 |
| rs289718 | 56976020 | Intron 10 | 0.304 | 0.023 | 0.043 | 0.133 | 0.438 | −0.4 | 0.035 | 0.129 | 4 |
| rs289719 | 56976029 | Intron 10 | 0.3 | 0.023 | 0.043 | 0.133 | 0.493 | 0.41 | 0.033 | 0.124 | 5 |
| rs1968905 | 56977036 | Intron 10 | - | - | - | - | 0.36 | 0.63 | 0.002 | 0.02 | 6 |
| rs12708980 | 56978467 | Intron 11 | - | - | - | - | 0.342 | −0.51 | 0.011 | 0.061 | 6 |
| rs12720889 | 56978651 | Intron 11 | 0.29 | 0.025 | 0.031 | 0.117 | - | - | - | - | No data |
| rs7195984 | 56981551 | Intron 12 | - | - | - | - | 0.169 | 0.72 | 0.005 | 0.035 | 4 |
| rs1800774 | 56981633 | Intron 12 | - | - | - | - | 0.217 | −0.51 | 0.025 | 0.103 | 4 |
| rs289740 | 56983038 | Intron 14 | - | - | - | - | 0.117 | 0.87 | 0.003 | 0.031 | 4 |
| rs1801706 | 56983750 | 3'UTR-Exon 16 | - | - | - | - | 0.176 | 0.57 | 0.021 | 0.088 | 4 |
| rs289743 | 56983884 | 3'flanking | - | - | - | - | 0.315 | −0.39 | 0.0480.015 | 0.165 | 2b |
| rs66495554 | 56984724 | 3'flanking | - | - | - | - | 0.193 | −0.58 | 0.071 | No data | |
dbSNPbuild 141
RefSeq: hg19, NC_000016.10 MAF: Minor allele frequency FDR; false discovery rate
RegulomeDB scores were generated by using http://regulome.stanford.edu/.
Scores represents; Category 1 (Likely to affect binding and linked to expression of a gene target): 1a- eQTL + TF binding + matched TF motif + matched DNase Footprint + DNase peak;1b- eQTL + TF binding + any motif + DNase Footprint + DNase peak;1c- eQTL + TF binding + matched TF motif + DNase peak;1d- eQTL + TF binding + any motif + DNase peak;1e- eQTL + TF binding + matched TF motif;1f- eQTL + TF binding / DNase peak; Category 2 (Likely to affect binding): 2a- TF binding + matched TF motif + matched DNase Footprint + DNase peak; 2b- TF binding + any motif + DNase Footprint + DNase peak; 2c- TF binding + matched TF motif + DNase peak; Category 3 (Less likely to affect binding) 3a- TF binding + any motif + DNase peak;3b- TF binding + matched TF motif; Category 4–6 (Minimal binding evidence): 4- TF binding + DNase peak; 5-TF binding or DNase peak; 6-Motif hit
Fig. 2. LD structure of 26 common CETP SNVs associated with HDL-C in 623 NHWs.
The degree of shades and values (r2 x 100) in each square represent the pairwise LD between 26 genotyped SNPs: black indicating complete LD (r2 = 1), white indicating no LD (r2 = 0), and shade intensity indicating the degree of LD (r2 between 0–1).
Fig. 3. LD structure of 31 common CETP SNVs associated with HDL-C in 788 African blacks.
The degree of shades and values (r2 x 100) in each square represent the pairwise LD between 31 genotyped SNPs: black indicating complete LD (r2 = 1), white indicating no LD (r2 = 0), and shade intensity indicating the degree of LD (r2 between 0–1).
