Abstract
KDM5 family proteins are critically important transcriptional regulators whose physiological functions in the context of a whole animal remain largely unknown. Using genome-wide gene expression and binding analyses in Drosophila adults, we demonstrate that KDM5 (Lid) is a direct regulator of genes required for mitochondrial structure and function. Significantly, this occurs independently of KDM5’s well-described JmjC domain-encoded histone demethylase activity. Instead, it requires the PHD motif of KDM5 that binds to histone H3 that is di- or trimethylated on lysine 4 (H3K4me2/3). Genome-wide, KDM5 binding overlaps with the active chromatin mark H3K4me3, and a fly strain specifically lacking H3K4me2/3 binding shows defective KDM5 promoter recruitment and gene activation. KDM5 therefore plays a central role in regulating mitochondrial function by utilizing its ability to recognize specific chromatin contexts. Importantly, KDM5-mediated regulation of mitochondrial activity is likely to play key roles in human diseases caused by dysfunction of this family of proteins.
Keywords: KDM5, Lid, transcription, PHD motif, histone, H3K4me3, mitochondria
Graphical Abstract

INTRODUCTION
Regulation of gene expression is essential for cell fate specification, developmental processes and to maintain cellular homeostasis. KDM5 family proteins are important transcriptional regulators that activate or repress gene expression in a context-dependent manner. Mammalian cells encode four KDM5 paralogs, KDM5A (Rbp2, JARID1A), KDM5B (Plu1, JARID1B), KDM5C (SMCX, JARID1C) and KDM5D (SMCY, JARID1D) while Drosophila has a single KDM5 ortholog (Lid). Evidence that KDM5 family proteins play key gene-regulatory roles comes from the observations that knockout of mouse KDM5B or Drosophila KDM5 results in lethality (Albert et al., 2013; Catchpole et al., 2011; Gildea et al., 2000). Moreover, dysregulation of KDM5 proteins in humans results in disease, with overexpression of KDM5 proteins being implicated in oncogenesis and their loss in cognitive impairment (Blair et al., 2011; Vallianatos and Iwase, 2015).
The most studied activity of KDM5 proteins is their Jumonji (JmjC) domain-encoded histone demethylase activity (Benevolenskaya, 2007a). In mammalian cells and Drosophila, KDM5 proteins demethylate trimethylated histone H3 (H3K4me3), a chromatin mark characteristic of promoter regions of actively transcribed genes (Santos-Rosa et al., 2002). To-date, most loss and gain of function KDM5 phenotypes described have been attributed exclusively to their demethylase activity. However, studies of mouse KDM5B and Drosophila KDM5 demonstrate that demethylase inactive animals develop normally to produce viable adults that are morphologically normal (Catchpole et al., 2011; Li et al., 2010). Consistent with this, we and others have shown demethylase-independent gene regulatory functions of KDM5 proteins (Benevolenskaya, 2007a; Cao et al., 2014; DiTacchio et al., 2011; Lee et al., 2009). While this leaves the in vivo significance of KDM5’s enzymatic activity uncertain, it emphasizes the functional importance of KDM5’s other gene regulatory activities. For example, KDM5 family proteins affect transcription through interactions with lysine deacetylases (HDACs), leading to changes in the acetylation of histones and other proteins (Barrett et al., 2007; DiTacchio et al., 2011; Lee et al., 2009; Liu et al., 2014; Nishibuchi et al., 2014). Importantly, because KDM5 proteins have additional motifs with defined in vitro functions, additional gene-regulatory mechanisms are likely to be crucial for KDM5 function in vivo. These include N- and C-terminal PHD motifs that bind to H3K4me0 and H3K4me2/3, respectively (Klein et al., 2014; Li et al., 2010; Torres et al., 2015; Wang et al., 2009). The binding activities of these two PHD motifs are particularly intriguing as they are the substrate and product of KDM5’s demethylase activity, and it remains unclear whether they act independently or in a coordinated manner to affect transcription.
Dysfunction of transcriptional regulators is well established to lead to a large number of diseases. Indeed, loss of function mutations in KDM5A, KDM5B or KDM5C are found in patients with intellectual disability, linking KDM5 function to cognition through unknown mechanism(s) (Vallianatos and Iwase, 2015). In addition, overexpression of KDM5A or KDM5B are implicated in the genesis and progression of several cancers, most notably melanoma, breast, gastric, lung and prostate cancers (reviewed by (Blair et al., 2011)). While the precise role of KDM5A and KDM5B in tumor development remains to be elucidated, their interaction with key tumorigenic factors such as the oncoprotein transcription factor Myc and the tumor suppressor pRB are likely to be important (Benevolenskaya, 2007b; Outchkourov et al., 2013; Secombe et al., 2007). Interestingly, recent data support the idea that KDM5 proteins promote the growth and survival of a more slowly proliferating subset of cancer cells (Roesch et al., 2010; Roesch et al., 2013; Sharma et al., 2010). Because standard therapies target rapidly dividing cells, this results in KDM5A and KDM5B overexpressing tumors being difficult to treat. Slow-growing KDM5 overexpressing tumor cells are also metabolically distinct from other tumor cells because they generate ATP through oxidative phosphorylation in the mitochondria and not via aerobic glycolysis (Roesch et al., 2013; Song et al., 2015). KDM5 proteins may therefore regulate fundamental metabolic processes related to energy production through unknown molecular mechanisms.
Key to understanding the biology of KDM5 proteins is defining their target genes and the mechanisms by which they regulate transcription. By combining genome-wide transcriptome and binding assays, we establish the repertoire of direct KDM5 target genes in Drosophila adults. Our analyses define KDM5 as a critical regulator of genes integral to mitochondrial structure and function. Significantly, the activation of critical mitochondrial genes does not require the JmjC domain-encoded demethylase activity of KDM5. Instead it relies on its C-terminal PHD motif that binds to di- and trimethylated H3K4. Recognition of a specific chromatin context is therefore a critical means by which KDM5 activates the transcription of genes essential to mitochondrial function in vivo. Because altered mitochondrial activity is a feature of cancer cells and is also implicated in cognitive dysfunction, our data provide key insights into the means by which loss or gain of KDM5 family genes leads to human disease.
RESULTS
KDM5 regulates distinct genes in larvae and adults
We previously demonstrated that larvae and adults with a hypomorphic combination of kdm5 alleles (kdm5K6801/10424) have 70% less KDM5 protein than wildtype and die more quickly than controls when exposed to the oxidizer paraquat (Liu et al., 2014). In kdm5 mutant larvae, paraquat sensitivity correlates with the downregulation of at least 15 target genes that are co-regulated by KDM5 and the stress response transcription factor Foxo (Liu et al., 2014). In contrast to our findings in larvae, analyses of those same 15 target genes in kdm5 mutant adults revealed that only six were downregulated, while six others were unaffected and three were upregulated (Figure S1A). KDM5 therefore regulates distinct targets during larval development and adulthood.
