Abstract
Neisseria gonorrhoeae is the etiological factor of the sexually transmitted gonorrhea disease that may lead, under specific conditions, to systemic infections. The gonococcal genome encodes many restriction modification (RM) systems, which main biological role is to defend the pathogen from potentially harmful foreign DNA. However, RM systems seem also to be involved in several other functions. In this study, we examined the effect of inactivation the N. gonorrhoeae FA1090 ngoAXmod gene encoding M.NgoAX methyltransferase on the global gene expression, biofilm formation, interactions with human epithelial host cells and overall bacterial growth. Expression microarrays showed at least a twofold deregulation of a total of 121 genes in the NgoAX knock-out mutant compared to the wild-type (wt) strain under standard grow conditions. Genes with changed expression levels encoded mostly proteins involved in cell metabolism, DNA replication and repair or regulating cellular processes and signaling (such as cell wall/envelop biogenesis). As determined by the assay with crystal violet, the NgoAX knock-out strain formed a slightly larger biofilm biomass per cell than the wt strain. Live biofilm observations showed that the biofilm formed by the gonococcal ngoAXmod gene mutant is more relaxed, dispersed and thicker than the one formed by the wt strain. This more relaxed feature of the biofilm, in respect to adhesion and bacterial interactions, can be involved in pathogenesis. Moreover, the overall adhesion of mutant bacterial cells to human cells was lower than adhesion of the wt gonococci [adhesion index = 0.672 (±0.2) and 2.15 (±1.53), respectively]; yet, a higher number of mutant than wt bacteria were found inside the Hec-1-B epithelial cells [invasion index = 3.38 (±0.93) × 105 for mutant and 4.67 (±3.09) × 104 for the wt strain]. These results indicate that NgoAX knock-out cells have lower ability to attach to human cells, but more easily penetrate inside the host cells. All these data suggest that the NgoAX methyltransferase, may be implicated in N. gonorrhoeae pathogenicity, involving regulation of biofilm formation, adhesion to host cells and epithelial cell invasion.
Keywords: Neisseria gonorrhoeae, restriction modification, NgoAX, adhesion and invasion of epithelial cells, biofilm, phase variation
Introduction
Gonorrhea, caused by Neisseria gonorrhoeae, is the second most common sexually transmitted disease in the world. Every year, this exclusively human pathogen infects about 106 million people (WHO, 2012). The disease is associated with low mortality but high prevalence, has serious socio-economic effects and is a public health problem worldwide. N. gonorrhoeae can infect the urogenital tract, anus, or throat. Gonococcal infections in men principally cause inflammation of the urethra, which can lead to such complications as urethral strictures, inflammation of the epididymis or the prostate gland (Edwards and Apicella, 2004; Marrazzo et al., 2010; Ison, 2011). The mucous membrane of the cervix is the most common site of infection of N. gonorrhoeae in women (Ison, 2011), in which, gonococci may cause asymptomatic or symptomatic cervical infections, or upper genital tract disease (Bozicevic et al., 2006; Edwards and Butler, 2011). Undetected or untreated gonorrhea can lead to very serious complications. These include: pelvic inflammatory disease, ectopic pregnancy, infertility in women and men, and systemic infections (Holmes, 1999; Ison, 2011; Unemo and Nicholas, 2012). In addition, gonorrhea may increase the risk of human immunodeficiency virus (HIV) transmission as suggested by many studies (for review see Jarvis and Chang, 2012). A recent study demonstrate that N. gonorrhoeae libarates a carbohydrate, heptose-monophosphate, that elicits an innate immune response and drives HIV-1 expression (Malott et al., 2013). Phase variation is a well-characterized mechanism by which numerous bacterial species, including N. gonorrhoeae, introduce phenotypic diversity within a population and increase pathogenicity (Snyder et al., 2001; Jordan et al., 2005; Srikhanta et al., 2009).
The main biological role of bacterial RM systems is to defend against the attack of potentially harmful foreign DNA. Yet, RM systems seem also to exhibit several other functions (for review see Vasu and Nagaraja, 2013). The role of RM systems in epigenetic signaling has been investigated by several research groups (Fox et al., 2007a; Srikhanta et al., 2009, 2010, 2011). RM systems have been shown to be involved in epigenetic regulation, which suggests their involvement in adaptive evolution by regulation of the global gene expression pattern (Low et al., 2001; Srikhanta et al., 2010). Type III RM system were also described affecting the interactions of pathogenic bacteria with host cells (Srikhanta et al., 2009). Through the modulation of genetic variation, RM may also affect the evolution rate (Arber, 2000). It seems that RM plays an important role in co-evolutionary interactions between mobile genetic elements and their hosts (Sneppen et al., 2015). RM systems are compared to toxin–antitoxin systems (Mruk and Kobayashi, 2014). It has been also shown that restriction products can stimulate their homologous recombination with the host genome (Price and Bickle, 1986; Arber, 2000). RM systems behave like selfish genetic elements that promote their own survival and increase their own turnout (Kobayashi, 2001; Vasu and Nagaraja, 2013). In addition, RM systems control the immigration of foreign DNA by restriction endonuclease function. This barrier allows controlling speciation of bacteria, which is associated with genetic isolation (Jeltsch, 2003).
Bacteria use DNA adenine methylation (Dam) as an epigenetic signal by which additional information is imparted to the DNA. Dam methyltransferases play, among others, a pivotal role in regulation of gene expression and phase variation (Marinus and Casadesus, 2009). However, as we previously demonstrated, N. gonorrhoeae FA1090 does not encode a Dam methyltransferase and biological functions of Dam are suggested to be taken over by another system (Kwiatek et al., 2014).
Restriction modification systems are ubiquitous in bacteria and include two enzymatic activities: endonucleolytic and methylating. Restriction endonucleases and methyltransferases recognize the same specific nucleotide sequence in double-stranded DNA, but differ in catalyzed reaction. Most endonucleases cut the foreign double-stranded DNA, when it is not specifically methylated, by hydrolysis of phosphodiester bonds. In contrast, methyltransferases modify nucleotides present in specific sequences of the host DNA by attaching to them a methyl group. Such modifications protect the host DNA from endonucleolytic restriction (Fox et al., 2007b; Rao et al., 2014).
Restriction and modification systems have been divided into four types: I, II, III, and IV, according to their mode of action and distribution of restriction, modification, and specificity functions within the enzyme subunits (Roberts et al., 2015).
Type III restriction-modification systems are the most poorly characterized among all types of RM systems (Williams, 2003; Rao et al., 2014). Rebase, the library of RM systems, revealed more than 155 confirmed enzymes belonging to type III RM (Roberts et al., 2015). Type III RM systems consist of two closely spaced genes, mod and res (Hümbelin et al., 1988). Mod genes encode the methyltransferase subunit Mod, responsible for sequence recognition and modification, while res genes encode the restriction endonuclease subunit. Methyltransferases are responsible for recognizing the asymmetric target sequence and methylation reaction catalysis. Enzymes of this type operate independently of the restriction endonuclease (Rao et al., 2014). Type III methyltransferases methylate adenine only on one strand of the recognized DNA sequence and convert it into an N6-methyladenine (Dryden et al., 2001). Restriction activity is performed only by a complex formed of Mod and Res subunit. Res subunit alone has no activity.
Few recent publications suggest that DNA methyltransferases belonging to type III RM systems play a role of epigenetic mechanisms, which control gene expression. Control of multiple gene expression by type III RM systems has been described in several human pathogenic bacteria (Srikhanta et al., 2005, 2009, 2011; Bayliss et al., 2006). Moreover, in Haemophilus influenzae and some other bacteria type I and III DNA methyltransferases undergo phase variation, which increases their potential role in controlling gene expression (Ryan and Lo, 1999; De Bolle et al., 2000; Srikhanta et al., 2005; Fox et al., 2007a; Broadbent et al., 2010; Seib et al., 2015).
Sixteen potential RM systems, in the chromosome of the N. gonorrhoeae FA1090 strain (accession no. AE004969), have been identified using bioinformatic analysis (Stein et al., 1995). However, only a few of them have been experimentally studied (Rebase). Among N. gonorrhoeae FA1090 RM systems, only two belong to the type III group (Stein et al., 1995). The first one is NgoAXII (encoded by the ngo0641 gene, also called modA13), for which the recognition sequence has been determined to be 5′ AGAAA 3′. Expression of the NgoAXII methyltransferase undergoes phase variation associated with the control of expression of many genes reassembled in a phasevarion (Srikhanta et al., 2009). The second type III RM system in N. gonorrhoeae FA1090 is NgoAX, encoded by two genes ngoAXmod (or ngo0545, also called modB1, Srikhanta et al., 2009) and ngoAXres (ngo0546), which respective products are a methyltransferase and restrictase. Recognition sequence of the NgoAX methyltransferase is: 5′-CCACC-3′ as was experimentally demonstrated. Like other type III methyltransferases, it methylates only one strand of the dsDNA (Adamczyk-Poplawska et al., 2009). We have previously observed that the restriction phenotype associated with NgoAX switches randomly when cloned in Escherichia coli. This phase variation was determined to be related to a variability in the number of 5′-CCAAC/G-3′, pentanucleotide repeats present at the 5′-end of the ngoAXmod coding region, which causes a frameshift of the ngoAXmod gene (Adamczyk-Poplawska et al., 2009). Such change resulted in switching the M.NgoAX methyltransferase (as well as the whole RM system) from ON to OFF state.
