Abstract
Rationale and Objectives
The discovery of germline genetic variants associated with breast cancer has engendered interest in risk stratification for improved, targeted detection and diagnosis. However, there has yet to be a comparison of the predictive ability of these genetic variants with mammography abnormality descriptors.
Materials and Methods
Our IRB-approved, HIPAA-compliant study utilized a personalized medicine registry in which participants consented to provide a DNA sample and participate in longitudinal follow-up. In our retrospective, age-matched, case-controlled study of 373 cases and 395 controls who underwent breast biopsy, we collected risk factors selected a priori based on the literature including: demographic variables based on the Gail model, common germline genetic variants, and diagnostic mammography findings according to BI-RADS. We developed predictive models using logistic regression to determine the predictive ability of: 1) demographic variables, 2) 10 selected genetic variants, or 3) mammography BI-RADS features. We evaluated each model in turn by calculating a risk score for each patient using 10-fold cross validation; used this risk estimate to construct ROC curves; and compared the AUC of each using the DeLong method.
Results
The performance of the regression model using demographic risk factors was not statistically different from the model using genetic variants (p=0.9). The model using mammography features (AUC = 0.689) was superior to both the demographic model (AUC = .598; p<0.001) and the genetic model (AUC = .601; p<0.001).
Conclusion
BI-RADS features exceeded the ability of demographic and 10 selected germline genetic variants to predict breast cancer in women recommended for biopsy.
Keywords: mammography, genetic variants, BI-RADS, risk estimation, predictive value
Introduction
Over the last several decades, predictive variables have been discovered and incorporated into risk prediction models [1–3] with the goal of personalizing breast cancer screening and diagnosis. One highly predictive source of information is abnormality level feature descriptors observed on mammography as described in the Breast Imaging Reporting and Data System (BI-RADS) [4–8]. Other emerging sources are the ever-growing genome-wide association studies (GWAS) that identify genetic variants (single nucleotide polymorphisms—SNPs). The SNPs discovered via recent GWAS are distinct from mutations in the BRCA1 and BRCA2 tumor suppressor genes [9]. While both are germ-line genetic risk factors (inherited from parental lineage), SNPs discovered in recent GWAS are single-base-pair DNA sequence variations conferring modest risk (low penetrance) but occurring commonly (high frequency) within the human population. Expansion of genetic risk prediction may depend on polygenic risk stratification, i.e. weighing many high frequency, low-penetrance SNPs at once [3, 10]. Early attempts to use such SNPs to predict breast cancer risk have demonstrated only modest improvements over conventional demographic risk factors, like those in the Gail model [11–13].
Breast cancer risk is determined by a combination of genetic and environmental factors. Intermediate phenotypes like imaging [14] can capture and convey these interactions of these risk factors and provide biomarkers that can augment comprehensive risk prediction. Since demographic risk factors, genetic variants, and imaging features will all likely have some level of predictive value, determining which variables provide the best predictive power in any given setting becomes extremely important. Investing limited resources in collection of the best predictive variables will provide the most benefit. Prior literature evaluated risk prediction with genetics and breast density [15, 16] and one paper added BI-RADS assessment category [17]. Despite the proven predictive ability of abnormality-level features described in the BI-RADS lexicon [4–8] (e.g. mass and calcification descriptors as well as associated findings like architectural distortion), comparison with demographic or genetic risk has been limited. We compare the performance of predictive models using distinct data elements: demographic risk factors, germline genetic variants, or mammography abnormality features to estimate breast cancer risk in women recommended for breast biopsy.
Materials and Methods
Subjects
The source of subjects for this project was the population-based __APMR_, details of which have been published previously [18]. Briefly, ___ Institution 1___ patients aged 18 years and older residing in one of 19 Zip codes surrounding ___City, State___ were invited to participate. After giving written informed consent, participants provided a blood sample from which DNA, plasma and serum was extracted and stored. Permission was given to link the biological samples with medical records and a brief questionnaire was completed.
We selected subjects from the __APMR_ using a retrospective case-control design. Women with a DNA sample available, a diagnostic mammogram, and a breast biopsy within 12 months after the mammogram were included. Cases were defined as women having a confirmed diagnosis of invasive breast cancer or ductal carcinoma in situ (DCIS) obtained from the __ Institution 1__ institutional cancer registry. Controls were confirmed through the electronic medical records (and absence from the cancer registry) as having a benign biopsy result, and never having had a breast cancer diagnosis.
To ensure a similar age distribution, we selected a control whose age was within five years of the age of each case. A total of 35 subjects were excluded from the statistical analysis. We excluded 3 cases with known BRCA1 mutation and 3 cases with BRCA2 mutation (because these mutations would likely dominate all other predictive variables). We excluded 8 non-white women from our study because GWAS variants can differ between races, and we did not have an appropriate number or distribution of non-whites to effectively consider race or to race-match cases and controls. Finally we excluded all instances in which BI-RADS features and breast density were all missing (21 cases). Some of the excluded subjects met more than one exclusion criterion.