To identify KDM5-regulated genes in adults we carried out RNA-seq analyses of kdm5K6801/10424 mutant flies and compared this to age-matched wildtype animals. (Figure S1B, C; Table S1). In unstressed conditions, kdm5 mutant flies had 8056 differentially expressed genes (DEGs). 4787 genes were significantly downregulated and thus require KDM5 for their activation while 3269 genes were upregulated so are normally repressed by KDM5 (p<0.05, FDR <0.05; Figure S1D, E). Similar to our previous microarray data from larval wing imaginal discs, many genes affected in kdm5 hypomorphic mutant adults were altered 2-fold or less (Figure S2A)(Liu et al., 2014). Analyses of KDM5-regulated genes (both up and downregulated) revealed enrichment for a number of diverse biological processes (GO categories), including glycosylation, lipid metabolism and cell division (Figure 1A). Differentially expressed genes were also implicated in the functioning of several subcellular compartments, including the mitochondria, ribosomes and lipid particles (Figure 1A).
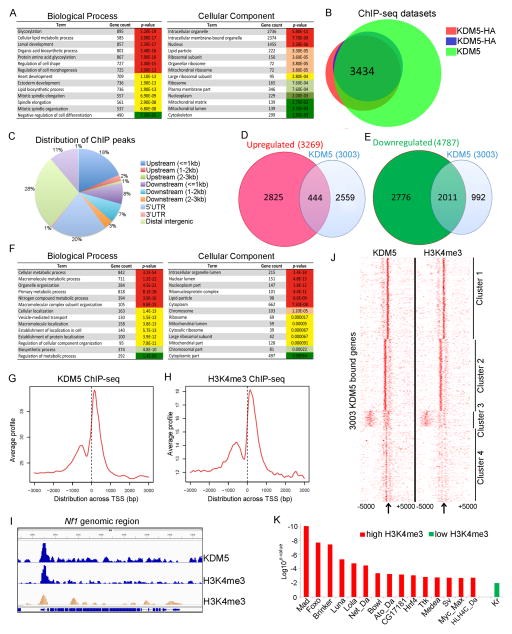
Figure 1. Genes directly regulated by KDM5 in adults.
(A) Analyses of RNA-seq data to identify enriched Biological Process and Cellular Component categories for genes with altered expression in kdm5 mutant flies (up and downregulated genes). (B) Overlap between anti-HA (two replicates) and KDM5 ChIP-seq datasets (q<0.05). (C) Genomic distribution of KDM5 ChIP-seq peaks using anti-HA. (D) Comparison of RNA-seq and ChIP-seq data showing directly repressed genes. (E) Directly activated genes. (F) Gene ontology analysis of directly regulated genes. (G) KDM5 ChIP-seq read density relative to the transcriptional start site (TSS) at differentially expressed genes. (H) H3K4me3 ChIP-seq read density relative to the TSS of KDM5 bound and regulated genes. (I) IGV genome browser view of Nf1 showing overlapping KDM5 and H3K4me3 peaks. Middle (blue) H3K4me3 trace is data from our analyses. Bottom (orange) H3K4me3 is published data (GSM400670). (J) Heat map showing distribution of KDM5 and H3K4me3 relative to TSS (clustered using k-means). (K) Transcription factor binding sites enriched within KDM5 peaks using MEME-ChIP. Abbreviations are Mad (Mothers against decapentaplegic), Lola (Longitudinals lacking), Da (Daughterless), Bowl (brother of odd with entrails limited), Ato (Atonal), Hnf4 (Hepatocyte nuclear factor 4), Ttk (Tramtrack), Kr (Krüppel).
Because kdm5 mutants are sensitive to the oxidizer paraquat we tested whether KDM5 was required for the transcriptional response to this stressor. To do this, we carried out RNA-seq analyses of wildtype and kdm5 mutant adults in oxidative stress conditions. Paraquat treatment of wildtype flies led to the upregulation of 2481, and downregulation of 3103, genes (≥1.5 fold, p≤0.05; Figure S2B, C). These included genes known to be activated by paraquat that are required to detoxify excess reactive oxygen species such as glutathione S transferase and cytochrome P450 genes (Zou et al., 2000). Paraquat-mediated changes to gene expression did not generally require KDM5 (Figure S2D–F). kdm5 mutant adult sensitivity to oxidative stress conditions is therefore due to defects that exist prior to stress conditions. We therefore focused on gene expression changes and phenotypes observed in kdm5 mutants in non-stressed conditions.
KDM5 binding correlates with the active chromatin mark H3K4me3
To determine which genes affected in kdm5 hypomorphic mutant adults were direct targets we carried out ChIP-seq analyses of wildtype adults using an anti-KDM5 antibody. To independently confirm these data we utilized a fly strain in which the sole source of KDM5 is from a transgene expressing an HA tagged form of KDM5 expressed under the control of its endogenous promoter (Liu et al., 2014). Analyses of two anti-HA and one KDM5 ChIP-seq datasets revealed a total of 3434 binding sites that were enriched at promoter regions (FDR<0.05; Figure 1B, C; Figure S3A–C). These 3434 KDM5 binding peaks mapped to 3003 genes that were used for subsequent integration with RNA-seq analyses from kdm5 mutant adults.
15% of KDM5-bound genes were upregulated in kdm5 mutant adults (444/3003; Figure 1D). A much higher proportion of KDM5-bound genes were downregulated (2011/3003, 67%), thus the primary function of KDM5 in adults is to activate gene expression (Figure 1E). Gene ontology analysis of these 2455 directly regulated KDM5 target genes revealed enrichment for diverse metabolic processes and cellular compartments, including those required for mitochondrial function (Figure 1F). The remaining 548 KDM5 binding sites (18%) occurred at genes whose expression was unaltered in kdm5 mutants. The function of KDM5 at these sites remains to be determined. Notably, consistent with our observation that paraquat-mediated changes to gene expression did not require KDM5, KDM5 binding was not altered by this stress condition (Figure S3D). These data confirm that the primary role of KDM5 is to regulate gene expression in normal, unstressed conditions.
The JmjC domain of KDM5 demethylates H3K4me3 and the C-terminal PHD motif (PHD3) binds to H3K4me2/3 (Li et al., 2010; Wang et al., 2009). We therefore investigated the link between KDM5 and H3K4me3 by carrying out anti-H3K4me3 ChIP-seq from wildtype adults and also utilizing a published modENCODE dataset (Negre et al., 2011). KDM5 and H3K4me3 showed a similar distribution, peaking near transcription start sites (TSS)(Figure 1G, H, I; Figure S3E). Additional genome-wide analyses of ChIP-seq peak data revealed four clusters with distinct patterns and degrees of enrichment for KDM5 binding and H3K4me3 (Figure 1J; Figure S3F). Genes within clusters 1 and 2 showed higher levels of H3K4me3 and KDM5 binding primarily surrounding the TSS and were more highly expressed. Genes in clusters 3 and 4 had lower levels of H3K4me3 and KDM5 binding and were expressed at low levels. At these genes, KDM5 and H3K4me3 were more dispersed and not limited to the TSS region. We also compared our KDM5 ChIP-seq data with three other published datasets that were generated using adult females: H3K9 acetylation (H3K9ac) that marks active promoters in addition to H3K4me1 and H3K27ac that are associated with enhancer elements. As expected based on the overlap between KDM5 and H3K4me3, KDM5 binding correlated with H3K9 acetylation (Figure S4A). In contrast, KDM5 binding was not significantly correlated with H3K27ac or H3K4me1 (Figure S4B–F). KDM5 therefore primarily localizes to promoters.
