Abstract
It is known since the discovery of DNA vaccines more than 20 years ago that DNA vaccines can function as adjuvants. Our recent study reported the involvement of Aim2 as the sensor of DNA vaccines in eliciting antigen-specific antibody responses. Our findings indicated the presence of previously unrecognized innate immune response pathways in addition to the TLR9 pathway, which is mainly activated by the CpG motifs of DNA vaccines. Our data further demonstrated the requirement of type I IFN in DNA vaccine-induced immune responses via the Aim2 pathway, but the exact downstream molecular mechanism was not characterized. In the current study, we investigated the roles of the putative DNA sensor cyclic GMP-AMP synthase (cGas), as well as the downstream interferon regulatory factors (Irf) 3 and 7 in type I IFN induction and antigen-specific immune responses elicited by DNA vaccination. Our results showed that DNA vaccine induced, Irf7-dependent signaling, as part of the Sting pathway, was critical for generation of both innate cytokine signaling and antigen-specific B and T cell responses. In contrast, Irf3 was not as critical as expected in this pathway and more surprisingly, immune responses elicited by DNA vaccines were not cGas dependent in vivo. Data presented in the current report provide more details on the innate immune mechanisms involved in DNA vaccination and further enrich our understanding on the potential utility of DNA vaccines in generating antigen-specific immune responses.
Introduction
Live attenuated vaccines are the most effective vaccines because they can elicit balanced T cell and antibody responses due to their in vivo replication potentials producing antigens inside the host. However, safety remains a concern for live attenuated vaccines (1). In contrast, many traditional inactivated or subunit vaccines, while safe when delivered as exogenous antigens, are not effective in eliciting T cell responses, and also lead to less optimal antibody responses due to the lack of strong T cell help. The discovery of DNA vaccines in the early 1990s provided a unique vaccination technology that is effective in delivering endogenous antigens to elicit both MHC-I and MHC-II restricted T cell responses and balanced humoral and cellular immunities (2–6). More recent progress in human studies with candidate HIV-1 and pandemic influenza vaccines has further confirmed that DNA vaccines are able to induce high quality and long lasting antibody responses in both animals and humans, particularly when used as a priming vaccination, followed by boosting with inactivated or recombinant protein vaccines (7–21). Studies on the underlying immunological mechanisms of DNA vaccination indicate that DNA priming is especially effective in guiding germinal center B cell development, possibly through the function of T follicular helper cells (22).
At the same time, it is long believed that DNA vaccines also induce innate immune signals, which may further contribute to the generation of acquired immunity. It has been known since the discovery of the DNA vaccine concept more than 20 years ago that the unmethylated CpG motifs encoded within the DNA vaccine plasmid could mediate immune responses via the endosomal DNA receptor, TLR9 (23). However, both TLR9 and MyD88-deficient mice mount immune responses comparable to wild-type mice, indicating antigen specific immune responses elicited by DNA vaccines are not TLR9 dependent (24–27).
More recent reports suggest that other innate signaling pathways are also involved in regulating the function of DNA vaccines (28, 29). We reported the role that Absent in Melanoma 2 (Aim2) plays in the induction of influenza HA antigen-specific antibody responses (30). In addition, the immunostimulatory double-stranded nature of the DNA plasmid itself, as cytosolic DNA, is a potent inducer of type I interferon (IFN-αβ) via the stimulator of interferon gene (Sting) and the noncanonical IκB kinase, TANK binding kinase-1 (Tbk1) (24, 31). Sting/Tbk1 activation triggers translocation of the interferon regulatory factor 3 (Irf3) transcription factor into the nucleus, driving IFN-αβ production through a positive feedback loop. Sting/Tbk1 mediated IFN-αβ production is required for DNA vaccine immunogenicity (24, 29, 32). However, the exact requirements of this pathway remain ambiguous as it has been reported that Irf3 deletion diminishes T cell immunity, but has little impact on B cell responses (33). Therefore, it was suggested that IFN-αβ does not play a significant role in generating high-level antibody responses following DNA vaccination (33) which is in stark contradiction to two other reports (29, 32). Additionally, the upstream DNA vaccine sensor has yet to be described, although multiple reports have identified the ubiquitously expressed cyclic GMP-AMP synthase (cGas) as a robust inducer of IFN-αβ capable of directly binding cytosolic dsDNA (27). As cGas is vital for immune responses to DNA viruses and bacterial infections (27, 34), we hypothesized that it would also be essential for DNA vaccine immunogenicity.
Here we investigated the role of multiple components of the DNA sensing pathway in immune responses elicited by a model DNA vaccine expressing the influenza HA antigen. We determined the deletion effects of cGas, Sting, Irf3, and the closely related Irf7, on both the innate and adaptive immune response. Our data identified the key modulator in the generation of antigen-specific immune responses downstream of Sting. Such information will provide much needed insight into the design and development of safer and more effective vaccines.
Materials and Methods
Mice and DNA Vaccination
C57BL/6 mice were obtained from Taconic Laboratories. Sting−/− mice were a gift from G. Barber (University of Miami) and were fully backcrossed to C57BL/6. Irf3−/− and Irf7−/− mice were from T. Taniguchi (University of Tokyo, Japan). Irf3−/−/Irf7−/− double knock out (DKO) mice and cGas−/− mice were generated in house by the K. Fitzgerald laboratory, at the University of Massachusetts Medical School (UMMS). All mice were maintained in the Department of Animal Medicine at UMMS according to IACUC-approved protocols. The H1HA DNA vaccine utilized in this study encodes a codon-optimized gene of full-length hemagglutinin (HA) protein from influenza A/Texas/04/09 as previously reported (30). Mice received 100 μg of H1HA DNA vaccine by intramuscular injection, at weeks 0 and 2 to study HA-specific antibody responses. A third boosting vaccination was delivered 1 week prior to sacrifice for the analysis of HA-specific T cell immunity. Serum was collected prior to vaccination and 6 hours post vaccination for cytokine assays, and at week 4 for antibody analysis. After collection, serum samples were stored at −80° C until the conclusion of the study. For in vivo immunogenicity studies involving Sting−/−, Irf3−/−, Irf7−/−, and DKO mice, an N=10 is reported. cGas−/− mouse studies are reported as an N=8. Immunogenicity data is representative of the average of two independently performed studies, where N value equals total number of mice/phenotype. Tissue punch biopsies were harvested at 0, 6, and 12 hours post vaccination for rt-PCR. Splenocytes were harvested at time of sacrifice. rt-PCR studies utilized an N=5.