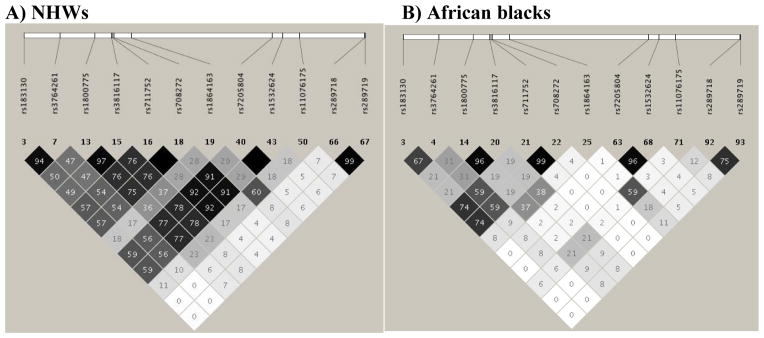
We identified twelve HDL-associated SNVs, which were significant in both populations, including rs183130, rs3764261 (GWAS significant), rs708272 (Taq1B), rs1800775 (-629C>A), rs3816117, rs711752, rs1864163, rs289718, rs289719, rs7205804, rs153624 and rs11076175 (see Table 4). The best SNV among the above-listed 12 HDL-associated SNVs was rs183130 (β=1.03; P=2.25E-06 in African blacks and β=0.040; P=1.91E-04 in NHWs) and this was in LD (r2=0.49–0.94) with 7 significant SNVs in NHWs (rs3764261, rs1800775, rs3816117, rs711752, rs708272, rs7205804, rs1532624) and in LD (r2=0.67–0.74) with 3 significant SNVs in African blacks (rs3764261, rs711752, rs708272) (see Fig. 4). The association of four remaining SNVs in NHWs (rs289719, rs289718, rs1864163 and rs11076175) and 8 remaining SNVs in African blacks (rs1800775, rs3816117, rs1864163, rs7205804, rs1532624, rs11076175, rs289718, rs289719) with HDL-C was independent of rs183130.
Fig. 4. LD structure of 12 common CETP SNVs associated with HDL-C in both populations. A) NHWs, B) African blacks.
The degree of shades and values (r2 x 100) in each square represent the pairwise LD between 12 genotyped SNPs: black indicating complete LD (r2 = 1), white indicating no LD (r2 = 0), and shade intensity indicating the degree of LD (r2 between 0–1).
3.6. Association of uncommon/rare variants (MAF < 0.05) with HDL-C
Three MAF thresholds, (MAF<0.05; MAF<0.02; and MAF<0.01) were used to separate the uncommon/rare variants (35 variants in NHWs, 45 variants in African blacks) in the rare variant association analyses (Table 5). Rare variants with MAF <0.01 were associated with HDL-C in both NHWs (P=0.024) and African blacks (P=0.009). We also checked the LD of significant rare variants (MAF <0.01) with common variants (MAF≥0.05) and found them not to be in LD with common variants (r2<0.20) that were associated with lipid traits in our NHW and African black samples.
Table 5.
Summary results of rare variant association analysis (SKAT-O) with HDL-C in NHWs and African blacks
| NHWs | African blacks | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| MAF threshold | N.RVa | N.Sample_RVb | N.Sample_NoRVc | Stat | P | N.RVa | N.Sample_RVb | N.Sample_NoRVc | Stat | P |
| Bin 1 (MAF <0.05) | 35 | 312 | 311 | 1.10E+05 | 0.030 | 45 | 598 | 190 | 2.16E+05 | 0.077 |
| Bin 2 (MAF <0.02) | 29 | 147 | 476 | 5.94E+04 | 0.072 | 17 | 195 | 593 | 2.27E+05 | 0.121 |
| Bin 3 (MAF <0.01) | 26 | 103 | 520 | 6.93E+04 | 0.024 | 6 | 30 | 758 | 3.47E+04 | 0.009 |
Abbreviation: SKAT-O, optimal sequencing Kernel association test. Bin 1 contained all variants with MAF<0.05, Bin 2 contained all variants with MAF<0.02 and Bin 3 contained all variants with MAF<0.01.
N.RV: Number of rare variants;
N_Sample_RV: Number of individuals carrying the rare allele with defined MAF cut offs.
N.Sample_NoRV: Number of individuals who do not carry any rare alleles with defined MAF cut offs.