In addition to identifying additional KDM5 target genes, our data support the biological importance of previously published KDM5 interactions with transcription-regulatory complexes. For example, MEME-ChIP (Machanick and Bailey, 2011) analyses of KDM5 ChIP peaks with high levels of H3K4me3 showed enrichment for Foxo and Myc binding sites that we and others have previously linked to KDM5 (Figure 1K) (Liu et al., 2014; Outchkourov et al., 2013; Secombe et al., 2007). Binding sites for a number of other transcription factors were also enriched within KDM5-bound regions, including those that mediate TGFβ/BMP signaling (e.g. Mad, Brinker, Medea) and neuronal development (e.g. Lola, Daughterless, Tramtrack). In contrast to KDM5 peaks that had high H3K4me3, KDM5 peaks with lower levels of this chromatin mark showed enrichment only for only one transcription factor binding site, the developmental regulator Kruppel (Figure 1K). KDM5 is therefore likely to be important for the regulation of several developmentally important pathways.
KDM5 directly regulates genes required for mitochondrial function
Our ChIP-seq and RNA-seq analyses revealed that KDM5 regulates genes involved in mitochondrial structure and function (Figure 2A). For example, among the direct targets that were downregulated in kdm5 mutants were genes required for mitochondrial morphology (deep orange (dor), twins (tws), Neurofibromin 1 (Nf1) and bicoid interacting protein 3 (bin3)), components of the inner or outer mitochondrial membrane (no mitochondrial derivative (nmd), Pmi and black pearl (blp)), the mitochondrial chaperone tumorous imaginal discs (l(2)tid), and genes essential to mitochondrial tRNA function (the Seryl tRNA synthase Slimp and the Leucyl-tRNA synthase Aats-leu). Other genes integral to mitochondrial function were bound by KDM5 but did not show changes to transcript levels in kdm5 mutants (e.g. mitochondrial assembly factor (Marf) and fission1 (Fis1)) (Figure 2A). These may be regulated in a more tissue-restricted manner, so changes to their expression were not detectable using whole animals.
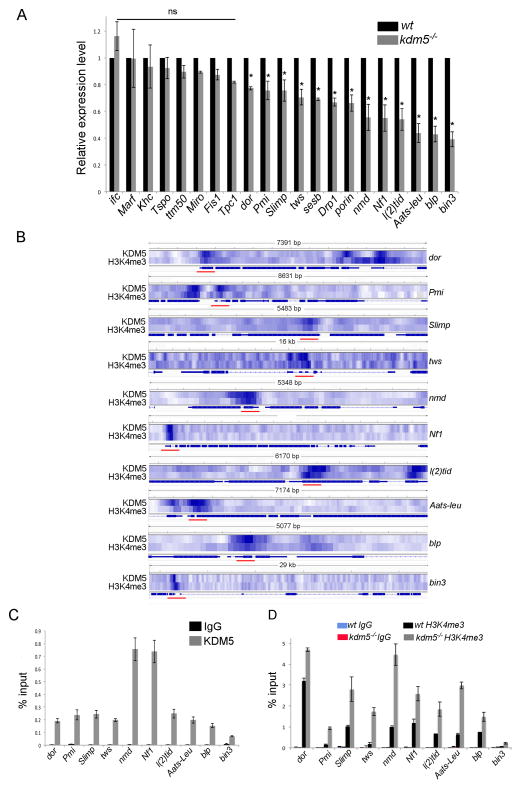
Figure 2. KDM5 directly regulates genes required for mitochondrial function.
(A) Real-time PCR of genes indicated in kdm5K6801/10424 adults shown relative to wildtype (w1118). Genes were normalized to rp49 and shown as relative expression in kdm5 mutants. * p<0.05. (B) Heatmap showing overlapping KDM5 and H3K4me3 peaks at target genes. Red bars show regions used for ChIP-PCR validation shown in (C) and (D). (C) ChIP-PCR using anti-HA (KDM5) and IgG control showing KDM5 enrichment at targets. (D) ChIP-PCR using anti-H3K4me3 or control IgG from wildtype (w1118) and kdm5K6801/10424 mutant adults.
Because of their established roles in mitochondrial function, we focused on KDM5-dependent activation of dor, Pmi, Slimp, tws, nmd, Nf1, l(2)tid, Aats-leu, blp and bin3, all of which showed KDM5 binding and the presence of H3K4me3 near the TSS (Figure 2B). To verify these ChIP-seq data, we confirmed KDM5 and H3K4me3 enrichment by ChIP-PCR (Figure 2C, D). We also examined H3K4me3 levels in kdm5 mutant flies and found that despite the decreased expression of these KDM5 target genes, promoter H3K4me3 levels were increased (Figure 2D). This is consistent with the histone demethylase activity of KDM5 and the observation by us and others that global levels of H3K4me3 are increased in kdm5 mutants (Eissenberg et al., 2007; Lee et al., 2007; Secombe and Eisenman, 2007). It is also consistent with the notion that while there is a correlation between H3K4me3 and increased transcription, there is not a causal relationship (Sims and Reinberg, 2006).
kdm5 mutants show mitochondrial defects, altered metabolism and increased oxidative stress
Because KDM5 directly regulates genes integral to mitochondrial structure and function, we examined kdm5 mutants for phenotypes in the highly metabolically active thoracic muscles. Mitochondria from wildtype animals were round and evenly distributed between the myofibrils (Figure 3A, B). In contrast, kdm5 mutant flies had irregularly shaped mitochondria that were larger than wildtype while other cellular structures appeared normal (Figure 3C, D). Consistent with the mitochondrial defects of kdm5 mutants interfering with the function of this organelle, total levels of ATP were decreased ~50% (Figure 3E). Interestingly, kdm5 mutants did not show altered levels of lactic acid, suggesting that they do not (or are unable to) generate ATP through anaerobic glycolysis to compensate for reduced mitochondrial ATP output (Figure 3F). The ability to climb (negative geotaxis) is often affected in flies with mitochondrial or ATP production defects (Fergestad et al., 2008). Consistent with this correlation, kdm5 mutant flies were significantly slower at climbing (Figure 3G).
