Abstract
Lethal disease caused by the fungus, Cryptococcus neoformans, is a consequence of the combined failure to control pulmonary fungal replication and immunopathology caused by induced type-2 helper T (Th2) cell responses in animal models. In order to gain incites into immune regulatory networks, we examined the role of regulatory T (Treg) cells in suppression of Th2 cells, using a mouse model of experimental cryptococcosis. Upon pulmonary infection with Cryptococcus, Treg cells accumulated in the lung parenchyma independently of priming in the draining lymph node. Using peptide-MHCII molecules to identify Cryptococcus-specific Treg cells combined with genetic fate-mapping, we noted that a majority of the Treg cells found in the lungs were induced during the infection. Additionally, we found that Treg cells utilized the transcription factor, Interferon Regulatory Factor 4 (IRF4), to dampen harmful Th2 cell responses, as well as mediate chemokine retention of Treg cells in the lungs. Taken together, induction and IRF4-dependent localization of Treg cells in the lungs allow Treg cells to suppress the deleterious effects of Th2 cells during cryptococcal infection.
Introduction
Cryptococcosis is an emerging infectious disease of humans caused by the fungus, Cryptococcus neoformans (1). Yeasts or spores are inhaled from the environment and enter the lower respiratory tract. Robust CD4+ helper T (Th) cell-mediated immunity controls this initial pulmonary infection, and as a result, immune replete individuals rarely experience overt disease. However, Th cell deficiencies associated with solid organ transplantation, cancer chemotherapy, and HIV/AIDS dramatically increases susceptibility to invasive cryptococcosis. C. neoformans emigrates from the lung, enters the bloodstream, and traverses the blood-brain barrier to cause cryptococcal meningitis. Despite access to standard antifungal and antiretroviral therapies, patients receiving treatment for cryptococcal meningitis exhibit a wide range of adverse clinical outcomes, including: infection relapse, immune reconstitution inflammatory syndrome due to excessive reaction to persistent antigen, and/or death (2). The reasons why some individuals recover without experiencing complications and others perish remains enigmatic.
Differences in the quality of the impaired Th cell responses in HIV patients stratify the spectrum of clinical outcomes (3–5). IFN-γ production by Type-1 helper T (Th1) cells defends against invasive cryptococcal disease and promotes fungal clearance (6–8). Dysregulated reconstitution of protective immunity in patients with recent cryptococcosis can also cause harmful inflammation (9). In addition, C. neoformans (serotype A) subverts protective immunity and exacerbates disease by driving Th2 cell production of interleukin IL-4, IL-5, and IL-13 (10, 11). Therefore, therapies that dampen detrimental Th cell responses could be used to ameliorate disease.
One mechanism the immune system uses to dampen Th cell responses is by employing regulatory T (Treg) cells. Treg cells are a distinct subset of Th cells that uniquely express the transcription factor forkhead box P3 (Foxp3), which stabilizes the suppressive function of Treg cells. Genetic aberrations in Foxp3 (i.e. IPEX syndrome) cause fatal Th cell-driven autoimmunity in humans, highlighting the importance of Foxp3 in immune homeostasis (12). Treg cells also inhibit effector Th cell responses to microbial infections (13). In particular, conditional depletion of Foxp3+ Treg cells in mice infected with C. neoformans increases Th2 cell abundance in the lungs, indicating Treg cells limit the proliferation of Th2 cells primed by cryptococcal infection (14, 15). Beyond these initial observations, little is known about the mechanism of Th2 cell suppression by Treg cells during cryptococcal infection.
Since Treg cell suppression of effector Th cells is contact dependent (16), Treg cells must colocalize with effector cells in order to function in tissues such as the lung (17). To accomplish this, Treg cells express chemokine receptors and integrins that allow them to home to and to be retained at sites of inflammation (18). Separate evidence indicates Treg cells that restrain mucosal Th cell responses exhibit highly specialized control of distinct Th cell subsets by expressing the same lineage-defining transcription factors as their effector Th cell counterpart (19–21). In particular, interferon regulatory factor 4 (IRF4) expression by Treg cells has been implicated in the suppression of Th2-driven autoimmunity (21).
Here, we utilized a mouse model of experimental cryptococcosis to investigate Treg cell responses to pulmonary fungal infection. Specifically, we explored the hypothesis that Treg cells utilize IRF4 and chemokine receptors to colocalize with Th2 cells in the lungs. While in proximity with Th2 cells, Treg cells are able to inhibit the expansion of deleterious Th2 cell responses to cryptococcal infection.
Materials and Methods
Mice
All mice used in this study were derived from a C57BL/6 background. B6.129P2-Ccr5tm1Kuz/J (22), B6.129P2(C)-Ccr7tm1Rfor/J (23), B6.Cg-Foxp3tm2Tch/J (24), B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J (25), Foxp3tm9(EGFP/cre/ERT2)Ayr/J (26), B6.129S1-Irf4tm1Rdf/J (27), B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, B6.PL-Thy1a/CyJ mice were purchased from Jackson Laboratories (Bar Harbor, ME). Foxp3-cre/GFP mice were a kind gift from Calvin Williams (28). Foxp3-eGFP mice were crossed with Thy1.1 mice to generate congenic marked mice for transfer experiements. Foxp3-cre ERT2 mice were crossed with tdTomato mice for Treg fate-mapping studies. All mice were housed in specific pathogen–free conditions.
Pathogen
Cryptococcus neoformans var. grubii strain KN99α was streaked on yeast peptone dextrose (YPD) agar plates and incubated for 2 days at 30°C. YPD broth was inoculated with colonies from the aforementioned plate and incubated for 16 hours at 30°C with gentle agitation. The inoculum was prepared by pelleting the culture, washing 3 times with phosphate buffered saline (PBS), and resuspending in PBS at a concentration of 2×106 cells/mL.
Infection
6–8 week old, sex-matched mice were anesthetized with pentobarbitol. 5×104 serotype A - KN99α (29) cryptococcal cells in 25 μL of PBS were placed on the nares of each mouse, and the mice aspirated the inoculum into the lower respiratory tract. Finally, the mice were suspended by their incisors for 5 minutes and subsequently placed upright in their cage until regaining consciousness. For survival studies, ten mice per group were infected as described above. Animals were monitored for morbidity and sacrificed when endpoint criteria were reached. Endpoint criteria were defined as 20% total body weight loss, loss of 2 grams of weight in 2 days, or symptoms of neurological disease.
Treatments
For intravital staining, 3 micrograms of anti-CD45.2 (104, BV421, Biolegend) was injected into the tail vein of mice or placed on the nares of sedated mice 3 minutes prior to sacrifice and whole blood/lung harvest (30). Foxp3-cre ERT2 tdTomato mice received 2mg/day tamoxifen IP for five consecutive days to induce endogenous fluorescence for Treg cell fate-mapping. For transfer studies, 1×106 negatively-selected CD4+ Th cells from naïve mice were injected via tail vein into congenic mice infected 7 days previously, and lungs were harvested at 14 days post-infection for leukocyte isolation and flow cytometric analysis. For CCR5 blockade experiments, mice were treated IP with 500 μg/day maraviroc (R&D Systems, Minneapolis, MN) from 9–14 days post-infection. Lastly, wildtype mice were treated 5 and 10 days post-infection with 1 mg of IL-10R antibody (1B1-3A, Bio-X-Cell) to block IL-10 signaling.
Pulmonary Leukocyte Preparation
Lung leukocytes were isolated as previously described (31). Briefly, lungs were excised and minced to generate approximately 1 mm3 pieces. The lung mince was incubated in HBSS (Invitrogen, Grand Island, NY) + 1.3 mM EDTA solution for 30 min at 37 °C with agitation, and then transferred to RPMI-1640 (Invitrogen) medium supplemented with 5% Fetal Bovine Serum (FBS) (Invitrogen) and 150 U/ml type I collagenase (Invitrogen) and incubated for 1 h at 37 °C with agitation. The cells were passed through a 70 μm filter, pelleted, and resuspended in 44% Percoll-RPMI medium (GE Life Sciences, Pittsburgh, PA). A percoll density gradient was created (44% top, 67% bottom), and the samples were centrifuged for 20 min at 650X g. The leukocytes at the interface were removed, washed 2 times with RPMI medium, and resuspended in PBS + FBS at a concentration of 107 cells/ml. CD4+ T cells were enriched using a Dynabeads CD4+ T Cell Negative Isolation Kit (Life Technologies, Grand Island, NY) per manufacture’s instructions. For intracellular cytokine analysis, ~106 CD4+ T cells were suspended in 200 μL of restimulation buffer (RPMI + 10% FBS + 1% penicillin/streptomycin + 5 μg brefeldin A) without (unstimulated) or with (stimulated) 10 ng phorbol myristate acetate (PMA) and 50 ng ionomycin. After 5 hours, the cells were washed and immediately prepared for flow cytometry.