In this work we studied the phenotypic effect of ngoAXmod inactivation in N. gonorrhoeae FA1090. The influence of the lack of DNA methylation by NgoAX methyltransferase on global gene expression was also evaluated. Moreover, inactivation of NgoAX on biofilm formation, interactions with human epithelial host cells and overall bacterial growth were examined.
Materials and Methods
Bacterial Strains Growth Conditions
Escherichia coli strain Top10 [F’[lacIq Tn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 AAA-] was grown in Luria-Bertani (LB) broth (Difco) or on LB agar plates at 37°C. When needed, medium was supplemented with kanamycin (30 μg/ml) or chloramphenicol (34 μg/ml) or ampicillin (100 μg/ml).
Neisseria gonorrhoeae strains were grown on GCB agar base (Difco) supplemented with 1% Kellogg’s supplement and 1% hemoglobin at 37°C in 5% CO2 or in GCB broth with 1% Kellogg’s supplement and 0.043% NaHCO3, according to Dillard (2006). The following antibiotics were used, when needed: kanamycin (30 μg/ml) or chloramphenicol (0.75 μg/ml).
Construction of N. gonorrhoeae Mutant Strains: The Knock-Out Mutant in NgoAX RM System (ngoAXmod::km) and the Complementation Mutant (igatrpb::ngoAXmod)
To construct the NgoAX knock-out mutant, we replaced the wt ngoAXmod gene on the FA1090 chromosome with a disrupted by antibiotic cassette allele.
The pNgoAXP plasmid encoding the NgoAX restriction-modification system from N. gonorrhoeae FA1090 (Adamczyk-Poplawska et al., 2009) was digested with MunI. In effect a deletion of a 600-bp fragment within the ngoAXmod gene encoding the ModNgoAX subunit of NgoAX was generated. The obtained fragment was ligated with the EcoRI fragment from the pDIY-km plasmid (Dziewit et al., 2011), containing a km cassette active in N. gonorrhoeae (Kwiatek et al., 2014). The resulting plasmid was linearized with MluI and used to transform piliated, i.e., competent N. gonorrhoeae cells. Linearization of the vector forced the replacement of the wt ngoAXmod gene by the ngoAXmod::km allele, by the occurrence of double crossing-over. N. gonorrhoeae FA1090 colonies lacking the NgoAX RM system (NgoAX knock-out mutants) were selected on kanamycin containing GCB plates.
To construct the N. gonorrhoeae complementation mutant, the ngoAXmod gene was amplified from chromosomal DNA of N. gonorrhoeae FA1090 (Gene Bank: AE004969, ATCC 700825) by PCR with Smamod and Nhemod primers (see Supplementary Table S1 for primer sequences), using PfuUltra II Fusion HS DNA polymerase (Agilent Technologies), according to the manufacturer’s protocol. The resulting fragment of 2094 bp was ligated with the PCR fragment (6578 bp) obtained by amplification using the pMPMigatrpBopaCM vector (pMPMA4Ω vector with cloned intergenic region iga-trpb and cm cassette) as template and primers Smatrpb and Nheiga (see Supplementary Table S1 for primer sequences). In the obtained construct, the ngoAXmod gene is under control of a strong constitutive promoter as previously described (Ramsey et al., 2012). The recombinant vector was linearized with EcoRV and used to transform the N. gonorrhoeae ngoAXmod::km strain. N. gonorrhoeae cells, in which the interrupted ngoAXmod gene was complemented with the wt ngoAXmod gene, inserted between iga and trpb genes, were selected on GCB plates containing 0.75 μg/ml chloramphenicol and 30 μg/ml kanamycin. The resulting recombinant strain was called N. gonorrhoeae igatrpb::ngoAXmod (complementation mutant).
Disruption of the ngoAXmod gene by the km cassette and introduction of the wt ngoAXmod gene into the region between iga and trpb genes in N. gonorrhoeae FA1090 was verified by PCR, sequencing and Southern Blot. For Southern Blot, isolated, from the wt N. gonorrhoeae, the knock-out and complementation mutants, 1.0 μg of chromosomal DNAs were digested with MluI restriction enzyme. Obtained fragments were separated on agarose gels (0.7%) at 130 V for 2 h and Southern alkali transfer, followed by hybridization were performed as described by Sambrook and Russell. We used non-radioactively labeled km or chloramphenicol cassette (DIG-High Prime, Roche) as probe (data not shown).
Field Emission Scanning Electron Microscopy (FE SEM)
For FE SEM, N. gonorrhoeae ngoAXmod::km, igatrpb::ngoAXmod and wt strains were harvested from 24 h GCB plates and suspended in GCB broth. OD600 were adjusted to 0.05, corresponding to 4 × 107 cfu/ml. Bacteria were then cultivated on cover glasses, placed in Petri dishes in GCB broth at 37°C in 5% CO2. After 24 h, samples for FE SEM were prepared as previously described (Kwiatek et al., 2014) and observed with MERLIN Carl Zeiss FE SEM Microscope.
Live Cell Confocal Microscopy (SCLM)
We quantified live biofilm development after 16 h growth on Glass Bottom Microwell Dishes (MatTek Corporation) as previously described by Kwiatek et al. (2014), using a Nikon Eclipse Ti (A1) microscope and the NIS-ELEMENTS interactive software.
Microtiter-Plate Adherence Assay
Microtiter-plate adherence assay was performed as described previously (Stepanovic et al., 2000; Kwiatek et al., 2014). Briefly, N. gonorrhoeae wt strains were harvested from 24 h GCB agar and OD600 = 0.05 suspensions were made in GCB broth supplemented with Kellogg’s supplement and MgSO4. (5 mM). Bacteria were then plated into 96-well microtiter plates and cultivated for 24 h at 37°C in 5% CO2. After the OD600 measurement, the supernatant was aspirated, and each well was extensively washed with PBS in order to remove all non-adherent bacteria. The attached bacteria were processed as previously described and stained with crystal violet (0.8%). After extensive washing and resolubilising the dye bound to biofilm-forming cells, OD570 was measured. Experiments were carried out three times and results were averaged. The significance of results was determined by Student’s t-test (P < 0.05).
RNA Isolation
RNA isolation was performed as described previously (Kwiatek et al., 2014). Total RNA was isolated from cells growing on one GCB agar plate (no pooled samples) using the High Pure RNA Isolation Kit (Roche) and DNA-freeTM, DNase Treatment and Removal (Ambion), according to the manufacturer’s recommendations. From one plate, approximately 20–30 μg of total bacterial RNA was purified. RNA quality was evaluated using and 2100 Bioanalyzer (Agilent Technologies) and NanoDrop 2000 (Thermo Scientific).
Microarray Experiments
Two color DNA microarray analysis was performed using the Agilent-034141 array as described in details by Kwiatek et al. (2014). The Cy5 dye was used to label the cRNA ngoAXmod::km strain and the Cy3 dye to label the cRNA N. gonorrhoeae FA1090 strain. Four wt and four mutant biological replicates were studied.
Microarray Analysis
Data files from the Agilent G2565CA Microarray Scanner System were loaded into the GeneSpring (Agilent Technologies, version 12.5) and analyzed as described previously (Kwiatek et al., 2014). Variations were presented as the ratio of gene expression of wt gonococcal strain over expression of the same gene for the NgoAXP knock-out mutant. Genes with divergent expression of ≥1.5 or 2-fold and a P-value < 0.05 were chosen. Microarray data have been deposited in the NCBI’s and are accessible through Gene Expression Omnibus series accession number GSE717031 (Edgar et al., 2002).
Real-Time qRT-PCR
Ten micro gram of total RNA (used for microarrays) were submitted to reverse transcription using the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific). Real-time PCR (5 × HOT FIREPol® EvaGreen® qPCR Mix Plus, Solis BioDyne) was carried out on The Applied Biosystems® StepOneTM Real-Time PCR Systems (Life Technologies). For each gene of interest we used HPLC-purified oligonucleotide primers (see Supplementary Table S1).
The comparative threshold cycle (Ct) method was used to relative quantification of gene transcription. The 16S rRNA gene was used as an internal reference to normalize the relative amount of target cDNA. Data represent averages of four independent samples.
Gonococcal Cell Viability Assay
Neisseria gonorrhoeae cells were harvested from GCB agar and cell suspensions were made in GCB broth with Kellogg’s supplement, 5 mM MgSO4, and 0.043% Na2CO3 (107 cells/ml). Bacterial cultures were incubated at 37°C with gentle shaking for 8 h. Every hour, samples were taken and the number of viable cells was determined using the BacTiter-Glo Microbial Cell Viability Assay (Promega) as previously described (Kwiatek et al., 2014). As negative control we used GCB broth with supplements. Signal detection was performed using the luminometer Glomax multiplus (Promega). All measurements were carried out three times and results were averaged. The significance of results was determined by Student’s t-test (P < 0.05).
Hec1-B Cell Culturing
The human adenocarcinoma endometrial cell line Hec-1-B, acquired from the ATCC (ATCC HTB113), was cultivated in DMEM (Sigma–Aldrich) supplemented with 10% fetal bovine serum (Cytogen), 2 mM glutamine (Cytogen) and 1 mM sodium pyruvate (Sigma–Aldrich). Cells were incubated at 37°C in 5% CO2 and passaged every 5 days.