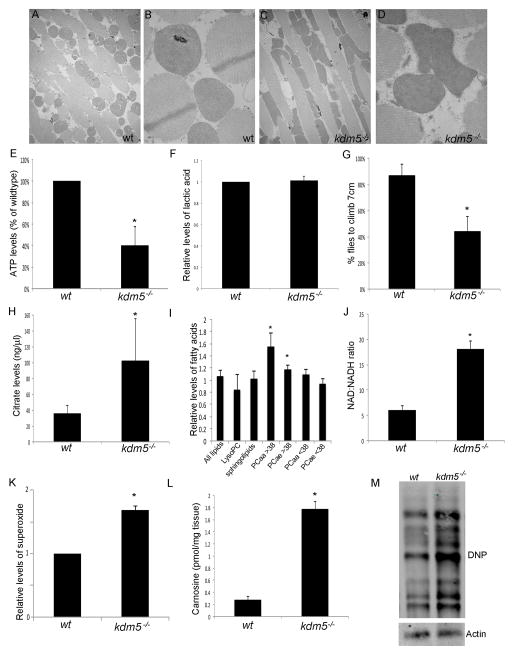
Figure 3. kdm5 mutant adults have mitochondrial and metabolic defects.
(A, B) TEM images showing 5K and 10K magnifications of mitochondria within thoracic muscles from wildtype (w1118) flies. (C, D) TEM images from kdm5K6801/10424 mutant flies. (E) ATP levels in kdm5K6801/10424 mutant flies shown as percentage of wildtype and normalized to the total protein levels. * p<0.01. (F) Lactic acid levels in kdm5K6801/10424 adults relative to w1118. (G) Quantitation the number of wildtype (w1118) or kdm5K6801/10424 flies that climb 7cm in 20 seconds after being tapped to the bottom of the vial. * p<0.001. (H) Citrate levels (ng/μl) in wildtype and kdm5K6801/10424 mutant flies. * p<0.01. (I) Levels of distinct lipid subtypes in kdm5K6801/10424 mutant flies relative to wildtype. LysoPC is lysophosphatidylcholine acyls (fatty acids generated as intermediate products), PCaa is phosphatidylcholine acyl molecules and PCae is phosphatidylcholine acyl-alkyl molecules. Numbers (<38 and >38) indicate number of carbon atoms in fatty acid chain. * p<0.01. (J) Ratio of NAD+ to NADH in wildtype (w1118) and kdm5K6801/10424 mutant flies. * p<0.001. (K) Levels of superoxide as assayed by MitoSOX in kdm5K6801/10424 mutant flies relative to wildtype. * p<0.05. (L) Levels of the antioxidant carnosine (pmol/mg tissue) in wildtype and kdm5K6801/10424 mutant flies. * p≪0.001. (M) Detection of oxidized proteins in wildtype and kdm5K680/104241 mutant fly heads using oxyblot (2-fold increase).
One key metabolic pathway that occurs in the mitochondria is the tricarboxylic acid (TCA) cycle that serves as a hub to integrate carbohydrate, fat and protein metabolism. To test if the TCA cycle was affected in kdm5 mutants we quantitated levels of citrate and found it was 2.5-fold higher than wildtype animals (Figure 3H). We also observed that several TCA cycle enzymes were upregulated in kdm5 mutants, but these changes to gene expression were mostly indirect (Table S2). In addition to being a TCA cycle intermediate, citrate is also a precursor for de novo synthesis of fatty acids that are necessary for membrane integrity and signaling. Examining the levels of 87 types of glycerophospholipids in kdm5 mutants revealed that levels of long chain glycerophospholipids were significantly increased (>38 carbons; Figure 3I). Shorter chain glycerophospholipids (<38 carbons), lysophosphatidylcholine lipids and sphingolipids were unaffected, suggesting that only a subset of lipids were altered in kdm5 mutants (Figure 3I). Other metabolic pathways that feed into or out of the TCA cycle, such as levels of amino acids and biogenic amines were also altered in kdm5 mutants (Table S3).
An important function of the TCA cycle is to convert NAD+ to NADH, which then serves as an electron donor in ATP production generated through the electron transport chain. A breakdown in the TCA cycle would be expected to decrease the conversion of NAD+ to NADH and alter the redox state of the cell. Consistent with this expectation, kdm5 mutants had a 3-fold increase in the ratio of NAD to NADH (Figure 3J). Through a number of different mechanisms, defective mitochondria can also lead to increased levels of potentially toxic reactive oxygen species (Cui et al., 2012). kdm5 mutants had superoxide levels that were 1.6-fold higher than wildtype, suggesting that they experience increased oxidative stress in the absence of any exogenous stress source (Figure 3K). Consistent with this, the antioxidant stress indicator carnosine was increased 8-fold and global levels of oxidized proteins 2-fold (Figure 3L, M). kdm5 mutants therefore have physical mitochondrial abnormalities and significant disruptions to metabolic processes related to energy production.
KDM5 activation of mitochondrial function genes is dependent on its C-terminal PHD motif and independent of its JmjC domain histone demethylase activity
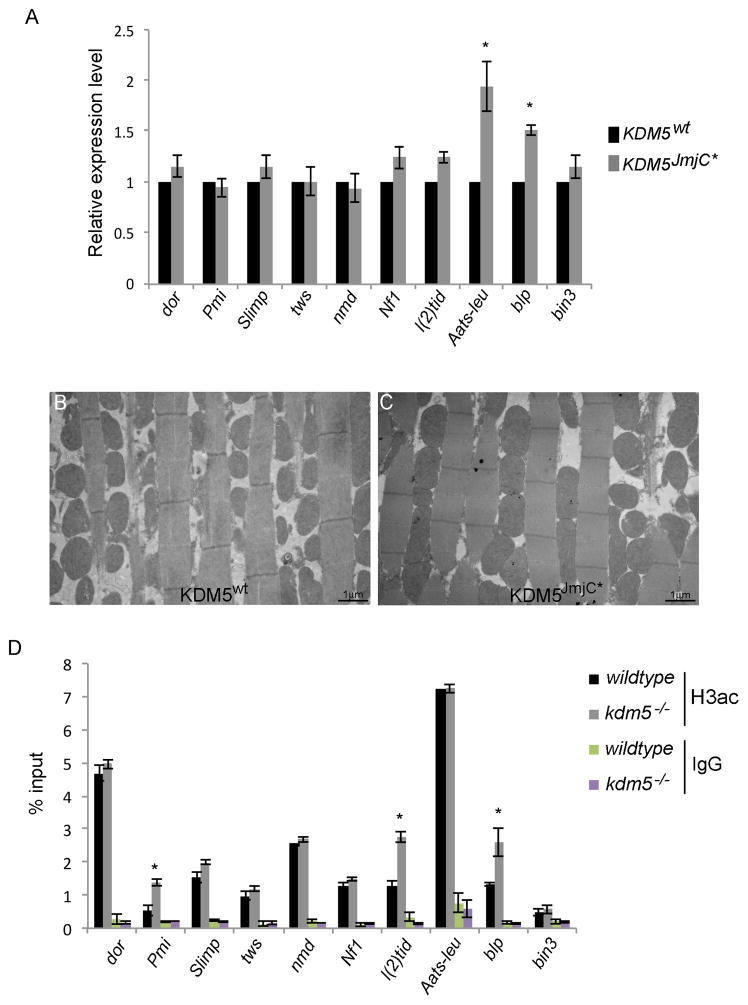
To test the extent to which the demethylase activity was required for the gene expression changes observed we examined flies specifically lacking KDM5-dependent enzymatic activity (Li et al., 2010). Of the 10 genes related to mitochondrial function tested, eight were unaffected in demethylase inactive flies, while the remaining two genes were slightly upregulated (Figure 4A). The histone demethylase activity of KDM5 is therefore not required for the activation of these target genes. Consistent with this, we do not observe mitochondrial abnormalities in the thoracic muscles of demethylase inactive flies (Figure 4B, C).