All epidemiologic, genetic, and mammographic risk factors were chosen a priori based on the literature [11–13, 19] to represent the variables most likely to influence breast cancer risk and these factors were included in analysis regardless of subsequent statistical significance.
Epidemiologic Risk Factors
Variables used in the current study that were collected at the time of enrollment into the __APMR_ included age and gender. Medical records were manually abstracted for the following information based on Gail risk factors: family history of breast cancer, age at menarche and number of biopsies (prior to the index biopsy qualifying each subject for inclusion). Age at first live birth was not available in our cohort so parity was instead used in our “DEMOGRAPHIC” model because of known association with breast cancer risk and correlation with age at first birth [20].
Genetic Variants
The __APMR_ was one of five initial biobanks in the eMERGE Network funded by the National Human Genome Research Institute [21]. We identified 10 genetic variants shown to predict breast cancer in large GWAS studies [22, 23] and tested for breast cancer risk prediction—Table 1 [11–13]. We sequenced these 10 SNPs on the Sequenom MassARRAY system. Because humans have two paired chromosomes with two chances to inherit the higher-risk (“risky”) allele, there are several accepted methods to quantify risky alleles for analysis. We enumerated the SNPs using two methods previously described in the literature [11]. The “variant count” method quantifies the number of risky alleles (one allele per SNP for heterozygotes and two alleles per SNP for homozygotes) aggregating them into categories of ≤ 6, 7–8, 9–10, 11–12, and ≥13. The “individual count” method, which we used in our “GENETIC” model, quantifies how many risky alleles are present (0, 1, or 2 risky alleles) for each individual SNP resulting in possible values of 0–30 inclusive.
Table 1.
| SNPs | Chromosome | Gene | High-Risk Allele | Low-Risk Allele |
|---|---|---|---|---|
| RS1045485 | 2q | CASP8 | G | C |
| RS13281615 | 8q | Unknown | G | A |
| RS13387042 | 2q | Unknown | A | G |
| RS2981582 | 10q | FGFR2 | T | C |
| RS3803662 | 16q | TOX3 | T | C |
| RS3817198 | 11p | LSP1 | C | T |
| RS889312 | 5q | MAP3K1 | C | A |
| RS10941679 | 5p | Unknown | G | A |
| RS999737 | 14q | RAD51L1 | C | T |
| RS11249433 | 1p | Unknown | C | T |
Mammography Features
In order to capture mammography abnormality level data, the biopsies of both cases and controls were matched with one diagnostic mammogram within 12 months prior to biopsy. If there were multiple mammograms within the year prior to biopsy we selected the mammogram with a most suspicious BI-RADS assessment category. If multiple diagnostic mammograms also had the same BI-RADS assessment category, we selected the mammogram closest in time but prior to the biopsy. If more than one mammogram were still candidates, with the same BI-RADS assessment category performed on the same day, we selected the one with the most extracted BI-RADS abnormality features. If the selected mammogram did not contain an assessment for breast density, each prior mammogram was checked for a breast density assessment and the most recent assessment was used.
Mammography features have been codified in the BI-RADS lexicon to standardize mammographic findings and recommendations [24] which contains three major categories: breast density, abnormality features (or “descriptors”) and BI-RADS assessment categories. We focused on breast density and abnormality features because most women receiving a biopsy recommendation will be assessed as BI-RADS 4 and less commonly BI-RADS 5; a uniformly assigned variable was unlikely to be contributory.
At the _____Clinic name____, mammography results were recorded as free text reports in the electronic health record. We use a parser based on the 3rd edition of BI-RADS [25], previously shown to outperform manual term identification [26, 27] to isolate BI-RADS mammography features. We used the BI-RADS 3rd edition rather than more recent version because this retrospective review analyzed mammograms from the era in which this edition was most commonly used, particularly keeping in mind that adoption of a lexicon typically lags dissemination. From these features, we selected from the most predictive abnormality descriptors, a priori, based on the literature [19] to use in our “MAMMOGRAPHIC” model: mass margin, microcalcification shape, microcalcification morphology, and architectural distortion. For microcalcification features we consolidated the suspicious morphology descriptors (linear, amorphous, and pleomorphic) and suspicious distribution descriptors (clustered, segmental, linear) into the “present” category; cases lacking any of these descriptors in their records were assigned “absent” calcifications. Breast density was discretized into the 4 values defined by BI-RADS: predominantly fatty, scattered fibroglandular, heterogeneously dense, or extremely dense. Importantly, the BI-RADS lexicon evolved from the 1st Edition to the 4th Edition over the time of our study. While the many of the descriptors were being used even before BI-RADS was released as a result of influential publications that codified predictive terms [28], the progressive development and dissemination of BI-RADS (1st Edition in 1993; 2nd Edition in 1995; 3rd Edition in 1998; and 4th Edition in 2003) [29] was undoubtedly accompanied by increasing uptake over time. Thus, in order to demonstrate how that evolution of BI-RADS might influence our results, we provide more detailed analysis over time (Appendix).