3.7. Haplotype analyses
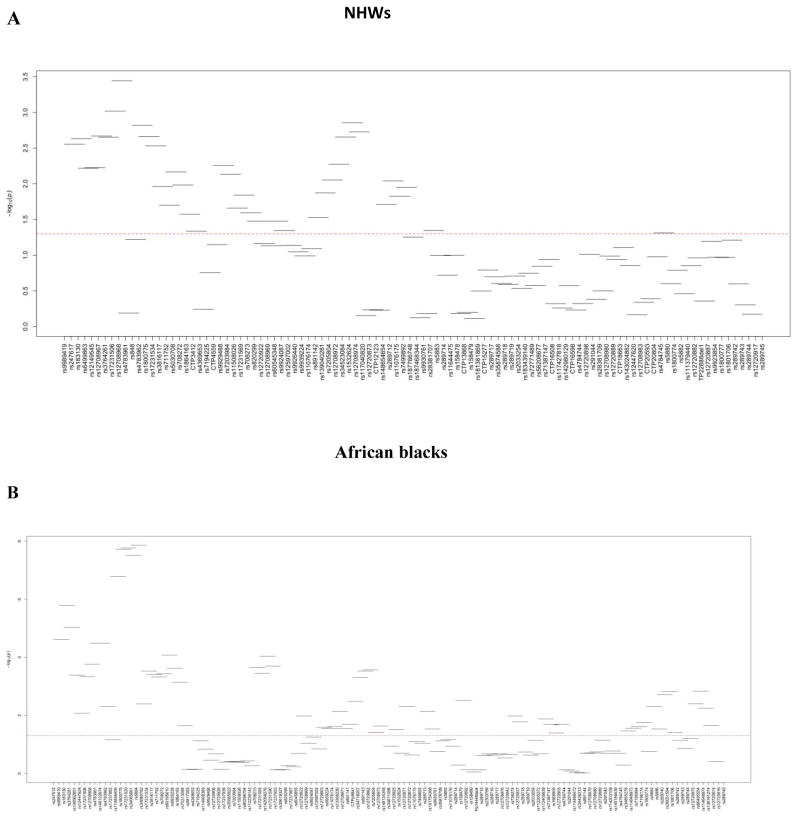
Sliding window approach was used to construct overlapping haplotypes of the genotyped SNVs and a global P-value was calculated for each 4-SNV window based on the comparison to the most common haplotype as a reference (R package-Haplostat). In both population groups, we observed multiple significant haplotype windows. Window 8 (rs17231506, rs12708968, rs4783961 and rs17245715) revealed the most significant signal for HDL-C (global P=3.60E-04) in NHWs, rs17231506 was the only significant SNV in single-site analysis in this haplotype window with a P-value of 1.25E-05. In African blacks, window 14 (rs180075, rs17231520, rs34065661 and rs5884) had the most significant association with HDL-C (global P=1.70E-08).
In both population groups, the haplotypes in the 5' of the CETP gene showed highly significant association with HDL-C. Fig. 5 shows the plots of the haplotype-based association test results for HDL-C. Details for the haplotype association analyses with HDL-C are shown in Supplementary Tables 11–12.
Fig. 5. CETP haplotype analysis with HDL-C levels in NHWs and African blacks.
Haplotype windows for NHWs (A) and for African blacks (B). The global (-log)10 P values are presented in the y-axis. Horizontal lines represent the window tested (101 haplotype windows in NHWs and 128 haplotype windows in African blacks). Variants are shown on the x-axis. Red line shows the threshold for statistical significance (P=0.05).
3.8. Functional annotation of significant SNVs
The RegulomeDB [37] scores for all SNVs identified in NHWs and African blacks are listed in Supplementary Tables 5 and 6, respectively. We identified 34 HDL-C-associated SNVs with a RegulomeDB score of less than 6, and among those, 3 (rs9989419, rs1800775 and rs289743) had a RegulomeDB score of less than or equal to 3 (Table 4). One of our independent signals with HDL-C in NHWs, rs9989419 (located ~8kb upstream of the CETP gene) had a RegulomeDB score of 1f (known cis-eQTL which lies within a TF binding site or a DNase peak based on the experimental evidence). This SNV has shown genome-wide significant association with HDL-C (18). Two other SNVs with a RegulomeDB score ≤3, [rs1800775 (Regulome DB score=3a); rs289743 (RegulomeDB score=2b)] were located near the CETP gene and they were highly correlated with other HDL-C significant SNVs (Fig. 2 and Fig. 3).