Figure 4. KDM5-mediated regulation of mitochondrial function genes is independent of its demethylase activity.
(A) Real-time PCR analyses of the genes indicated in demethylase inactive adults (described in (Li et al., 2010); kdm5JmjC*) relative to wildtype (w1118). Each gene was normalized to rp49 and shown as relative expression in kdm5JmjC mutants. * p<0.05. (B, C) TEM images showing 5K magnifications of mitochondria within thoracic muscles from kdm5K6801 mutant flies rescued by a wildtype KDM5 transgene (KDM5WT; B) and kdm5JmjC mutant flies (C). (D) ChIP-PCR showing levels of histone H3 acetylation using a pan-acetyl antibody in wildtype and kdm5K6801/10424 flies. IgG is a negative control. ChIP-PCR fragments are indicated in Figure 2B. * p<0.05.
One means by which KDM5 can activate transcription is by increasing promoter histone H3 acetylation by inhibiting HDACs (DiTacchio et al., 2011; Lee et al., 2009). To test whether this mechanism plays a role in the activation of mitochondrial function genes, we tested whether levels of promoter H3 acetylation were decreased in kdm5 mutant flies. Seven of the 10 genes examined had no change in promoter histone H3 acetylation and the remaining three showed a slight increase (Figure 4D). KDM5-mediated activation of these genes therefore does not occur through changes to histone acetylation.
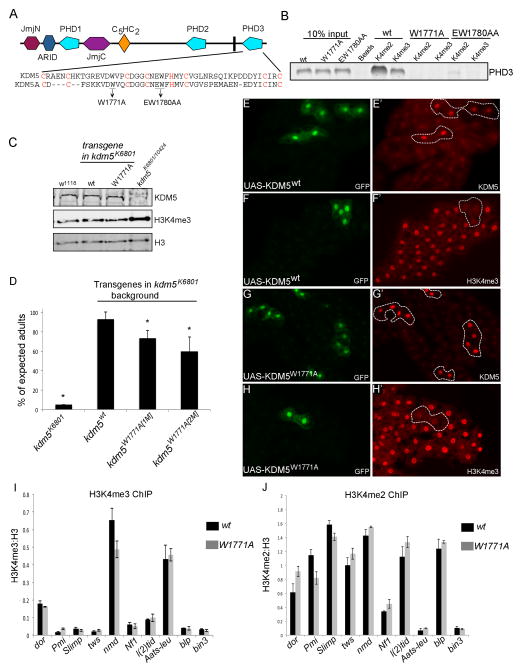
Because KDM5 binding correlated with H3K4me3, a chromatin mark bound by the C-terminal PHD3 motif of KDM5, we next tested the functional significance of this activity. To abolish H3K4me2/3 binding we made two different mutations in PHD3 based on structural data from human KDM5A (Wang et al., 2009) and tested their ability to bind histone peptides in vitro (Figure 5A). Mutating tryptophan 1771 to alanine (W1771A) eliminated PHD3 binding to both di- and trimethylated H3K4 (Figure 5B). Altering glutamate and tryptophan at amino acid positions 1780 and 1781 to alanine (EW1780A) reduced binding to H3K4me2 and H3K4me3, but not as dramatically as W1771A. We therefore generated flies endogenously expressing full length KDM5W1771A using a genomic rescue transgene approach that we used previously (Li et al., 2010). The only source of KDM5 in kdm5W1771A flies harbors the PHD3 mutation and is expressed at physiological levels using its endogenous promoter (Figure 5C). Flies harboring a wildtype version of the KDM5 genomic transgene served as a control for these analyses.
Figure 5. kdm5 mutant flies unable to bind H3K4me2/3 retain demethylase activity.
(A) Schematic of the PHD3 region of KDM5. Amino acids required for KDM5A binding to H3K4me2/3 are underlined and for zinc coordination in red (Wang et al., 2009). (B) in vitro binding between biotinylated histone H3 peptides di- or trimethylated at lysine 4 (H3K4me2 and H3K4me3) and GST-PHD3. The interaction between KDM5 and H3K4me2 and H3K4me3 is abolished by GST-PHD3W1771A and reduced by GST-PHD3EW1780AA. (C) Western from adult heads showing levels of KDM5, H3K4me3 and histone H3. H3 and H3K4me3 are two different channels from the same Western blot. The H3K4me3:H3 ratio is increased in kdm5K6801/10424 flies (1.6+/−0.3-fold). (D) Number of kdm5K6801 homozygous flies observed without a genomic rescue transgene (kdm5K6801), with a wildtype rescue transgene (kdm5wt) and two independent insertions of a kdm5W1771A transgene (1M and 5M). * p<0.01. (E–H) Clones of fat body cells marked by the presence of GFP that express wildtype KDM5 or KDM5W1771A. hs-FLP; UAS-KDM5 (or UAS-KDM5W1771A) females were crossed to actin>CD2>Gal4, UAS-GFP males. Transgene-expressing cells are shown by the co-expression of GFP (E, F, G, H). Anti-KDM5 is shown in E′ and G′ and anti-H3K4me3 in F′ and H′. (I) ChIP-PCR analyses of H3K4me3 levels at KDM5 target genes in wildtype flies (kdm5K6801 mutant flies rescued by a wildtype genomic rescue construct (wt)) and kdm5W1771A mutant flies. No significant changes were observed. (J) ChIP-PCR analyses of H3K4me2 levels in wildtype and kdm5W1771A mutant flies.
kdm5W1771A homozygous flies eclosed at a slightly reduced Mendelian ratio, but surviving adults appeared morphologically normal (Figure 5D). Interestingly, in contrast to kdm5 mutants that show increased global levels of H3K4me3 due to the loss of its demethylase activity, kdm5W1771A animals did not have altered global levels of H3K4me3 (Figure 5C). Because PHD3 binds to the target substrate of KDM5’s demethylase activity, we took two additional approaches to examine whether the PHD3 motif influenced the enzymatic activity of KDM5. Firstly, we overexpressed KDM5W1771A in fat body cells using the UAS/Gal4 system and found that was able to demethylate H3K4me3 in a similar manner to wildtype KDM5 (Figure 5E–H). Secondly, we examined promoter proximal H3K4me3 by ChIP in kdm5W1771A mutant flies and found that in contrast to loss of KDM5 that results in increased H3K4me3, mutating the PHD3 motif had no effect on promoter H3K4me3 (Figure 5I). H3K4me2, which is also bound by PHD3, is also unaltered in kdm5W1771A mutant flies (Figure 5J). The PHD3 motif of KDM5 therefore acts independently of the JmjC domain-encoded demethylase activity.