Statistical Analysis
We used logistic regression to model risk of malignancy. Odds ratios and 95% confidence intervals were obtained for each predictor in a model that contained all of the variables. We adjusted for age at biopsy in each model. We built 3 models each containing variables from single data types: demographic risk factors (the DEMOGRAPHIC model), SNPs (the GENETIC model), and imaging features (the MAMMOGRAPHIC model). In order to comprehensively summarize odds ratios for all variables, we created a single model—Table 2 (using the variant count method because it is more concise). Prior to building our models we imputed age at menarche, number of pregnancies and breast density to populate missing values [30, 31] as implemented in the R mice package [32–34].
Table 2.
Demographic variables, genetic factors, and mammographic features in the predictive models*
| Variable | Cases (N = 373) | Controls (N = 395) | OR | 95% CI¥ | p-value¥ |
|---|---|---|---|---|---|
| Age at menarche | |||||
| ≥14 | 105 (28.2%) | 84 (21.2%) | Referent | ||
| 12–13 | 189 (50.7%) | 224 (56.6%) | 0.63 | (0.33,1.21) | 0.15 |
| 7–11 | 79 (21.1%) | 88 (22.2%) | 0.73 | (0.29,1.83) | 0.46 |
| Number of biopsies | |||||
| 0 | 303 (81.2%) | 337 (85.6%) | Referent | ||
| 1 | 60 (16.0%) | 52 (13.1%) | 1.35 | (0.84,2.16) | 0.21 |
| ≥2 | 10 (2.6%) | 6 (1.5%) | 2.89 | (0.95,8.75) | 0.061 |
| Number of pregnancies | |||||
| ≥6 | 52 (14.0%) | 47 (12.0%) | Referent | ||
| 3–5 | 164 (43.9%) | 175 (44.4%) | 1.09 | (0.63,1.89) | 0.76 |
| 1–2 | 126 (33.8%) | 129 (32.7%) | 1.18 | (0.66,2.13) | 0.57 |
| 0 | 31 (8.3%) | 43 (10.9%) | 0.72 | (0.34,1.51) | 0.38 |
| Number. of first-degree relatives with breast cancer | |||||
| 0 | 268 (71.8%) | 325 (82.2%) | Referent | ||
| 1 | 91 (24.4%) | 57 (14.4%) | 2.05 | (1.34,3.15) | 0.0010 |
| ≥2 | 14 (3.7%) | 13 (3.2%) | 1.68 | (0.70,4.04) | 0.24 |
| Number of risk-conferring variant alleles | |||||
| 0–6 | 26 (6.9%) | 58 (14.6%) | Referent | ||
| 7 or 8 | 97 (26.0%) | 126 (31.9%) | 1.92 | (1.04,3.55) | 0.037 |
| 9 or 10 | 135 (36.1%) | 126 (31.9%) | 2.69 | (1.46,4.95) | 0.0016 |
| 11 or 12 | 90 (24.1%) | 73 (18.4%) | 2.74 | (1.41,5.30) | 0.0029 |
| ≥13 | 25 (6.7%) | 12 (3.0%) | 4.95 | (1.89,12.96) | 0.0012 |
| Breast density | |||||
| Fatty | 24 (6.5%) | 26 (6.5%) | Referent | ||
| Scattered | 30 (8.1%) | 58 (14.6%) | 0.46 | (0.12,1.80) | 0.24 |
| Heterogeneous | 293(78.6%) | 298 (75.4%) | 0.99 | (0.25,3.92) | 0.98 |
| Extremely dense | 25 (6.8%) | 14 (3.4%) | 2.47 | (0.27,22.27) | 0.37 |
| Mass margin | |||||
| Circumscribed | 16 (4.2%) | 36 (9.1%) | 0.42 | (0.21,0.85) | 0.016 |
| Obscured | 8 (2.1%) | 14 (3.5%) | 0.59 | (0.22,1.58) | 0.29 |
| Ill-defined | 47 (12.6%) | 48 (12.1%) | 1.42 | (0.87,2.31) | 0.16 |
| Spiculated | 82 (21.9%) | 4 (1.0%) | 27.56 | (9.64,78.78) | < 0.001 |
| Suspicious microcalcification shape | 63 (16.8%) | 79 (20.0%) | 1.49 | (0.82,2.72) | 0.19 |
| Suspicious microcalcification distribution | 79 (21.1%) | 117 (29.6%) | 0.56 | (0.33,0.94) | 0.029 |
| Architectural distortion | 50 (13.4%) | 21 (5.3%) | 2.32 | (1.25,4.28) | 0.0073 |
Sample sizes are shown as an average across the 5 imputed datasets
Bold signifies statistically significant variables
Assessment of Model Performance
Ten-fold cross-validation was used to fit and evaluate the models. We calculated the areas under the ROC curve (AUC) with 95% confidence intervals and did statistical comparisons using the DeLong method [35]. A two-sided p-value of < 0.05 was the criterion for statistical significance. All statistical analyses and graphics were done in R version 3.0.1.[36]
IRB approvals
The ________ Institutional Review Board reviewed and approved the data collection of the __Anonymized Personalized Medicine Registry___ (_APMR_), which required informed consent. The __Institution 1___ and the __ Institution 2 ___ IRBs also approved this retrospective study and waived any further specific informed consent. All study activities were HIPAA compliant.