Cell Culture Stimulation and Cytokine Measurement
Cells were treated under several conditions. LPS stimulation was performed at a concentration of 200 ng/ml. Poly (deoxyadenylic-deoxythymidylic) [poly(dA:dT)] (Sigma Aldrich) DNA and HA-expressing DNA vaccine plasmid were added to the cells in the presence of Lipofectamine 2000 at a concentration of 1.5 μg/ml. Cultures were incubated 16–18 hours at 37°C, and supernatants were harvested. Murine IFN-β sandwich ELISA was used as previously described (35). IFN-α and IFN-β rt-PCR was performed on RNA isolated from immortalized BMDM. rt-PCR reactions were performed on a Bio-Rad CFX-96 cycler. Primer sequences are designed based on GenBank information.
Serum cytokine analysis
Cytokines were quantified in sera collected from individual mice prior to vaccination at hour 0, and 6 hours post the first DNA vaccination using the Mouse Th1/Th2/Th17 CBA kit (BD Biosciences) according to manufacturer’s instructions. The panel of cytokines included: IFN-γ, IL-2, IL-4, IL-6, TNF, IL-10, and IL-17. All serum samples from each time point of interest were run in a single CBA experiment. Prior to assay, serum samples were diluted 1:2 in sample diluent. Samples were read on a LSRII flow cytometer and analyzed with FCAP Array Software version 3.0 (BD Biosciences).
ELISA
Microtiter plates were coated with standard H1HA antigen at 1.0 μg/mL in PBS for 1 hour at room temperature and assayed as previously described (36). For sodium thiocyanate, displacement was performed at a serum dilution of 1:100. After washing of serum samples, sodium thiocyanate was added at various (0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 M) concentrations in PBS for 15 min followed by 5 washes in ELISA wash buffer. The assay was then completed as above.
ELISPOT assay
Splenocyte T and B cell ELISPOT reagents were obtained from Mabtech. H1HA specific T cells were quantified per manufactures instructions. Positive controls were stimulated with 20 ng/ml PMA (Sigma-Aldrich) and 500 ng/ml ionomycin (Sigma-Aldrich). The H1HA relevant peptide used was a CD8+ T cell epitope (IYSTVASSL). H1HA-specific antibody secreting cells were detected by coating of MAIPSWU (Millipore) plates with the standard H1HA antigen utilized for ELISA (1.0 μg/mL). Positive spots were visualized on a CTL Imager and counting was performed with Immunospot software (Cellular Technology Ltd.)
Quantitative real time PCR
C57BL/6 and various knockout mice were shaved and vaccinated with 100 μg H1HA DNA vaccine plasmid intramuscularly in the hind quad muscle. Punch biopsies were performed from the injection site at 6 hours and 12 hours post vaccination and snap frozen. RNA was isolated from tissue biopsies using TRIzol Reagent (Life Technologies), and cDNA was generated using the Bio Rad iScript cDNA synthesis kit (Bio-Rad). cDNA was then used for rt-PCR reactions on a Bio-Rad CFX-96 cycler. Primer sequences were designed based on GenBank HA sequences.
Statistical analysis
All data is presented as the mean of individual mice +/− standard error of the mean (SEM). Statistical analysis was performed using a Student’s t test, a one-way ANOVA followed by a Tukey post-test, or a two-way ANOVA followed by a Bonferonni post-test.
Results
Sting is required for induction of both innate and adaptive immune responses to DNA vaccination
It was recently reported, using a model ovalbumin encoding vaccine, that Sting-deficient mice exhibited severe defects in the adaptive immune responses generated by DNA vaccination (24). To confirm these results with a DNA vaccine expressing a viral immunogen, we vaccinated mice with DNA vaccine pH1HA expressing the HA antigen of the influenza A virus which was responsible for the H1N1 pandemic in 2009. As HA is highly immunogenic and is the major protective antigen in clinically licensed inactivated and live-attenuated influenza vaccines, HA DNA vaccines provide a sensitive study model (15, 16, 18, 30, 36–38) to detect the impact of innate immunity on a clinically relevant immunogen.
