Abstract
Background & Aims
Inflammatory bowel disease (IBD) has familial aggregation in African Americans (AAs), but little is known about the molecular genetic susceptibility. Mapping studies using the Immunochip genotyping array expand the number of susceptibility loci for IBD in Caucasians to 163, but the contribution of the 163 loci and European admixture to IBD risk in AAs is unclear. We performed a genetic mapping study using the Immunochip to determine whether IBD susceptibility loci in Caucasians also affect risk in AAs and identify new associated loci.
Methods
We recruited AAs with IBD and without IBD (controls) from 34 IBD centers in the US; additional controls were collected from 4 other immunochip studies. Association and admixture loci were mapped for 1088 patients with Crohn's disease (CD), 361 with ulcerative colitis (UC), 62 with IBD type-unknown (IBDU), and 1797 controls; 130,241 autosomal single-nucleotide polymorphisms (SNPs) were analyzed.
Results
The strongest associations were observed between UC and HLA rs9271366 (P=7.5e–6), CD and 5p13.1 rs4286721 (P=3.0e–6), and IBD and KAT2A rs730086 (P=2.3e–6). Additional suggestive associations (P<4.2e-5) were observed between CD and IBD and African-specific SNPs in STAT5A and STAT3; between IBD and SNPs in IL23R, IL12B, and C2 open reading frame 43; and between UC and SNPs near HDAC11 and near LINC00994. The latter 3 loci have not been previously associated with IBD, but require replication. Established Caucasian associations were replicated in AAs (P<3.1e-4) at NOD2, IL23R, 5p15.3, and IKZF3. Significant admixture (P<3.9e–4) was observed for 17q12-17q21.31 (IZKF3 through STAT3), 10q11.23-10q21.2, 15q22.2–15q23, and 16p12.2–16p12.1. Network analyses showed significant enrichment (false discovery rate <1e–5) in genes that encode members of the JAK–STAT, cytokine, and chemokine signaling pathways, as well those involved in pathogenesis of measles.
Conclusions
In a genetic analysis of 3308 AA IBD cases and controls, we found that many variants associated with IBD in Caucasians also showed association evidence with these diseases in AAs; we found evidence for loci and variants not previously associated with IBD. The complex genetic factors that determine risk for or protection from IBD in different populations require further study.
Keywords: race, ethnicity, genetic variant, intestinal inflammation
Introduction
Inflammatory bowel disease (IBD) is a complex genetic disorder of immune dysregulation causing chronic idiopathic inflammation of the gastrointestinal tract, estimated to affect about 1.4 million Americans. It comprises two major, genetically related phenotypes: Crohn's disease (CD) and ulcerative colitis (UC).
IBD shares many clinical and immunological characteristics with other complex genetic immune-mediated diseases (IMDs), especially with seronegative auto-inflammatory diseases1, such as ankylosing spondylitis, psoriasis and primary immune-deficiencies2, indicating overlapping etiological factors. To facilitate genetic studies on IMDs a custom Illumina array of ~200,000 SNPs, the Immunochip, was designed based on genome wide association (GWA) analyses on Caucasian populations of 12 IMDs including IBD3. The main purposes were to fine map established associations and to replicate suggestive, but not yet proven, associations3. The Immunochip also contains ancestry informative markers (AIMs) allowing for genome-wide admixture estimates and adjustment for population stratification.
IBD GWA studies, including those performed using the Immunochip, have expanded the number of IBD susceptibility loci to 163 (including 30 CD- and 23 UC-specific loci)2, and have enhanced our understanding of IBD immunopathogenesis by identifying key cellular pathways, both known – such as barrier function, the role of T cell subsets and cytokine–cytokine receptor signaling – and unknown – such as autophagy, regulation of interleukin 23 (IL23) signaling, and host defense4. However, compared to hundreds of IBD genetic studies in Caucasian populations including massive GWA mega-analyses and replication studies like the Caucasian Immunochip Study (CIS)2, only a handful of IBD-associated gene variations have been evaluated in African Americans (AAs), in relatively small sample sizes of a few hundred cases and controls, and only for CD, not UC5-7.
AAs are a recently admixed population derived from an average of approximately 80% West African and 20% European ancestries8. IBD prevalence is lower in AAs than Caucasian Americans (CAs) possibly as a result of both genetic and environmental differences9-11. IBD sibling risk in AA IBD patients is relatively high (2.5%), suggesting underlying genetic risk factors are in-part responsible for IBD in AAs, albeit lower than that observed for Caucasians (4.6%)12.
An admixed population is one where two or more previously separated populations have interbred. Loci that have different allele frequencies in the founder populations become correlated because of gene flow, a phenomenon known as admixture linkage disequilibrium (ALD). AAs are a typical example of an admixed population, where there has been recent introduction of European genetic lineages into a West-African-derived population. A powerful technique called mapping by ALD (MALD) can be used on admixed populations such as AAs and the underlying assumption is that the difference in disease frequency is due in part to differences in allele frequencies of causal variants between populations.
We therefore undertook an evaluation of the Immunochip in ~4000 AA IBD cases and controls, primarily to determine the importance of the 163 established CIS IBD loci in the understudied AA population and to identify novel IBD loci, including loci identified by MALD.
Patients and Methods
Study Population and Phenotyping
The study population included unrelated self-identified non-Hispanic AA volunteers recruited from three coordinating centers: (1) Johns Hopkins: Multicenter African American IBD Study (MAAIS) coordinated by Johns Hopkins IBD Genetics Research Center (GRC) of the NIDDK IBD Genetics Consortium (IBDGC) with recruitment from 13 collaborating IBD centers and 4 other IBDGC GRCs5, and additional AA control samples from the controls from rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and type 1 diabetes (T1D) Immunochip studies; (2) Emory: GENESIS AA cohort, an ancillary study of the NIDDK IBDGC, coordinated by Emory University with recruitment of IBD cases and matched controls from 12 of their collaborating IBD centers; and (3) Cedars: Cedars-Sinai Medical Center recruited IBD cases and controls with additional controls from the Pharmacogenetics and Risk of Cardiovascular disease Study Group (PARC) from San Francisco General Hospital and University of California, Los Angeles13.
All subjects gave informed consent to participate in genetics research studies in protocols approved by each sites institutional review board. Cases were confirmed as CD, UC, or IBD type undetermined (IBDU) in accordance with the NIDDK IBDGC phenotyping manual14. Details regarding controls are described in the Supplement. See Acknowledgements for listing of all recruitment centers.
Genotyping and Quality Control (QC)
DNA samples were derived from whole blood. All DNA samples were genotyped using the Immunochip3 and genotype determinations (allele calls) were made using GenomeStudio version 2011.1 and Genotyping Module Version 1.9.4. All MAAIS samples and the RA, SLE, and T1D controls were genotyped at Feinstein Institute for Medical Research. All Emory, Cedars-Sinai and PARC samples were genotyped at Cedars-Sinai Medical Center Genetics Institute.