Results
We collected a total of 373 cases and 395 controls with a biopsy date between 1/29/1989 – 12/15/2010. The majority of mammograms were performed between 1993–2005 with approximately equal proportions of cases and controls in these years (Figure 1). The age range (i.e. age at biopsy) was 29 to 90 years of age (mean = 62, SD = 12.8). Of the cases, 77% were invasive and 23% were in situ. The variables included in the predictive models are shown in Table 2. Some of our predictors had a high percentage of missing observations. Menarche was missing 239 (60.5%) for cases and 64 (17.2%) for controls; number of pregnancies was missing 1 (0.03%) for cases and 16 (4.0%) for controls, and breast density was missing 157 (42.1%) for cases and 179 (45.3%) for controls.
Figure 1.
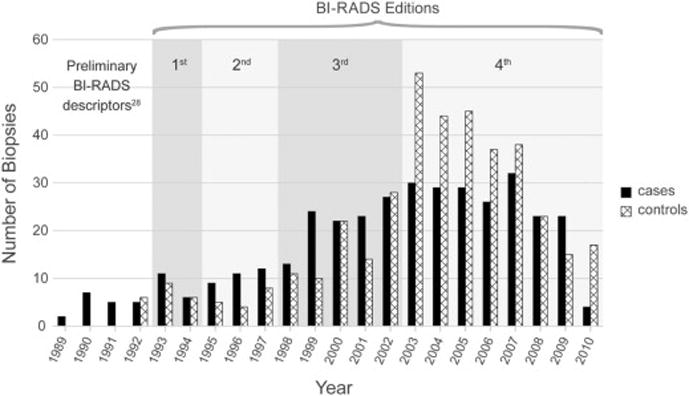
Plot shows the year that included biopsies (cases and controls) were performed, from which corresponding diagnostic mammography examinations were identified. Date ranges of the sequential editions of the BI-RADS lexicon are demarcated to illustrate the evolution of the lexicon noting that utilization invariably lags dissemination. Prior to the publication of the BI-RADS lexicon, standardized descriptors were also available in the scientific literature.28
Demographic variables were associated with risk of a malignant diagnosis; associations were not statistically significant for age at menarche, number of biopsies, and parity, but risk estimates were in the expected directions. As expected, women with a family history of breast cancer had a two-fold risk of a malignant diagnosis (OR 2.05, 95% CI 1.34, 3.15). Greater numbers of risky genetic alleles were also significantly associated with greater risk of a malignant diagnosis.
Mammographic features (Table 3) were also significantly associated with risk of a malignant diagnosis (Table 2). Spiculated mass margin, p < 0.001, and architectural distortion, p < 0.01 were significant predictors of malignancy amongst imaging features while circumscribed mass margin decreased the risk of malignancy, p = 0.016. We observed that suspicious calcification shape predicted malignancy but not significantly. However suspicious calcification distribution significantly decreased the risk of malignancy, p = 0.029.
Table 3.
Frequency of microcalcification descriptors in cases and controls
| Control | Case | Total | |
|---|---|---|---|
| Microcalcification Shape | |||
| Benign descriptor | 33 (53%) | 29 (47%) | 62 |
| Amorphous | 10 (83%) | 2 (17%) | 12 |
| Pleomorphic | 70 (54%) | 59 (46%) | 129 |
| Fine linear | 0 (0%) | 5 (100%) | 5 |
| Total | 113 | 95 | 208 |
| Microcalcification Distribution | |||
| Benign descriptor | 11 (52%) | 10 (48%) | 21 |
| Grouped | 115 (62%) | 71 (38%) | 186 |
| Segmental | 0 (0%) | 6 (100%) | 6 |
| Linear | 18 (46%) | 21 (54%) | 39 |
| Total | 144 | 108 | 252 |
We found that use of BI-RADS descriptors in our study population did increase over time (Appendix).