Infection of Hec-1-B Cells with N. gonorrhoeae
Bacteria were plated on GCB agar plates 2 days before infection and grown at 37°C, in a 5% CO2-containing atmosphere. Then, colonies were checked under a light microscope for pilus and Opa expression status by colony morphology (Swanson, 1978). Appropriate clones were picked and streaked onto new GCB agar plates. The plates were then grown for no more than 16 h to avoid autolysis of bacteria. Bacteria were scraped from GCB agar plates in GCB broth and used to infect the epithelial cells at a multiplicity of infection of 100.
Adhesion Assay of N. gonorrhoeae to Human Cells
Human Hec-1-B cells were seeded in 24-well plates (Cellstar) at a density of 105 cells/well. After incubation for 5 days at 37°C in 5% CO2, bacteria were added. Four hour post infection, the medium was discarded from one set of wells, and human cells were washed extensively with pre-warmed PBS and then lysed with 0.5% saponin (Sigma–Aldrich). Released bacterial cells were plated for enumeration of bacteria associated to human cells: human cell-associated colony forming units (CFU). At the same time, the supernatant was collected from a parallel set of infected cultures and bacteria present in this supernatant were plated on GCB agar for enumeration of bacteria not associated to human cells. Hec-1-B cells from this parallel set of wells were also lysed and released bacteria plated on GCB agar for CFU enumeration. The total CFU was defined as the sum of CFU of bacteria not associated to human cells and bacteria released from human cells. The adhesion index was the quotient of dividing the cell-associated CFU by the total CFU (Hopper et al., 2000; Kwiatek et al., 2014). Tests were carried out at least three times and results were averaged. The significance of results was determined by the Student’s t-test (P < 0.05).
Invasion Assay of N. gonorrhoeae into Human Cells
Hec-1-B cells were seeded and infected as described above for adhesion assays. Invasion assays were performed essentially as described by Hopper et al. (2000). Four hour after infection, sets of wells were washed with pre-warmed PBS and further incubated for 60 min with DMEM containing 200 μg/ml of gentamicin (Gibco) in order to kill extracellular bacteria. Subsequently, cultures were washed extensively again to remove gentamicin and then lysed as described above. Released bacteria were plated for enumeration of intracellular CFU. At the same time point, separate sets of cultures were processed for calculation of adhesion index. The invasion index was calculated by dividing the number of intracellular bacteria by the adhesion index. All infections were performed in triplicate. The significance of results was determined by the Student’s t-test (P < 0.05).
Enzymes, Oligonucleotides and Chemicals
Enzymes were acquired from Thermo Scientific. Kits for chromosomal DNA purification, DNA clean-up and plasmid DNA extraction were obtained from A&A Biotechnology, Gdansk, Poland. Unless otherwise noted, other chemicals were acquired from Sigma–Aldrich. Primers for DNA amplification were purchased from the Institute of Biochemistry and Biophysics (Poland) or from Sigma–Aldrich (HPLC-purified primers for qRT-PCR).
Other Methods
Routine methods were carried out as described by Sambrook and Russell (2001).
Computer Analysis
DNA and protein sequences were analyzed with BLAST, Genebank, KEGG and Uniprot databases. COG were assigned using http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. Predicted outer membrane proteins were checked using http://db.psort.org/.
Results
Expression of Type III Methyltransferases in N. gonorrhoeae FA1090
According to Rebase (Roberts et al., 2015) and literature data (Stein et al., 1995; Srikhanta et al., 2009), N. gonorrhoeae FA1090 encodes two type III RM systems: NgoAX, encoded by the ngo0545 gene, also called ngoAXmod (encoding the Mod subunit) and ngo0546 (coding the Res subunit) and the NgoAXII system, encoded by the ngo0641 gene, called ngoAXIImod (Mod) and ngo0640 (Res). By qRT-PCR, we determined the relative level of expression of genes encoding methyltransferases in comparison to the level of expression of a housekeeping gene, encoding 16S rRNA. Our results indicated that the ngoAXmod gene is expressed at 4.488 (±1.11) copies/million 16S rRNA copies and ngoAXIImod at 0.621 (±0.067) copies/million.
Construction of the N. gonorrhoeae NgoAX Knock-Out Mutant and Complementation Strain
To study gene expression and selected physiological aspects of neisserial pathogenesis in the presence or absence of NgoAX methyltransferase activity, two gonococcal mutants were obtained. Mutant strains were constructed using suicide plasmids, by the gene replacement method. A plasmid with the ngoAXmod gene with cloned km cassette was used for interruption of the chromosomal ngoAXmod gene, resulting in the N. gonorrhoeae ngoAXmod::km strain (otherwise called the NgoAX knock-out strain).
To obtain the N. gonorrhoeae complementation mutant, we cloned the wt ngoAXmod gene into the intergenic region between iga and trpb locus present on the pMPMigatrpBopaCM plasmid. In this vector, the cloned ngoAXmod gene was under control of a constitutive promoter as described previously and a chloramphenicol cassette was used for recombinant vector selection (Ramsey et al., 2012). This plasmid was transformed into the N. gonorrhoeae ngoAXmod::km strain, generating N. gonorrhoeae igatrpb:: ngoAXmod. The obtained N. gonorrhoeae complementation strain was both, kanamycin and chloramphenicol resistant: the km cassette disrupted the original ngoAXmod locus and the wt ngoAXmod copy was inserted into the chromosome, followed by the chloramphenicol cassette for recombinant cell selection.
To confirm that the constructed mutant strains contain only the desired insertions PCR, gene sequencing and Southern Blot were performed. With Smamod and Nhemod primers (Supplementary Table S1), we performed PCR on genomic DNA isolated from the wt and mutants strains. As expected, a PCR product of 2095 bp for wt gene or a 2552-bp long PCR for interrupted ngoAXmod:: km were obtained, while both products were generated in case of the igatrpb::ngoAXmod mutant. With AXPleft and AXPright primers (Supplementary Table S1), we obtained a PCR product of 1435 bp for the wt strain, 1893-bp DNA fragment for the knock-out mutant in the NgoAX RM system, and both products for the complementation mutant. PCR analysis performed with Trpb and Iga primers (Supplementary Table S1) on genomic DNA isolated from the wt strain and complementation mutant resulted, as expected, in a 1134-bp fragment for the wt strain or a 4286-bp long PCR for the igatrpb::ngoAXmod mutant, corresponding to the intergenic region and inserted ngoAXmod gene (data not shown).
Absence of the M.NgoAX Methyltransferase Enhances N. gonorrhoeae Growth
Neisseria gonorrhoeae cells were grown in liquid medium with gentle agitation at 37°C. To measure the viability of ngoAXmod::km mutant and control gonococcal strains, we measure a luminescent signal, proportional to the number of viable cells by using the Microbial Cell Viability Assay (BacTiter-GloTM).
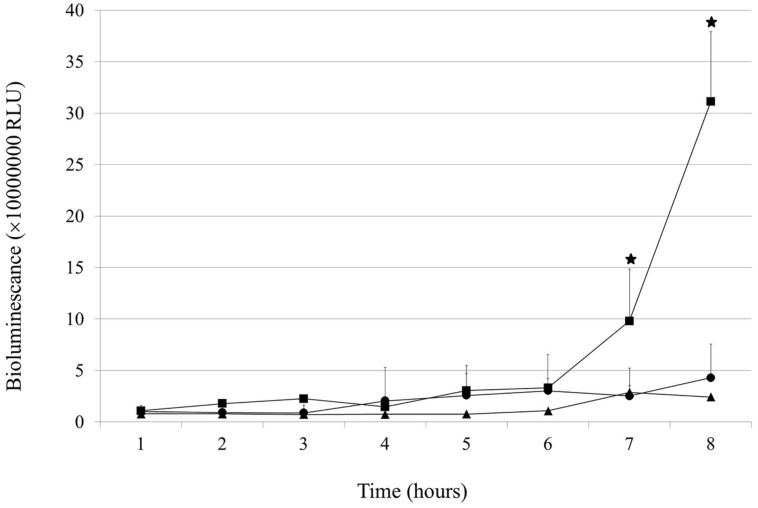
Under the applied conditions, the growth of the wt gonococcal cells was very slow and a plateau-like phase was achieved after approximately 5 h (Figure 1). After 8 h of growth, wt cells reached the level of 4.28 (±3.3) × 106 RLU (relative light units). In contrast, NgoAX knock-out cells kept growing exponentially during 5 h and after 7 and 8 h reached the level of 9.79 (±5.05) and 31.1 (±6.79) × 106 RLU, respectively (which is statistically different from the level of the wt cells: P < 0.05 at both laps times). Complementation of the ngoAXmod gene mutation (igatrpb::ngoAXmod mutant) restored the growth to a similar level as for the wt gonococci (P > 0.05).
FIGURE 1.
Effect of ngoAXmod disruption on Neisseria gonorrhoeae growth. Growth and viability of the wt N. gonorrhoeae (triangle), ngoAXmod::km mutant (square) and igatrpb::ngoAXmod mutant (circle) were measured for 8 h using the BacTiter-GloTM Microbial Cell Viability Assay kit. The amount of viable cells was directly proportional to the produced bioluminescence. The asterisk represents the statistical difference in comparison to the wt strain (P < 0.05).
Impact of the ngoAXmod Gene Inactivation on Gonococcal Gene Expression
In the NgoAX knock-out mutant, a total of 121 genes were deregulated (38 down and 83 up) at least twofold as compared to the wt strain under standard growth conditions. This group of genes represented 5.61% of the total gene pool. When the cut-off was positioned at 1.5 fold, the expression of 249 genes (11.54% of total genes) was changed in the ngoAXmod::km mutant: 106 genes were downregulated and 143 upregulated (Supplementary Table S2). Expression of randomly chosen genes was examined by qRT-PCR (Supplementary Table S2).