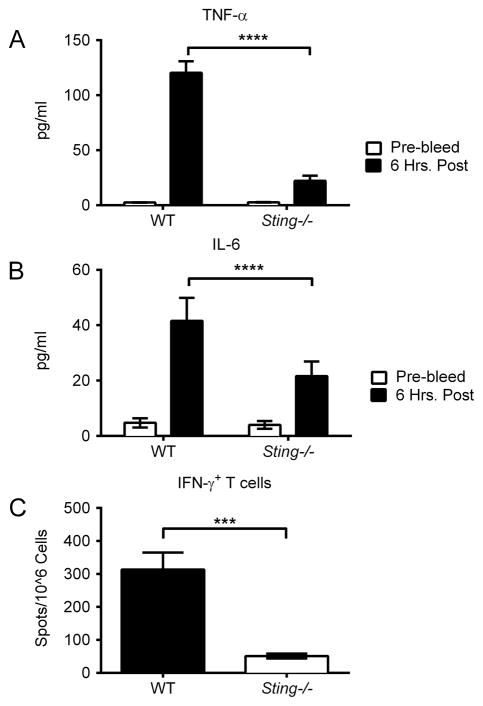
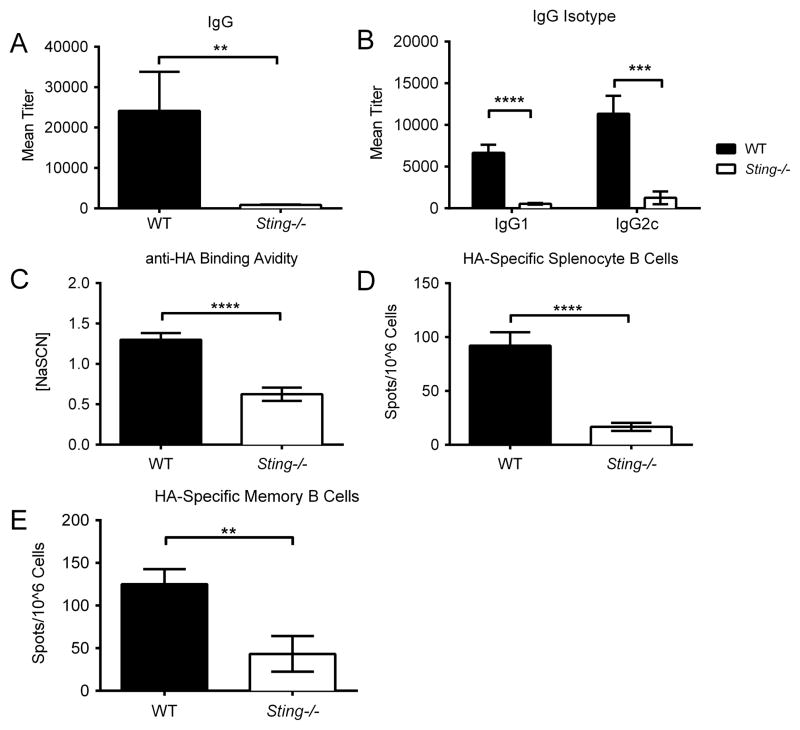
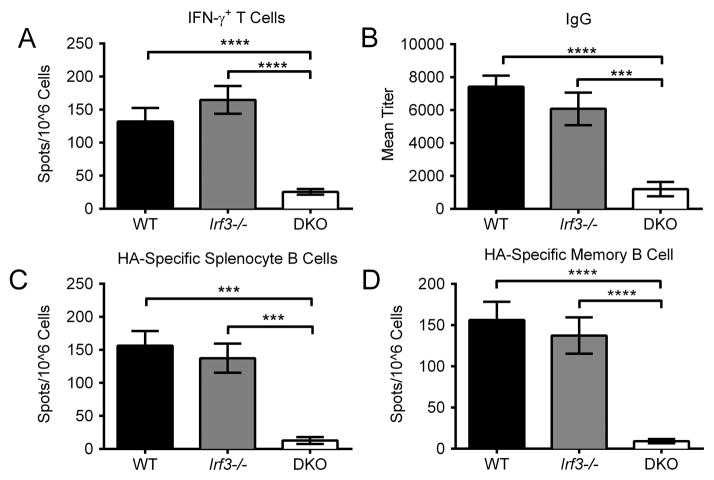
To gain a more complete understanding of the requirement for Sting signaling in innate immune responses, wild-type C57BL/6 mice (WT) and Sting−/− mice were vaccinated with the DNA vaccine pH1HA and serum cytokines were measured 6 hours post vaccination. Quantification of cytokine levels revealed that Sting−/− mice had a marked decrease in TNF-α and IL-6 production (Figure 1A–B). Sting-deletion also negatively impacted the generation of antigen-specific adaptive immunity. While WT mice yielded high levels of HA-specific IFN-γ secreting CD8+ T cells following stimulation with an MHC-I peptide encoded within the pH1HA vaccine, Sting−/− splenocytes failed to respond to peptide stimulation (Figure 1C). Furthermore, humoral responses were also impaired because vaccinated WT, but not Sting−/−, mice elicited robust anti-HA IgG responses (Figure 2A–B). Correspondingly, the HA-specific B cell population was decreased in both the circulating (spleen) and memory (bone marrow) compartments of Sting−/− mice, as was the avidity of anti-HA binding antibodies (Figure 2C–E).
FIGURE 1. Sting is required for DNA vaccine immunogenicity.
Sera were collected pre-vaccination and 6 hours post DNA vaccination. Cytokines were quantified in the serum of individual mice at a 1:2 dilution using a BD Th1/Th2/Th17 cytokine kit. Shown are significant decreases in cytokines after vaccination with pH1HA encoding DNA vaccine: (A) TNF-α and (B) IL-6. WT and Sting−/− mice were vaccinated intramuscularly with pH1HA at weeks 0, 2, and 8. Splenocytes were harvested at termination 7 days following the third vaccination. (C) Frequency of HA peptide-specific IFN-γ+ T cells in mice immunized with pH1HA. Data are the averages ± SEM of 2 independent studies of 5 mice per group (Total N=10 per phenotype). ***p<0.001, ****p<0.0001 versus control group.
FIGURE 2. DNA vaccine induced antibody responses are Sting dependent.
HA-specific IgG titers (A), including their (B) isotypes, were analyzed 14 days post second vaccination of WT and Sting−/− mice. Anti-HA binding avidity was quantified via ELISA and reported as molar concentration of sodium thiocyanate required to displace anti-HA serum antibodies to 2x pre-bleed levels (C). Splenocytes and bone marrow were harvested at termination 7 days following the third boosting vaccination. (D) Splenocyte B cells were plated immediately following isolation, while (E) bone marrow cells were plated after 5 days of culturing in non-specific stimulation to promote clonal expansion. Data are the averages ± SEM of 2 independent studies of 5 mice per group (Total N=10 per phenotype). **p<0.01, ***p<0.001, ****p<0.0001 versus control group.