Model Performance
The “baseline” model with age alone had an area under the ROC curve (AUC) of 0.547 (95% CI 0.506, 0.587). When we added the Gail demographic factors to the baseline model—i.e. the DEMOGRAPHIC model—AUC increased to 0.598 (95% CI 0.558, 0.638; p = 0.012; Figure 2). When we added the ten SNPs to the baseline model—i.e. the GENETIC model—the AUC increased to 0.601 (95% CI 0.562, 0.641; p < 0.01). There was no statistically significant difference between the AUCs of the DEMOGRAPHIC model and the GENETIC model (p = 0.90). When we added mammographic features to the baseline model—i.e. the MAMMOGRAPHIC model—the AUC significantly increased to 0.689 (95% CI 0.652, 0.727; p < 0.001). The AUC of the MAMMOGRAPHIC model was statistically significantly better than the DEMOGRAPHIC (p < 0.001) and the GENETIC (p < 0.001) models respectively. We also show that if we limit our study to cases after a year when a new BI-RADS edition became available (e.g. after 1993 when BI-RADS 1st Edition became available or after 1998 when BI-RADS 3rd Edition became available), the predictive ability of our BI-RADS models increase in keeping with improved utilization and consistency (Appendix).
Figure 2.
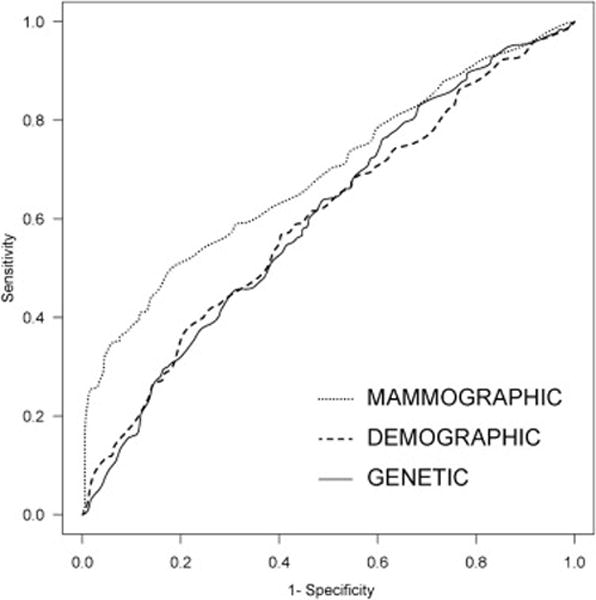
Receiver operating characteristic curves for the three models that were compared.
Discussion
Determining the predictive ability of variables that confer risk for any given population of patients is important even if these variables (or combinations of these variables) are not sufficiently well-understood to influence clinical decision-making. We show that mammographic findings are significantly better at predicting malignancy in a population of women recommended for breast biopsy as compared to risk factors based on the Gail model or 10 selected SNPs. We found no significant difference between our GENETIC and DEMOGRAPHIC models in this population. By consolidating a unique data set of demographic risk factors, genetic variants, and mammographic findings, our regression models revealed both expected and novel statistically significant predictors of malignancy among women recommended for biopsy.
Our results are comparable to, yet extend, previous literature, which to date has not considered finding level imaging variables.[11–13] The predictive ability of genetic variants and demographic risk factors (without mammography features) has been investigated in the literature before. Two studies [11, 12] extracted a case-control cohort from longitudinal observational studies and used a similar logistic regression modeling technique to demonstrate that demographic (Gail) risk factors achieved an AUC of 0.607 and 0.534 respectively (comparable to our DEMOGRAPHIC model AUC of 0.598). With regard to genetic variants, one of these studies [12] analyzed seven SNPs (a subset of those used in our model) revealing an AUC of 0.607 while the other [11] used 10 SNPs (almost the exact list used in our model) demonstrating an AUC of 0.597. These results are again comparable to our GENETIC model AUC of 0.601). Our contribution is to demonstrate that mammography features have superior predictive ability in this population as shown by our MAMMOGRAPHIC model AUC of 0.689, statistically significantly better than either the DEMOGRAPHIC OR GENETIC models. Our findings reflect an early indication as to the comparative value of each variable type at this point of care (recommendation for breast biopsy). Further research in this area has the potential to determine the wisest investment of resources in order to accurately predict breast cancer risk and optimize management.