4. DISCUSSION
Previous studies have examined the role of common variants and haplotypes in the CETP gene in relation to dysplidemia, atherosclerosis and coronary heart disease with inconsistent outcomes. To our knowledge, this is first study that has resequenced the CETP gene and its flanking regions in selected individuals with extreme HDL-C levels from two racial groups and then examined the association of identified common and rare variants with variation in HDL-C levels in the total samples of NHWs and African blacks.
The complete resequencing of the CETP gene in 95 NHWs and 95 African blacks revealed a total of 279 variants, of which 80 were found in both populations. We identified all but 16 common variants (8 in NHWs and 8 in African blacks) present in dbSNP (build 137) in populations of European and African descent and also in 1000 genome project. The 16 known variants were missed probably due to technical issues in resequencing and so these SNVs were genotyped in the entire sample of both populations in order to cover the full range of variation in this gene. In addition, we identified 25 low-frequency variants that were not reported in any public databases. In tandem with SNV discovery effort, 200 SNVs including tagSNPs, several low frequency variants (MAF <0.005) and significantly reported variants in and around the CETP were successfully genotyped in 623 NHWs and 788 African blacks. A total of 184 QC-passed variants (104 in NHWs and 131 in African blacks) were included in the subsequent association analyses.
In single-site association analysis, we identified 11 and 16 FDR-significant HDL-associated SNVs (FDR<0.05) in NHWs and African blacks, respectively (1.03E-07≤FDR≤0.041). This is consistent with published studies where CETP polymorphisms have been reported to influence mostly HDL-C levels (6, 39–40). Notably, 12 SNVs (rs183130, rs3764261, rs1800775, rs3816117, rs711752, rs708272, rs1864163, rs7205804, rs1532624, rs11076175, rs289718, rs289719) were associated with HDL-C levels in both populations (P<0.05; 7.37E- 05≤FDR≤0.133). In NHWs, majority of these 12 SNVs were in strong LD with each other and with the GWAS significant variant, rs3764261 as well as with -629C>A (rs1800775) and Taq1B (rs70272), whereas little LD were observed among these SNVs among African blacks (Fig. 4A–4B).
Previously, three SNVs, TaqIB (rs708272; located in intron 1), -629C>A (rs1800775; located in promoter) and GWAS significant rs3764261 (5' flanking region), have been shown to be associated with lipid profile and/or CETP mass/activity in several studies [9,42]. It has been reported that up to 10% of the variation in the HDL-C can be explained by the TaqIB polymorphisim [9] and that Taq1B is a marker of promoter variant -629C>A (rs1800775) which is located in Sp2/SP2 binding sites of the proximal promoter [6, 43–46]. Although there is a strong LD between these two SNVs, their association with HDL-C has not been consistent in all studies [2,9]. In our study, -629C>A was strongly correlated with Taq1B in NHWs (r2=0.75) but not in African blacks (r2=0.19). Despite this, both Taq1B (β=0.033; P=2.42E-03 for NHWs; β=0.88; P=6.33E-05 for African blacks) and -629C>A (β=−0.027, P=1.25E-02 for NHWs; β=−0.53; P=0.006 for African blacks) were associated with plasma HDL-C levels in both populations. Recently, Lu et al. (2013) investigated the association of Taq1B and -629C>A polymorphisms with coronary heart disease and lipid levels in a multiethnic population of Singapore [47]. They found a weak correlation between the two polymorphisms and Taq1B showed stronger association with HDL-C and ApoA1 levels than -629C>A. Hence, they concluded that there could be additional functional sites, other than -629C>A, within or around the CETP gene that is in LD with Taq1B (47). In our study, rs183130 (located in ~3kb upstream) was the most significant HDL-associated SNV in both populations and that was also in LD with Taq1B and -629C>A. This has also shown consistent association with HDL-C across multiple ethnic groups including Europeans, Asians, and African Americans and it appears to be functional since it alters the consensus transcription binding site [42, 48].
Furthermore, an uncommon non-synonymous SNV, rs1800777 (R451Q), located in exon 15, that was previously reported to be associated with HDL-C levels [39] was found in only NHWs with a MAF of 0.035 (MAF= 0.00 % for both HapMAP YRI and our African blacks) and was associated with HDL-C (P=0.023) levels (see Supplementary Table 9).