cGas does not mediate the immune response to DNA vaccination
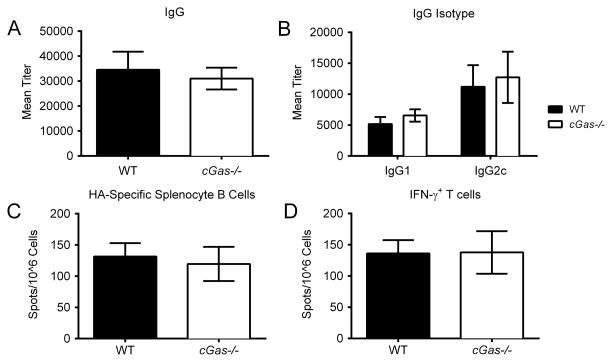
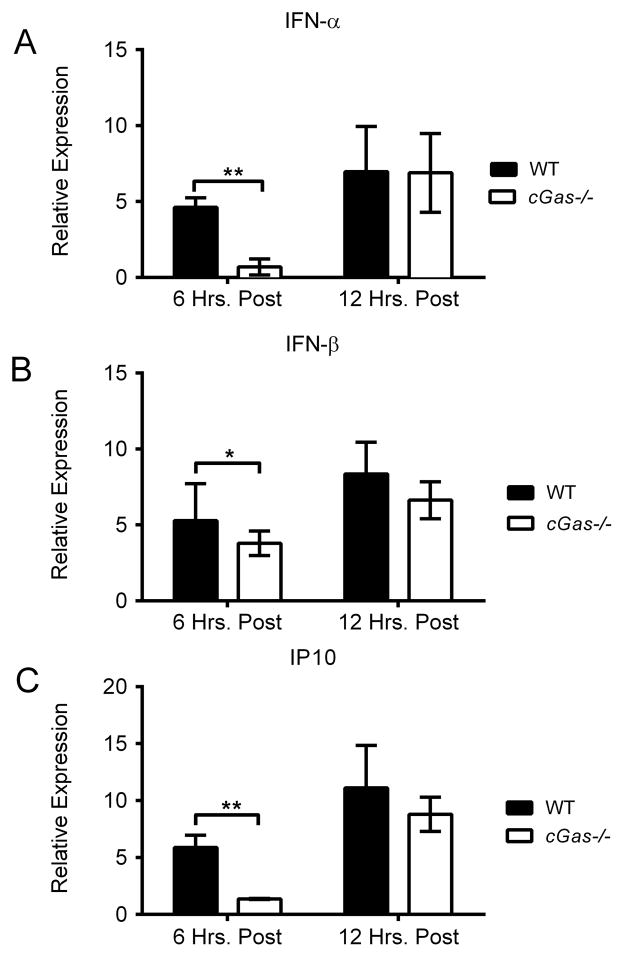
Previous attempts to identify the upstream sensor for DNA vaccination have been unsuccessful (28, 29). It is widely believed that a cytosolic DNA sensor regulates IFN-αβ production in response to DNA vaccination. Studies over the last few years have identified cytosolic DNA binding proteins important in the immune response to intracellular dsDNA. Among these, cyclic GMP-AMP synthase (cGas) is a powerful activator of the Sting/Tbk1 pathway, providing a strong candidate for the unknown DNA vaccine sensor. cGas is a nucleotidyl transferase enzyme which catalyzes the conversion of ATP and GTP into a novel cyclic dinucleotide, cGMP-AMP (cGAMP) which in turn binds and activates Sting. Hence, we evaluated cGas’s role in DNA vaccination. Contrary to expectations, no significant change in the magnitude of the innate or adaptive responses were seen in cGas−/− mice, as they had comparable levels of serum cytokines, anti-HA antibodies, and cytotoxic T cell responses to WT mice (Figure 3A–D and Supplemental 1). The ability to generate high-level adaptive immune responses following DNA vaccination suggested that IFN-αβ production was not dependent on cGas. This result was surprising to us, given the importance of cGas in sensing cytosolic dsDNA. We next quantified local IFN-αβ production at the site of injection by taking punch biopsies at 6 and 12 hours post vaccination. This ensured that we only measured pH1HA-induced IFN-αβ locally. IFN-αβ production was attenuated in cGas−/− mice at the initial 6 hour time point. However, the levels of IFN-αβ in vaccinated cGas−/− mice recovered by the 12 hour time point, approaching those seen in wild type mouse controls (Figure 4A–C). These findings indicate that although cGas contributes to the initial innate immune response upon detection of the DNA vaccine plasmid itself, this effect was transient, and dispensable for sustained immunity. There may be an alternative DNA sensing, IFN-αβ inducing pathway in cGas−/− mice for DNA vaccine induced immune responses.
FIGURE 3. DNA vaccine immunogenicity is cGas independent.
WT and cGas−/− mice were vaccinated intramuscularly with pH1HA at weeks 0, 2, and 8. (A) HA-specific total IgG and (B) IgG isotype titers were analyzed 14 days post second vaccination. Either (C) anti-HA antibody secreting cells or (D) the frequency of HA peptide-specific IFN-γ+ T cells were quantified in mice immunized with pH1HA 1 week post third vaccination. Data are the averages ± SEM of 2 independent studies of 4 mice per group (Total N=8 per phenotype).
FIGURE 4. Decreases in cGas−/− IFN-αβ production following DNA vaccination are transient.
WT and cGas−/− mice were vaccinated intramuscularly with the pH1HA vaccine, and the site of injection was harvested either 6 or 12 hours later. Total RNA was isolated from tissue biopsies and subjected to rt-PCR for (A) IFN-α, (B) IFN-β, and (C) IP10. Data are the averages ± SEM of 5 mice per group. Values are relative to naïve mouse gene expression. *p<0.05, **p<0.01 versus control group.
DNA vaccine induced adaptive immune responses are Irf3 independent
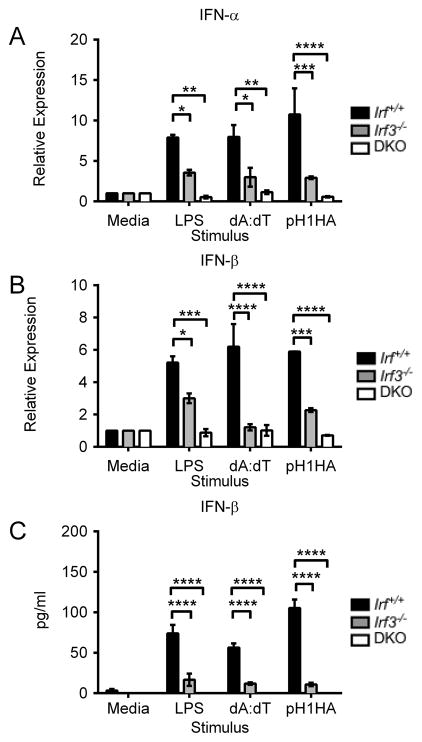
Although the effects of Sting deletion on DNA vaccine immunogenicity were previously reported, the mechanism by which Sting promotes IFN-αβ production is not. Therefore, we also dissected the downstream signaling molecules to elucidate the IFN-αβ pathway. Irf3 is a critical downstream mediator of Sting driven type I IFN responses. As Irf3 is endogenously expressed at high levels and is required for initiating the IFN-αβ cascade, we analyzed its effects on DNA vaccine immunogenicity. In vitro stimulation of immortalized BMDM cultures showed a clear, negative effect of Irf3 deletion on IFN-β production (Figure 5A–C). As expected, the synthetic B-form dsDNA poly(dA:dT) and pH1HA induced robust IFN-β levels in Irf+/+ BMDM as measured by both rt-PCR and cell culture ELISA. Conversely, IFN-β production was limited in Irf3−/− and, to a greater extent, BMDM lacking both Irf3 and Irf7 (DKO). Surprisingly, however, vaccination of Irf3−/− mice did not result in impaired adaptive immunity. WT and Irf3−/− mice had equivalent levels of IFN-γ producing CD8+ T cells, in contrast to previously reported results (33) (Figure 6A). Furthermore, Irf3−/− mice exhibited high levels of anti-HA IgG titers, a result not seen in DKO mice (Figure 6B–D). Overall, while IFN-αβ is required for DNA vaccine immunogenicity, Irf3 alone does not play a substantial role in impacting the acquired immune response.