Flow Cytometry
Samples were incubated for 5 minutes with CD16/32 antibody (Biolegend) and LIVE/DEAD Fixable Far Red stain (Invitrogen) to prevent nonspecific antibody binding, as well as mark dead cells. 25nM Cda2-tetramer was added to the sample and incubated at 25°C for 1 hour in the dark. CCR3 (J07E35, PE, Biolegend), CCR4 (2G12, PE, Biolegend), and CCR5 (HM-CCR5, PE, Biolegend) were added 1:50 during tetramer staining when appropriate. Samples were surface-stained at 4°C for 30 minutes with the following antibodies: CD4 (RM4-5, BV605, Biolegend), CD11b (M1/70, PE-Cy5, eBioscience, San Diego, CA), CD11c (N418, PE-Cy5, eBioscience), B220 (RA3-6B2, PE-Cy5, eBioscience), CD25 (3C7, BV650, Biolegend), CD44 (IM7, Alexa Fluor 700, Biolegend) and/or Siglec F (E50-2440, PE, BD Biosciences). When applicable, the cells were then incubated in Foxp3 Transcription Factor Buffer (eBioscience) at 4°C for 30 minutes. The cells were labeled with antibodes against the following intracellular antigens: Foxp3 (FJK-16s, FITC, eBioscience), IL-5 (TRFK5, APC, Biolegend), IL-13 (eBio13A, eFluor 450, eBioscience), GATA3 (L50-823, PE-Cy7, BD Biosciences), and/or IRF4 (3E4, eFluor 450, eBioscience). 1:200 antibody concentrations were used for most surface staining, and 1:100 antibody concentrations were used for intracelluar staining. For data acquisition, events from the entire sample (500,000–1,000,000) were collected on a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA), and the data were analyzed with FlowJo X (Tree Star Inc., Ashland, OR).
Naïve Cda2+ Th cell Enrichment
Analysis of antigen-specific Th cells within the pre-immune repertoire was performed, as previously published (32). Briefly, thymi and secondary lymphoid organs were collected from uninfected Foxp3-GFP mice. Cell suspensions were labeled with Cda2-tetramer and enriched using anti-PE MACS cell isolation kits (Miltenyi, San Diego, CA). Flow cytometry was performed as described above.
Lung Cytokines
Lungs from naïve mice or mice 14-days post-infection were excised, snap frozen in liquid nitrogen, and homogenized in 3 mL of T-PER (Thermo Fisher Scientific) with Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN). The lung homogenate was pelleted, and the supernatant was collected and stored at −80°C until analysis. Samples were diluted 1:4 in assay buffer immediately before processing. Cytokines were quantified using Luminex technology according to manufacturer instructions (Bio-Rad, Hercules, CA).
Lung Histology
Lungs were removed from mice 14 days post-infection, perfused via the right ventricle with cold PBS, inflated with 10% formalin (Thermo Fisher Scientific, Rockford, IL), and placed in a container of 10% formalin. Tissues were dried with organic solvent, embedded in parafin, sectioned, and stained with hematoxylin and eosin, before images were captured.
Statistics
P-values for pairwise comparisions were by Mann-Whitney U with Bonferroni adjustments for multiple comparisons. Global tests were by Kruskal-Wallis ANOVA. Surival curves were compared with log-rank tests. Power calculations were performed to assess appropriate sample size for all experiments. P-values ≤ 0.05 were considered statistically significant. All statistics and graphs were processed with Prism 6 (GraphPad Software, La Jolla, CA).
Study Approval
All animal experiments were done in concordance with the Animal Welfare Act, U.S. federal law, and NIH guidelines. Mice were handled in accordance with guidelines defined by the University of Minnesota Institutional Animal Care and Use Committee protocol numbers 1010A91133 and 1207A17286.
Results
Regulatory and effector Th cells coexist in the lung parenchyma
Upon pulmonary infection with the pathogenic fungus, Cryptococcus neoformans (KN99α), mice develop lethal disease that results from a combination of unabated fungal replication and Th2-driven immunopathology. Importantly, these detrimental Th2 cells are primed and accumulate in the lungs (15). Since Treg cell suppression of effector cells requires these cells to be in close proximity (16), we hypothesized that Treg/effector cells colocalize within Cryptococcus-infected lungs.
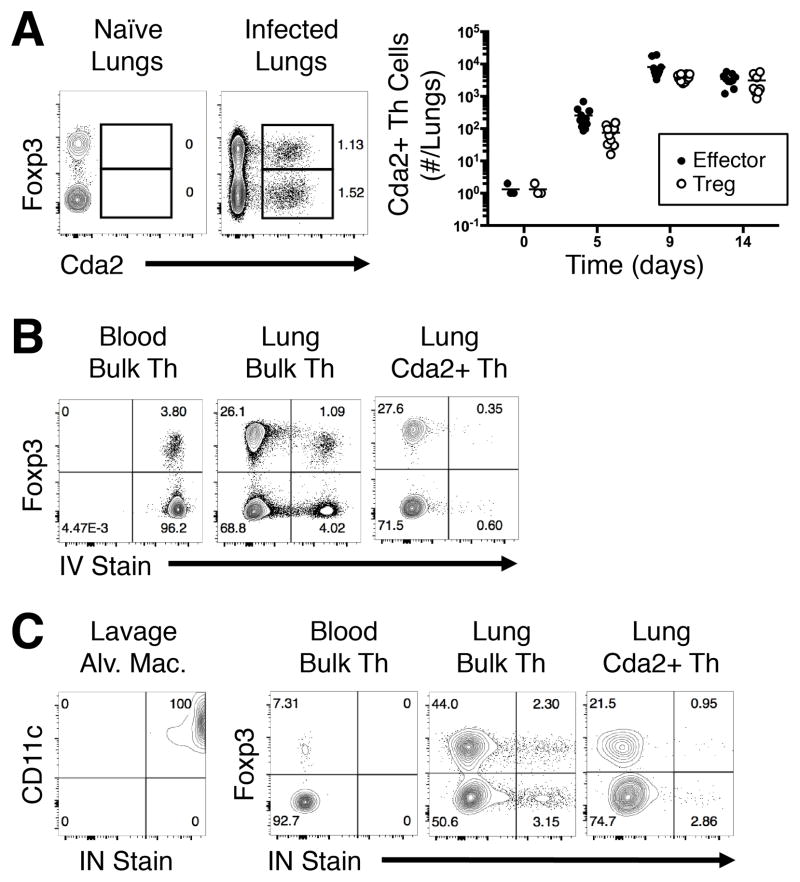
Th cells are highly heterogeneous with respect to their T cell receptor and cognate functions. Thus, the use of Cryptococcus-specific reagents to track antigen-specific Th cells within a polyclonal repertoire facilitates direct comparisons of Th cell subsets responding to infection. We used a peptide from chitin deacetylase 2 (Cda2), an immunogenic cryptococcal protein (33), to construct a recombinant peptide-MHCII tetramer to track Cryptococcus-specific Th cells by flow cytometry (15). The Cda2-MHCII tetramer not only identified Cda2+ Foxp3− effector Th cells in the lungs of infected mice, but the tetramer also labeled a sizable population of Cda2+ Foxp3+ Treg cells (Fig. 1A).
Figure 1. Antigen-specific Treg and effector Th cells colocalize in the lung parenchyma.
(A) Flow plots (left) and composite graph (right) of antigen-specific Cda2+ CD4+ Treg and effector Th cells in naïve and infected mice. (B) Cytometry plots of CD4+ Th cells in peripheral blood (left) or whole lung (center), and antigen-specific CD4+ Th cells in whole lungs (right) all treated with intravital/intravenous (IV) fluorescent CD45 antibody 3 minutes prior to euthanasia and tissue harvest. (C) Cytometry plots of CD11c+ Siglec F+ alveolar macrophages (far left), polyclonal Th cells in the blood (left), or CD4+ polyclonal Th cells (left) and antigen-specific Th cells (right) from whole lung digests after intravital/intranasal (IN) instillation of fluorescent CD45 antibody 3 minutes prior to euthanasia and tissue harvest. Plots represent two independent experiments.