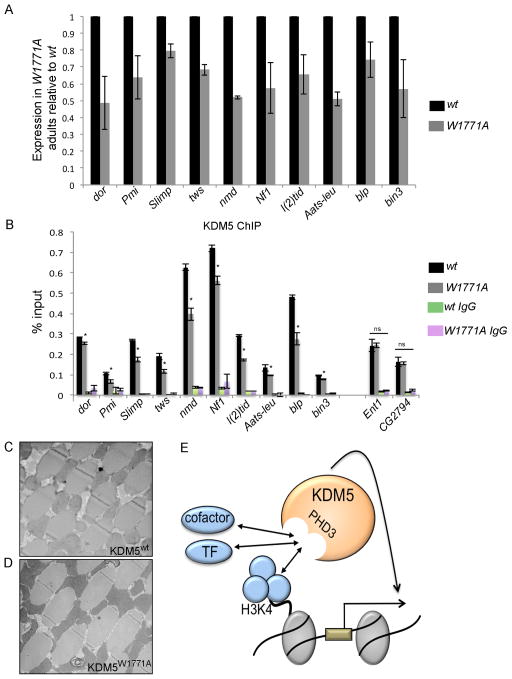
To determine the extent to which H3K4me3 binding was required for transcriptional regulation by KDM5, we tested target gene expression levels in kdm5W1771A flies. All 10 mitochondrial-related genes examined were downregulated in kdm5W1771A flies (Figure 6A). KDM5 promoter recruitment was also significantly attenuated at dor, Pmi, Slimp, tws, nmd, Nf1, l(2)tid, Aats-leu, blp and bin3 (Figure 6B). To test if H3K4me3 recognition contributed to KDM5 recruitment at a target gene not related to mitochondrial activity we examined binding to the promoter of the gene Equilibrative nucleoside transporter 1 (Ent1). Interestingly, KDM5 binding to this gene was unaltered in kdm5W1771A mutant flies (Figure 6B). We also tested KDM5 binding to the promoter of the putative kinase CG2794, which has lower H3K4me3 levels, and found it to be unaffected in kdm5W1771A mutants (Figure 6B). Consistent with the gene expression changes observed, kdm5W1771A mutant flies had deformed mitochondria in their thoracic muscles (Figure 6C, D). H3K4me3 recognition by KDM5 therefore plays an important role in the regulation of gene expression, and abrogating this activity results in mitochondrial abnormalities.
Figure 6. The PHD3 motif of KDM5 is important for gene activation.
(A) Real-time PCR comparing mRNA levels of direct KDM5 genes in flies lacking H3K4me2/3 binding (kdm5W1771A; W1771A) and wildtype (kdm5 mutant flies rescued by a wildtype transgene). All genes shown are significantly downregulated in kdm5W1771A flies (p<0.05). (B) anti-KDM5 ChIP-PCR analyses of binding at the target genes indicated in wildtype flies and kdm5W1771A mutant flies. * p<0.05. (C, D) TEM images showing 5K magnifications of mitochondria within thoracic muscles from wildtype flies (KDM5WT; C) and kdm5W1771A mutant flies (KDM5W1771A; D). (E) Model for KDM5 function. KDM5 recruitment to its target mitochondrial function genes is regulated in part through recognition of the chromatin mark H3K4me3, but PHD3-mediated interaction(s) with as yet unidentified transcription factor(s) (TF) and transcriptional co-factors are also likely to be crucial.
To determine whether the effect on KDM5 recruitment was specific to PHD3 or a general feature of PHD motifs, we examined the function of the N-terminal PHD motif (PHD1) that binds to histone H3 that is unmethylated at lysine 4 (H3K4me0)(Li et al., 2010; Torres et al., 2015). Similar to our analyses of PHD3, we generated flies expressing a mutant form of KDM5 unable to bind H3K4me0 at endogenous levels (kdm5W490A; Figure S5A–C). We also overexpressed KDM5W490A using a UAS transgene and found that in contrast to deleting the PHD1 motif (Li et al., 2010), specifically abrogating H3K4me0 binding did not affect the demethylase activity of KDM5 (Figure S5D–G). Importantly, ChIP analyses demonstrated that KDM5W90A was recruited to mitochondrial function genes in a similar manner to wildtype KDM5 (Figure S5H). The PHD1 and PHD3 motifs therefore have distinct functions and only PHD3 is involved in recruiting KDM5 to the promoters of mitochondrial function genes.
Because abrogating KDM5-dependent recognition of H3K4me2/3 attenuated but did not abolish promoter binding (Figure 6B), chromatin-recognition is not the sole means by which KDM5 is recruited to its targets. The PHD3 motif of KDM5 is therefore likely to interact with additional factors critical for transcriptional activation. One possibility for this is that the PHD3 of KDM5 interacts with transcription factor(s) necessary for gene activation. We therefore used MEME-ChIP to determine whether KDM5-regulated mitochondrial function genes were enriched for DNA motifs that could contribute to gene activation. Three DNA motifs were identified by these analyses, none of which have been associated with a known transcription factor (Figure S6A–D). All 10 mitochondrial function genes contained motif 3 (TGGAAA), seven contained motif 2 (GGAACANGGA) and six contained motif 1 (GGGGCAGA), and four promoters had all three motifs. Additionally, based on the observation that the H3K4me3-binding PHD motif of KMT2A also interacts with the transcriptional cofactor Cyp33 (Ali et al., 2014), the PHD3 of KDM5 may be required for the recruitment of cofactor(s) that are limiting for gene activation. These possibilities are summarized in Figure 6E.
DISCUSSION
Based on genome-wide transcriptome and ChIP data, we demonstrate a direct, critical role for KDM5 in the activation of genes required for the integrity and functioning of mitochondria. Focusing on 10 mitochondrial function genes, we found that KDM5-dependent target gene activation was independent of its well-established histone demethylase activity. Taken with previous reports from us and others (Catchpole et al., 2011; Li et al., 2010; Secombe et al., 2007), these data further support the view that KDM5 has essential gene regulatory functions that lie outside of its JmjC domain-encoded enzymatic activity. Consistent with non-demethylase functions playing important roles, we observed a striking correlation between KDM5 binding and the presence of the active chromatin mark H3K4me3. Further support for a significant connection between KDM5 binding and the presence of H3K4me3 comes from studies of mammalian KDM5 proteins (Kidder et al., 2014; Lopez-Bigas et al., 2008; Ram et al., 2011) and of Drosophila KDM5 in larval wing imaginal disc cells (Lloret-Llinares et al., 2012). While the target genes are distinct in different tissues and at different developmental stages, the binding of KDM5 proteins to H3K4me3-enriched regions of the genome appears to be highly conserved. Importantly, we extend these correlative data by specifically addressing the functional importance of PHD3-mediated recognition of H3K4me2/3 by KDM5 in vivo. Flies harboring a mutation in the PHD3 of KDM5 that abrogated H3K4me2/3 binding showed decreased binding to, and expression of, mitochondrial function genes. Moreover, this was specific to PHD3, as a mutation in the PHD1 motif of KDM5 that binds H3K4me0 did not alter KDM5 recruitment. These data demonstrate that the C-terminal PHD motif (PHD3) is a vital contributor to promoter recruitment and to gene activation by KDM5. Importantly, because both the PHD1 and PHD3 domains behaved independently of the JmjC demethylase domain, our data reinforce the notion that KDM5 proteins regulate transcription by distinct mechanisms that utilize the activity of different domains.