The value of mammography features has been well-established in the literature [4–8], but has not yet been directly compared to new predictors like germline genetic variants. We find that abnormality features such as spiculated mass margins and architectural distortion were strong predictors of breast cancer in this population. While mammographic breast density has been used to effectively predict risk in concert with demographic risk factors [37] in screening populations, breast density was in fact a weak a predictor in our population recommended for breast biopsy. Both suspicious morphologic and distribution descriptors for microcalcification yielded surprising results. Morphology was not significantly predictive (but tended in the expected direction, i.e. to predict malignancy) however distribution was significantly predictive of benignity. This result is counter to prior literature [5, 38, 39] showing both of these variables to be highly predictive of malignancy (usually DCIS). The reasons for the poor and unexpected predictive direction of microcalcification descriptors is likely multifactorial. First, microcalcification descriptors extracted from free text reports were dominated by two descriptors: pleomorphic morphology and clustered distribution (Table 3). The heavy use of these descriptors likely diminishes their predictive power. Linear morphology and segmental distribution were noted only in malignant cases and thus may have been predictive had our numbers been larger. Second, our small sample size is likely at least partially responsible for these results, because only a small proportion (22.3%) of our already small number of cases were DCIS, those most likely to be manifest by microcalcifications on mammography. Third, though these descriptors are shown to be predictive of malignancy in previous literature, these results were found in consecutive biopsy cases including only microcalcifications rather than a case-control design, such as ours, which includes all imaging findings.
Limitations of this study should be considered when interpreting results. Our predictive models are undoubtedly limited by the challenges of medical record and self-reported data that are inherently imperfect. Prior comparable studies [11–13] used genetic and demographic data from high-quality cohort studies conducted using prospective and systematic collection of risk factors resulting in few missing values. We, on the other hand, found it necessary to extract many risk factors from an existing electronic health record, resulting in a higher rate of missing values. For this reason, we imputed three variables: age at menarche, number of pregnancies, and breast density. By assessing our level of convergence diagnostics, comparing the distribution of observed and imputed variables, and comparing results with prior literature, we are confident that our imputed dataset are robust. The small size of the study limited precision in estimates, yet associations were observed in the expected directions. In order to compensate for small sample size we needed to include clinical encounters over a substantial time range (1989–2010). This study design has the disadvantage that mammography descriptors were used more consistently by interpreting radiologist in the latter years included in our study due to increased adoption of the BI-RADS lexicon and adherence with the lexicon descriptors. In fact, increasing utilization of BI-RADS appears to result in increased predictive performance of logistic regression models based on descriptors as more recent mammograms are included. Therefore, the results of the larger data set, across the entire study timeframe, may underestimate the predictive ability of the models using BI-RADS descriptors (Appendix), likely making our results, i.e. the magnitude by which BI-RADS descriptors outperform genetics and demographic risk factors, conservative. Finally, our patient population was limited to white women recommended for biopsy, so results cannot be generalized beyond this group.
By constructing a unique data set, we demonstrate that mammography features according to BI-RADS provide discriminative performance that is statistically significantly superior to demographic and genetic models in women recommended for breast biopsy. Because BI-RADS descriptors predict the outcome of a given finding (the target for biopsy) in the immediate diagnostic setting, while demographic and genetic variables (as well as breast density) predict longer term risk of malignancy, our results make intuitive sense. However, our study quantifies this differential value without claiming that any of these variables or combinations are ready to influence clinical decision-making at this point in time. These results provide a necessary step toward quantifying the value of these variables knowing that personalizing risk is a promising direction in the future. For example, now over 100 SNPs have been discovered as GWAS consortium datasets grow [40]. Improved codification and collection of mammography variables will likely result from the new BI-RADS lexicon 5th edition and the growth of the National Mammography Database. Continued research like ours has the potential to guide prioritization of variable collection for optimal risk prediction.
Acknowledgments
We thank Jacqueline Bohne for their help with data extraction and consolidation from the ____Clinic name____ EMR. We also thank Elizabeth A. Simcock for figure development and graphic design.
Funding*: Grants: The authors acknowledge the support of the Wisconsin Genomics Initiative from the state of Wisconsin and support from the National Institutes of Health (grants: R01CA127379, R01CA127379-03S1, R01GM097618, R01LM011028, R01ES017400). We also acknowledge support from the eMERGE Network (U01HG004608), the UW Institute for Clinical and Translational Research (UL1TR000427) and the UW Carbone Comprehensive Cancer Center (P30CA014520).
Appendix
In order to analyze the use of BI-RADS descriptors over the date range of our study we plotted the use of BI-RADS descriptor per individual for the whole study population (Figure A1), cases (Figure A2) and controls (Figure A3). These figures all demonstrate increasing utilization of the BI-RADS descriptors over time. We then took our imputed data and created two new logistic regression models subsets of the data: 1) after 1993 when BI-RADS 1st Edition became available and 2) after 1998 when BI-RADS 3rd Edition became available (Table A1). These AUC values demonstrate a trend of improvement for models using mammography reports collected in more recent timeframes (Table A.1 and Figure 1—main manuscript).
Figure A1.