Several SNP-wise and sample-wise quality filters were applied (Supplementary Figure 1). Samples were excluded if they had <99% data completeness, differed by >3 standard deviations from the mean heterozygosity for the study, had discrepant gender, or were unexpectedly the first-degree relative of any other sample in the study. Matching of cases and controls was done by determining principal components (PC) using the software EIGENSTRAT15, plotting PC1 against PC2, followed by visual inspection and elimination of outlier samples. Three successive rounds were necessary until a satisfactory matching of cases/controls (Supplementary Figure 2) was obtained.
Local and Global Ancestry Estimation
Because AAs are well-modeled as linear combinations of West African (YRI) and European (CEU) ancestries16, to estimate the locus-specific local ancestry, we chose the WINPOP model in LAMP package17 for its fast computation with low error rate18. Average West African (global YRI) ancestry was estimated by using ADMIXTURE19.
Study-wide Association and Admixture, and Replication of 163 SNPs Established in Caucasian Immunochip Study (CIS)
For all analyses, we compared three phenotypes, all IBD, CD, and UC, against the same set of AA controls. We performed association and admixture mapping both under generalized linear models. Sex, recruitment coordinating center (Hopkins, Emory or Cedars), GRC (Feinstein or Cedars), global YRI ancestry, and the first 10 PCs were included in multiple regression with IBD affection status, and those significantly associated with IBD were used as covariates in association and admixture mapping.
We estimated the testing burdens empirically by assessing the autocorrelation (of genotypes for association or local ancestry proportions for admixture) of all the SNPs on each autosomal chromosome for each individual, and then summing over the 22 chromosomes and averaging across individuals20.
The testing burdens of independent SNPs or admixture regions, specific to our dataset, were 23,639.4 and 128.4 for association and admixture mapping respectively. Association peaks with p<2.1e-6 (5% false positive rate corrected for number of independent SNPs, i.e. 0.05/23,639.4) were marked as significant, while those with 2.1e-6≤p<4.2e-5 were marked as suggestive (i.e. one false positive per study-wide test burden). Admixture peaks with p<3.9e-4 (0.05/128.4) were marked as significant.
For chromosomes with multiple significant/suggestive association SNPs, conditional regression was performed to determine number of independent signals. Specifically for m SNPs, all possible m*(m-1)/2 pairwise combinations were tested. For each pair if the target SNP remained significant in the presence of the conditional SNP then they are independent of each other; otherwise, they have arisen from the same signal.
To evaluate the 163 loci with reported genome-wide significance in the CIS, we tested the most statistically significant SNPs at each locus and present on the Immunochip (Maximal-CIS SNPs). Criteria for replication was p<3.1e-4 (0.05/163).
Gene Ontology was utilized to functionally annotate the genes within each significant admixture region, using the PANTHER Classification System (http://pantherdb.org). Genes with immunological or gastrointestinal-relevant functions were considered to be suggestive candidate genes within the region.
eQTL Evaluation
For significant/suggestive association SNPs we also performed local (cis-) and distant (trans-) expression quantitative trait locus (eQTL) mapping with R package MatrixEQTL21 on peripheral blood mononuclear cells (PBMCs) drawn from a different set of 85 unrelated AA study subjects profiled with Illumina HumanHT-12 v4 Expression BeadChips and genotyped on Illumina HumanOmni1 or HumanOmni2.5 BeadChips22. We defined gene-SNP pairs within 1Mb of another as local, otherwise distant, and those with false discovery rate (FDR) <0.05 were considered significant.
Network Analysis
We first selected genes with association p<0.005, and then the identified genes annotated with multiple biologically functional databases including Reactome23, human protein reference databases (www.hprd.org), and NCI/Nature Pathway Interaction Database. The networks were then constructed from the known interactions in those databases. In addition, we identified the top KEGG pathways associated with each network using the enrichment analysis tool in STRING (http://string-db.org/). See Supplement for additional details.
Results
After QC measures were performed, 3,308 samples (1,511 IBD cases and 1,797 controls) and 130,241 unique autosomal SNPs remained for association and admixture analyses (Supplementary Figures 1 and 2). Average West African ancestry was higher in controls than cases (81.7±9.7% vs. 80.0±10.3%, p<1e-5), and also differed by coordinating centers (p<1e-10), with YRI proportion lowest for AAs from the American West Coast, similar to other genetic studies24 (Supplementary Table 1).
Five variables were significantly associated with IBD (p<0.05) and therefore were included as covariates for all subsequent analyses: sex (p=5e-7), recruitment coordinating center (p=2e-5), GRC (p=4e-4), PC2 (p=0.015), and global ancestry (p=0.03). Quantile-quantile plots for association mappings and genomic inflation factors are shown in Supplementary Figure 3.
Replication of Maximal-CIS SNPs
152 of the 163 Maximal-CIS SNPs passed QC. The remaining 11 SNPs did not have any tagging (r2<0.6) SNPs that could be used as alternative markers. Five SNPs met criteria for replication (p<3.1e-4): rs5743289 that tags the three common NOD2 mutations, rs11209026 that encodes the IL23R R381Q protective variant, rs1801274 that encodes the FCGR2A H167R risk variant, rs11742570 in the 5p13.1 gene desert near PTGER4 and rs12946510 on 17q21 just 3’ of IKZF3 (Table 1).
Table 1.
Significant association for replication (p<3.1e-4) for SNPs established in Caucasian Immunochip Study (CIS)
| SNP | Chr | Mbc | Nearest Gene |
A1d | A2d | YRIe | CEUe | Casee | Ctrle | CISe | Sigf | pf | ORf | L95CIf | U95CIf | CIS Sigg |
CIS ORg |
CIS L95CIg |
CIS U95CIg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11209026a,h | 1p31.3 | 67.71 | IL23R | A | G | 0.009 | 0.041 | 0.007 | 0.014 | 0.067 | CD | 7.53E-05 | 0.219 | 0.103 | 0.464 | IBD | 0.497 | 0.465 | 0.531 |
| rs1801274a | 1q23 | 161.48 | FCGR2A | A | G | 0.496 | 0.491 | 0.506 | 0.453 | 0.525 | IBD | 2.60E-04 | 1.211 | 1.093 | 1.343 | UC | 1.192 | 1.15 | 1.234 |
| rs11742570a | 5p13.1 | 40.41 | PTGER4 | A | G | 0.385 | 0.354 | 0.342 | 0.390 | 0.39 | IBD | 1.29E-04 | 0.812 | 0.729 | 0.903 | CD | 0.773 | 0.747 | 0.799 |
| rs5743289b,h | 16q21 | 50.76 | NOD2 | A | G | 0 | 0.226 | 0.052 | 0.032 | 0.178 | CD | 6.12E-05 | 1.763 | 1.336 | 2.326 | CD | 1.557 | 1.497 | 1.618 |
| rs12946510a | 17q21 | 37.91 | IKZF3 | A | G | 0.119 | 0.500 | 0.233 | 0.201 | 0.467 | CD | 9.31E-05 | 1.319 | 1.148 | 1.516 | IBD | 1.157 | 1.124 | 1.19 |
IBD-general SNP in CIS;
Crohn's disease-specific SNP in CIS;
SNP position in mega base-pair (Mb, Genome Build 37);
Minor (A1) and major (A2) allele in current dataset respectively;
Frequency corresponding to minor allele (A1) for Hapmap African samples (YRI), Hapmap European samples (CEU), current African American IBD case (Case) and control (Ctrl) samples, and Immunochip case/control samples used in CIS respectively;
Most significant phenotype (Sig), corresponding p-value (p), minor allele odds ratio (OR) with upper (U95CI) and lower (L95CI) 95% confidence interval.