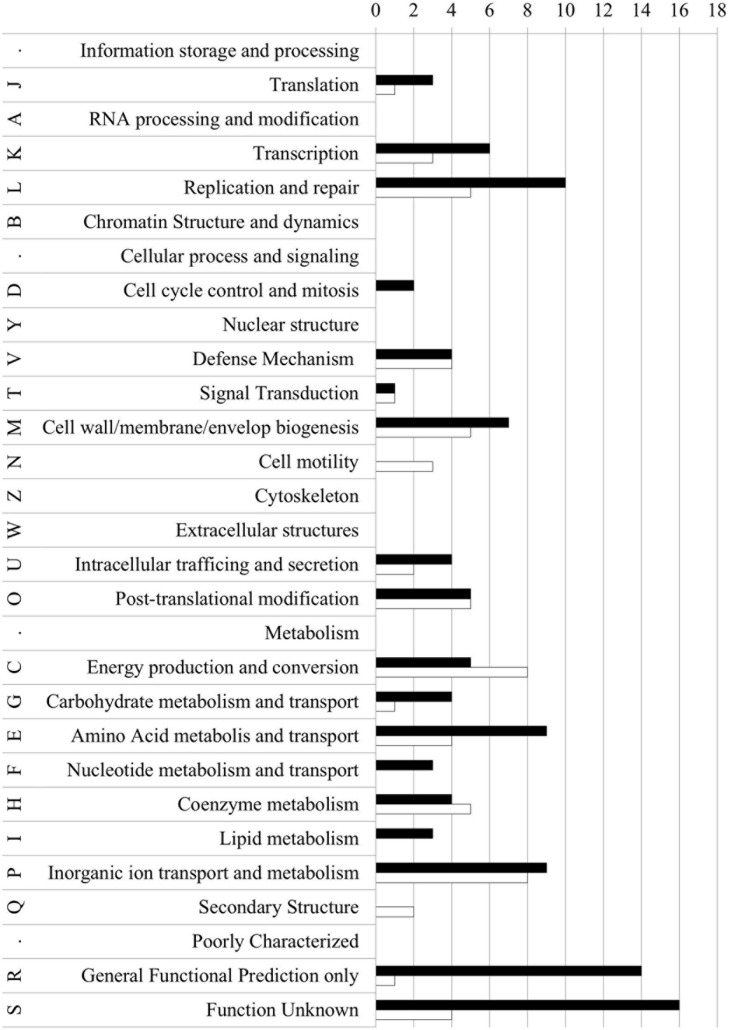
For each gene with deregulated expression, we determined the COG category (cluster of the orthologous gene, Natale et al., 2000; Tatusov et al., 2000) category (Supplementary Table S2; Figure 2). Most genes with affected expression encoded hypothetical proteins with unknown function (S or R COG category: 35 gene products) or proteins in which no conserved domains (87 gene products). Sixty five genes with changed expression levels encoded proteins involved in cell metabolism (mostly categories C and P). Fifteen genes encoded proteins involved in DNA replication and repair (category L). Forty three genes encoded proteins regulating cellular processes and signaling, among them 12 genes encoded proteins involved in cell wall/envelop biogenesis (M category) and 10 – proteins engaged in posttranslational modifications (O category).
FIGURE 2.
Clusters of orthologous groups classification of proteins encoded by up- and down-regulated genes in the NgoAX knock-out N. gonorrhoeae versus the wt strain. Letters represent COGs categories; numbers – the number of proteins in each category; black bars – proteins encoded by up-regulated genes; white bars – proteins encoded by down-regulated genes.
Among the affected genes, encoding proteins with unknown functional COG, many were involved in biofilm formation and/or adhesion. For example, the operon containing ngo0095–ngo0098 genes, involved in pilus formation, was downregulated. Other downregulated genes involved in adhesion were those encoding adhesins MafA and MafB (ngo1393 and ngo41068) or OpaD, encoded by ngo1513. In contrast, expression of the ngo0180 gene, encoding a Maf-like protein, was upregulated.
Detailed information about deregulated genes can be found in Supplementary Table S2.
Biofilm Formation by N. gonorrhoeae FA1090 is Modulated by M.NgoAX Methyltransferases
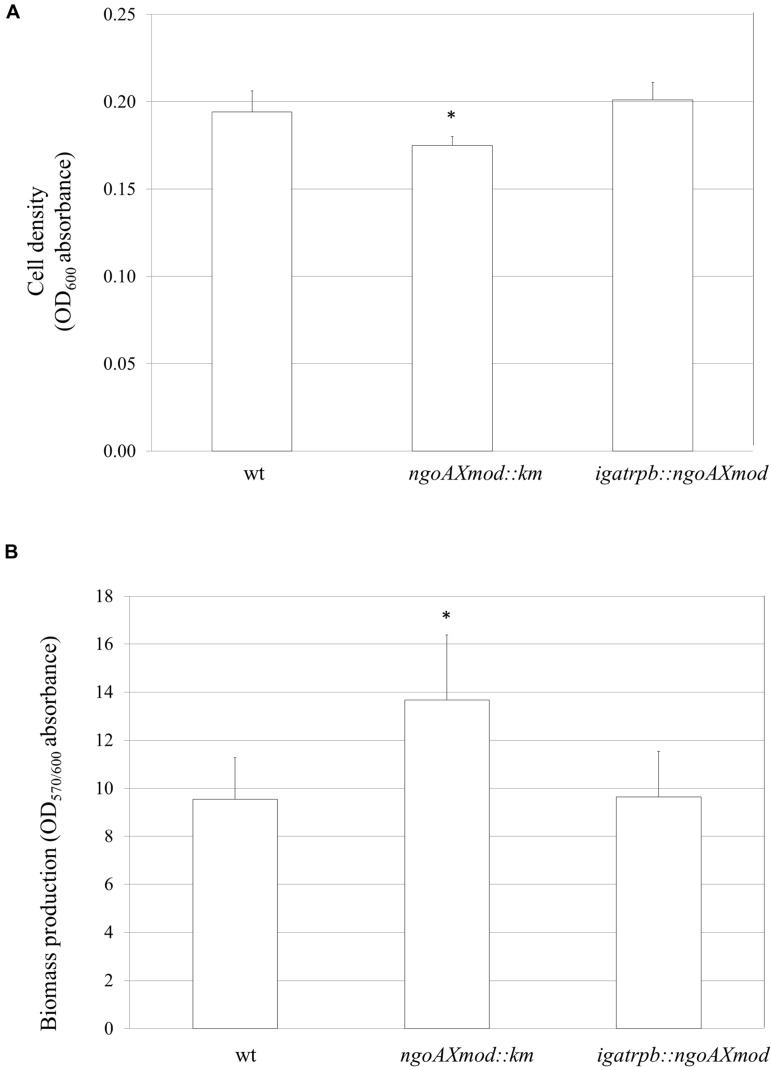
Gonococci under study were examined for their growth and adherence abilities to polystyrene surfaces. After a 24-h growth without shaking, the overall density OD600, reflecting both planktonic and biofilm-engaged gonococcal cells, was determined. The amount of N. gonorrhoeae ngoAXmod::km cells was slightly yet significantly lower (OD600 = 0.175 ± 0.005) than gonococcal wt cells (OD600 = 0.194 ± 0.012; P < 0.05). Reintroduction of the wt copy of ngoAXmod resulted in a similar amount of complementary mutant cells (OD600 = 0.201 ± 0.010) as compared to the wt strain (P > 0.05) (Figure 3A). Subsequently, after intensive washing of cells not engaged in biofilm formation, we measured the proportion of biofilm-forming cells by staining them with crystal violet. The NgoAX knock-out strain formed a slightly larger biofilm biomass per cell than the wt strain (OD570/600 = 13.8 ± 2.24 and 9.35 ± 2.06 respectively; P < 0.05) as determined by comparing the biofilm biomass to the total amount of cells (Figure 3B). Introduction of the wt copy of ngoAXmod reduced biofilm formation at the level of wt strain: OD570/600 = 9.64 ± 1.89.
FIGURE 3.
Growth and biofilm biomass of ngoAXmod::km and igatrpb::ngoAXmod N. gonorrhoeae mutants compared to the wt strain. Measurements were performed by crystal violet staining after a 24-h growth without shaking. (A) Biofilm and planktonic cell growth was measured at absorbance OD600; (B) Biomass production: ratio of cells that forms biofilm (OD570) versus total grown cells (OD600). Asterisks represent statistical significant differences in comparison to the wt strain (P < 0.05).
The ability of N. gonorrhoeae FA1090 wt, ngoAXmod::km and complementation mutant to form biofilm was also evaluated using FE SEM and SCLM.