The lung consists of three physically separate compartments that may contain Th cells: blood vessels, airways, and lung parenchyma. To determine whether the Treg/effector cells were circulating in the blood vasculature or residing in the airways/lung parenchyma, we performed intravital antibody staining. Anti-CD45 antibody administered i.v. was used to label polyclonal Foxp3+ Treg cells and Foxp3− effector Th cells retrieved from the peripheral blood. The complete labeling of all Foxp3+ and Foxp3− cells in the blood indicates that the antibody completely penetrated the entirety of the blood vasculature (Fig. 1B). Cells collected from whole lung digests had a mixture of i.v. stained and unstained cells, showing the lung was composed of circulating and lung-resident Treg/effector Th cells (Fig. 1B). In contrast, nearly all antigen-specific cells were unstained, showing that infection-induced Th cells resided outside of the blood vasculature (Fig. 1B). To distinguish Treg/effector Th cells in the lungs from the airways, we sedated infected mice and instilled fluorescent-coupled anti-CD45 antibody into the nares. Airway-resident alveolar macrophages collected by lavage were fully labeled by this intranasal antibody treatment, showing this method of antibody delivery was effective (Fig 1C). Conversely, Th cells in the peripheral blood remained unlabeled, indicating the antibody did not leak from the airways into blood circulation (Fig 1C). A majority of polyclonal and antigen-specific Treg/effector Th cells obtained from lung digests did not stain with this inhaled antibody (Fig. 1C). Therefore, most of the Treg/effector Th cells were not resident in the airways. By extension, these data collectively demonstrate that Treg and effector Th cells responding to infection coexist in the lung parenchyma.
Treg cell induction does not depend on lymphoid priming
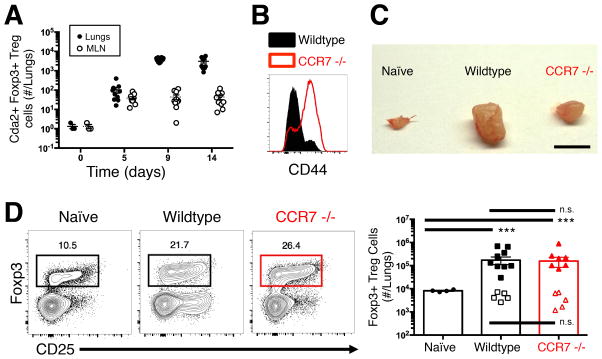
We next sought to determine the location of Treg cell induction. The mediastinal lymph node (MLN) is the principle origin of Th cell priming to most microbes that breech the airway mucosa. However, we previously showed that effector Th cells gather in the lungs in the absence of dendritic cell trafficking and subsequent T cell activation in the lung-draining lymph node (15). Thus, we compared the presence of Treg cells at the location of traditional Th cell priming in the MLN and the site of C. neoformans infection in the lungs. Cryptococcus-specific Foxp3-expressing Treg cells existed in the MLN and spleen, yet these populations remained relatively small compared to the large population of Cryptococcus-specific Treg cells that accumulated in the lungs (Fig. 2A, Fig. S1).
Figure 2. Lymph node priming is dispensable for Treg cell accumulation in the lungs.
(A) Cryptococcus-specific (Cda2) Foxp3+ Treg cells in the lungs and mediastinal lymph nodes (MLN). (B) Histogram of CD44 expression by CD4+ Th cells from lymph nodes of wildtype and CCR7 −/− mice 14 days post-infection. (C) MLN of naïve wildtype, as well as 14 days post-infection wildtype and chemokine receptor 7 (CCR7) deficient mice. Scale bar = 2 mm (D) Cytometry plot (left) and composite graph (right) of Foxp3+ Treg cells from lungs of naïve wildtype and 14 days post-infection wildtype and CCR7 −/− mice. Filled symbols indicate polyclonal Treg cells and open symbols are Cryptococcus-specific Treg cells. Pairwise comparisons were made by Man-Whitney U with Bonferoni adjustments for multiple comparisons. *** = P<0.0005, n.s. = non-significant. All data are presented as the mean +/− standard error of the mean and represent two independent experiments.
The relatively small Treg cell response in the MLN suggests that either Treg cells immediately migrate after activation in the lymph node to the site of infection or Treg cell induction occurs autonomously in the lungs. We used CCR7 −/− mice (23) to answer this question. CCR7 is required for naïve T cell entry into lymph nodes, thus CCR7 deficiency expectedly inhibited naïve Th cells (i.e. CD44 low) from accumulating in the MLN (Fig. 2B). Likewise, the MLN of infected CCR7 −/− mice exhibited decreased swelling compared to wildtype mice (Fig. 2C), further indicating dysfunctional Th cell priming in the MLN of CCR7-deficient mice. Despite the aberrant MLN response, bulk and Cryptococcus-specific Treg cells accumulated in the lungs of CCR7 −/− mice similar to levels in wildtype mice after C. neoformans infection (Fig. 2D). Thus, Treg cell induction and accumulation in the lungs does not require mediastinal lymph node priming during pulmonary cryptoccocal infection.
Cryptococcus-specific Treg cells are induced in the lungs upon pulmonary infection
Treg cells develop along two ontologically distinct lineages: “peripheral” Treg (pTreg) cells and “thymic” Treg (tTreg) cells. Upon receiving secondary cues of excessive inflammation, naïve Th cells can differentiate in the periphery into pTreg cells. Conversely, tTreg cells become regulatory cells during thymic selection based on T cell receptor affinity for self-antigens (34). Of note, tTreg cells emigrate from the thymus with full suppressive potency and do not need to undergo further activation in lymphoid tissues (35). Therefore, we asked whether the Treg cells in cryptococcal infected lungs are tTreg cells that populate the lungs independently of lymph node priming or pTreg cells autonomously induced in the lungs.
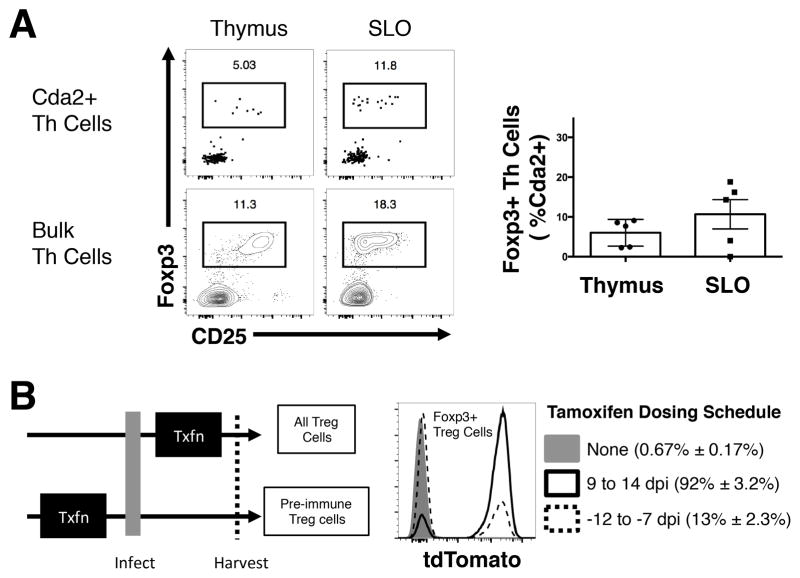
Cryptococcus-specific tTreg cells must exist in the pre-immune, naïve Th cell repertoire, if these cells are the dominant source of Treg cells in the lungs of infected mice. Therefore, we examined the thymus and secondary lymphoid organs of uninfected mice for the presence of Cda2+ Foxp3+ Treg cells. Cda2-specific Treg cells were present in the pre-immune repertoire contained in the thymus and secondary lymphoid organs, albeit at lower Treg/effector proportions compared with polyclonal Th cells (Fig. 3A). Therefore, a small number of Cryptococcus-specific tTreg cells can be found in uninfected mice, and these cells could migrate to the lung and proliferate in response to cryptococcal infection.