The most significant phenotype (Sig) and the corresponding odds ratio (OR), lower (L95CI) and upper (U95CI) bounds for 95% confidence interval for odds ratio in CIS;
SNP without expression data available in eQTL evaluation.
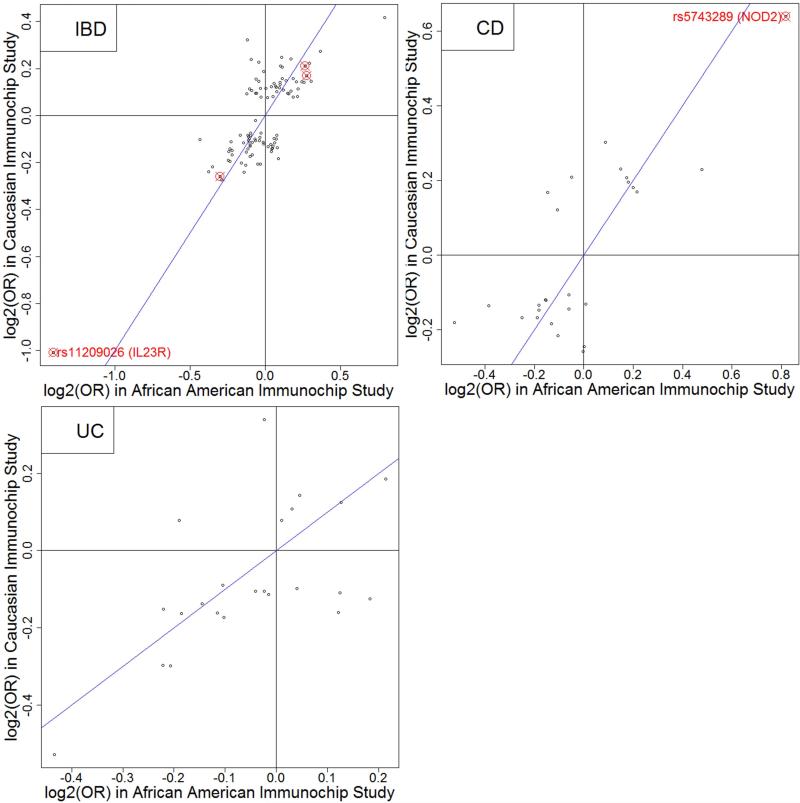
For most of the 152 SNPs, the AA odds ratios are in the same direction as those from the CIS (correlation=0.78, Figure 1). 81 SNPs had AA ORs within the 95% CIS confidence intervals (CI), including one replicated SNP (rs11742570). Nominal levels of significance (p<0.05 and same risk allele) were observed for 41 of the 152 (27%) variants (Supplementary Table 2).
Figure 1.
Comparison of log2(Odds ratios) estimated in this study with those in Caucasian Immunochip Study (CIS) for 110 IBD-general (correlation=0.78), 26 CD-specific (correlation=0.77) and 23 UC-specific (correlation=0.61) Maximal-CIS SNPs.
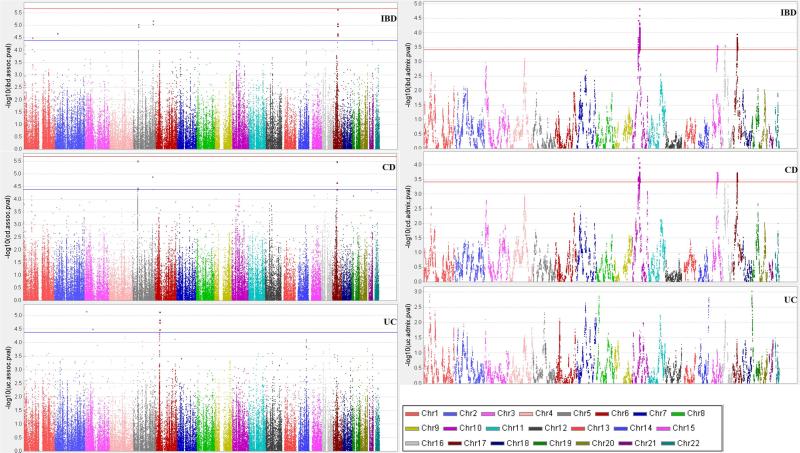
Study-wide Association
Association results for IBD, CD and UC are shown in Figure 2 (left panels). Suggestive association for IBD was observed at the following 7 loci for 15 SNPs (Table 2): 1p at IL23R; 2p near the p-telomere and 4 Mb upstream of an associated locus in the CIS near C2orf43; 5p13.1 gene desert (5 SNPs in tight LD, pairwise r2 ranging from 0.52 to 1.00); 5q 70kb telomeric of IL12B (2 SNPs in tight LD, r2=0.87); and 17 in the STAT5A/STAT3 region (6 SNPs in 3 LD blocks). Conditional regression revealed 3 independent signals from the latter region: three STAT5A SNPs (rs7220367, rs7217884 and rs13380828, all monomorphic in CEU, r2 ranging from 0.46 to 0.99) in one block; two SNPs 200 kb apart (rs730086 in KAT2A and rs1053004 in STAT3, r2=0.44); and rs7224339. IBD, being the phenotype with the largest sample size had greatest power to detect association evidence. However, 72% of cases had CD, and not surprisingly SNPs at 5p15.3 (PTGER4), IL12B, KAT2A and STAT5A also showed suggestive association for CD, with CD consistently showing greater association evidence at 5p15.3 than all IBD (Table 2).
Figure 2.
Association mapping (left) and admixture (right) mapping p-values (in −log10) for IBD (top panel), CD (middle panel) and UC (bottom panel).
Table 2.