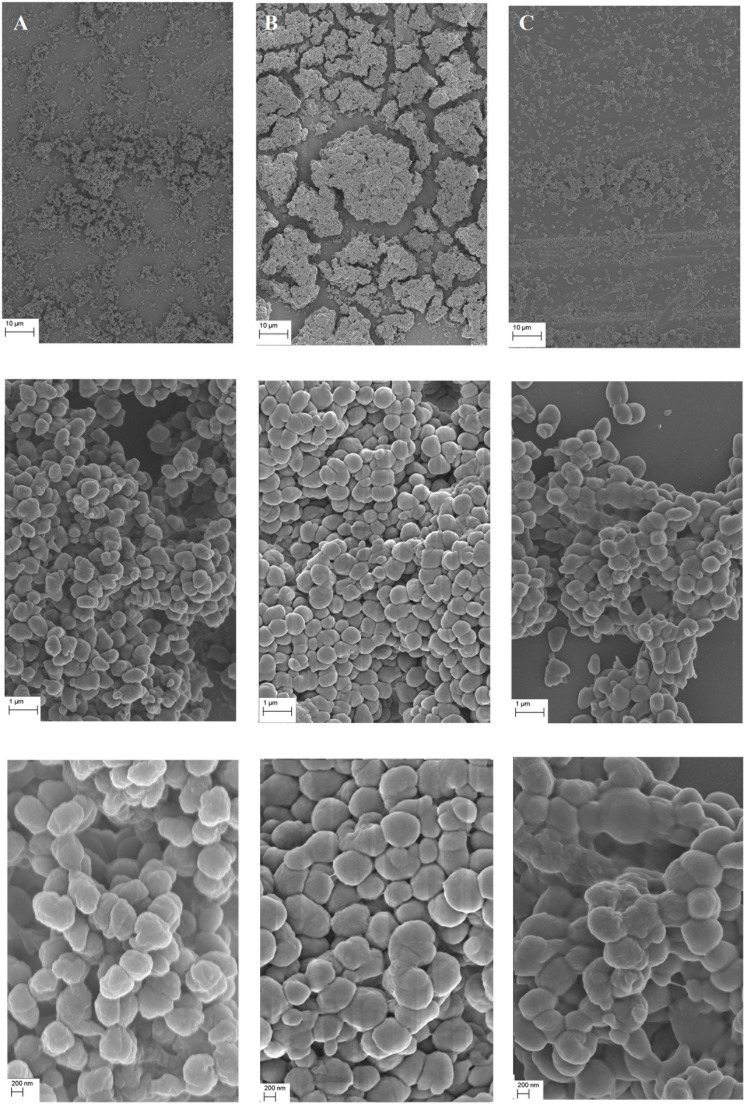
For FE SEM, bacteria were grown under standard growth conditions on a cover slip glass for 24 h. As shown on Figure 4A, biofilm formed by the wt N. gonorrhoeae was evenly spread on the whole area of the glass plates with cells associated in layers. The biofilm was observed to have a structural roughness. Gonococcal cells seemed to be attached to each other in a dispersed, relaxed way and exhibited characteristic for gonococci, membrane vesicles (blebbings) on their surface and had coffee-bean shape. These observations were in consistency with those from our previous work (Kwiatek et al., 2014).
FIGURE 4.
Biofilm produced by N. gonorrhoeae on cover glass after 24 h visualized by Field Emission Scanning Electron Microscopy. (A) Biofilm formed by the wt FA1090 strain; (B) Biofilm formed by the N. gonorrhoeae ngoAXmod::km mutant and (C) Biofilm formed by the N. gonorrhoeae igatrpb::ngoAXmod mutant. Experiments were triplicated and representative photographs are shown.
In contrast, biofilm formed by the N. gonorrhoeae NgoAX knock-out mutant was more compact with closely associated cells forming compact clumps on the glass surface (Figure 4B). The cell shape seemed similar to that of the wt, yet slightly more rounded. However, the surface of the cells was different: mutant cells with interrupted ngoAXmod gene were smooth, without blebbings characteristic for the wt Neisseria. Moreover, the mutant cells seemed to be positioned more closely to each other than wt cells. The formed biofilm was more opulent. Complementation of the ngoAXmod-encoded function restored the wt feature of the biofilm: it was more dispersed than the one formed by the ngoAXmod::km mutant. Moreover, cells presented blebbings on their surface, characteristic for the wt strain (Figure 4C).
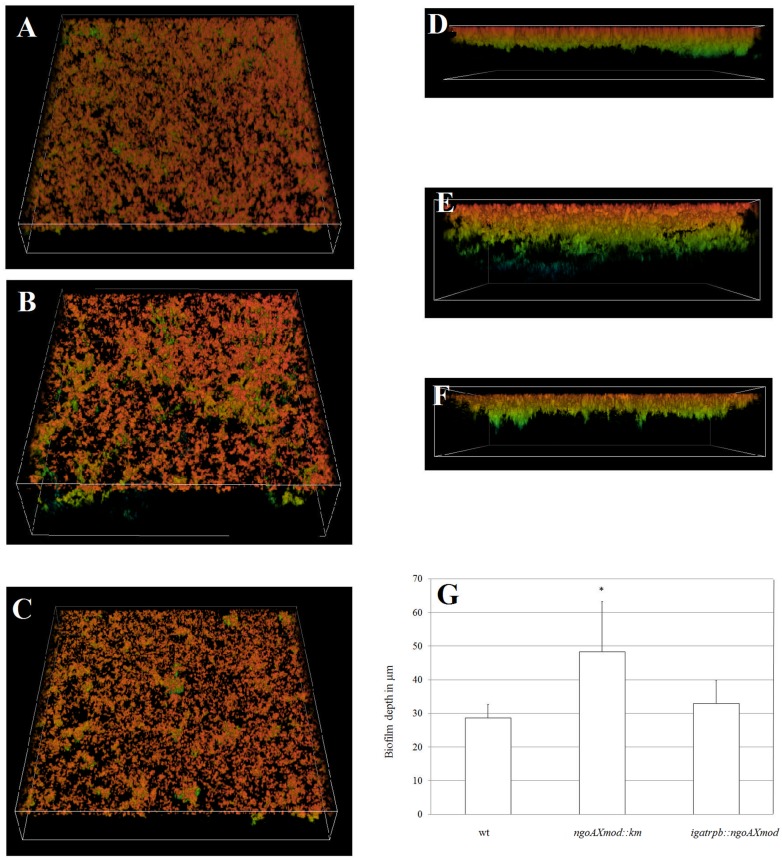
To study live gonococcal biofilm SCLM was applied. For this purpose, N. gonorrhoeae was cultivated on glass plates and after a 24-h growth cells were stained by acridine orange. As shown on Figures 5A,D biofilm formed by the wt strain was dense and uniformly distributed on the glass surface. On the other side, the biofilm formed by the gonococcal mutant in the gene encoding M.NgoAX was more relaxed, dispersed but thicker (Figures 5B,E). The wt feature of the biofilm was recovered in the complementation mutant (Figures 5C,F). We also measured the thickness of each biofilm. This examination revealed that the thickest biofilm was in case of the ngoAXmod::km strain: 48.3 (±14.9) μm (Figure 5G), while the wt strain produced a biofilm with 28.6 (±4.0) μm. Finally, the biofilm produced by the N. gonorrhoeae complementation mutant in the ngoAXmod gene resembled the one formed by the wt strain: 32.9 (±6.9) μm. The difference in thickness of the biofilm formed by the NgoAX knock-out mutant and wt strain was statistically significant (P <0.05).
FIGURE 5.
Three-dimensional biofilm structures visualized by SCLM produced by N. gonorrhoeae after 24 h. (A–C) Three-dimensional biofilm structures and density; (D–F) biofilm thickness. (A) and (D) biofilm produced by the wt strain; (B) and (E) biofilm produced by the gonococcal ngoAXmod::km mutant; (C) and (F) biofilm produced by the gonococcal igatrpb::ngoAXmod mutant (G) biofilm thickness in μm as measured by the NIS-ELEMENTS software. Experiments were triplicated and representative images are shown.