Most of the significant SNVs identified in this study were located in the upstream and 5' half of the gene that were strongly correlated with each other and majority of these associations have been reported previously [9,38,41,48–49, 50–57]. However, we also observed two novel independent associations (rs1968905 and rs289740) with HDL-C in African blacks.
We also performed haplotype-based association analysis to evaluate whether combinations of CETP SNVs are more strongly associated with HDL-C than individual SNVs. We found that HDL-C associated significant haplotypes were mainly located in the 5' region, including the promoter and first ~12 kb of the CETP gene, and all these significant haplotypes included the HDL-C associated SNVs in single-site analysis. Similar results were obtained by a recent study in Latvians where a haploblock including SNVs located in ~0.6kb upstream of the CETP gene, promoter and first ~10kb of the CETP was associated with extreme HDL-C levels [41].
In addition to the single-site and haplotype association analyses, rare variant analyses (MAF<0.01) also revealed significant association with HDL-C levels. Although previous studies have shown the contribution of rare variants in other lipid genes in relation to lipid levels [12–15,38] to our knowledge, this is first study indicating that rare CETP variants make a substantial contribution to the variation of HDL-C levels in the general white and black populations. Our results are in concordance with a recent study published by ENGAGE consortium that highlights the aggregate effects of low-frequency/rare variants on inter-individual variation in lipid traits [59].
There are some limitations in our study. First, our sequencing sample in extreme HDL-C groups was relatively small in both ethnic groups and thus we may have missed some functional rare variants. Second, resequencing of the CETP failed to capture ~2kb in intron 2. Although we genotyped all reported significant and common SNVs in this un-sequenced region, some uncommon SNVs may have been missed and thus could not be analyzed.
In conclusion, this study provides credence to the "common disease-common variant" and "common disease-rare variant" hypotheses as both common and rare CETP variants were associated with HDL-C levels in American white and African black populations, and it further establishes the pivotal role of CETP in HDL metabolism.
Supplementary Material
Acknowledgments
SOURCES OF FUNDING
This study was supported by the National Heart, Lung and Blood Institute (NHLBI) grant, HL084613.
Abbreviations
- CETP
cholesteryl ester transfer protein
- SNP
single-nucleotide polymorphism
- QC
quality control
- TG
triglyceride
- MAF
minor allele frequency
- HDL-C
HDL cholesterol
- LD
linkage disequilibrium
- FDR
false discovery rate
Footnotes
DISCLOSURES
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993;34:1255–1274. [PubMed] [Google Scholar]
- 2.Dullaart RP, Groener JE, Dikkeschei BD, Erkelens DW, Doorenbos H. Elevated cholesteryl ester transfer protein activity in IDDM men who smoke. Possible factor for unfavorable lipoprotein profile. Diabetes Care. 1991;14:338–341. doi: 10.2337/diacare.14.4.338. [DOI] [PubMed] [Google Scholar]
- 3.de Grooth GJ, Zerba KE, Huang SP, Tsuchihashi Z, Kirchgessner T, Belder R, Vishnupad P, Hu B, Klerkx AH, Zwinderman AH, Jukema JW, Sacks FM, Kastelein JJ, Kuivenhoven JA. The Cholesteryl Ester Transfer Protein (CETP) TaqIB Polymorphism in the Cholesterol and Recurrent Events Study: No Interaction with the Response to Pravastatin Therapy and No Effects on Cardiovascular Outcome: a Prospective Analysis of the CETP TaqIB Polymorphism on cardiovascular outcome and interaction with cholesterol-lowering therapy. J Am Coll Cardiol. 2004;43:854–857. doi: 10.1016/j.jacc.2003.08.056. [DOI] [PubMed] [Google Scholar]
- 4.Bruce C, Sharp DS, Tall AR. Relationship of HDL and coronary heart disease to a common amino acid polymorphism in the cholesteryl ester transfer protein in men with and without hypertriglyceridemia. J Lipid Res. 1998;39:1071–1078. [PubMed] [Google Scholar]