Suggestive Study-wide Association (p<4.2e-5)
| SNP | Chr | Mba | A1b | A2b | YRIb | CEUb | Caseb | Ctrlb | Nearest Gene | Location | Sigc | pc | ORc | L95CIc | U95CIc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7515029g | 1 | 67.60 | G | A | 0.093 | 0.049 | 0.075 | 0.101 | IL23R | intergenic | IBD | 3.17E-05 | 0.670 | 0.555 | 0.809 |
| rs3072g | 2 | 20.88 | G | A | 0.027 | 0.354 | 0.100 | 0.130 | C2orf43 | intergenic | IBD | 2.15E-05 | 0.697 | 0.590 | 0.823 |
| rs10043340g | 5 | 40.49 | C | A | 0.466 | 0.275 | 0.361 | 0.434 | PTGER4 | intergenic | CDd | 3.25E-06 | 0.754 | 0.669 | 0.849 |
| rs4286721 | 5 | 40.50 | G | A | 0.385 | 0.252 | 0.319 | 0.386 | PTGER4 | intergenic | CDd | 2.97E-06 | 0.748 | 0.662 | 0.845 |
| rs6866402 | 5 | 40.52 | A | G | 0.442 | 0.257 | 0.368 | 0.428 | PTGER4 | intergenic | CDd | 4.05E-05 | 0.782 | 0.695 | 0.879 |
| rs1876141 | 5 | 40.53 | G | A | 0.442 | 0.267 | 0.367 | 0.428 | PTGER4 | intergenic | CDd | 4.00E-05 | 0.782 | 0.695 | 0.879 |
| rs1505994 | 5 | 40.53 | A | G | 0.442 | 0.267 | 0.368 | 0.429 | PTGER4 | intergenic | CDd | 3.65E-05 | 0.781 | 0.694 | 0.878 |
| rs35990859g | 5 | 158.80 | G | A | - | 0.092e | 0.014 | 0.027 | IL12B | intergenic | IBDd | 6.62E-06 | 0.393 | 0.261 | 0.590 |
| rs36048684g | 5 | 158.82 | A | T | - | 0.050e | 0.013 | 0.027 | IL12B | intergenic | IBDd | 9.04E-06 | 0.398 | 0.265 | 0.598 |
| rs730086 | 17 | 40.27 | G | A | 0.097 | 0.664 | 0.238 | 0.177 | KAT2A | intron | IBDd | 2.26E-06 | 1.373 | 1.204 | 1.567 |
| rs7220367 | 17 | 40.44 | G | C | 0.392 | 0 | 0.285 | 0.349 | STAT5A | intergenic | CD | 2.23E-05 | 0.758 | 0.667 | 0.862 |
| rs7217884g | 17 | 40.45 | G | A | 0.270 | 0 | 0.211 | 0.267 | STAT5A | intron | IBDd | 2.21E-05 | 0.765 | 0.675 | 0.866 |
| rs13380828g | 17 | 40.45 | A | T | 0.297 | 0 | 0.210 | 0.266 | STAT5A | intron | IBD | 2.57E-05 | 0.766 | 0.677 | 0.867 |
| rs1053004 | 17 | 40.47 | A | G | 0.119f | 0.611 | 0.169 | 0.118 | STAT3 | utr-3 | IBD | 1.08E-05 | 1.409 | 1.209 | 1.641 |
| rs7224339g | 17 | 40.49 | A | G | 0 | 0 | 0.033 | 0.058 | STAT3 | intron | IBD | 8.76E-06 | 0.552 | 0.425 | 0.717 |
| rs2655211 | 3 | 13.49 | G | A | 0.179 | 0.431 | 0.190 | 0.265 | HDAC11 | intergenic | UC | 6.97E-06 | 0.619 | 0.503 | 0.763 |
| rs254855 | 3 | 64.07 | A | G | 0.261 | 0.301 | 0.287 | 0.214 | LINC00994 | coding | UC | 3.13E-05 | 1.491 | 1.235 | 1.799 |
| rs2395178g | 6 | 32.41 | C | G | 0.533 | 0.602 | 0.415 | 0.500 | HLA-DRA | intergenic | UC | 1.49E-05 | 0.692 | 0.586 | 0.818 |
| rs9270986g | 6 | 32.57 | A | C | 0.173 | 0.204 | 0.218 | 0.156 | HLA-DRB1 | intergenic | UC | 1.85E-05 | 1.559 | 1.272 | 1.91 |
| rs9271366g | 6 | 32.59 | G | A | 0.168 | 0.192 | 0.202 | 0.141 | HLA-DQA1 | intergenic | UC | 7.47E-06 | 1.627 | 1.315 | 2.013 |
| rs2097431g | 6 | 32.59 | A | G | 0.332 | 0.394 | 0.445 | 0.364 | HLA-DQA1 | intergenic | UC | 3.34E-05 | 1.436 | 1.210 | 1.703 |
SNP position in mega base-pair (Mb, Genome Build 37);
Frequency corresponding to minor allele (A1) for Hapmap African (YRI), Hapmap European (CEU), and current African American case (Case) and control (Ctrl) samples respectively;
The most significant phenotype (Sig) and corresponding p-values (p), odds ratio (OR) with lower (L95CI) and upper (U95CI) bounds for 95% confidence intervals;
SNP with suggestive association (p<4.2e-5) for both IBD and CD;
Minor allele frequency (MAF) for samples from 1000 Genomes pilot 1 CEU low coverage panel;
MAF for samples from the human variation panel of African Americans;
SNP without expression data available in eQTL evaluation.
For UC, the major peak located in the HLA region, with greatest association for rs9271366 (p=7e-6). Conditional analysis revealed that all suggestive HLA SNPs could be accounted for by rs9271366 (Supplementary Figure 4). Two SNPs showed suggestive associations on chromosome 3, at 13Mb and at 64Mb (near HDAC11 and LINC00994), neither within 5Mb of any established IBD loci.
Raw genotyping intensities were visually examined for all replicated/suggestive SNPs and only rs35990859 (at IL12B) had sub-optimal cluster separation (data not shown).
Admixture Association
We observed four chromosomal regions of significant admixture association (Figure 2, right panels and Table 3) with all having significant evidence for both IBD and CD except for 16p12 (IBD-only). For all of these regions, CEU (European) ancestry increases risk and YRI (West African) ancestry was protective. Just below significance was an area with increased CD risk from CEU, maximal at rs2111112 (p=4.9e-4) and 3 Mb from NOD2 (with increased CEU ALD extending through NOD2). Conditioning on NOD2 genotype rs5743289 weakened this ALD evidence (p=0.017). A list of annotated genes with immune-related functions that map to each region is included in Supplementary Table 3.
Table 3.
Significant (p<3.9e-4) admixture association regions
| Maximal SNP | Chromosomala | Positiona | Phenotypeb | pb | ORb | L95CIb | U95CIb | Candidate Genesd |
|---|---|---|---|---|---|---|---|---|
| rs1684909 | 10q11.23-10q21.2 | 49.94-63.25 | IBDc | 1.47E-05 | 0.542 | 0.472 | 0.715 | ZNF365, REEP3, MBL2 |
| rs603439 | 15q22.2-15q23 | 62.76-70.75 | CDc | 1.80E-04 | 0.549 | 0.484 | 0.751 | TRIP4, SMAD6, SMAD3, MAP2K1, IGDCC4, IGDCC3 |
| rs1423086 | 16p12.2-16p12.1 | 21.74-28.39 | IBD | 2.67E-04 | 0.586 | 0.465 | 0.781 | IL4R, IL21R, SLC5A11, PRKCB, ITFG1 |
| rs2227322 | 17q12-17q21.31 | 37.39-40.93 | IBDc | 1.10E-04 | 0.580 | 0.519 | 0.765 | ORMDL3, CSF3, CCR7, Keratin cluster |
Window containing SNPs with suggestive admixture (p<7.8e-4) by chromosomal banding and physical positions in Mb (Genome Build 37).
The most significant phenotype (Phenotype) and the corresponding: p-value (p); odds ratio with each additional African ancestry (YRI) allele (OR); lower (L95CI) and upper (U95CI) bounds for 95% confidence interval for YRI odds ratio.
SNP with significant admixture (p<3.9e-4) for both IBD and CD.