Figure 3. Treg cells in the preimmune repertoire are not the dominant source of Treg cells that accumulate in the lungs of fungal infected mice.
(A). Foxp3+ Treg cells as a proportion of Th cells in the thymus or secondary lymphoid tissue of naïve mice. Representative cytometry plots of polyclonal and antigen-specific Th cells (left) and composite graph of antigen-specific Th cells (right). (B) In vivo genetic fate-mapping strategy of Treg cells using Foxp3-cre ERT2 x Rosa26 stop-floxed tdTomato mice (left). Cytometry plots of fluorescent reporter activity in antigen-specific Treg cells from lungs with or without tamoxifen (Txfn) (right). All data are presented as the mean ± standard error of the mean and represent two independent experiments.
To further address the question of whether the Treg cells accumulating in lungs of infected mice migrated from the thymus or were induced in the lungs, we developed a genetic fate-mapping system to distinguish where these cells developed. Mice containing a Foxp3-cre Estrogen Receptor 2 (ERT2) transgene (26) were crossed with mice that had a Rosa26 stop codon-floxed tdTomato allele to make Foxp3-i-cre tdTomato mice. Effectively, the combination of these transgenes allows for inducible fluorescent marking in vivo of Treg cells and all of the progeny of these cells. Similarly, when tamoxifen is removed, Treg cells produced de novo will not have any detectable fluorescence reporter activity. Ultimately, this allowed us to label tTreg cells (and all cells derived from this progenitor) within the pre-immune repertoire, halt new reporter induction by stopping tamoxifen administration, and determine whether the lung-resident Treg cell progenitors existed prior to infection (i.e. tdTomato+) or were produced post-infection (i.e. tdTomato−) (Fig. 3B). Less than 1% of Treg cells from Foxp3-i-cre tdTomato mice that did not receive tamoxifen were fluorescent (Figure 3B), and tamoxifen administered during the peak Th cell response, 9–14 days post-infection, induced fluorescence in more than 90% of the Treg cells (Figure 3B). Thus, the genetic fate-mapping system is not leaky and suitably penetrant. When tamoxifen was given 12–7 days prior to infection to label the pre-immune Treg cells, a minor fraction of Treg cells retained fluorescence when harvested at 14 days post-infection (Figure 3B). Therefore, a small proportion of Treg cells in the lungs came from tTreg cells in the pre-immune repertoire, and instead, the majority were pTreg cells that acquire a regulatory phenotype as a consequence of fungal infection.
Interferon Regulatory Factor 4 expression by Treg cells is required to efficiently suppress the pathologic Th2 cell response to pulmonary fungal infection
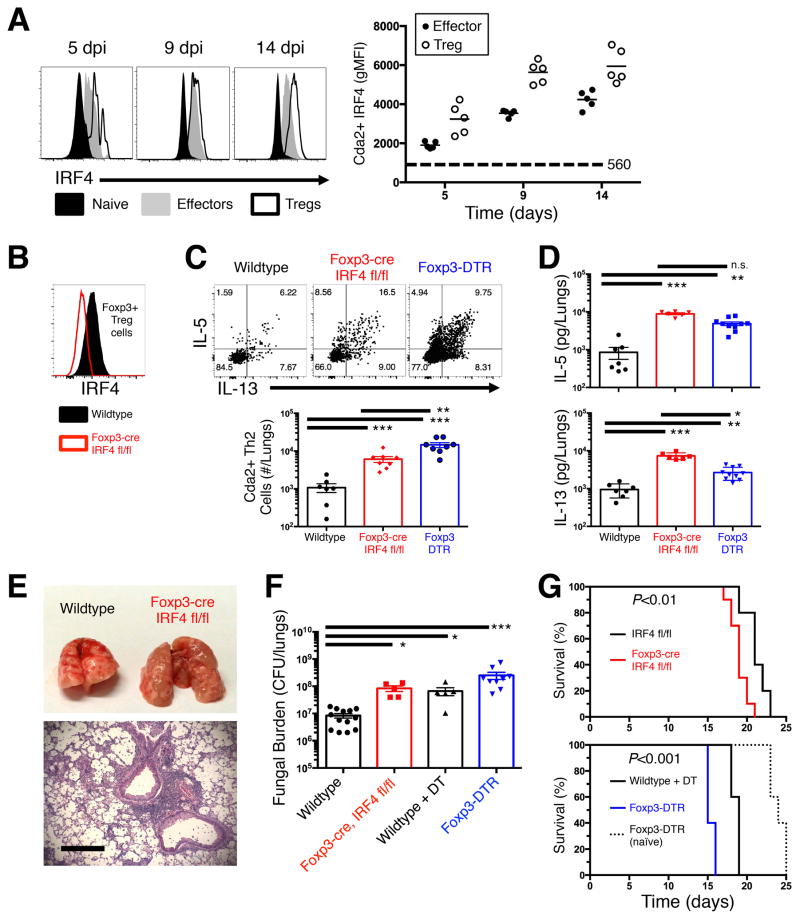
Treg cells generated during cryptococcal infection are poised to uniquely suppress Th2 cells (14, 15). Additionally, our data indicating that Treg cells are induced and reside in the lungs of infected mice led us to investigate features consistent with Treg cells that develop extrathymically, accumulate at mucosal surfaces, and target Th2 cells for suppression (36–38). A prominent feature of pTreg cells that suppress distinct Th cell subsets in mucosal tissues is the expression of transcription factors that mirror the lineage of the effector Th cell populations targeted for suppression (19, 21). Therefore, we examined the expression kinetics of the Th2 cell transcription factor, IRF4, by both antigen-specific Foxp3+ Treg cells and cognate Foxp3− effector Th2 cells from mice infected with C. neoformans. As hypothesized, IRF4 expression increased in Treg cells and effector Th cells throughout the course of infection (Fig. 4A). This raised the possibility that IRF4 is utilized by Treg cells to suppress the Th2 cell response to pulmonary fungal infection.
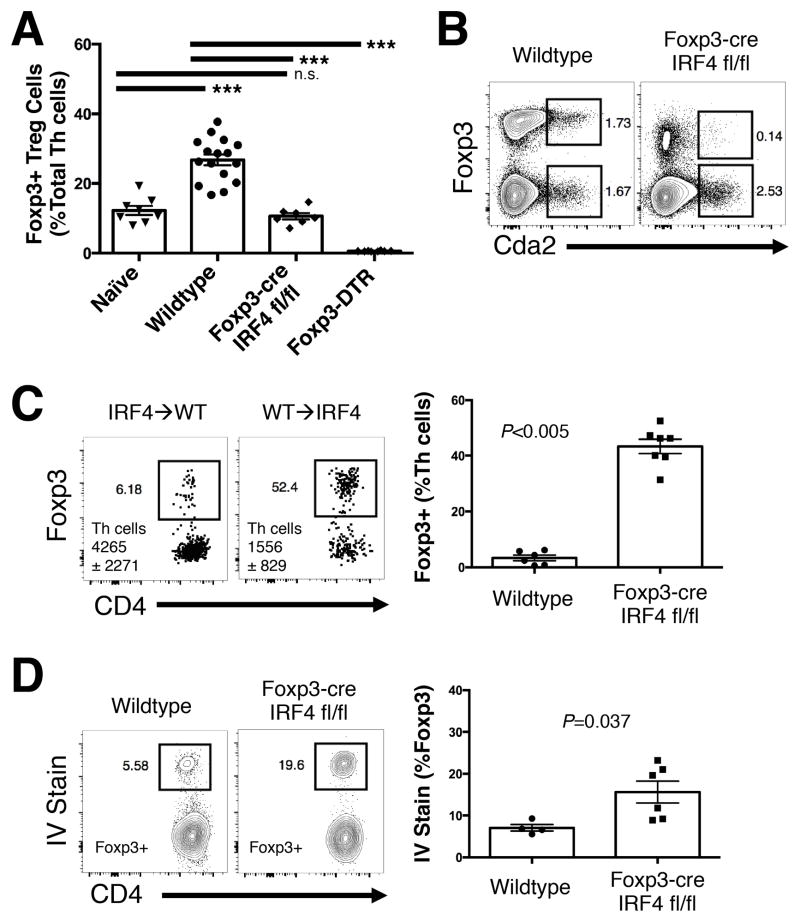
Figure 4. IRF4 is required by Treg cells to efficiently suppress the detrimental Th2 cell response to pulmonary cryptococcal infection.