- 5.Vasan RS, Pencina MJ, Robins SJ, Zachariah JP, Kaur G, D'Agostino RB, Ordovas JM. Association of circulating cholesteryl ester transfer protein activity with incidence of cardiovascular disease in the community. Circulation. 2009;120:2414–2420. doi: 10.1161/CIRCULATIONAHA.109.872705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schierer A, Been LF, Ralhan S, Wander GS, Aston CE, Sanghera DK. Genetic variation in cholesterol ester transfer protein, serum CETP activity, and coronary artery disease risk in Asian Indian diabetic cohort. Pharmacogenet Genomics. 2012;22:95–104. doi: 10.1097/FPC.0b013e32834dc9ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brousseau ME, Goldkamp AL, Collins D, Demissie S, Connolly AC, Cupples LA, Ordovas JM, Bloomfield HE, Robins SJ, Schaefer EJ. Polymorphisms in the gene encoding lipoprotein lipase in men with low HDL-C and coronary heart disease: the Veterans Affairs HDL Intervention Trial. J Lipid Res. 2004;45:1885–1891. doi: 10.1194/jlr.M400152-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Barter P, Gotto AM, LaRosa JC, Maroni J, Szarek M, Grundy SM, Kastelein JJ, Bittner V, Fruchart JC. Treating to New Targets Investigators. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357:1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- 9.Boekholdt SM, Thompson JF. Natural Genetic Variation as a Tool in Understanding the Role of CETP in Lipid Levels and Disease. J Lipid Res. 2003;44:1080–1093. doi: 10.1194/jlr.R200018-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Clarke AJ, Cooper DN. GWAS: heritability missing in action? Eur J Hum Genet. 2010;18:859–861. doi: 10.1038/ejhg.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 12.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 13.Evans DJ, Arzer J, Aberle J, Beil FU. Rare Variants in the Lipoprotein Lipase (LPL) Gene Are Common in Hypertriglyceridemia but Rare in Type III Hyperlipidemia. Atherosclerosis. 2011;214:386–390. doi: 10.1016/j.atherosclerosis.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Johansen CT, Wang J, McIntyre AD, et al. Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ Cardiovasc Genet. 2012;5:66–72. doi: 10.1161/CIRCGENETICS.111.960864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Cao H, Ban MR, Kennedy BA, Zhu S, Anand S, Yusuf S, Pollex RL, Hegele RA. Resequencing genomic DNA of patients with severe hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2007;27:2450–2455. doi: 10.1161/ATVBAHA.107.150680. [DOI] [PubMed] [Google Scholar]
- 16.Khovidhunkit W, Chartyingcharoen P, Siriwong S, Limumpornpetch P, Plengpanich W. Resequencing CETP, LIPC and LIPG genes in Thai subjects with hyperalphalipoproteinemia. Am J Cardiol. 2012;110:62–66. doi: 10.1016/j.amjcard.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 17.Hamman RF, Marshall JA, Baxter J, Kahn LB, Mayer EJ, Orleans M, Murphy JR, Lezotte DC. Methods and prevalence of non-insulin-dependent diabetes mellitus in a biethnic Colorado population. The San Luis Valley Diabetes Study. Am J Epidemiol. 1989;129:295–311. doi: 10.1093/oxfordjournals.aje.a115134. [DOI] [PubMed] [Google Scholar]
- 18.Rewers M, Shetterly SM, Hoag S, Baxter J, Marshall J, Hamman RF. Is the risk of coronary heart disease lower in Hispanics than in non-Hispanic whites? The San Luis Valley Diabetes Study. Ethnicity Dis. 1993;3:44–54. [PubMed] [Google Scholar]
- 19.Demirci FY, Dressen AS, Hamman RF, Bunker CH, Kammerer CM, Kamboh MI. Association of a Common G6PC2 Variant with Fasting Plasma Glucose Levels in Non-diabetic Individuals. Ann Nutr Metab. 2010;56:59–64. doi: 10.1159/000268019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunker CH, Ukoli FA, Matthews KA, Kriska AM, Huston SL, Kuller LH. Weight Threshold and Blood Pressure in a Lean Black Population. Hypertension. 1995;26:616–623. doi: 10.1161/01.hyp.26.4.616. [DOI] [PubMed] [Google Scholar]