Candidate genes highlighted based on literature review of genes located in physical window.
eQTL Evaluation
We conducted PBMC eQTL mapping for 26 (5 significant replication and 21 suggestive study-wide association) SNPs with eQTL data. After controlling for multiple comparisons, 0 distant and 6 local gene-SNP pairs showed significant association (Supplementary Table 4): rs1053004 on chromosome 17 was associated with KRT19 (p=1.7e-4) and with TTC25 (p=8.0e-4) expression; 3 SNPs (rs1876141 [p=5.5e-4], rs6866402 [p=7.5e-4], and rs1505994 [p=7.5e-4] in tight LD (r2>0.99) at 5p13.1 with Complement Component 6 (C6) expression; and rs1801274 with CD84 expression (p=1.2e-3). PTGER4 expression was evaluated but was not associated with any SNP evaluated (p>0.05).
Network Analysis
We found multiple networks demonstrating significant influence for IBD, CD and UC in AAs even after FDR and Bonferroni corrections (Table 4). Cytokine-cytokine receptor interaction was the dominant pathway for CD and IBD overall, but for UC was second in effect to Measles (which also played a significant but less prominent role in IBD and CD). Jak-STAT and Chemokine signaling pathways were included in top 3 networks for IBD and CD (and Jak-STAT alone for UC). In addition, T cell receptor signaling pathway was one of the top pathways in UC and IBD, but not CD.
Table 4.
Significant KEGG pathways for IBD, CD and UC
| Top KEGG Pathways | Phenotypea | Genesb | p-valuec | FDRc | Bonferronic |
|---|---|---|---|---|---|
| Cytokine-cytokine receptor interaction | IBD | 12 | 1.55E-12 | 3.66E-10 | 3.66E-10 |
| Jak-STAT signaling pathway | IBD | 8 | 4.07E-09 | 4.82E-07 | 9.64E-07 |
| Chemokine signaling pathway | IBD | 8 | 1.59E-08 | 1.26E-06 | 3.77E-06 |
| Measles | IBD | 7 | 3.32E-08 | 1.97E-06 | 7.88E-06 |
| Pathways in cancer | IBD | 9 | 8.65E-08 | 4.10E-06 | 2.05E-05 |
| T cell receptor signaling pathway | IBD | 5 | 6.33E-06 | 2.50E-04 | 1.50E-03 |
| Focal adhesion | IBD | 6 | 9.10E-06 | 3.08E-04 | 2.16E-03 |
| Gap junction | IBD | 4 | 5.97E-05 | 1.77E-03 | 1.42E-02 |
| Fc gamma R-mediated phagocytosis | IBD | 4 | 7.47E-05 | 1.97E-03 | 1.77E-02 |
| Vascular smooth muscle contraction | IBD | 4 | 1.74E-04 | 4.13E-03 | 4.13E-02 |
| Cytokine-cytokine receptor interaction | CD | 18 | 3.11E-21 | 7.37E-19 | 7.37E-19 |
| Chemokine signaling pathway | CD | 10 | 4.13E-11 | 4.89E-09 | 9.79E-09 |
| Jak-STAT signaling pathway | CD | 9 | 2.29E-10 | 1.81E-08 | 5.42E-08 |
| Measles | CD | 6 | 1.15E-06 | 6.84E-05 | 2.74E-04 |
| Pathways in cancer | CD | 8 | 1.76E-06 | 8.32E-05 | 4.16E-04 |
| Measles | UC | 7 | 1.97E-09 | 4.67E-07 | 4.67E-07 |
| Cytokine-cytokine receptor interaction | UC | 8 | 1.03E-08 | 1.22E-06 | 2.43E-06 |
| Jak-STAT signaling pathway | UC | 6 | 2.05E-07 | 1.62E-05 | 4.87E-05 |
| T cell receptor signaling pathway | UC | 5 | 8.88E-07 | 5.26E-05 | 2.11E-04 |
| African trypanosomiasis | UC | 3 | 2.33E-05 | 1.11E-03 | 5.53E-03 |
| NOD-like receptor signaling pathway | UC | 3 | 1.15E-04 | 4.56E-03 | 2.74E-02 |
| Adipocytokine signaling pathway | UC | 3 | 1.88E-04 | 6.38E-03 | 4.47E-02 |
Associated phenotype;
Number of genes in the pathway;
Nominal p-value, false discovery rate (FDR), and Bonferroni-corrected p-value respectively.
Discussion
In this study of IBD genetics in the AA population, we assembled the largest set of cases and controls (more than 4 times larger than any prior AA IBD study and the first genetic study of AA UC), evaluated for replication the majority of established IBD associations and interrogated the majority of known IBD loci for novel associations, and performed pathway and eQTL analyses to further inform about the nature of AA IBD genetics. Although our genotyping platform (not being GWA) limited association mapping primarily to replicating known associations and interrogating established, immunologically related loci, we also used the more powerful method of MALD to identify novel loci throughout the autosomal genome and to complement the association mapping.
We replicated, using stringent criteria (p<3.1e-4), five CIS loci. As our study has more CD than UC cases, most of our replications were for CD. Not surprisingly we replicated loci with greater impact in other populations. Three of the five replications, NOD2, IL23R and 5p13.1, account for the greatest degree of CD variance estimated in CIS. The two other replicated loci also have relatively high OR for Caucasian IBD: FCGR2A and IKZF3 (17th and 47th ranked ORs of the 163 CIS loci).
Two replicated CIS SNPs are more frequent in, and hence IBD risk is more likely to arise from, CEU than YRI genome: rs5743289 that tags the 3 common NOD2 CD-associated mutations rs12946510 at IKZF3 (Table 1). In contrast, the two other replicated SNPs, rs11742570 at 5p13.1 and the rs1801274 at FCGR2A with functional variant H167R, have similar presence in CEU and YRI genomes, whereas the R381Q wildtype albeit risk variant of IL23R at rs11209026 is more common in YRI than CEU genomes.
Given our sample size, we had 80% power to replicate Maximal-CIS SNPs with OR>1.3 and MAF>0.2. Supplementary Figure 5 illustrates our power at different ORs/MAFs at three levels of significance. Although our study was underpowered a-priori for genome-wide association (and obtaining enough AA individuals for genetic studies, as statically powerful as those in Caucasians, may be impractical for years to come), we have identified SNPs in the AA population at a level of association for which to target for independent replication.
Two replicated loci also had additional SNPs with suggestive association: 5p13.1 and IL23R. At 5p13.1 rs4286721 showed greatest association with CD (p=2.97e-6), only marginally below study-wide significance (p<2.1e-6). The suggestive SNPs are in LD, are protective and more frequent in YRI than CEU, and, in our PBMC eQTL analysis, correlated with complement component 6 (C6) instead of PTGER4, the associated gene in the original discovery in lymphoblastoid cell lines25. C6 is a reasonable candidate gene, given that C2 and other complement-related genes are associated with numerous IMDs. For rs7515029 at IL23R the protective allele is almost twice as frequent in YRI (9%) than CEU (5%) indicating another African-derived protective variant, in addition to the well-established R381Q replication.