FIGURE 5. Irf3 is required for DNA vaccine-induced IFN-αβ production in vitro.
Irf+/+, Irf3−/−, and DKO BMDMs were transfected with poly (dA:dT) or pH1HA plasmid for 18 hours. (A) IFN-α and (B) IFN-β levels in transfected BMDM were quantified by rt-PCR. (C) Secreted IFN-β in the culture supernatants was analyzed by ELISA. Data are presented as mean ± SEM from 3 independent experiments. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 versus control group.
FIGURE 6. Irf3 is dispensable for DNA vaccine immunogenicity.
WT, Irf3−/−, and DKO mice were vaccinated intramuscularly with a pH1HA encoding DNA vaccine at weeks 0, 2, and 8. Splenocytes were harvested at termination 7 days following the third boosting vaccination. (A) Frequency of HA peptide-specific IFN-γ+ T cells in mice immunized with pH1HA. (B) HA-specific IgG titers were analyzed 14 days post second vaccination. (C) HA-specific splenocyte B cells were plated immediately following isolation. (D) Memory B cells were isolated from bone marrow and plated after 5 days of culturing in non-specific stimulation to promote clonal expansion. Data are the averages ± SEM of 2 independent studies of 5 mice per group (Total N=10 per phenotype). ***p<0.001, ****p<0.0001 versus control group.
Irf7 regulates DNA vaccine-induced innate cell signaling
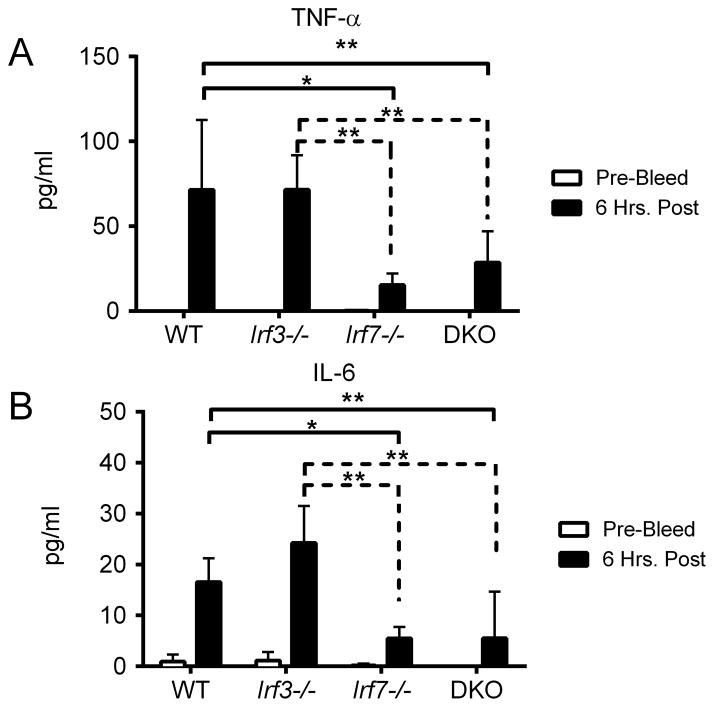
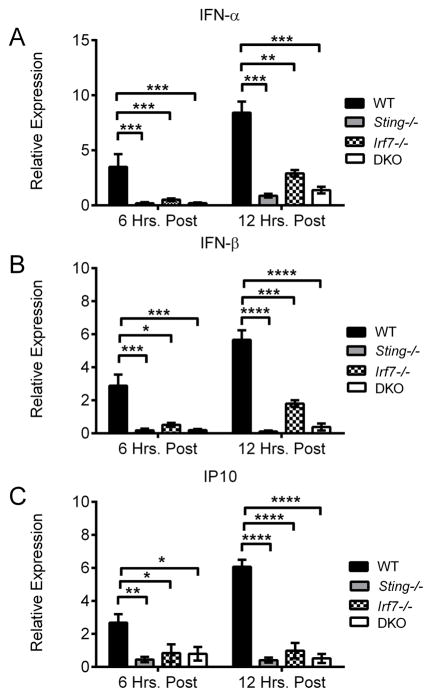
We next investigated Irf7’s role in the context of DNA vaccination. As shown in Figure 7, both WT and Irf3−/− mice yielded commensurate levels of serum cytokines 6 hours post vaccination. However, Irf7−/− and DKO mice showed clear defects in TNF-α and IL-6 production, at levels comparable to Sting−/− mice. The similar cytokine profiles seen in Sting−/− and Irf7−/− mice indicated that both are required for innate signaling following DNA vaccination. Furthermore, both molecules have been identified as key regulators of IFN-αβ production following DNA virus infection (24, 39). As DNA vaccine immunogenicity is IFN-αβ dependent (29), we performed a more detailed analysis on the effect of Sting and Irf7 deletion on IFN-αβ expression. To ensure we only measured pH1HA-induced IFN-αβ, we limited our measurements to the site of vaccination. WT, Sting−/−, Irf7−/−, and DKO mice were vaccinated with the pH1HA vaccine, and punch biopsies of the injection site were harvested 6 or 12 hours later. rt-PCR analysis of mRNA illustrates that Sting−/− and DKO mice lack significant IFN-α and IFN-β expression compared to WT controls (Figure 8A–B). Interestingly, Irf7−/− mice exhibited a similar decrease in IFN-αβ production, although to a lesser degree. Consistent with a decrease in IFN-αβ, both Sting−/− and Irf7−/− mice had a corresponding decrease in the IFN stimulated gene IP10, illustrating the wide-ranging effect of Sting and Irf7 deletion on the innate immune response (Figure 8C).