Interaction of N. gonorrhoeae with Human Epithelial Cells Hec-1-B is Modulated by M.NgoAX Expression
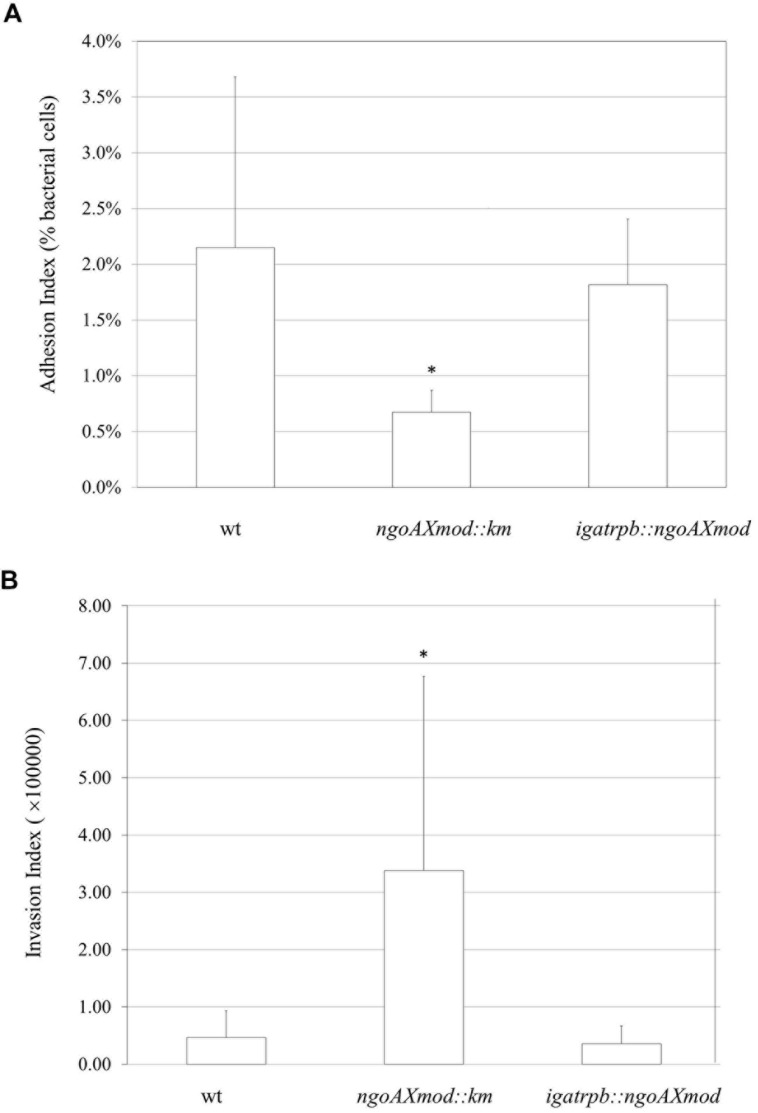
After demonstrating that the gonococcal NgoAX knock-out mutant strain differs from the wt strain in adherence to glass or polystyrene surfaces, we investigated its ability to attach and invade epithelial cells. Monolayers of human Hec-1-B cells were infected with gonococci for 4 h. Then, bacteria that were attached to or have invaded into the epithelial cells were released. We also enumerated total bacterial cells added to the Hec-1-B cells (total CFU). Under our experimental conditions, the adhesion index of the wt strain was 2.15 (±1.53) (Figure 6A). The gonococcal ngoAXmod::km mutant had almost a three-times lower adhesion compared to the wt strain (Figure 6A). The adhesion index determined for the ngoAXmod::km strain was 0.672 (±0.2; P < 0.05). Adhesion of the complementation mutant seemed similar to that of the wt type with the adhesion index 1.82 (±0.59). Yet, the difference between the igatrpb::ngoAXmod strain and wt strain was not statistically significant (P > 0.05).
FIGURE 6.
Neisseria gonorrhoeae adhesion and invasion of human Hec-1-B cells. Human epithelial cells were infected for 4 h with the gonococcal ngoAXmod::km mutant, wt strain or igatrpb:: ngoAXmod mutant. (A) The adhesion index was calculated as a ratio between the numbers of bacteria that adhered to human cells to the total number of cells used for infection. (B) The invasion index was calculated by dividing the number of gentamicin resistant CFU by the adhesion index. The asterisk represents the statistical difference in comparison to the wt strain (P < 0.05).
Using the gentamycin resistance assay we also established the invasion index, which allows to determine the amount of gonococcal cells that penetrate into the host human cells. As seen on Figure 6B, in our experimental conditions, the invasion index for wt gonococcal cells was 4.67 (±3.09) × 104. Inactivation of the M.NgoAX methyltransferase enhanced almost 10-fold the penetration of mutant gonococci cells into human host cells: 3.38 (±0.93) × 105 (P < 0.05). Complementation of the ngoAXmod gene restored the level of invasion to 3.58 (±1.89) × 104, which was not statistically different from that noted for the wt cells (P > 0.05).
Discussion
We studied the impact of M.NgoAX methyltransferase inactivation on the overall gene expression and fitness of N. gonorrhoeae FA1090. For this purpose, we constructed two mutant strains: one, with inactivated ngoAXmod gene by insertion of the km cassette (the NgoAX knock-out mutant) and a second, in which this mutation was complemented by insertion of the wt ngoAXmod allele into a non-coding intergenic region between iga and trpb chromosomal genes (Ramsey et al., 2012). Inactivation of the M.NgoAX methyltransferase mimics phase variation of the ngoAXmod gene in N. gonorrhoeae cells. Indeed, the ngoAXmod gene was designated as a candidate for phase variation (Snyder et al., 2001; Srikhanta et al., 2009) and we have previously observed that the restriction phenotype associated with the NgoAX RM system can switch randomly (Adamczyk-Poplawska et al., 2009). This transition may be involved in a mechanism that increases gonococcal evasion strategies or environmental adaptation.
Inactivation of type III M.NgoAX methyltransferase studied in this work had a huge impact on cell fitness and resulted in deregulation of expression of 121 genes at least twofold compared to the wt strain under standard growth conditions. In contrast, it was demonstrated that inactivation of the M.NgoAXII type III methyltransferase encoded by ngoAXIImod did not influence gene expression under standard gonococcal growth conditions and the cut-off threshold set at 1.5 fold (Srikhanta et al., 2009). A small pool of 54 genes was deregulated by M.NgoAXII inactivation when N. gonorrhoeae FA1090 was cultivated under iron-limiting conditions (Srikhanta et al., 2009). Different effects of methyltransferases on gene expression may be explained by variations in their expression levels. As demonstrated by qRT-PCR, the ngoAXmod gene, encoding M.NgoAX, is sevenfold more highly expressed in the wt N. gonorrhoeae FA1090 than ngoAXIImod, encoding M.NgoAXII. Srikhanta et al. (2009) have compared the amino acid sequences of type III methyltransferases present in N. gonorrhoeae and meningitidis strains. Phylogenetic studies revealed that Neisseriaceae have two distinct mod genes: modA and modB. All studied N. gonorrhoeae strains possess the same modB1 gene but different modA alleles. N. gonorrhoeae FA1090 has modB1 gene (ngoAXmod), encoding M.NgoAX and modA13 (ngoAXIImod), encoding M.NgoAXII. The conservation of modB1 gene among strains suggests that proteins encoded by this gene have a general role. Due to, we postulated, that M.NgoAX is the main epigenetic regulator in N. gonorrhoeae FA1090. Phase variation of gene encoding M.NgoAXII plays a role in regulation of phasevarion in particular growth conditions such as iron-depleted conditions. Our hypothesis is reinforced by the fact that methylation by M.NgoAXII is not detected in N. gonorrhoeae FA1090 genome (PacBio results, Rebase).
Lack of methylation by NgoAX affected not only gene expression. Also changes in bacterial cell growth were observed. Mutant cells with inactivated M.NgoAX methyltransferase were able to grow more rapidly when cultivated in liquid medium with shaking and without interaction with a solid support. During growth without shaking, the number of NgoAX knock-out gonococci increased at a lower rate than wt Neisseria. This suggested a role of M.NgoAX methyltransferase in growth control of N. gonorrhoeae FA1090 depending of growth conditions. Differences in growth abilities of the studied strains were also reflected in biofilm formation.
Biofilms provide a number of advantages when it comes to bacterial survival and pathogenesis. Biofilm formation by gonococci may promote their ability to persist as an asymptomatic infection in the female genital tract (Greiner et al., 2005; Bozicevic et al., 2006). Adhesion studies of the NgoAX knock-out strain to polystyrene plates or human epithelial cells indicated that the mutant strain adhered more poorly to both supports than the wt FA1090. Surprisingly, however, a greater amount of mutant gonococci were engaged in biofilm formation as determined by microplate assay and biofilm biomass/cell measurements or by FE SEM or SCLM observations. All assays confirmed an increase of mutant gonococcal cells involved in biofilm structure compared to the wt strain. However, there seemed to be fewer cell to cell interactions in case of the NgoAX knock-out mutant than for the wt gonococci. The observation of live bacterial biofilm by confocal microscopy indicated that the biofilm formed by NgoAX knock-out cells was thicker and more relaxed compared to the wt strain. Similar changes in biofilm structure were observed for mutants in which the other type III methyltransferase, M.NgoAXII, was deleted. Three-dimensional images of the biofilms showed that the ngoAXIImod::km (modA13::kan) FA1090 strain formed a thick and dense biofilm, while the wt biofilm was thinner with few sparse patches of cells scattered across the attachment surface (Srikhanta et al., 2009).
Biofilm FE SEM observations, which demanded different sample preparations, showed distortion of the biofilm, most probably due to a large cell-free space at the base of the biofilm structure formed by the NgoAX knock-out mutant cells, and collapsing of the biofilm structure. Altogether, the gathered data suggest that NgoAX methylation on one side increases the planktonic cell division rate and by this augments the number of cells that may be engaged in biofilm formation, and on the other, decreases the cell-cell or cell-support adhesion. In in vivo conditions phase variation of NgoAX methyltransferase expression may tightly regulate this process.
Phillips et al. (2012) used stable isotope labeling by amino acids in cell culture (SILAC method) to compare gene expression in biofilm-forming and planktonic N. gonorrhoeae 1291 strain. In this study, nitrite reductase was detected as the key enzyme required for anaerobic growth – conditions favoring biofilm formation. The ngo1276 (aniA) gene, encoding this enzyme, was shown to be upregulated in the N. gonorrhoeae FA1090 NgoAX knock-out mutant as demonstrated by expression microarrays assay. The biofilm of the mutant was shown to consist of a greater number of bacteria than wt. In comparison to proteomes of planktonic organisms, Phillips et al. (2012) demonstrated that 73 proteins were upregulated and 54 downregulated. Nearly a third of the upregulated proteins were involved in energy metabolism, while cell envelope proteins represented the second largest group. In our study, we observed in the mutant strain deregulation of 13 genes encoding proteins directly involved in energy production and conversion, such as the previously mentioned aniA gene or genes coding for NADH-quinone oxidoreductase (ngo1737 and ngo1748, respectively).