kdm5 mutant flies had physically abnormal mitochondria in addition to metabolic defects that led to decreased ATP production, altered lipid metabolism and increased levels of oxidative stress. These phenotypes were likely to be caused by the combined dysregulation of numerous KDM5 targets, as individual genes showed relatively mild (~2-fold) change to transcript levels. Significantly, we expect that our observed regulation of mitochondrial function genes is a conserved function of KDM5 proteins, as KDM5A knockdown in SAOS-2 cells that lack the KDM5 interacting protein pRB show a “long rod” mitochondrial phenotype (Lopez-Bigas et al., 2008; Varaljai et al., 2015). However, in contrast to our findings in flies, KDM5A knockdown cells did not show any change to the levels of reactive oxygen species, possibly due to compensation from other KDM5 family proteins. In the case of KDM5A, the only characterized direct target was Mitofusin 2 (Mnf2), whose expression was repressed by KDM5A (Lopez-Bigas et al., 2008). The Drosophila paralog of Mnf2, Marf (Mitochondrial assembly regulatory factor), was bound by KDM5 in our ChIP-seq data, but its expression was not altered in whole animals. KDM5 may therefore regulate Marf in a tissue specific manner in flies.
Our observation that KDM5 affects mitochondrial function is particularly relevant to our understanding of the link between overexpression of KDM5A or KDM5B and the genesis and progression of a number of different types of cancers (Blair et al., 2011). KDM5A and KDM5B overexpressing tumors are often resistant to traditional chemotherapy treatments (Blair et al., 2011). This effect has been characterized in some detail in KDM5B overexpressing melanomas where drug resistance is linked to the survival of a small population of slowly dividing cells (Roesch et al., 2010; Roesch et al., 2013). Proteomic analyses of KDM5B overexpressing cells revealed an increase in mitochondrial bioenergy that correlated with resistance to chemotherapy drugs (Roesch et al., 2013). However, no gene expression changes were reported to account for the link between KDM5B and changes to mitochondrial activity. Based on our data, we suggest that KDM5B may directly regulate genes required to increase mitochondrial activity, and that this is key to the survival of this subset of slow growing cells that can cause therapy resistance. We further propose that the effects of KDM5B on mitochondrial activity may be PHD3-dependent. In this regard it is interesting to note that the PHD3 motif has been previously linked to cancer, as this domain of KDM5A forms part of a leukemia-causing fusion protein (van Zutven et al., 2006; Wang et al., 2009). In addition, our previous studies showed that the PHD3 motif of KDM5 is required for induction of cell growth in concert with the oncoprotein transcription factor Myc (Li et al., 2010). Together, these data show the importance of chromatin recognition by KDM5, and suggest a direct role for dysregulation of PHD motif-mediated H3K4me3 recognition in cancer.
Mitochondrial dysfunction is also an emerging theme among conditions that affect neuronal functioning, including Fragile X-related syndromes, Down syndrome, Alzheimer’s disease, Rett syndrome and some autism spectrum disorders (De Felice et al., 2012; Pagano and Castello, 2012; Yan et al., 2013). Mutations in KDM5A and KDM5B are found in patients with autosomal recessive intellectual disability, and mutations in KDM5C are found in those with the X-linked form of this disease (Vallianatos and Iwase, 2015). Reduced mitochondrial function and/or increased oxidative stress may contribute to the neuronal phenotypes observed in these patients. It is also noteworthy that our analyses of directly regulated KDM5 target genes revealed significant enrichment for several transcription factors critical for neuronal development and function. In particular, factors such as Lola (Longitudinals lacking) and Ttk (Tramtrack) are required for dendrite development (Iyer et al., 2013; Parrish et al., 2006). Because dendrite abnormalities are found in KDM5C knockdown cells (Iwase et al., 2007), the dysregulation of KDM5-Lola and KDM5-Ttk target genes may contribute to the cognitive pathologies seen in patients with mutation in KDM5 family genes.
MATERIALS AND METHODS
Fly strains
Fly strains were obtained from Bloomington stock center. Individually kdm510424 and kdm5K6801 homozygotes survive at less than 5%, but kdm510242/kdm5K6801 is a hypomorphic allelic combination in which adults survive at ~50% expected frequency (30% of wildtype protein levels) (Li et al., 2010; Liu et al., 2014; Secombe et al., 2007). The wildtype untagged, HA-tagged kdm5 and demethylase inactive (kdm5JmjC*) genomic rescue transgenes are published (Li et al., 2010; Liu et al., 2014). The PHD3 and PHD1 mutant genomic rescue transgenes (gKDM5W1771A and gKDM5W490A) were generated by site directed mutagenesis. gKDM5W1771A transgene insertion numbers 1M, 2M and 5M were tested and behaved indistinguishably. gKDM5W490A 8M or 9M were used. UAS-KDM5 was generated using the pUASt vector. UAS-KDM5W1771A and UAS-KDM5W490A were generated by site-directed mutagenesis. Transgenic flies were generated by The Best Gene (TheBestGene.com).
RNA-seq and ChIP-seq analyses
RNA-seq analyses were carried out comparing wildtype (w1118) flies and kdm5K6801/10424 flies that were 1–3 days old. ChIP-seq using anti-KDM5 (using w1118 flies) and anti-HA (using genomic tagged KDM5:HA flies) were carried out using 1–3 day old flies. Details of RNA-seq and ChIP-seq analyses are described in supplementary methods. Datasets are publically available (Accession number GSE70591).
Real-time PCR
1ug of total RNA was reverse transcribed using Verso cDNA kit (Thermo Scientific) with oligo (dt) primer. qRT-PCR reactions were performed in triplicate as described previously (Liu et al., 2014). Primer sequences are provided in Table S4.
Chromatin immunoprecipitation
ChIP-PCR binding signals were calculated as a percentage of input DNA as described previously (Liu et al., 2014). IgG was used as a negative control. Primers used for ChIP-PCR are shown in Table S4.
Quantitation of ATP, citrate, lactic acid levels and the ratio of NAD to NADH
ATP concentration was determined using the ATP bioluminescence assay kit HS II (Invitrogen). Values were normalized to protein concentration. Citrate and lactic acid concentrations were determined using kits from Sigma (MAK057 and MAK064, respectively). The ratio of NAD/NADH was determined using the NAD/NADH assay kit II (Sigma MAK037).
MitoSOX (superoxide) quantitation
MitoSOX (Invitrogen) fluorescence was quantitated from control (w1118) and kdm5K6801/10424 mutant flies using isloated mitochondria as described previously (Tang et al., 2009).
Histone binding
Histone binding assays were carried out as described previously (Li et al., 2010).