FIGURE 7. Irf7 is required for immune cytokine production.
WT, Irf3−/−, Irf7−/−, and DKO sera were collected pre-vaccination and 6 hours post DNA vaccination. Serum cytokines were quantified from individual mice at a 1:2 dilution using a BD Th1/Th2/Th17 cytokine kit. Shown are significant decreases in cytokines after vaccination with pH1HA encoding DNA vaccine: (A) TNF-α and (B) IL-6. Data are the averages ± SEM of 2 independent studies of 5 mice per group (Total N=10 per phenotype). *p<0.05, **p<0.01 versus control group.
FIGURE 8. IFN-αβ production following DNA vaccination is Irf7 dependent.
WT, Sting−/−, Irf7−/−, and DKO mice were vaccinated intramuscularly with pH1HA vaccine, and the site of injection was harvested 6 or 12 hours later. Total RNA was isolated from tissue biopsies and subjected to rt-PCR for (A) IFN-α, (B) IFN-β, and (C) IP10. Data are the averages ± SEM of 5 mice per group. Values are relative to naïve mouse gene expression. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 versus control group.
Irf7 is required for generation of antigen-specific immune responses following DNA vaccination
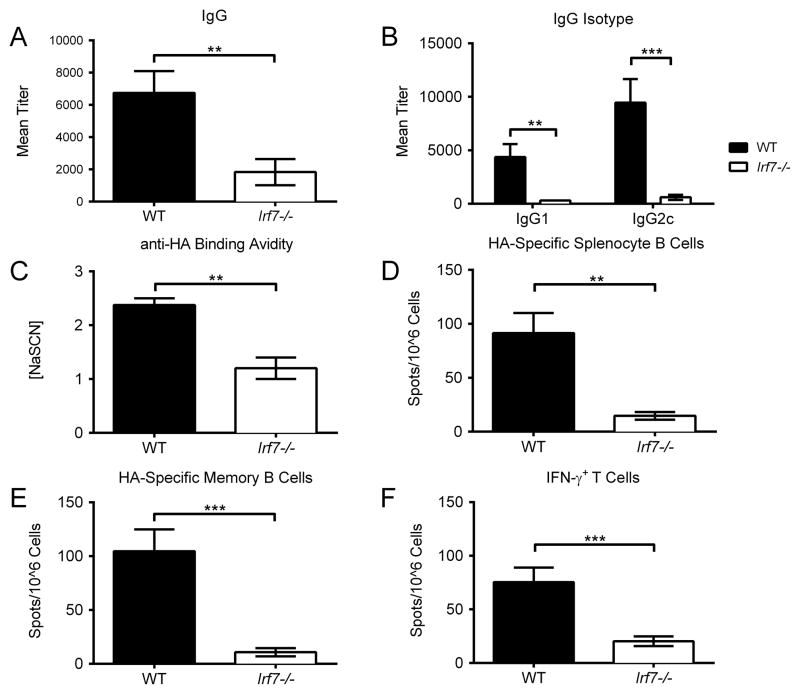
We next tested whether Irf7 deletion would inhibit development of antigen-specific immunity. pH1HA vaccination of WT mice elicited strong antigen-specific humoral responses to the encoded HA antigen, whereas Irf7−/− mice failed to generate the same levels of anti-HA IgG after two vaccinations (Figure 9A–B). Likewise, antibody-binding avidity was decreased approximately 3 fold compared to WT controls (Figure 9C), consistent with the underdevelopment of HA-specific B cells in both the spleen and bone marrow (Figure 9D–E). Vaccination of Irf7−/− mice also failed to induce significant numbers of IFN-γ+ CD8+ splenocytes (Figure 9F). Altogether, these results strongly support the view that Irf7 plays a broad role in the activation of both T and B cell subsets following DNA vaccination.
FIGURE 9. Irf7 is required for generation of anti-pH1HA adaptive immune responses.
WT and Irf7−/− mice were vaccinated intramuscularly with pH1HA. (A) HA-specific IgG titers, including their isotypes (B), were analyzed 14 days post second vaccination. Anti-HA binding avidity was quantified via ELISA and reported as molar concentration of sodium thiocyanate required to displace anti-HA serum antibodies to 2x pre-bleed levels (C). Splenocytes and bone marrow were harvested at termination 7 days following the third boosting vaccination. (D) Splenocyte B cells were plated immediately following isolation, while (E) bone marrow cells were plated after 5 days of culturing in non-specific stimulation to promote clonal expansion. (F) Frequency of HA peptide-specific IFN-γ+ T cells in mice immunized with pH1HA. Data are the averages ± SEM of 2 independent studies of 5 mice per group (Total N=10 per phenotype). **p<0.01, ***p<0.001 versus control group.
Discussion
The complicated interplay between DNA vaccination and the innate immune system is just beginning to be elucidated, as the canonical TLR pathways have little influence on DNA vaccine immunogenicity. Instead, previous reports have shown that sensing of intracytoplasmic DNA plasmid governs DNA vaccine immunogenicity via the Sting/Tbk1/IFN-αβ pathway. However, the processes involved are still unclear. In particular, the requirement for IFN-αβ in generating high-level antibody responses has yielded contradictory results (29, 32, 33). Similarly, the necessary transcription factors downstream of Sting and Tbk1 remain uncertain. The current report investigates these factors and provides an updated understanding of the requirement for such factors in DNA vaccine immunogenicity.