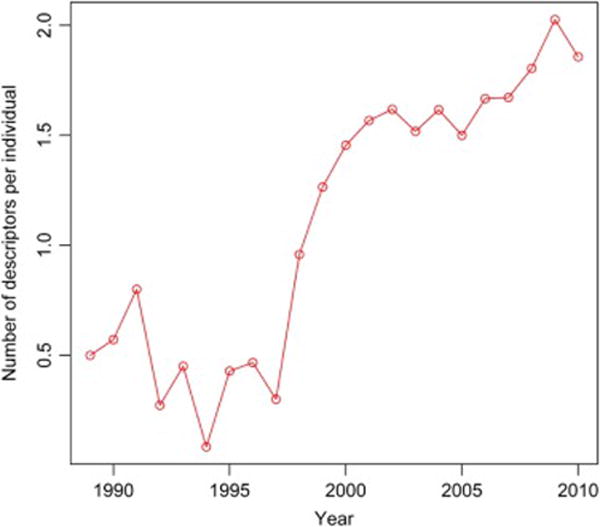
Utilization of BI-RADS descriptors for cases and controls over the time of study
Figure A2.
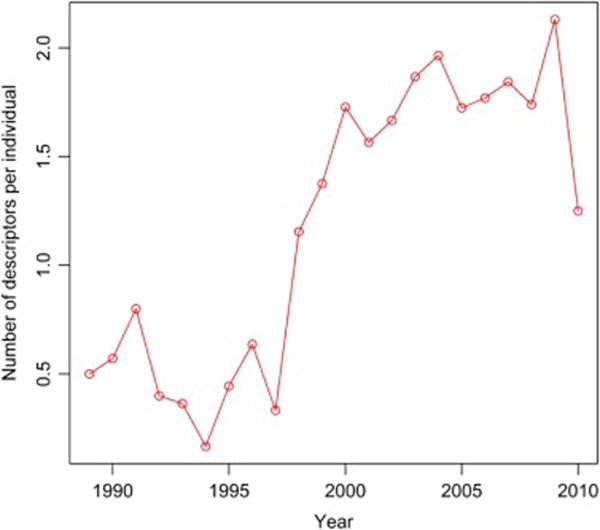
Utilization of BI-RADS descriptors for cases over the time of study
Figure A3.
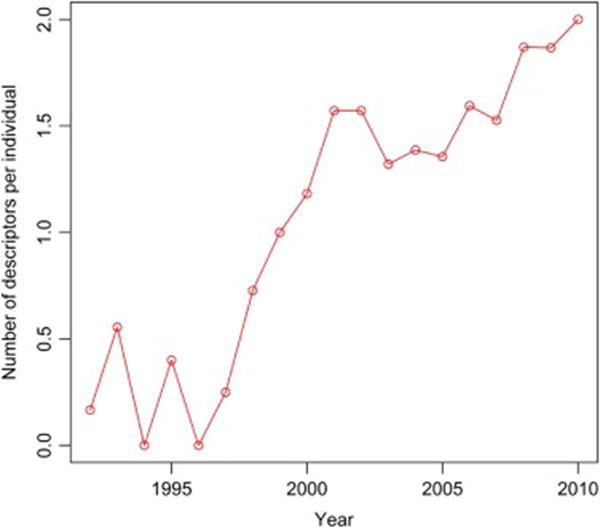
Utilization of BI-RADS descriptors for controls over the time of study
Table A1.
Performance of MAMMOGRAPHIC models using progressively more recent subsets of the data
| Timeframe | n | AUC |
|---|---|---|
| 1989–2010 | 768 | .693 |
| 1993–2010 | 743 | .701 |
| 1998–2010 | 662 | .711 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Of note, the sponsors/funders of this study had no role in: 1) study design; 2) collection, analysis or interpretation of data, 3) writing of the report; or 4) decision to submit the article for publication.