To study the mechanism of gonococcal biofilm formation, Falsetta et al. (2009) compared gene expression in biofilms and planktonic N. gonorrhoeae 1291 cells. The study showed that 3.8% of the genome of the biofilm-forming strain was differentially regulated at least twofold compared to planktonic cells. Genes that were highly upregulated in biofilms included, among others, aniA, and ccp genes. These genes encode enzymes involved in the anaerobic respiratory metabolism and stress tolerance. It was also noted that aniA and ccp insertional mutants were attenuated for biofilm development on glass, and on transformed primary human cervical cells. We found that expression of these genes (ngo1276 and ngo1769, respectively) was also upregulated in the NgoAX knock-out mutant. These data suggest that biofilm formation during infection of epithelial cells by N. gonorrhoeae may be associated with the control of nitric oxide steady-state levels (Falsetta et al., 2009).
The changes in biofilm structure and the ability to penetrate into human epithelial cells can reflect the facility of N. gonorrhoeae to efficient adaptation to different environments during the gonococcal infection. Indeed, the overall adhesion of mutant bacteria to the human host was lower than adhesion of wt gonococci; yet, ngoAXmod::km cells were more numerous inside human epithelial Hec-1-B cells than wt bacteria. These results suggest that NgoAX knock-out cells have less ability to attach to human cells, but have a greater facility to penetrate inside the host cells. Adherence to and invasion of host epithelial cells are the first steps of infection initiated by N. gonorrhoeae in humans. Therefore, it is suggested that phase variation of the ngoAXmod gene may have a physiological role in dissemination of N. gonorrhoeae. Also functional studies using N. gonorrhoeae FA1090 confirmed that ngoAXIImod ON and OFF strains have distinct phenotypes in biofilm formation, in antimicrobial resistance or in infection of primary human cells (Srikhanta et al., 2009).
Interaction of gonococci to the epithelial cells has been extensively studied and shown to be based on the interaction of several bacterial virulence factors, for example pili, porin or Opa, to their respective receptors (Edwards and Apicella, 2004). In the N. gonorrhoeae FA1090 ngoAXmod knock-out, we found many genes encoding proteins with predicted outer membrane functions2 or which belong to the COG M (cell wall/membrane/envelop biogenesis) category. Among them, expression of genes, encoding proteins with predicted outer membrane localization, ngo1513 (encoding OpaD), ngo2093 (encoding FetA), ngo2121(encoding lipoprotein VacJ), ngo0021, ngo0510, ngo0555 and ngo0868 (encoding OpcA), was deregulated both in the studied NgoAX knock-out mutant (Supplementary Table S2) and biofilm-forming gonococci versus planktonic cells (Phillips et al., 2012). Moreover, several other proteins, which gene expression was affected in the ngoAXmod::km mutant cells, are suspected to be involved in cell adhesion. This includes ngo1068, ngo1393 and ngo0180 genes, which encode Maf adhesins. The Maf family comprises adhesins with multiple silent loci, which bind to glycolipids on host cell genes (Paruchuri et al., 1990). LgtE (ngo2159), encoding lacto-N-neotetraose biosynthesis glycosyl transferase, and rfaC (ngo1934), encoding ADP-heptose–LPS heptosyltransferase, genes encode enzymes that are involved in lipooligosacharride (LOS) synthesis. Their expression is downregulated in the ngoAXmod::km gonococci. Oligosaccharide extension of LOS can be involved in receptor-mediated interactions. It was demonstrated that LOS containing lacto-N-neotetraose facilitates invasion but not the adherence of N. gonorrhoeae to the human cervical epidermal carcinoma ME180 cell line (Song et al., 2000). Moreover, the terminal lactosamine of lacto-N-neotetraose play an important role in invasion of primary urethral epithelial cells (Harvey et al., 2001). The downexpression of the lgtE gene in NgoAXP knock-out mutant cells may result in control of the lacto-N-neotetraose biosynthesis, which may be necessary for Hec-1-B cell invasion by gonococci. A naturally occurring variation of the terminal carbohydrates on the LOS molecule correlates with altered disease states and may reflect the adaptation of Neisseria to different steps of infection (John et al., 1999). Moreover, the LOS transport across the periplasmic space and their exposition at the cell surface seemed to be enhanced in NgoAX knock-out cells by a more intensive expression of lgtF (ngo1228) and lgtG (ngo1229) genes, encoding LgtF and LgtG transporters, respectively.
Our results also showed downregulation of genes involved in pili formation. The expression of ngo0095 (pilP), ngo0096 (pilO), ngo0097 (pilN) and ngo0098 (pilM) was deregulated in the NgoAX knock-out mutant. Type IV pili are another element responsible for the first steps of bacterial adhesion, but account also for bacterial aggregation (i.e., microcolony formation). Weaker expression of the respective genes may contribute to a more relaxed feature of the biofilms. For N. meningitidis, immunofluorescent staining with antipilus antibodies showed that meningococcal piliation dramatically reduces at later time points of bacterial cell interaction with human cells (Pujol et al., 1997).
The most downregulated gene determined in our study, excluding ngo0545 (ngoAXmod) was ngo0574, coding for carbonic anhydrase (Cah), which catalyzes the conversion of carbon dioxide to bicarbonate and protons. Deregulation of this gene was demonstrated also by proteomic analysis of the biofilm (Phillips et al., 2012), and transcriptomic analysis of type III RM mutant (Srikhanta et al., 2009), but not by transcriptomic analysis of the biofilm (Falsetta et al., 2009). The role of this enzyme in biofilm formation should be investigated as N. gonorrhoeae FA1090 possesses two Cah genes and the second one (ngo2079) was not deregulated in NgoAX knock-out cells. Bacterial Cah proteins are speculated to be involved in the critical steps of bacterial life cycle, including stages important for survival, invasion, and pathogenicity (Supuran, 2011). It seems that Cah activity may be necessary for biofilm formation and may also be involved in cell to host adhesion.
We also observed deregulation of genes encoded by prophages present in the N. gonorrhoeae chromosome. N. gonorrhoeae FA1090 contains several prophages in its genome (Piekarowicz et al., 2006, 2007). Five of them are dsDNA phages (Φ1–5) and 4 are ssDNA phages (Φ6–9). In the NgoAX knock-out mutant, 14 genes of Φ1 phage, 4 genes from Φ2 phage and two from Φ5 phage were deregulated. Moreover, 2 genes from Φ8 ssDNA phage were shown to be downregulated. The role of proteins encoded by these deregulated genes remains unknown. Yet, it is known that viral proteins maybe involved in bacterial fitness and pathogenesis. For example, proteins encoded by prophages found in Streptococcus mitis (Bensing et al., 2001) or Enterococcus faecalis (Matos et al., 2013) were necessary for adhesion of these bacteria to human platelets, which is the first step toward the development of infective endocarditis. Further studies should be performed to investigate the role of prophage-encoded proteins in N. gonorrhoeae pathogenesis.
Regulation of gene expression by methylation of specific DNA sequences by Dam or type III methyltransferases has been reviewed (Wion and Casadesús, 2006; Marinus and Casadesus, 2009; Srikhanta et al., 2010). As in case of Dam methyltransferase, the target site methylation state may affect DNA binding by the regulatory protein, which directly regulates transcription or even acts on RNA polymerase affinity to DNA. The widespread distribution of phase variable RM systems in host-adapted pathogenic bacteria, such as H. influenzae or N. gonorrhoeae, suggests the existence of a mechanism coordinating the switch of multiple gene expression. The interactions between RNA polymerase with DNA are randomly affected by a different methylation pattern. Such strategy may be used to generate cells specialized distinctly in human cell invasion or biofilm formation (Srikhanta et al., 2009).
Our data suggest that the M.NgoAX methyltransferase may be involved in N. gonorrhoeae pathogenicity, specifically in control of biofilm formation, adhesion to host cells and invasion of epithelial cells. Phase variation of ngoAXmod gene expression may be necessary for changing the gonococcal abilities in the first step of human cell invasion.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Ministry of Science and Higher Education in Poland (grant no. NN303816940) and the National Science Center in Poland (2014/15/B/NZ6/02514) and the CePT Project No.: POIG. 02.02.00-14-024/08-00. We also thank Aleksandra Szymanska and Ada Kuka for technical support.