(A) Flow cytometry histogram (left) and composite graphs (right) of Interferon Regulatory Factor 4 (IRF4) expression in CD44 low naïve, Cryptococcus-specific (Cda2+) Foxp3+ Treg, Cda2+ Foxp3− effector cells collected from the lungs of mice. (B) Histogram of IRF4 expression by lung Treg cells in wildtype and Foxp3-cre IRF4 fl/fl mice 14 days post infection. (C) Cda2+ Th2 cells producing interleukin-5 (IL-5) and/or IL-13 in the lungs of wildtype, Foxp3-cre, and Foxp3-DTR mice 14 days post-infection. (D) IL-5 and IL-13 secreted in lung homogenates from wildtype, Foxp3-cre, and Foxp3-DTR mice 14 days post-infection. (E) Photograph of gross-level pathology of lungs from mice infected 14 days previously (top). Hematoxylin and eosin staining of lung sections from Foxp3-cre mice 14 days post-infection (bottom). Bar = 200 μm. (F) Colony forming units (CFU) in the lungs of wildtype +/− diphtheria toxin (DT), Foxp3-cre IRF4 fl/fl, or Foxp3-DTR + DT. (G) Survival curve of IRF4 fl/fl and Foxp3-cre IRF4 fl/fl infected mice (top). Survival curves of naïve Foxp3-DTR, as well as infected wildtype and Foxp3-DTR mice – all groups treated every other day with 200 ng DT beginning at 5 days post-infection (bottom). Survival curves include 10 mice per group, and P-values calculated by log rank test. Pairwise comparisons were made by Man-Whitney U with Bonferoni adjustments for multiple comparisons. *** = P<0.0005, ** = P<0.005, * = P<0.05, n.s. = non-significant. All data are presented as the mean ± standard error of the mean and represent two independent experiments.
To test whether IRF4 expression by Tregs was important for Th2 suppression, we bred Foxp3-cre mice (28) with IRF4 floxed mice (27) to generate mice with a conditional IRF4 gene deletion in Treg cells (Foxp3-cre IFR4 fl/fl) (Fig. 4B). Cryptococcus-specific Th2 cells increased >5-fold in the lungs of Foxp3-cre IRF4 fl/fl mice, and this impaired suppression of Th2 cells resembled the situation observed with complete Treg abrogation (25) (Fig. 4C). Consistent with the increase in Th2 cell numbers, Foxp3-cre IRF4 fl/fl mice also had significantly elevated amounts of IL-5 and IL-13 from infected lung homogenates compared with both wildtype animals and Treg cell depleted mice (Fig. 4D). Importantly, Foxp3-cre IRF4 fl/fl mice did not experience a concomitant increase in Th17 and Th1 cell cytokine production (Fig. S2). Altogether, IRF4 is utilized by Treg cells to suppress Th2 cell responses to pulmonary cryptococcal infection.
The failure to efficiently suppress Th2 cell proliferation and effector function in mice with IRF4-deficient Treg cells also correlated with exacerbation of Th2-mediated disease. IRF4-deficiency in Treg cells aggravated gross lung pathology (Fig 4E), as well as enhanced pulmonary accumulation of macrophages, multinucleate giant cells, and polymorphonuclear cells (Fig. 4E). Foxp3-cre IRF4 fl/fl mice had elevated fungal burden and accelerated cryptococcal disease (Fig. 4F&G). While lethal disease onset appeared to be more rapid in Foxp3-DTR mice relative to Foxp3-cre IRF4 fl/fl mice, naïve Foxp3-DTR mice receiving DT experienced fatal autoimmunity (Fig. 4G). DT treatment also negatively impacted the survival of infected wildtype mice independently of Treg cell ablation by potentially augmenting fungal burden (Fig. 4F&G). Thus, DT treatment and autoimmunity additionally contributed to the faster disease experienced by Foxp3-DTR mice. Taken together, IRF4-deficient Treg cells exhibited a profound Th2 suppression defect that was comparable to complete Treg cell deficiency.
Interferon Regulatory Factor 4 is needed for localization of Treg cells in cryptococcal infected lungs
How Treg cells utilize IRF4 to suppress Th2 cells remains incompletely understood. Existing evidence suggests that IRF4 may dictate expression of suppressive factors employed by Treg cells. For example, IRF4 interacts with Blimp-1 to mediate transcription of the suppressive cytokine, IL-10 (39). Chromatin immunoprecipitation of IRF4 confirms that IRF4 binds to the IL-10 locus, and IRF4 has been shown to mediate IL-10 production by Th2 cells (40, 41). However, IL-10 in lung homogenates of cryptococcal infected mice was unaffected by IRF4-deficiency in Treg cells and actually increased in mice with complete Treg cell abrogation (Fig. S3A). Furthermore, blockade with substantial quantities (i.e. 2 mg/mouse over the course of 9 days) of anti-IL-10R antibody did not alter Th2 cell production (Fig. S3B) or IL-5 and IL-13 secretion in lungs of infected mice (Fig. S3C). Finally, in other systems, IRF4-deficient Treg cells still suppress effector Th cells in an in vitro assay, indicating that IRF4 is dispensable for the direct suppression of effector Th cells by Treg cells (21).
An alternative hypothesis is IRF4 promotes the retention of Treg cells at the site of inflammation. Although IRF4-deficiency in Treg cells did not alter the proportion of Foxp3+ Treg cells among total CD4+ Th cells in the spleen and MLN (Fig. S4A), it significantly decreased Treg cell proportions in the lungs of infected mice (Fig. 5A). Furthermore, Foxp3-cre IRF4 fl/fl mice had substantially fewer antigen-specific Treg cells in the lungs in comparison to wildtype mice (Fig. 5B). However, these studies could not determine whether the decreased Treg cell proportions were due to biased effector Th cell accumulation or defective Treg cell retention in the lungs.
Figure 5. Pulmonary retention of Treg cells is maintained by IRF4.
(A) Frequency of Foxp3+ Treg cells in the lungs of naïve wildtype mice and wildtype, Foxp3-cre, and Foxp3-DTR mice 14 days post-infection. (B) Flow cytometry plots of antigen-specific Treg and effector cells in the lungs of mice 14 days post-infection. (C) Flow cytometry plots and composite graphs of Foxp3-IRF4 fl/fl or widltype donor Th cells collected from lungs of wildtype or Foxp3-cre IRF4 mismatched recipients at 14 days post-infection. Cytometry plots show congenic marked donor Th cells collected from the recipient with quantities of Th cells deposited also indicated in the plot. (D) Flow plots and composite graphs of pulmonary Treg cells from wildtype and Foxp3-cre IRF4 mice infected 14 days previously and treated with intravenous anti-CD45 antibody 3 minutes before lung harvest. Pairwise comparisons were made by Man-Whitney U with Bonferoni adjustments for multiple comparisons. *** = P<0.0005, n.s. = non-significant. All data are presented as the mean ± standard error of the mean and represent two independent experiments.
To test the hypothesis that IRF4 promotes Treg cell localization in the lungs, we performed a set of adoptive transfers. Naïve CD4+ Th cells from Foxp3-cre IRF4 fl/fl mice were transferred into congenic wildtype recipients, as well as the reciprocal transfer of wildtype Th cells into Foxp3-cre IRF4 fl/fl mice. After resting in infected mice for 5 days (9–14 days post-infection of recipient), the donor cells were identified using the congenic markers. Transferred Th cells from Foxp3-cre IRF4 and wildtype mice parked equivalently in the lungs of their respective hosts (Fig. 5C), yet the transferred cells remained vastly outnumbered by the recipient cells. This allowed us to observe the behavior of transferred cells in the context of a recipient dominated inflammatory milieu. Despite a fully competent wildtype suppressive response that should suppress Th2 cell proliferation, we still observe a blunted Treg cell response skewed towards effector Th cells in Foxp3-cre IRF4 fl/fl transferred cells (Fig. 5C), indicating Treg cells lacking IRF4 were inefficiently retained in the lungs. IRF4-deficient Treg cells deposited normally in the MLN and spleens of infected mice (Fig. S4B). Thus, a generalized defect in Treg induction of the Foxp3-cre IRF4 cells was not apparent. Finally, Treg cells from Foxp3-cre IRF4 fl/fl mice were over-represented in the blood vasculature of infected lungs (Fig. 5D), correlating the decrease in pulmonary retention of Treg cells with the diffusion of these cells into the local bloodstream. These data demonstrate that IRF4 intrinsically regulates Treg cell localization in the lungs of cryptococcal infected mice.