References
- 1.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 2.Barlow WE, White E, Ballard-Barbash R, et al. Prospective breast cancer risk prediction model for women undergoing screening mammography. J Natl Cancer Inst. 2006;98:1204–1214. doi: 10.1093/jnci/djj331. [DOI] [PubMed] [Google Scholar]
- 3.Pashayan N, Duffy SW, Chowdhury S, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104:1656–1663. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker JA, Kornguth PJ, Lo JY, Williford ME, Floyd CE., Jr Breast cancer: prediction with artificial neural network based on BI-RADS® standardized lexicon. Radiology. 1995;196:817–822. doi: 10.1148/radiology.196.3.7644649. [DOI] [PubMed] [Google Scholar]
- 5.Liberman L, Abramson AF, Squires FB, Glassman JR, Morris EA, Dershaw DD. The breast imaging reporting and data system: positive predictive value of mammographic features and final assessment categories. AJR Am J Roentgenol. 1998;171:35–40. doi: 10.2214/ajr.171.1.9648759. [DOI] [PubMed] [Google Scholar]
- 6.Burnside ES, Rubin DL, Fine JP, Shachter RD, Sisney GA, Leung WK. Bayesian network to predict breast cancer risk of mammographic microcalcifications and reduce number of benign biopsy results: initial experience. Radiology. 2006;240:666–673. doi: 10.1148/radiol.2403051096. [DOI] [PubMed] [Google Scholar]
- 7.Chhatwal J, Alagoz O, Lindstrom MJ, Kahn CE, Jr, Shaffer KA, Burnside ES. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol. 2009;192:1117–1127. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmers JM, Verbeek AL, IntHout J, Pijnappel RM, Broeders MJ, den Heeten GJ. Breast cancer risk prediction model: a nomogram based on common mammographic screening findings. Eur Radiol. 2013;23:2413–2419. doi: 10.1007/s00330-013-2836-8. [DOI] [PubMed] [Google Scholar]
- 9.Linger RJ, Kruk PA. BRCA1 16 years later: risk-associated BRCA1 mutations and their functional implications. The FEBS journal. 2010;277:3086–3096. doi: 10.1111/j.1742-4658.2010.07735.x. [DOI] [PubMed] [Google Scholar]
- 10.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 11.Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100:1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101:959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devilee P, Rookus MA. A tiny step closer to personalized risk prediction for breast cancer. N Engl J Med. 2010;362:1043–1045. doi: 10.1056/NEJMe0912474. [DOI] [PubMed] [Google Scholar]
- 15.Tamimi RM, Cox D, Kraft P, Colditz GA, Hankinson SE, Hunter DJ. Breast cancer susceptibility loci and mammographic density. Breast Cancer Res. 2008;10:R66. doi: 10.1186/bcr2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darabi H, Czene K, Zhao W, Liu J, Hall P, Humphreys K. Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement. Breast Cancer Res. 2012;14:R25. doi: 10.1186/bcr3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong K, Handorf EA, Chen J, Bristol Demeter MN. Breast cancer risk prediction and mammography biopsy decisions: a model-based study. American journal of preventive medicine. 2013;44:15–22. doi: 10.1016/j.amepre.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarty CA, Wilke RA, Giampietro PF, Wesbrook SD, Caldwell MD. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Alagoz O, Ayvaci MU, et al. A comprehensive methodology for determining the most informative mammographic features. Journal of digital imaging. 2013;26:941–947. doi: 10.1007/s10278-013-9588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi S, Sugiura H, Ando Y, et al. Reproductive history and breast cancer risk. Breast cancer (Tokyo, Japan) 2012;19:302–308. doi: 10.1007/s12282-012-0384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breast Imaging Reporting And Data System (BI-RADS®) 4th. Reston VA: American College of Radiology; 2003. [Google Scholar]
- 25.Breast Imaging Reporting And Data System (BI-RADS) 3rd. Reston VA: American College of Radiology; 1998. [Google Scholar]
- 26.Houssam N, Ryan W, Elizabeth B, Mehmet A, Jude S, David P. Proceedings of the 2009 IEEE International Conference on Data Mining Workshops. IEEE Computer Society; 2009. Information Extraction for Clinical Data Mining: A Mammography Case Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19:913–916. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 29.Burnside ES, Sickles EA, Bassett LW, et al. The ACR BI-RADS experience: learning from history. J Am Coll Radiol. 2009;6:851–860. doi: 10.1016/j.jacr.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DB R. Multiple Imputation for Nonresponse in Surveys. John Wiley & Sons; New York: 1987. [Google Scholar]
- 31.van Buuren S. Flexible imputation of missing data CRC/Chapman & Hall Interdisciplinary Statistics Series. Chapman and Hall/CRC; 2012. [Google Scholar]
- 32.vB S, Groothuis-Oudshoorn mice: Multivariate Imputation by Chained Equations. R package version 2.9. 2011 [Google Scholar]
- 33.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software. 2011;45:1–67. [Google Scholar]
- 34.Van Buuren S, Oudshoorn CGM. Multivariate Imputation by Chained Equations MICE V1.0 User’s manual. TNO Prevention and Health, Leiden; Jun, 2000. [Google Scholar]
- 35.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 36.R: A language and environment for statistical computing. Vienna, Austria: R Core Team: R Foundation for Statistical Computing; 2013. [3.0.1] In. [Google Scholar]
- 37.Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148:337–347. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg WA, Arnoldus CL, Teferra E, Bhargavan M. Biopsy of amorphous breast calcifications: pathologic outcome and yield at stereotactic biopsy. Radiology. 2001;221:495–503. doi: 10.1148/radiol.2212010164. [DOI] [PubMed] [Google Scholar]
- 39.Burnside ES, Ochsner JE, Fowler KJ, et al. Use of Microcalcification Descriptors in BI-RADS 4th Edition to Stratify Risk of Malignancy. Radiology. 2007;242:388–395. doi: 10.1148/radiol.2422052130. [DOI] [PubMed] [Google Scholar]
- 40.Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. 361e351–352. [DOI] [PMC free article] [PubMed] [Google Scholar]