- 21.Bunker CH, Ukoli FA, Okoro FI, Olomu AB, Kriska AM, Huston SL, Markovic N, Kuller LH. Correlates of Serum Lipids in a Lean Black Population. Atherosclerosis. 1996;123:215–225. doi: 10.1016/0021-9150(96)05810-8. [DOI] [PubMed] [Google Scholar]
- 22.Richmond W. Preparation and properties of a cholesterol oxidase from Nocardia wp. and its application to the enzymatic assay of total cholesterol in serum. Clin Chem. 1973;19:1350–1356. [PubMed] [Google Scholar]
- 23.Harris MR, Clareann HB, Richard FH, Dharambir KS, Christopher EA, Kamboh MI. Racial Differences in the Distribution of a Low Density Lipoprotein Receptor-related Protein (LRP) Polymorphism and Its Association with Serum Lipoprotein, Lipid and Apolipoprotein Levels. Atherosclerosis. 1998;137:187–195. doi: 10.1016/s0021-9150(97)00230-x. [DOI] [PubMed] [Google Scholar]
- 24.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 25.Willer CJ, Sanna S, Jackson AU, et al. Newly Identified Loci That Influence Lipid Concentrations and Risk of Coronary Artery Disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boes E, Coassin S, Kollerits B, Heid IM, Kronenberg F. Genetic-epidemiological Evidence on Genes Associated with HDL Cholesterol Levels: a Systematic In-depth Review. Exp Gerontol. 2009;44:136–160. doi: 10.1016/j.exger.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spirin V, Schmidt S, Pertsemlidis A, Cooper RS, Cohen JC, Sunyaev SR. Common Single-nucleotide Polymorphisms Act in Concert to Affect Plasma Levels of High-density Lipoprotein Cholesterol. Am J Hum Genet. 2007;81:1298–1303. doi: 10.1086/522497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teslovich TM, Musunuru K, Smith AV, et al. Biological, Clinical and Population Relevance of 95 Loci for Blood Lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edmondson AC, Braund PS, Stylianou IM, et al. Dense Genotyping of Candidate Gene Loci Identifies Variants Associated with High-density Lipoprotein Cholesterol. Circ Cardiovasc Genet. 2011;4:145–155. doi: 10.1161/CIRCGENETICS.110.957563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridker PM, Paré G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP Gene Region, HDL Cholesterol, and Risk of Future Myocardial Infarction: Genomewide Analysis Among 18 245 Initially Healthy Women from the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bansal A, van den Boom D, Kammerer S, Honisch C, Adam G, Cantor CR, Kleyn P, Braun A. Association testing by DNA pooling: an effective initial screen. Proc Natl Acad Sci U S A. 2002;99:16871–16874. doi: 10.1073/pnas.262671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 34.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2002;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of Functional Variation in Personal Genomes Using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chasman DI, Paré G, Zee RY, et al. Genetic Loci Associated with Plasma Concentration of Low-density Lipoprotein Cholesterol, High-density Lipoprotein Cholesterol, Triglycerides, Apolipoprotein A1, and Apolipoprotein B Among 6382 White Women in Genome-wide Analysis with Replication. Circ Cardiovasc Genet. 2008;1:21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoblauch H, Bauerfeind A, Toliat MR, Becker C, Luganskaja T, Günther UP, Rohde K, Schuster H, Junghans C, Luft FC, Nürnberg P, Reich JG. Haplotypes and SNPs in 13 Lipid-relevant Genes Explain Most of the Genetic Variance in High-density Lipoprotein and Low-density Lipoprotein Cholesterol. Hum Mol Genet. 2004;13:993–1004. doi: 10.1093/hmg/ddh119. [DOI] [PubMed] [Google Scholar]
- 41.I, Fridmanis D, Vaivade I, Nikitina-Zake L, Klovins J. The association of common SNPs and haplotypes in CETP gene with HDL cholesterol levels in Latvian population. PloS One. 2013;8:e64191. doi: 10.1371/journal.pone.0064191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson JF, Wood LS, Pickering EH, Dechairo B, Hyde CL. High-density genotyping and functional SNP localization in the CETP gene. J Lipid Res. 2007;48:434–443. doi: 10.1194/jlr.M600372-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Klerkx AH, Tanck MW, Kastelein JJ. Haplotype analysis of the CETP gene: not TaqIB, but the closely linked -629C->A polymorphism and a novel promoter variant are independently associated with CETP concentration. Hum Mol Genet. 2003;12:111–123. doi: 10.1093/hmg/ddg013. [DOI] [PubMed] [Google Scholar]