Treg cell accumulation in the lungs is dependent on Chemokine Receptor 5
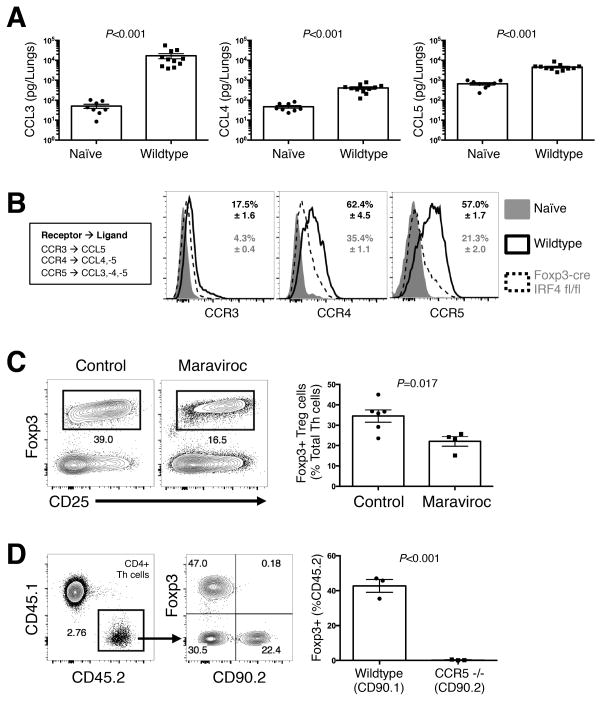
Th cells follow chemokine gradients to traffic to the site of inflammation (18). Therefore, we investigated chemotactic signals that may influence pulmonary localization of Treg cells. CCL3, CCL4, and CCL5 are involved in type-2 immunity (42), and these chemokines increased 5–100-fold in the lungs of infected mice compared with naïve controls (Fig. 6A). To determine if the Treg cells could recognize these chemokines, we examined expression of the cognate chemokine receptors by Treg cells in the lungs of infected mice (Fig. 6B). CCR4 and CCR5 were highly expressed by Treg cells, and expression of these receptors decreased in IRF4-deficient Treg cells (Fig. 6B). In contrast, CCR3 was minimally expressed by Treg cells (Fig. 6B). Thus, CCL3, CCL4, and CCL5 were highly abundant in the lungs of infected mice, and the ability to detect these chemokine signals by Treg cells would require IRF4-dependent expression of CCR4 and CCR5.
Figure 6. Treg cell accumulation is mediated by CCR5 via IRF4.
(A) Chemokine Ligands (CCL) measured in lung homogenates from wildtype mice 14 days post-infection. (B) Cytometry histograms of Chemokine Receptor (CCR) expression on CD44 low naïve cells, as well as Treg cells from wildtype and Foxp3-cre IRF4 fl/fl mice 14 days post-infection. (C) Flow plots (left) and composite graphs (right) of Treg cells in the lungs of mice 14 days post-infection with and without maraviroc treatment. (D) CD45.2/CD90.1 wildtype and CD45.2/CD90.2 CCR5 −/− naïve Th cells transferred into a CD45.1/CD90.2 Foxp3-DTR mouse infected and Treg cell-depleted 7 days previously. Pairwise comparisons were made by Man-Whitney U. All data are presented as the mean ± standard error of the mean and represent 2 independent experiments.
Due to the elevated expression of CCR5, abundance of cognate chemokine ligands, and the high dependence of CCR5 on IRF4, we tested the causal relationship between CCR5 and pulmonary retention of Treg cells during fungal infection. Maraviroc is a selective inhibitor of CCR5 that is used in HIV patients to block CCR5-mediated entry of HIV into leukocytes (43). Mice that received 500 micrograms of maraviroc every day from 9 days to 14 days post-infection had significantly reduced accumulation of pulmonary Treg cells compared to similarly infected, vehicle treated controls (Fig. 6C). To further test the requirement of CCR5 for Treg localization in the lungs. CD45.1/CD90.2 congenic Foxp3-DTR mice were infected, and Treg cells were eliminated at 7 days post-infection by administering DT. 1 million naïve CD4+ Th cells from uninfected CD45.2/CD90.1 wildtype and CD45.2/CD90.2 CCR5 −/− mice (22) were transferred into the Foxp3-DTR mice. At 14 days post-infection the lungs were harvested and analyzed for Treg accumulation in the lungs. Strikingly, while wildtype Tregs readily accumulated in the lungs, the Tregs transferred from CCR5 −/−, mice were absent from the lungs of infected mice (Fig. 6D). Thus, Treg cell induction and retention in the lungs requires CCR5.
Discussion
Th cells are central to immunity and immunopathology associated with cryptococcal infection. While Th1 cells correlate with protection, Th2 cells exacerbate cryptococcal disease. Therefore, a deeper understanding of how the diseased host regulates Th cell responses could lead to development of interventions that ameliorate disease in predisposed individuals. One promising target of immune modulation is the Foxp3+ Treg cell population. Previously, Treg cells were found to counterbalance pathologic Th2 cell inflammation following pulmonary cryptococcal infection (14, 15). Yet, the mechanism behind this suppression was largely unexplored. Herein, we tracked Cryptococcus-specific Th cell responses with multi-parameter flow cytometry and manipulated host immune responses to unravel the mechanism of Treg-mediated suppression of Th2 cells during cryptococcal infection. We showed that Treg cells are induced in the tissues and utilize CCR5 and IRF4 to colocalize with and suppress Th2 effector cells in the lung parenchyma.
Many fungal pathogens elicit Treg cell responses. In most cases, Treg cells control the axis of Th17 cell responses and fungal clearance (44–47). In contrast, the primary function of Treg cells generated during cryptococcal infection is Th2 cell suppression. This clearly benefits the host, as disease is enhanced when Treg cells fail to adequately control Th2 cell proliferation. The signals the host uses to detect host damage and elicit Treg cell induction are unknown in the case of pulmonary cryptococcal infection. Additional insight into these processes could lead to the identification of potent biomarkers to predict immune dysfunction in patients stricken with cryptococcal disease. Furthermore, therapeutic targeting of these pathways could be used to prompt the host to dampen harmful Th2 cell production.
We report that Treg cells are induced in substantial quantities during cryptococcal infection, even in the absence of CCR7-mediated entry of naïve Th cells into the MLN. Th2 cells also accumulate in the lungs independently of dendritic cell trafficking to the MLN and subsequent Th cell priming (15). This begs the question as to the precise location of T cell priming during cryptococcal infection. Bronchus-associate lymphoid tissue (BALT) is comprised of stochastically distributed clusters of lymphocytes in proximity to high endothelial venules and tissue-resident dendritic cells (48). These structures exist under homeostatic conditions and readily increase in size and number (known as “inducible” BALT) in individuals with chronic inflammatory conditions (49, 50). BALT sufficiently supports T cell priming in the absence of canonical lymphoid responses (51, 52). It is plausible that the BALT is responsible for T cell priming in cryptococcal infection. Fungi may not freely diffuse through the lymphatics to reach the lymph nodes due to the relatively large size of individual fungal cells. Consequently, during early cryptococcal infection in mice, an overwhelming majority of antigen is contained in the lungs. The lung is a high blood flow organ, so circulating naïve Th cells have consistent access to the depot of cryptococcal antigen. Naïve Th cells could be coaxed into the lungs via high endothelial venules in a chemokine/integrin-mediated process, and the BALT could direct naïve Th cell activation, proliferation, and differentiation.
Treg cells are required for the suppression of Th2 cells in this model of pulmonary cryptococcal infection. However, the mechanism of Treg cell-mediated suppression was unknown. IL-10 production by Treg cells is a well-known pathway by which Treg cells inhibit pulmonary Th cell responses (53). IL-10 signaling reduces the proliferative potential of Th cells (54), as well as amplifies the suppressive potency of Treg cells (55). However, IL-10 blockade had minimal impact on Th2 cell responses to cryptococcal infection. Previous studies have shown IL-10-independent Treg cell suppression of effector Th cells involves close contact (56). Thus, mechanistically, the colocalization of Treg cells with effector Th2 cells during cryptococcal infection is an important observation, and the ability of Treg cells to inhibit effector Th cell niches affords unique functional opportunities.