The necessity for Irf3 in cellular mediated immune responses was previously demonstrated (33). Supporting these findings, multiple reports have shown that Irf3-dependent IFN-β can augment the production of Th1 and Th2 cytokines both in vitro and in vivo (40, 41). However, our results show a more limited impact for Irf3, with reductions in IFN-γ+ T cell numbers seen only in Irf3−/−/Irf7−/− DKO mice. Still, in agreement with previous reports (33), we did not see a substantial effect on humoral immunity, confirming that Irf3 is not required for B cell activation. The discrepancy in CD8+ T cell responses may be attributed to the inclusion of more than one vaccination in our study. Another possible explanation may be our choice of a more clinically relevant antigen, which is highly immunogenic, providing a more sensitive system to detect the impact of innate immunity. In this study system, Irf3 alone plays a limited role in DNA vaccine induced antigen-specific immune responses.
In contrast to Irf3, our results unexpectedly identified the important role played by Irf7 in DNA vaccine immunogenicity. Irf7−/− mice exhibited a similar immune phenotype to Sting−/− mice in that Irf7 deficiency resulted in significantly diminished T and B cell responses, indicating a broad contribution by Irf7 to the induction of adaptive immunity. Moreover, Irf7−/− mice had impaired TNF-α and IL-6 production, characteristic of Sting dependent signaling. The lack of IFN-αβ production in Irf7−/− mice further suggests that the defects in DNA vaccination seen in Sting−/− mice may be due to a failure to initiate the Irf7-dependent IFN-αβ feedback loop. This is in accordance with previous reports that Irf7 is the main regulator of immunity with regards to DNA virus infection (39, 42). Altogether, our results indicate that Irf7 is the key driving force behind sustained DNA vaccine-induced IFN-αβ production, implying that the temporary defect in immune priming provided by Irf3 deletion is overcome by the induction of Irf7, allowing for rescue of DNA vaccine immunogenicity.
Our attempt to identify the upstream DNA vaccine sensor yielded surprising results, as cGas deletion did not significantly limit DNA vaccine immunogenicity. In particular, the ability of cGas−/− mice to generate wild type IFN-αβ levels in vivo was somewhat unexpected. Our results suggest that while cGas is engaged at the site of vaccination and required for the early induction of IFN-αβ, it is not necessary for sustained IFN-αβ production in response to DNA vaccines. Previous studies have suggested that cGas is a non-redundant cytosolic DNA sensor (27, 43), but our data implies that at least one additional sensor functions in this context. Several potential candidates have been defined in the literature, including Ifi16 and Ddx41, amongst others. The engagement of one or more of these sensors and their signaling via Sting provides a possible explanation for the ability of cGas−/− mice to generate adaptive immune responses following DNA vaccination.
In summary, our results further extend previous knowledge on the importance of the Sting pathway by adding new information both up- and downstream of the Sting molecule. Deletion of cGas did not dramatically impact the immune response, pointing to the need for searching additional upstream mechanisms or alternate cytosolic DNA receptors that control the Sting pathway. Realizing that Irf3 is not essential for DNA vaccine immunogenicity led to the finding that Irf7 is the key signaling molecule downstream of Sting in DNA vaccination. Of note, the necessity of both the Irf3 and Irf7 pathways appears to be independent of route of vaccination, as gene gun delivery did not affect data trends (results not shown).
In addition to obtaining a clearer understanding of the molecular mechanisms governing DNA vaccine immunogenicity, our findings may be relevant to clinical applications as well. Much attention has been focused recently on the inclusion of molecular adjuvants in DNA vaccine formulations (44, 45). The identification of the necessary signaling components for DNA vaccine immunogenicity may provide for additional adjuvant targets that can prime the immune system more efficiently. Overall, our results provide further insight on the cellular mechanisms through which DNA vaccines link both the innate and adaptive immune pathways to maximize protective, antigen-specific immune responses.
Supplementary Material
Footnotes
Funding: This study was supported in part by NIH grants 5 U19 AI082676, 5 P01 AI082274 and 5R33AI087191, and Bill and Melinda Gates Foundation grant OPP1033112.
References
- 1.Cann AJ, Stanway G, Hughes PJ, Minor PD, Evans DM, Schild GC, Almond JW. Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine. Nucleic acids research. 1984;12:7787–7792. doi: 10.1093/nar/12.20.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 3.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 4.Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 5.Fuller DH, Haynes JR. A qualitative progression in HIV type 1 glycoprotein 120-specific cytotoxic cellular and humoral immune responses in mice receiving a DNA-based glycoprotein 120 vaccine. AIDS research and human retroviruses. 1994;10:1433–1441. doi: 10.1089/aid.1994.10.1433. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly JJ, Martinez D, Jansen KU, Ellis RW, Montgomery DL, Liu MA. Protection against papillomavirus with a polynucleotide vaccine. The Journal of infectious diseases. 1996;173:314–320. doi: 10.1093/infdis/173.2.314. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy JS, Co M, Green S, Longtine K, Longtine J, O’Neill MA, Adams JP, Rothman AL, Yu Q, Johnson-Leva R, Pal R, Wang S, Lu S, Markham P. The safety and tolerability of an HIV-1 DNA prime-protein boost vaccine (DP6-001) in healthy adult volunteers. Vaccine. 2008;26:4420–4424. doi: 10.1016/j.vaccine.2008.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, Shen S, Green S, Rothman AL, Ennis FA, Arthos J, Pal R, Markham P, Lu S. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26:3947–3957. doi: 10.1016/j.vaccine.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khurana S, Wu J, Dimitrova M, King LR, Manischewitz J, Graham BS, Ledgerwood JE, Golding H. DNA priming prior to inactivated influenza A(H5N1) vaccination expands the antibody epitope repertoire and increases affinity maturation in a boost-interval-dependent manner in adults. The Journal of infectious diseases. 2013;208:413–417. doi: 10.1093/infdis/jit178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledgerwood JE, Hu Z, Gordon IJ, Yamshchikov G, Enama ME, Plummer S, Bailer R, Pearce MB, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS Vrc, and V. R. C. S. Teams. Influenza virus h5 DNA vaccination is immunogenic by intramuscular and intradermal routes in humans. Clinical and vaccine immunology : CVI. 2012;19:1792–1797. doi: 10.1128/CVI.