- 44.Ordovas JM, Cupples LA, Corella D, Otvos JD, Osgood D, Martinez A, Lahoz C, Coltell O, Wilson PW, Schaefer EJ. Association of cholesteryl ester transfer protein-TaqIB polymorphism with variations in lipoprotein subclasses and coronary heart disease risk : the Framingham study. Arterioscler Thromb Vasc Biol. 2000;20:1323–1329. doi: 10.1161/01.atv.20.5.1323. [DOI] [PubMed] [Google Scholar]
- 45.Dachet C, Poirier O, Cambien F, Chapman J, Rouis M. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20:507–515. doi: 10.1161/01.atv.20.2.507. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Bai H, Liu R, Liu Y, Liu BW. Analysis of cholesterol ester transfer protein gene TaqIB and 629 C/A polymorphisms in patients with endogenous hypertriglyceridemia in Chinese population. Zhonghua Yi Xue Yi Chuan XueZaZhi. 2006;23:640–646. [PubMed] [Google Scholar]
- 47.Lu Y, Tayebi N, Li H, Saha N, Yang H, Heng CK. Association of CETP Taq1B and -629C > A Polymorphisms with Coronary Artery Disease and Lipid Levels in the Multi-ethnic Singaporean Population. Lipids Health Dis. 2013;12:85. doi: 10.1186/1476-511X-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spirin V, Schmidt S, Pertsemlidis A, Cooper RS, Cohen JC, Sunyaev SR. Common single-nucleotide polymorphisms act in concert to affect plasma levels of high-density lipoprotein cholesterol. Am J Hum Genet. 2007;81:1298–1303. doi: 10.1086/522497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boes E, Coassin S, Kollerits B, Heid IM, Kronenberg F. Genetic-epidemiological Evidence on Genes Associated with HDL Cholesterol Levels: a Systematic In-depth Review. Exp Gerontol. 2009;44:136–160. doi: 10.1016/j.exger.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heid IM, Boes E, Müller M, et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ Cardiovasc Genet. 2008;1:10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- 51.Coram MA, Duan Q, Hoffmann TJ, et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am J Hum Genet. 2013;92:904–916. doi: 10.1016/j.ajhg.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weissglas-Volkov D, Pajukanta P. Genetic causes of high and low serum HDL-cholesterol. J Lipid Res. 2010;51:2032–2057. doi: 10.1194/jlr.R004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds CA, Gatz M, Pedersen NL, Prince JA. An assessment of CETP sequence variation in relation to cognitive decline and dementia risk. Int J Mol Epidemiol Genet. 2011;2:122–129. [PMC free article] [PubMed] [Google Scholar]
- 54.Lettre G, Palmer CD, Young T, et al. Genome-wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project. PLoS Genet. 7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridker PM, Paré G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterworth DM, Ricketts SL, Song K, et al. Genetic Variants Influencing Circulating Lipid Levels and Risk of Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edmondson AC, Braund PS, Stylianou IM, et al. Dense Genotyping of Candidate Gene Loci Identifies Variants Associated with High-density Lipoprotein Cholesterol. Circ Cardiovasc Genet. 2011;4:145–155. doi: 10.1161/CIRCGENETICS.110.957563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Razzaghi H, Aston CE, Hamman RF, Kamboh MI. Genetic screening of the lipoprotein lipase gene for mutations associated with high triglyceride/low HDL-cholesterol levels. Hum Genet. 2000;107:257–267. doi: 10.1007/s004390000367. [DOI] [PubMed] [Google Scholar]
- 59.Surakka I, Horikoshi M, Mägi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–97. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.