Perhaps, the most interesting regulatory pathway concerns the potential ability of Treg cells to mediate suppression by starving Th cells of local growth factors. In particular, Treg cells scavenge IL-2 via their high affinity IL-2 receptor (57). This competition limits IL-2 growth factor availability and restricts Th cell proliferation (58). There is some evidence to suggest this might occur in the context of pulmonary cryptococcal infection. First, IL-2 complexes administered to infected mice massively augment Th2 cell accumulation (15). Thus, IL-2 is not only an important signal for Th2 cell proliferation, but IL-2 is also a limited resource in this setting. Additionally, Signal Transducer and Activator 6 (STAT6) and IRF4 are each individually required for Th2 cell generation during cryptococcal infection (unpublished observations). However, the requirement for these transcription factors can be bypassed by treating infected knockout mice with IL-2 complexes (unpublished observations). Local IL-2 starvation by Treg cells leading to Th2 cell suppression is an intriguing, but still untested hypothesis in this model of pulmonary fungal infection.
Collectively, our data unify several emerging concepts regarding Treg cell suppression of Th2 cells. Peripherally induced Treg cells inhibit Th2 cells at mucosal surfaces (38), and Treg cells utilize effector cell programs like IRF4 to mediate specific suppression of Th2 cells (21). IRF4 functions as a rheostat for T cell receptor signaling (59), and TCR signaling is required to maintain a portion of the suppressive program of Treg cells (60). Additionally, chemokines promote the migration and retention of Treg cells in inflamed tissues (18), and CCR5 is important for Treg cells to suppress Th cell responses to pulmonary fungal infections (61). In our model, Treg cells were induced in the periphery and IRF4 expression by Treg cells was required for efficient Th2 suppression. Treg cells in the lungs of cryptococcal infected mice expressed high levels of CCR5, and the few remaining IRF4-deficient Treg cells in the lungs had significantly decreased expression of CCR5. IRF4 does not directly interact with the promoter region of CCR5 (41) and does not likely influence CCR5 gene transcription. Thus, we favor a model where diminished T cell receptor signaling in Treg cells due to IRF4 deficiency reduces CCR5 expression. This prevents Treg/effector cell colocalization and hinders Treg suppression of Th2 cells. Thus, our data provide a logical connection between the hitherto disjointed observations of extrathymically induced Treg cells, IRF4-dependent suppression, chemokine-mediated localization, and Th2-specific inhibition.
Skewed type-2 cytokine responses in the peripheral blood and cerebral spinal fluid of patients with cryptococcal meningitis are associated with early mortality and onset of immune reconstitution inflammatory syndrome (5, 62). CCR5+ T cells are recruited to the CSF of patients experiencing cryptococcal meningitis and increased presence of CCR5+ T cells is associated with poor clinical outcome (63). HIV infects and lyses CCR5+ and CXCR4+ Th cells equally (64), and the in vivo evolution of CXCR4-tropic virus is assisted by efficient elimination of CCR5+ Th cells (65). Moreover, maraviroc treatment selectively eliminates Treg cells in HIV patients (66). Taken together with our findings that Treg cells require CCR5 to colocalize and suppress detrimental Th2 cell responses, these observations unveil a novel potential etiology of cryptococcal pathogenesis. Perhaps, HIV-directed lysis of CCR5+ Treg cells and/or therapeutic targeting of CCR5 in people living with HIV could exacerbate Th2-driven disease experienced by patients with cryptococcal meningitis.
Supplementary Material
Acknowledgments
We thank Dr. Calvin Williams (Medical College of Wisconsin) for kindly providing Foxp3-cre mice, as well as Dr. Marc Jenkins for helpful discussions. We are also grateful to the University of Minnesota Flow Cytometry Core Facility for instrumentation, and the University of North Carolina Lineberger Comprehensive Cancer Center Animal Histopathology Core Facility.
This work was supported by NIH grant AI080275 to KN. DLW received support from NIH T32 training grant AI007313, University of Minnesota Doctoral Dissertation Fellowship, and Dennis W. Watson Fellowship.
Abbreviations used in this article
- BALT
Bronchus associated lymphoid tissue
- Cda2
Chitin deacetylase 2
- DTR
Diptheria toxin receptor
- ERT2
Estrogen receptor 2
- Foxp3
Forkhead box P3
- Th
Helper T
- IRF4
Interferon regulatory factor 4
- MLN
Mediastinal lymph node
- pTreg
Peripheral regulatory T
- Treg
regulatory T
- STAT6
Signal transducer and activator 6
- tTreg
Thymic regulatory T
Footnotes
Author Contributions
DLW conceived and performed experiments and wrote the manuscript. KDS, DIK, and JNK performed experiments and edited the manuscript. PRB and KN helped conceive experiments and edited the manuscript.
References
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Wiesner DL, Boulware DR. Cryptococcus-Related Immune Reconstitution Inflammatory Syndrome(IRIS): Pathogenesis and Its Clinical Implications. Current fungal infection reports. 2011;5:252–261. doi: 10.1007/s12281-011-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bicanic T, Meintjes G, Wood R, Hayes M, Rebe K, Bekker LG, Harrison T. Fungal burden, early fungicidal activity, and outcome in cryptococcal meningitis in antiretroviral-naive or antiretroviral-experienced patients treated with amphotericin B or fluconazole. Clin Infect Dis. 2007;45:76–80. doi: 10.1086/518607. [DOI] [PubMed] [Google Scholar]
- 4.French N, Gray K, Watera C, Nakiyingi J, Lugada E, Moore M, Lalloo D, Whitworth JA, Gilks CF. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS. 2002;16:1031–1038. doi: 10.1097/00002030-200205030-00009. [DOI] [PubMed] [Google Scholar]
- 5.Boulware DR, Meya DB, Bergemann TL, Wiesner DL, Rhein J, Musubire A, Lee SJ, Kambugu A, Janoff EN, Bohjanen PR. Clinical features and serum biomarkers in HIV immune reconstitution inflammatory syndrome after cryptococcal meningitis: a prospective cohort study. PLoS medicine. 2010;7:e1000384. doi: 10.1371/journal.pmed.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarvis JN, Meintjes G, Rebe K, Williams GN, Bicanic T, Williams A, Schutz C, Bekker LG, Wood R, Harrison TS. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. Aids. 2012;26:1105–1113. doi: 10.1097/QAD.0b013e3283536a93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decken K, Kohler G, Palmer-Lehmann K, Wunderlin A, Mattner F, Magram J, Gately MK, Alber G. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infection and immunity. 1998;66:4994–5000. doi: 10.1128/iai.66.10.4994-5000.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wormley FL, Jr, Perfect JR, Steele C, Cox GM. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infection and immunity. 2007;75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulware DR, Meya DB, Muzoora C, Rolfes MA, Huppler Hullsiek K, Musubire A, Taseera K, Nabeta HW, Schutz C, Williams DA, Rajasingham R, Rhein J, Thienemann FW, Lo M, Nielsen K, Bergemann TL, Kambugu A, Manabe YC, Janoff EN, Bohjanen PR, Meintjes G CT Team. Timing of antiretroviral therapy after diagnosis of cryptococcal meningitis. The New England journal of medicine. 2014;370:2487–2498. doi: 10.1056/NEJMoa1312884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller U, Stenzel W, Kohler G, Polte T, Blessing M, Mann A, Piehler D, Brombacher F, Alber G. A gene-dosage effect for interleukin-4 receptor alpha-chain expression has an impact on Th2-mediated allergic inflammation during bronchopulmonary mycosis. The Journal of infectious diseases. 2008;198:1714–1721. doi: 10.1086/593068. [DOI] [PubMed] [Google Scholar]
- 11.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, Sonstein J, Curtis JL, Huffnagle GB, Toews GB, Olszewski MA. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. The American journal of pathology. 2009;174:932–943. doi: 10.2353/ajpath.2009.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verbsky JW, Chatila TA. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and IPEX-related disorders: an evolving web of heritable autoimmune diseases. Current opinion in pediatrics. 2013;25:708–714. doi: 10.1097/MOP.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe JH, Ertelt JM, Way SS. Foxp3(+) regulatory T cells, immune stimulation and host defence against infection. Immunology. 2012;136:1–10. doi: 10.1111/j.1365-2567.2011.03551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze B, Piehler D, Eschke M, von Buttlar H, Kohler G, Sparwasser T, Alber G. CD4(+) FoxP3(+) regulatory T cells suppress fatal T helper 2 cell immunity during pulmonary fungal infection. European journal of immunology. 2014;44:3596–3604. doi: 10.1002/eji.201444963. [DOI] [PubMed] [Google Scholar]