Our strongest UC signal was in the HLA region, similar to that observed in all examined populations. The most significant SNP, rs9271366, known to tag the HLA class II allele DRB1*1502 in other populations, is the same SNP found most statistically significant in both Japanese26 and Korean27 UC GWASs (p=1e-70 and p=1e-18 respectively). Hence, our rs9271366 UC association (p=7.47e-6) is justified as replication of an established UC SNP. It also accounted for nearby associations according to conditional analysis (Supplementary Figure 4). Interestingly, it was found to be the most associated HLA SNP in AA SLE, a disease 4-fold more frequent in AAs than CAs28.
The strongest IBD association was observed for rs730086, with OR=1.4 (p=2.26e-6, just below study-wide significance), at an intron of KAT2A, a histone acetyltransferase gene recently linked to repression of IFN-beta and innate antiviral immunity through inhibition of TKB129. In LD with rs730086 is rs1053004 (r2=0.44), located in the 3’utr of STAT3, which is eQTL for two genes (FDR=0.038): KRT19, a gene that produces a major type 1 keratin expressed in ileal and colonic epithelium with a broad distribution in simple and stratified epithelia; and TTC25, a gene important for celiogenesis expressed in bone marrow but not in intestine. The risk variants are much more frequent in CEU than YRI.
We observed two other independent signals for IBD and CD on chromosome 17 centered about STAT5A/STAT3 adjacent genes. These two signals are protective and African-specific (monomorphic in CEU). We found no association (p>0.1) for rs12942547, the CIS SNP located in STAT3 intron with nearly identical frequencies in CEU and YRI. Complementing these findings is significant ALD that overlaps this chromosome 17 region (and extends through IKZF3 and STAT3) with the CEU genome producing IBD risk and YRI being protective. Conditioning on the IKZF3 replicated SNP (rs11742570) and the 3 independent STAT5A/STAT3 associations eliminated this ALD (p=0.8). An important implication of our association and admixture results is that the common CEU STAT5A/STAT3 haplotype likely contributes to IBD risk in the Caucasians – a finding made possible by studying an admixed population. Sequencing studies in AAs will be important to identify potential functional variants in LD with the YRI-specific protective alleles.
We found three other regions with significant ALD. Although ALD regions should be unbiased to known loci (as Immunochip contained AIMs throughout the genome) all four regions overlap CIS loci. The most highly associated region is on chromosome 10, includes the IPMK/CISD1/UBE2D1/TFAM locus and extends to within 800 kb of ZNF365 (using a genome-wide admixture p=0.10 cut-off). A region with ALD associated with CD and IBD on chromosome 15 contains SMAD3, a gene associated with need for multiple surgeries in CD30. A region on chromosome 16 contains PRKCB, IL4R and IL21R, and extends centromeric just to SBK1. Each of the ALD regions contains additional potential candidate genes, as noted in Supplementary Table 3.
The top three KEGG pathways in AA IBD (cytokine, Jak-STAT and chemokines) were among the top four KEGG pathways found in CIS. However, not observed in AA IBD was the Leishmania infection pathway – second most significant pathway in the CIS; whereas AA UC, IBD, and CD all showed strong evidence for measles pathway and UC also showed significant evidence for African trypanosomiasis – both pathways not observed in CIS.
The high frequency of genes associated with infectious disease supports the hypothesis that IBD genetic susceptibility may be a by-product of evolutionary adaption to human pathogens. Hence the differential infectious disease pathways associated with AA and Caucasian IBD may be related to adaptation to geographically distinct infectious diseases. In total, these findings suggest that, while major pathways are similar across populations, some are distinct and may be targets for personalized IBD therapies.
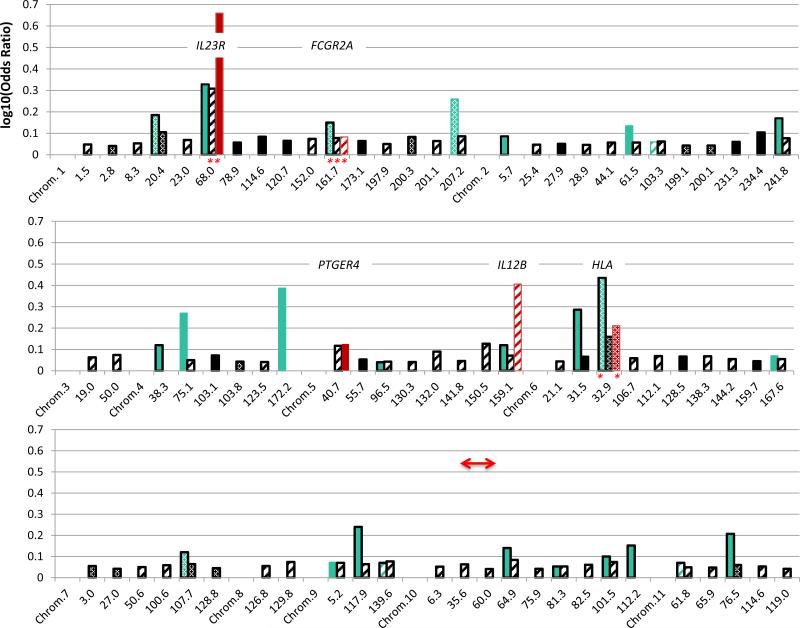
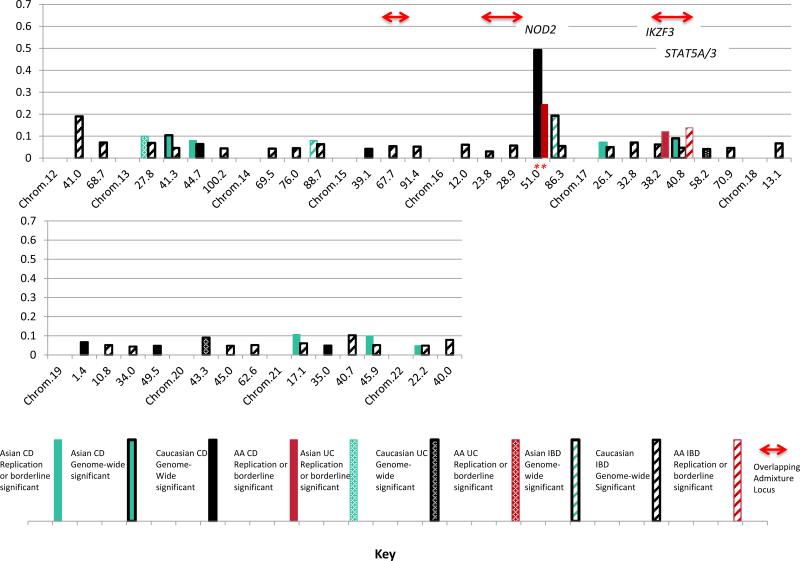
This first, in-depth characterization of AA IBD genetics provides a basis to compare IBD genetics with Caucasian2 and East Asian26,27,31-36 populations where multiple GWAS, replication and Immunochip studies have been performed (Figure 3). Five of the AA loci with suggestive/replication evidence are associated in all three populations: IL23R, FCGR2A, IL12B, HLA and STAT5A/3, with the functional SNPs for IL23R and FCGR2A the same. The higher OR loci tend to be observed in more than one population. The STAT5A/STAT3 locus stands out in AAs: the ORs for the three independent associations are 0.76, 1.40 and 0.55, suggesting that this region likely has a stronger influence in AA than Caucasian IBD (OR=1.1 for rs12942547 in CIS). A Japanese CD GWAS found relatively strong influence for STAT333, maximal at rs9891119, but we found no evidence in AAs for this SNP equally frequent in YRI, CEU and East Asians.