Westerns and immunofluorescence
The KDM5 antibody has been described previously (Secombe et al., 2007). Anti-H3K4me2, H3K4me3, and H3 were obtained from Active Motif. Western analysis was carried out using standard protocols, infrared conjugated secondary antibodies (LiCOR) and band intensity quantitated using LiCOR odyssey v3.0 software. Immunofluorescence of larval tissue was carried out as described in (Secombe et al., 2007).
Metabolite analyses
Wildtype (w1118) or kdm5K6801/10424 mutant adults were homogenized in 2.5mM ammonium acetate in methanol and analyzed using UPLC-MS/MS (Acquity UPLC- Xevo TQ MS). Assay procedures and metabolite nomenclature have been previously described (Gieger et al., 2008; Illig et al., 2010). Six biological replicates of pooled QC samples (for each genotype) were analyzed to calculate the %CV for each metabolite.
Electron microscopy
Thoraxes from 1–3 day old flies were fixed in 2% paraformaldehyde, 2.5% glutaraldehyde, 5 mM CaCl2, 0.1 mM sodium cacodylate for 24hours at 4°C. This was followed by 2-hour post-fixation in 2.5% glutaraldehyde, 0.8% osmium tetroxide, 0.1 mM sodium cacodylate at 4°C. Ultrathin sections were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate and viewed on a JEOL 1200EX transmission electron microscope at 80kv.
Negative geotaxis assays
Flies were tapped down to the bottom of an empty standard plastic food vial and the number of flies that climbed 7 cm within 20 seconds quantified. 80 females (10 flies per vial; 8 biological replicates) were scored for each genotype.
Statistical analyses
Unless otherwise indicated, data are presented as mean ± SEM from at least three independent biological replicates (each done in triplicate). Statistical significances were evaluated using Student’s t-test and one-way ANOVA analyses using GraphPad Prism Software. Statistical analyses of mendelian ratios was carried out using chi-squared test.
Supplementary Material
Acknowledgments
The authors thank Secombe lab members and Hardik Shah and Yongping Qui from the Einstein Diabetes Research and Training Center (DRTC) Metabolomics Core Facility. This work was supported by NIH grant R01GM112783 to J.S.
Footnotes
Author Contributions: Conceived and designed the experiments: X.L and J.S. Performed the experiments: X.L and J.S. Analyzed the data: X.L and J.S. Wrote the paper: J.S. and X.L.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 2013;9:e1003461. doi: 10.1371/journal.pgen.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Hom RA, Blakeslee W, Ikenouye L, Kutateladze TG. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta. 2014;1843:366–371. doi: 10.1016/j.bbamcr.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A, Santangelo S, Tan K, Catchpole S, Roberts K, Spencer-Dene B, Hall D, Scibetta A, Burchell J, Verdin E, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV. Histone H3K4 demethylases are essential in development and differentiation. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2007a;85:435–443. doi: 10.1139/O07-057. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV. Retinoblastoma binding protein 2 (RBP2) and differentiation. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2007b;85:523–523. doi: 10.1139/O07-057. [DOI] [PubMed] [Google Scholar]
- Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel) 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Liu Z, Cheung WK, Zhao M, Chen SY, Chan SW, Booth CJ, Nguyen DX, Yan Q. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell reports. 2014;6:868–877. doi: 10.1016/j.celrep.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG, Burchell JM, Taylor-Papadimitriou J. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38:1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct. 2012;2012:646354. doi: 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice C, Signorini C, Leoncini S, Pecorelli A, Durand T, Valacchi G, Ciccoli L, Hayek J. The role of oxidative stress in Rett syndrome: an overview. Annals of the New York Academy of Sciences. 2012;1259:121–135. doi: 10.1111/j.1749-6632.2012.06611.x. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Olson L, Patel KP, Miller R, Palladino MJ, Ganetzky B. Neuropathology in Drosophila mutants with increased seizure susceptibility. Genetics. 2008;178:947–956. doi: 10.1534/genetics.107.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmuller G, Kato BS, Mewes HW, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Iyer EP, Iyer SC, Sullivan L, Wang D, Meduri R, Graybeal LL, Cox DN. Functional genomic analyses of two morphologically distinct classes of Drosophila sensory neurons: post-mitotic roles of transcription factors in dendritic patterning. PLoS One. 2013;8:e72434. doi: 10.1371/journal.pone.0072434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome biology. 2014;15:R32. doi: 10.1186/gb-2014-15-2-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, Wen H, Larson C, Zhang X, Zheng X, et al. The Histone-H3K4-Specific Demethylase KDM5B Binds to Its Substrate and Product through Distinct PHD Fingers. Cell reports. 2014 doi: 10.1016/j.celrep.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The H3K4 Demethylase Lid Associates with and Inhibits the Histone Deacetylase Rpd3. Mol Cell Biol. 2009;29:1401–1410. doi: 10.1128/MCB.01643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Zhang JY, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genet. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Greer C, Secombe J. KDM5 interacts with Foxo to modulate cellular levels of oxidative stress. PLoS Genet. 2014;10:e1004676. doi: 10.1371/journal.pgen.1004676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M, Perez-Lluch S, Rossell D, Moran T, Ponsa-Cobas J, Auer H, Corominas M, Azorin F. dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, DeWaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi G, Shibata Y, Hayakawa T, Hayakawa N, Ohtani Y, Sinmyozu K, Tagami H, Nakayama J. Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2014;289:28956–28970. doi: 10.1074/jbc.M114.573725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outchkourov NS, Muino JM, Kaufmann K, van Ijcken WF, Groot Koerkamp MJ, van Leenen D, de Graaf P, Holstege FC, Grosveld FG, Timmers HT. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell reports. 2013;3:1071–1079. doi: 10.1016/j.celrep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- Pagano G, Castello G. Oxidative stress and mitochondrial dysfunction in Down syndrome. Advances in experimental medicine and biology. 2012;724:291–299. doi: 10.1007/978-1-4614-0653-2_22. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1B(high) Cells. Cancer cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases - The Myc connection. Cell Cycle. 2007;6:1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos LS, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Gene Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Song IS, Jeong JY, Jeong SH, Kim HK, Ko KS, Rhee BD, Kim N, Han J. Mitochondria as therapeutic targets for cancer stem cells. World J Stem Cells. 2015;7:418–427. doi: 10.4252/wjsc.v7.i2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Le PK, Tse S, Wallace DC, Huang T. Heterozygous mutation of Opa1 in Drosophila shortens lifespan mediated through increased reactive oxygen species production. PLoS One. 2009;4:e4492. doi: 10.1371/journal.pone.0004492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat Commun. 2015;6:6204. doi: 10.1038/ncomms7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos CN, Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics. 2015;7:503–519. doi: 10.2217/epi.15.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, Veronese A, Mecucci C, Negrini M, de Greef GE, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- Varaljai R, Islam AB, Beshiri ML, Rehman J, Lopez-Bigas N, Benevolenskaya EV. Increased mitochondrial function downstream from KDM5A histone demethylase rescues differentiation in pRB-deficient cells. Genes Dev. 2015;29:1817–1834. doi: 10.1101/gad.264036.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan MH, Wang X, Zhu X. Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic Biol Med. 2013;62:90–101. doi: 10.1016/j.freeradbiomed.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.