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS, Team VRCS. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. The Lancet infectious diseases. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21:346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, Lu S. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. Journal of virology. 2008;82:7369–7378. doi: 10.1128/JVI.00562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaine M, Wang S, Hackett A, Arthos J, Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine. 2010;28:2999–3007. doi: 10.1016/j.vaccine.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Hackett A, Jia N, Zhang C, Zhang L, Parker C, Zhou A, Li J, Cao WC, Huang Z, Li Y, Lu S. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PloS one. 2011;6:e28757. doi: 10.1371/journal.pone.0028757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Parker C, Taaffe J, Solorzano A, Garcia-Sastre A, Lu S. Heterologous HA DNA vaccine prime--inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine. 2008;26:3626–3633. doi: 10.1016/j.vaccine.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal R, Yu Q, Wang S, Kalyanaraman VS, Nair BC, Hudacik L, Whitney S, Keen T, Hung CL, Hocker L, Kennedy JS, Markham P, Lu S. Definitive toxicology and biodistribution study of a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 (HIV-1) vaccine in rabbits. Vaccine. 2006;24:1225–1234. doi: 10.1016/j.vaccine.2005.07.112. [DOI] [PubMed] [Google Scholar]
- 18.Suguitan AL, Jr, Cheng X, Wang W, Wang S, Jin H, Lu S. Influenza H5 hemagglutinin DNA primes the antibody response elicited by the live attenuated influenza A/Vietnam/1203/2004 vaccine in ferrets. PloS one. 2011;6:e21942. doi: 10.1371/journal.pone.0021942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S. Combination DNA plus protein HIV vaccines. Springer seminars in immunopathology. 2006;28:255–265. doi: 10.1007/s00281-006-0028-1. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, Liu Q, Whitney S, Keen T, Nair BC, Kalyanaraman VS, Markham P, Lu S. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350:34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 21.Vaine M, Wang S, Liu Q, Arthos J, Montefiori D, Goepfert P, McElrath MJ, Lu S. Profiles of human serum antibody responses elicited by three leading HIV vaccines focusing on the induction of Env-specific antibodies. PloS one. 2010;5:e13916. doi: 10.1371/journal.pone.0013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristin Hollister YC, Wang Shixia, Wu Hao, Mondal Arpita, Clegg Ninah, Lu Shan, Dent Alexander. The role of follicular helper T cells and the germinal center in HIV-1 gp120 DNA prime and gp120 protein boost vaccination. Human Vaccines & Immunotherapeutics. 2014:10. doi: 10.4161/hv.28659. epub: ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tudor D, Dubuquoy C, Gaboriau V, Lefevre F, Charley B, Riffault S. TLR9 pathway is involved in adjuvant effects of plasmid DNA-based vaccines. Vaccine. 2005;23:1258–1264. doi: 10.1016/j.vaccine.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babiuk S, Mookherjee N, Pontarollo R, Griebel P, van Drunen Littel-van den Hurk S, Hecker R, Babiuk L. TLR9−/− and TLR9+/+ mice display similar immune responses to a DNA vaccine. Immunology. 2004;113:114–120. doi: 10.1111/j.1365-2567.2004.01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 30.Suschak JJ, Wang S, Fitzgerald KA, Lu S. Identification of aim2 as a sensor for DNA vaccines. Journal of immunology. 2015;194:630–636. doi: 10.4049/jimmunol.1402530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature immunology. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 32.Tudor D, Riffault S, Carrat C, Lefevre F, Bernoin M, Charley B. Type I IFN modulates the immune response induced by DNA vaccination to pseudorabies virus glycoprotein C. Virology. 2001;286:197–205. doi: 10.1006/viro.2001.0957. [DOI] [PubMed] [Google Scholar]
- 33.Shirota H, Petrenko L, Hattori T, Klinman DM. Contribution of IRF-3 mediated IFNbeta production to DNA vaccine dependent cellular immune responses. Vaccine. 2009;27:2144–2149. doi: 10.1016/j.vaccine.2009.01.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. The Journal of experimental medicine. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Taaffe J, Parker C, Solorzano A, Cao H, Garcia-Sastre A, Lu S. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. Journal of virology. 2006;80:11628–11637. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Jia N, Li J, Han Y, Cao W, Wang S, Huang Z, Lu S. Optimal designs of an HA-based DNA vaccine against H7 subtype influenza viruses. Hum Vaccin Immunother. 2014:10. doi: 10.4161/hv.28795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almansour I, Chen H, Wang S, Lu S. Cross reactivity of serum antibody responses elicited by DNA vaccines expressing HA antigens from H1N1 subtype influenza vaccines in the past 30 years. Hum Vaccin Immunother. 2013;9:2049–2059. doi: 10.4161/hv.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 40.Shirota H, Petrenko L, Hong C, Klinman DM. Potential of transfected muscle cells to contribute to DNA vaccine immunogenicity. Journal of immunology. 2007;179:329–336. doi: 10.4049/jimmunol.179.1.329. [DOI] [PubMed] [Google Scholar]
- 41.Oganesyan G, Saha SK, Pietras EM, Guo B, Miyahira AK, Zarnegar B, Cheng G. IRF3-dependent type I interferon response in B cells regulates CpG-mediated antibody production. The Journal of biological chemistry. 2008;283:802–808. doi: 10.1074/jbc.M704755200. [DOI] [PubMed] [Google Scholar]
- 42.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 43.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalams SA, Parker S, Jin X, Elizaga M, Metch B, Wang M, Hural J, Lubeck M, Eldridge J, Cardinali M, Blattner WA, Sobieszczyk M, Suriyanon V, Kalichman A, Weiner DB, Baden LR, Network NHVT. Safety and immunogenicity of an HIV-1 gag DNA vaccine with or without IL-12 and/or IL-15 plasmid cytokine adjuvant in healthy, HIV-1 uninfected adults. PloS one. 2012;7:e29231. doi: 10.1371/journal.pone.0029231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JJ, Simbiri KA, Sin JI, Dang K, Oh J, Dentchev T, Lee D, Nottingham LK, Chalian AA, McCallus D, Ciccarelli R, Agadjanyan MG, Weiner DB. Cytokine molecular adjuvants modulate immune responses induced by DNA vaccine constructs for HIV-1 and SIV. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 1999;19:77–84. doi: 10.1089/107999099314441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.