- 15.Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, Lee CG, Elias JA, Nielsen JN, Boulware DR, Bohjanen PR, Jenkins MK, Levitz SM, Nielsen K. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS pathogens. 2015;11:e1004701. doi: 10.1371/journal.ppat.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T. Regulatory T cells: how do they suppress immune responses? International immunology. 2009;21:1105–1111. doi: 10.1093/intimm/dxp095. [DOI] [PubMed] [Google Scholar]
- 17.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nature immunology. 2013;14:1007–1013. [Google Scholar]
- 18.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nature reviews Immunology. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program coopts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuziel WA, Dawson TC, Quinones M, Garavito E, Chenaux G, Ahuja SS, Reddick RL, Maeda N. CCR5 deficiency is not protective in the early stages of atherogenesis in apoE knockout mice. Atherosclerosis. 2003;167:25–32. doi: 10.1016/s0021-9150(02)00382-9. [DOI] [PubMed] [Google Scholar]
- 23.Hopken UE, Droese J, Li JP, Joergensen J, Breitfeld D, Zerwes HG, Lipp M. The chemokine receptor CCR7 controls lymph node-dependent cytotoxic T cell priming in alloimmune responses. European journal of immunology. 2004;34:461–470. doi: 10.1002/eji.200324690. [DOI] [PubMed] [Google Scholar]
- 24.Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. Journal of immunology. 2007;178:2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- 25.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature immunology. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 26.Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nature immunology. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. The Journal of experimental medicine. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infection and immunity. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D. Intravascular staining for discrimination of vascular and tissue leukocytes. Nature protocols. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Dong Z, Zhou R, Luo D, Wei H, Tian Z. Isolation of lymphocytes and their innate immune characterizations from liver, intestine, lung and uterus. Cell Mol Immunol. 2005;2:271–280. [PubMed] [Google Scholar]
- 32.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levitz SM, Nong S, Mansour MK, Huang C, Specht CA. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10422–10427. doi: 10.1073/pnas.181331398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. The Journal of experimental medicine. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Zheng Y. Regulatory T cell identity: formation and maintenance. Trends in immunology. 2015;36:344–353. doi: 10.1016/j.it.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends in immunology. 2013;34:74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S, Fagarasan S, Mielke LA, Afshar-Sterle S, Masters SL, Nakae S, Saito H, Wentworth JM, Li P, Liao W, Leonard WJ, Smyth GK, Shi W, Nutt SL, Koyasu S, Kallies A. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nature immunology. 2015;16:276–285. doi: 10.1038/ni.3085. [DOI] [PubMed] [Google Scholar]
- 38.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature immunology. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 40.Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. Journal of immunology. 2009;183:1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sallusto F, Lanzavecchia A, Mackay CR. Chemokines and chemokine receptors in T-cell priming and Th1/Th2-mediated responses. Immunology today. 1998;19:568–574. doi: 10.1016/s0167-5699(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 43.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrobial agents and chemotherapy. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroetz DN, Deepe GS., Jr CCR5 dictates the equilibrium of proinflammatory IL-17+ and regulatory Foxp3+ T cells in fungal infection. Journal of immunology. 2010;184:5224–5231. doi: 10.4049/jimmunol.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinley L, Logar AJ, McAllister F, Zheng M, Steele C, Kolls JK. Regulatory T cells dampen pulmonary inflammation and lung injury in an animal model of pneumocystis pneumonia. Journal of immunology. 2006;177:6215–6226. doi: 10.4049/jimmunol.177.9.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Netea MG, Sutmuller R, Hermann C, Van der Graaf CA, Van der Meer JW, van Krieken JH, Hartung T, Adema G, Kullberg BJ. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. Journal of immunology. 2004;172:3712–3718. doi: 10.4049/jimmunol.172.6.3712. [DOI] [PubMed] [Google Scholar]
- 47.Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422–434. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foo SY, Phipps S. Regulation of inducible BALT formation and contribution to immunity and pathology. Mucosal immunology. 2010;3:537–544. doi: 10.1038/mi.2010.52. [DOI] [PubMed] [Google Scholar]
- 49.Richmond I, Pritchard GE, Ashcroft T, Avery A, Corris PA, Walters EH. Bronchus associated lymphoid tissue (BALT) in human lung: its distribution in smokers and non-smokers. Thorax. 1993;48:1130–1134. doi: 10.1136/thx.48.11.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suda T, Chida K, Hayakawa H, Imokawa S, Iwata M, Nakamura H, Sato A. Development of bronchus-associated lymphoid tissue in chronic hypersensitivity pneumonitis. Chest. 1999;115:357–363. doi: 10.1378/chest.115.2.357. [DOI] [PubMed] [Google Scholar]
- 51.Halle S, Dujardin HC, Bakocevic N, Fleige H, Danzer H, Willenzon S, Suezer Y, Hammerling G, Garbi N, Sutter G, Worbs T, Forster R. Induced bronchus-associated lymphoid tissue serves as a general priming site for T cells and is maintained by dendritic cells. The Journal of experimental medicine. 2009;206:2593–2601. doi: 10.1084/jem.20091472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, Hartson L, Sprague F, Goodrich S, Woodland DL, Lund FE, Randall TD. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nature medicine. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 53.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, Muller W, Rudensky AY. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Taga K, Tosato G. IL-10 inhibits human T cell proliferation and IL-2 production. Journal of immunology. 1992;148:1143–1148. [PubMed] [Google Scholar]
- 55.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Bruning JC, Muller W, Rudensky AY. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Garra A, Vieira PL, Vieira P, Goldfeld AE. IL-10-producing and naturally occurring CD4+ Tregs: limiting collateral damage. The Journal of clinical investigation. 2004;114:1372–1378. doi: 10.1172/JCI23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sitrin J, Ring A, Garcia KC, Benoist C, Mathis D. Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. The Journal of experimental medicine. 2013;210:1153–1165. doi: 10.1084/jem.20122248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leon B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nature communications. 2014;5:3495. doi: 10.1038/ncomms4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nature immunology. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 60.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nature immunology. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreira AP, Cavassani KA, Massafera Tristao FS, Campanelli AP, Martinez R, Rossi MA, Silva JS. CCR5-dependent regulatory T cell migration mediates fungal survival and severe immunosuppression. Journal of immunology. 2008;180:3049–3056. doi: 10.4049/jimmunol.180.5.3049. [DOI] [PubMed] [Google Scholar]
- 62.Boulware DR, Bonham SC, Meya DB, Wiesner DL, Park GS, Kambugu A, Janoff EN, Bohjanen PR. Paucity of initial cerebrospinal fluid inflammation in cryptococcal meningitis is associated with subsequent immune reconstitution inflammatory syndrome. The Journal of infectious diseases. 2010;202:962–970. doi: 10.1086/655785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang CC, Omarjee S, Lim A, Spelman T, Gosnell BI, Carr WH, Elliott JH, Moosa MY, Ndung’u T, French MA, Lewin SR. Chemokine levels and chemokine receptor expression in the blood and the cerebrospinal fluid of HIV-infected patients with cryptococcal meningitis and cryptococcosis-associated immune reconstitution inflammatory syndrome. The Journal of infectious diseases. 2013;208:1604–1612. doi: 10.1093/infdis/jit388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grivel JC, Margolis LB. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nature medicine. 1999;5:344–346. doi: 10.1038/6565. [DOI] [PubMed] [Google Scholar]
- 65.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, Fenyo EM, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nature medicine. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 66.Pozo-Balado MM, Martinez-Bonet M, Rosado I, Ruiz-Mateos E, Mendez-Lagares G, Rodriguez-Mendez MM, Vidal F, Munoz-Fernandez MA, Pacheco YM, Leal M. Maraviroc reduces the regulatory T-cell frequency in antiretroviral-naive HIV-infected subjects. The Journal of infectious diseases. 2014;210:890–898. doi: 10.1093/infdis/jiu180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.