Figure 3.
Comparison of log10(Odds Ratios) at the risk alleles by chromosomal position (in Mb, Genome Build 37) between Caucasian (black), East Asian (green) and African American (red) loci for IBD (hatched), CD (solid) and UC (stippled). Bars with black outlines represent loci with reported genome-wide significant association evidence in the CIS (black, limited to OR≥1.1) or in East Asian studies (green). Bars without outlines are present for AA or East Asian replications (at p<3.1e-4) of CIS-Maximal SNPs, or for SNPs evaluated in these populations with suggestive evidence for association at established CIS loci (p<4.2e-5) or borderline of genome-wide significance for novel loci (p≤1e-6). Asterisks denote the same associated SNPs in AA and additional populations. Red arrows denote overlaps between significant admixture regions and association loci.
In summary, our study reveals that in AAs HLA SNPs demonstrate the dominant signal for UC; STAT5A/STAT3 shows disproportionate association with IBD and CD including novel African protective haplotypes and common European risk haplotypes that together with an IKZF3 CD replication 3 Mb centromeric likely gives risk to significant chromosome 17 regional ALD. We identified three other chromosomal regions contributing significant IBD risk from European admixture. We replicated Maximal-CIS SNPs for NOD2, IL23R, 5p13.1, and FCGR2A, identified additional risk variants at 5p13.1and IL23R. We observed a strong correlation in ORs for Maximal-CIS SNPs between our study and the CIS suggesting that additional Caucasian established loci likely play a role in AAs. We also demonstrated other known (IL12B) and novel (C2orf43, HDAC11 and LINC00994) suggestive areas of association interest. The new suggestive associations will need to be validated by independent replication. Finally a network analysis showed that, as in other studies, cytokines, Jak-STAT signaling and chemokine pathways play major roles, but also suggests that measles and African trypanosomiasis pathways may be important for further investigation. This study has yielded vital information not only on the etiopathogenesis of IBD specifically for AAs, but also about risk variants and ancestral chromosomal regions that may also contribute to IBD pathogenesis in Caucasians, as we continue our exploration of AA IBD genetics.
Supplementary Material
Acknowledgements
We are grateful to all of the patients and the controls that volunteered to join this study. This study is dedicated to Dr. Linda Kao who helped plan and direct the study and passed away just prior to the study's completion.
IBDGC sample recruitment was from the Meyerhoff Inflammatory Bowel Disease Center at The Johns Hopkins Hospital and Johns Hopkins University School of Medicine, Departments of Medicine and Surgery (Patricia Ushry, Sharon Dudley-Brown, Theodore M. Bayless, Christina Ha, Jonathon Efron; Susan Gearhart, and Michael Marohn); and the Division of Pediatric Gastroenterology and Nutrition at The Johns Hopkins Children's Center (Maria Oliva-Hemker and Carmen Cuffari); and funded satellite centers at Henry Ford Health System (Martin Zonca, Qiana Samuels and Aref Araya); University of North Carolina Department of Medicine (Dolly Walkup); Baylor College of Medicine; Virginia Commonwealth University (Kasiah Banks; Alisa Maibauer; Amy Newcombe); Washington Hospital Center (Michael S. Gold and Averell Sherker); University of Florida; Howard University (Duane T. Smoot); University of Alabama (Toni Seay; Tajuanna Lucious); Perelman School of Medicine at the University of Pennsylvania (James D. Lewis); University of Maryland Department of Pediatrics; University of Chicago (Lici Shen); Cornell University; and Columbia University (Arun Swaminath). IBDGC Genetic Research Centers at Yale University, University of Pittsburgh, University of Toronto, and University of Montreal (John D. Rioux holds a Canada Research Chair). The database was developed with assistance from Phil Schumm (University of Chicago). Additional DNA samples on African American IBD cases were provided by Material Transfer Agreements from Washington University Inflammatory Bowel Disease Program with assistance from Rodney Newberry and Ellen Li.
Emory samples were recruited from: Emory University School of Medicine and funded satellite centers at The Children's Hospital of Philadelphia, Cincinnati Children's Hospital Medical Center, Case Western Reserve University, University of Maryland, Vanderbilt Children's Hospital, University of Texas Southwestern Medical Center, University of North Carolina Chapel Hill Department of Pediatrics, University of Chicago Comer Children's Hospital (Thomas Mangatu; Kathleen Van’t Hof, The Barnett and Alscher Families), Louisiana State University Health Sciences Center, Cook Childrens Medical Center, and Willis-Knighton Physician Network.
Cedars samples were recruited from: Cedars-Sinai Medical Center F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute (Stephan Targan, Andrew Ippoliti, Eric Vasiliauskas, David Shih, Gil Melmed, Marla Dubinsky), Charlotte Gastroenterology and Hepatology PLLC; and the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center Institute for Translational Genomics and Population Sciences.
We thank Suna Onengut-Gumuscu for assistance with extraction of control data from RA, SLE and T1D Immunochip studies. We thank Feng Zhou for assistance with Figure 3 and compiling IBD association information in East Asian populations.
Grant Support: NIH Grants DK062431 (S.R.B.), DK087694 (S.K.), DK062413 (D.P.B.M and K.T), DK046763-19, AI067068 and U54DE023789-01 (D.P.B.M.), DK062429 and DK062422 (J.H.C.), DK062420 (R.H.D.), DK062432 (J.D.R.), and DK062423 (M.S.S.). Additional support from Harvey M. and Lynn P. Meyerhoff Inflammatory Bowel Disease Center, the Morton Hyatt Family, the Buford and Linda Lewis family (S.R.B.); Endowed professorship from Marcus foundation (SK); The Joshua L and Lisa Z Greer Endowed Chair, HS021747 from the Agency for Healthcare Research and Quality, grant 305479 from the European Union, and The Leona M. and Harry B. Helmsley Charitable Trust (D.P.B.M.); from the Veterans Administration HSR&D Center for Innovations in Quality, Effectiveness and Safety (#CIN 13-413) and the Michael E. DeBakey VA Medical Center (J.K.H.). PARC control samples supported by NIH/NHLBI grant HL06957 (Ronald M. Krauss, PI). RA control samples recruited and supported by the Consortium for the Longitudinal Evaluation of African-Americans with Early Rheumatoid Arthritis (CLEAR), NIH grants N01-AR-02247 and AR-6-2278 (S.L.B.) and the University of Alabama GCRC (M01-RR-00032); SLE control samples recruited and supported by the PROFILE Study group coordinated at the University of Alabama Birmingham and supported by NIH grants P01-AR49084 (R.P.K.) and M01-RR-00032; T1D control samples recruited and supported by the Type 1 Diabetes Genetics Consortium U01 DK062418 (S.S.R.), and sponsored by NIDDK, NIAID, NHGRI, NICHD and the Juvenile Diabetes Research Foundation (JDRF).
Abbreviations
- ALD
admixture linkage disequilibrium
- AAs
African Americans
- AIMs
ancestry informative markers
- CAs
Caucasian Americans
- CIS
Caucasian Immunochip study
- CD
Crohn's disease
- CEU
European ancestry
- eQTL
expression quantitative trait locus
- FDR
false discovery rate
- GRC
genetics research center
- GWA
genome wide association
- IMD
immune-mediated diseases
- IBD
inflammatory bowel disease
- IBDU
inflammatory bowel disease type undetermined
- OR
odds ratio
- PBMCs
peripheral blood mononuclear cells
- QC
quality control
- RA
rheumatoid arthritis
- Maximal-CIS SNPs
the most statistically significant SNPs at each locus in the Caucasian Immunochip study and present on the Immunochip genotyping array
- SLE
systemic lupus erythematosus
- T1D
type 1 diabetes
- UC
ulcerative colitis
- YRI
West African ancestry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: No authors have conflicts of interest to declare.
Author Contributions: Study concept and design: SRB SK DPBM LWHK JHC CH DJC. Analysis and interpretation of data: CH TH DTO DJC MEZ KDT JCM ZL CLS LWHK DPBM SRB SK. Data acquisition and material support: LWD JSA RNB JHC RKC TD TAD RHD JSH JKH SZH KLI KEK HK MK MDK JK RK BSK JFK AK JHK ML PM DEM BOO AP JDR JIR SS EJS MSS AS SRT JV MHW SLB RPK SSR ADR DPBM SRB SK. Drafting manuscript: CH SRB. Critical revision of the manuscript for important intellectual content: CH TH DTO HK CLS MHW SSR ADR JHC LWHK DPBM SRB SK.
Author names in bold designate shared co-first authors
References
- 1.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang MH, Okazaki T, Kugathasan S, et al. Contribution of higher risk genes and European admixture to Crohn's disease in African Americans. Inflamm Bowel Dis. 2012;18:2277–2287. doi: 10.1002/ibd.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adeyanju O, Okou DT, Huang C, et al. Common NOD2 risk variants in African Americans with Crohn's disease are due exclusively to recent Caucasian admixture. Inflamm Bowel Dis. 2012;18:2357–2359. doi: 10.1002/ibd.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanaan Z, Ahmad S, Roberts H, et al. Crohn's disease in Caucasians and African Americans, as defined by clinical predictors and single nucleotide polymorphisms. J Natl Med Assoc. 2012;104:420–427. doi: 10.1016/s0027-9684(15)30195-4. [DOI] [PubMed] [Google Scholar]
- 8.Patterson N, Hattangadi N, Lane B, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YR, Loftus EV, Jr, Cangemi JR, Picco MF. Racial/Ethnic and regional differences in the prevalence of inflammatory bowel disease in the United States. Digestion. 2013;88:20–25. doi: 10.1159/000350759. [DOI] [PubMed] [Google Scholar]
- 10.Betteridge JD, Armbruster SP, Maydonovitch C, Veerappan GR. Inflammatory bowel disease prevalence by age, gender, race, and geographic location in the U.S. military health care population. Inflamm Bowel Dis. 2013;19:1421–1427. doi: 10.1097/MIB.0b013e318281334d. [DOI] [PubMed] [Google Scholar]
- 11.Malaty HM, Hou JK, Thirumurthi S. Epidemiology of inflammatory bowel disease among an indigent multi-ethnic population in the United States. Clin Exp Gastroenterol. 2010;3:165–170. doi: 10.2147/CEG.S14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen GC, Torres EA, Regueiro M, et al. Inflammatory bowel disease characteristics among African Americans, Hispanics, and non-Hispanic Whites: characterization of a large North American cohort. Am J Gastroenterol. 2006;101:1012–1023. doi: 10.1111/j.1572-0241.2006.00504.x. [DOI] [PubMed] [Google Scholar]
- 13.Simon JA, Lin F, Hulley SB, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 14.Dassopoulos T, Nguyen GC, Bitton A, et al. Assessment of reliability and validity of IBD phenotyping within the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) IBD Genetics Consortium (IBDGC). Inflamm Bowel Dis. 2007;13:975–983. doi: 10.1002/ibd.20144. [DOI] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasaniuc B, Sankararaman S, Kimmel G, Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–21. doi: 10.1093/bioinformatics/btp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol. 2011;7:e1002325. doi: 10.1371/journal.pcbi.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maranville JC, Baxter SS, Witonsky DB, Chase MA, Di Rienzo A. Genetic mapping with multiple levels of phenotypic information reveals determinants of lymphocyte glucocorticoid sensitivity. Am J Hum Genet. 2013;93:735–743. doi: 10.1016/j.ajhg.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews L, Gopinath G, Gillespie M, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–22. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen GK, Millikan RC, John EM, et al. The potential for enhancing the power of genetic association studies in African Americans through the reuse of existing genotype data. PLoS Genet. 2010;6:e1001096. doi: 10.1371/journal.pgen.1001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada Y, Yamazaki K, Umeno J, et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn's disease. Gastroenterology. 2011;141:864–871. e1–5. doi: 10.1053/j.gastro.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 27.Yang SK, Hong M, Zhao W, et al. Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm Bowel Dis. 2013;19:954–966. doi: 10.1097/MIB.0b013e3182802ab6. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Narvaez EA, Fraser PA, Palmer JR, et al. MHC region and risk of systemic lupus erythematosus in African American women. Hum Genet. 2011;130:807–815. doi: 10.1007/s00439-011-1045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Q, Zhuang L, Lai B, et al. Gcn5 and PCAF negatively regulate interferon-beta production through HAT-independent inhibition of TBK1. EMBO Rep. 2014;15:1192–1201. doi: 10.15252/embr.201438990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowler SA, Ananthakrishnan AN, Gardet A, et al. SMAD3 gene variant is a risk factor for recurrent surgery in patients with Crohn's disease. J Crohns Colitis. 2014;8:845–851. doi: 10.1016/j.crohns.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano K, Matsushita T, Umeno J, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. 2009;41:1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- 32.Hirano A, Yamazaki K, Umeno J, et al. Association study of 71 European Crohn's disease susceptibility loci in a Japanese population. Inflamm Bowel Dis. 2013;19:526–533. doi: 10.1097/MIB.0b013e31828075e7. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki K, Umeno J, Takahashi A, et al. A genome-wide association study identifies 2 susceptibility Loci for Crohn's disease in a Japanese population. Gastroenterology. 2013;144:781–788. doi: 10.1053/j.gastro.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 34.Hong SN, Park C, Park SJ, et al. Deep resequencing of 131 Crohn's disease associated genes in pooled DNA confirmed three reported variants and identified eight novel variants. Gut. 2015;0:1–9. doi: 10.1136/gutjnl-2014-308617. [DOI] [PubMed] [Google Scholar]
- 35.Yang SK, Hong M, Choi H, et al. Immunochip analysis identification of 6 additional susceptibility Loci for Crohn's disease in koreans. Inflamm Bowel Dis. 2015;21:1–7. doi: 10.1097/MIB.0000000000000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015 doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.