Abbreviations
- COG
clusters of orthologous groups
- FE SEM
Field Emission Scanning Electron Microscopy
- km
kanamycin cassette
- RM
restriction modification
- SCLM
Scanning Confocal Laser Microscopy
- wt
wild-type
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01426
References
- Adamczyk-Poplawska M., Lower M., Piekarowicz A. (2009). Characterization of the NgoAXP: phase-variable type III restriction-modification system in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 300 25–35. 10.1111/j.1574-6968.2009.01760.x [DOI] [PubMed] [Google Scholar]
- Arber W. (2000). Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24 1–7. 10.1111/j.1574-6976.2000.tb00529.x [DOI] [PubMed] [Google Scholar]
- Bayliss C. D., Callaghan M. J., Moxon E. R. (2006). High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 34 4046–4059. 10.1093/nar/gkl568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B. A., Siboo I. R., Sullam P. M. (2001). Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect. Immun. 69 6186–6192. 10.1128/IAI.69.10.6186-6192.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozicevic I., Fenton K. A., Martin I. M., Rudd E. A., Ison C. A., Nanchahal K., et al. (2006). Epidemiological correlates of asymptomatic gonorrhea. Sex. Transm. Dis. 33 289–295. 10.1097/01.olq.0000194582.44222.c9 [DOI] [PubMed] [Google Scholar]
- Broadbent S. E., Davies M. R., van der Woude M. W. (2010). Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 77 337–353. 10.1111/j.1365-2958.2010.07203.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle X., Bayliss C. D., Field D., van de Ven T., Saunders N. J., Hood D. W., et al. (2000). The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35 211–222. 10.1046/j.1365-2958.2000.01701.x [DOI] [PubMed] [Google Scholar]
- Dillard J. P. (2006). Genetic manipulation of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 4A2.1–4A2.19. 10.1002/9780471729259.mc04a02s00 [DOI] [PubMed] [Google Scholar]
- Dryden D. T., Murray N. E., Rao D. N. (2001). Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 29 3728–3741. 10.1093/nar/29.18.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Adamczuk M., Szuplewska M., Bartosik D. (2011). DIY series of genetic cassettes useful in construction of versatile vectors specific for Alphaproteobacteria. J. Microbiol. Methods 86 166–174. 10.1016/j.mimet.2011.04.016 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. L., Apicella M. A. (2004). The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin. Microbiol. Rev. 17 965–981. 10.1128/CMR.17.4.965-981.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. L., Butler E. K. (2011). The pathobiology of Neisseria gonorrhoeae lower female genital tract infection. Front. Microbiol. 2:102 10.3389/fmicb.2011.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L., Bair T. B., Ku S. C., Vanden Hoven R. N., Steichen C. T., McEwan A. G., et al. (2009). Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect. Immun. 77 3522–3532. 10.1128/IAI.00036-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. L., Dowideit S. J., Erwin A. L., Srikhanta Y. N., Smith A. L., Jennings M. P. (2007a). Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 35 5242–5252. 10.1093/nar/gkm571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. L., Srikhanta Y. N., Jennings M. P. (2007b). Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol. Microbiol. 65 1375–1379. 10.1111/j.1365-2958.2007.05873.x [DOI] [PubMed] [Google Scholar]
- Greiner L. L., Edwards J. L., Shao J., Rabinak C., Entz D., Apicella M. A. (2005). Biofilm formation by Neisseria gonorrhoeae. Infect. Immun. 73 1964–1970. 10.1128/IAI.73.4.1964-1970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H. A., Jennings M. P., Campbell C. A., Williams R., Apicella M. A. (2001). Receptor-mediated endocytosis of Neisseria gonorrhoeae into primary human urethral epithelial cells: the role of the asialoglycoprotein receptor. Mol. Microbiol. 42 659–672. 10.1046/j.1365-2958.2001.02666.x [DOI] [PubMed] [Google Scholar]
- Holmes K. K. (1999). Sexually Transmitted Diseases. New York, NY: McGraw-Hill Health Professions Division Publisher. [Google Scholar]
- Hopper S., Wilbur J. S., Vasquez B. L., Larson J., Clary S., Mehr I. J., et al. (2000). Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68 896–905. 10.1128/IAI.68.2.896-905.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümbelin M., Suri B., Rao D. N., Hornby D. P., Eberle H., Pripfl T., et al. (1988). Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 200 23–29. 10.1016/0022-2836(88)90330-0 [DOI] [PubMed] [Google Scholar]
- Ison C. A. (2011). “Biology of Neisseria gonorrhoeae and the clinical picture of infection,” in Sexually Transmitted Infections and Sexually Transmitted Diseases, eds Gross G. E., Tyring S. K. (Heidelberg: Springer; ), 77–90. 10.1007/978-3-642-14663-3_6 [DOI] [Google Scholar]
- Jarvis G. A., Chang T. L. (2012). Modulation of HIV transmission by Neisseria gonorrhoeae: molecular and immunological aspects. Curr. HIV Res. 10 211–217. 10.2174/157016212800618138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. (2003). Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene 317 13–16. 10.1016/S0378-1119(03)00652-8 [DOI] [PubMed] [Google Scholar]
- John C. M., Schneider H., Griffiss J. M. (1999). Neisseria gonorrhoeae that infect men have lipooligosaccharides with terminal N-acetyllactosamine repeats. J. Biol. Chem. 274 1017–1025. 10.1074/jbc.274.2.1017 [DOI] [PubMed] [Google Scholar]
- Jordan P. W., Snyder L. A., Saunders N. J. (2005). Strain-specific differences in Neisseria gonorrhoeae associated with the phase variable gene repertoire. BMC Microbiol. 5:21 10.1186/1471-2180-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. (2001). Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29 3742–3756. 10.1093/nar/29.18.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek A., Bacal P., Wasiluk A., Trybunko A., Adamczyk-Poplawska M. (2014). The dam replacing gene product enhances Neisseria gonorrhoeae FA1090 viability and biofilm formation. Front. Microbiol. 5:712 10.3389/fmicb.2014.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D. A., Weyand N. J., Mahan M. J. (2001). Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69 7197–7204. 10.1128/IAI.69.12.7197-7204.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott R. J., Keller B. O., Gaudet R. G., McCaw S. E., Lai C. C., Dobson-Belaire W. N., et al. (2013). Neisseria gonorrhoeae-derived heptose elicits an innate immune response and drives HIV-1 expression. Proc. Natl. Acad. Sci. U.S.A. 110 10234–10239. 10.1073/pnas.1303738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Casadesus J. (2009). Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33 488–503. 10.1111/j.1574-6976.2008.00159.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrazzo J. M., Handsfield H. H., Sparling P. F. (2010). “Neisseria gonorrhoeae,” in Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, eds Mandell G. L., Bennett J. E., Dolin R. (Philadelphia, PA: Churchill Livingstone/Elsevier; ), 2753–2770. 10.1016/B978-0-443-06839-3.00212-5 [DOI] [Google Scholar]
- Matos R. C., Lapaque N., Rigottier-Gois L., Debarbieux L., Meylheuc T., Gonzalez-Zorn B., et al. (2013). Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9:e1003539 10.1371/journal.pgen.1003539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I., Kobayashi I. (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 42 70–86. 10.1093/nar/gkt711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D. A., Galperin M. Y., Tatusov R. L., Koonin E. V. (2000). Using the COG database to improve gene recognition in complete genomes. Genetica 108 9–17. 10.1023/A:1004031323748 [DOI] [PubMed] [Google Scholar]
- Paruchuri D. K., Seifert H. S., Ajioka R. S., Karlsson K. A., So M. (1990). Identification and characterization of a Neisseria gonorrhoeae gene encoding a glycolipid-binding adhesin. Proc. Natl. Acad. Sci. U.S.A. 87 333–337. 10.1073/pnas.87.1.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N. J., Steichen C. T., Schilling B., Post D. M., Niles R. K., Bair T. B., et al. (2012). Proteomic analysis of Neisseria gonorrhoeae biofilms shows shift to anaerobic respiration and changes in nutrient transport and outermembrane proteins. PLoS ONE 7:e38303 10.1371/journal.pone.0038303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A., Kłyz A., Majchrzak M., Adamczyk-Popławska M., Maugel T. K., Stein D. C. (2007). Characterization of the dsDNA prophage sequences in the genome of Neisseria gonorrhoeae and visualization of productive bacteriophage. BMC Microbiol. 7:66 10.1186/1471-2180-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A., Majchrzak M., Kłyz A., Adamczyk-Popławska M. (2006). Analysis of the filamentous bacteriophage genomes integrated into Neisseria gonorrhoeae FA1090 chromosome. Pol. J. Microbiol. 55 251–260. [PubMed] [Google Scholar]
- Price C., Bickle T. A. (1986). A possible role for DNA restriction in bacterial evolution. Microbiol. Sci. 3 296–299. [PubMed] [Google Scholar]
- Pujol C., Eugène E., de Saint Martin L., Nassif X. (1997). Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65 4836–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey M. E., Hackett K. T., Kotha C., Dillard J. P. (2012). New complementation constructs for inducible and constitutive gene expression in Neisseria gonorrhoeae and Neisseria meningitidis. Appl. Environ. Microbiol. 78 3068–3078. 10.1128/AEM.07871-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D. N., Dryden D. T., Bheemanaik S. (2014). Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res. 42 45–55. 10.1093/nar/gkt616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Vincze T., Posfai J., Macelis D. (2015). REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43 D298–D299. 10.1093/nar/gku1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. A., Lo R. Y. (1999). Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res. 27 1505–1511. 10.1093/nar/27.6.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn, Vol. 1 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Seib K. L., Jen F. E., Tan A., Scott A. L., Kumar R., Power P. M., et al. (2015). Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N(6)-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 43 4150–4162. 10.1093/nar/gkv219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneppen K., Semsey S., Seshasayee A. S., Krishna S. (2015). Restriction modification systems as engines of diversity. Front. Microbiol. 6:528 10.3389/fmicb.2015.00528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L. A., Butcher S. A., Saunders N. J. (2001). Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147 2321–2332. 10.1099/00221287-147-8-2321 [DOI] [PubMed] [Google Scholar]
- Song W., Ma L., Chen R., Stein D. C. (2000). Role of lipooligosaccharide in Opa-independent invasion of Neisseria gonorrhoeae into human epithelial cells. J. Exp. Med. 191 949–960. 10.1084/jem.191.6.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikhanta Y. N., Dowideit S. J., Edwards J. L., Falsetta M. L., Wu H. J., Harrison O. B., et al. (2009). Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 5:e1000400 10.1371/journal.ppat.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikhanta Y. N., Fox K. L., Jennings M. P. (2010). The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 8 196–206. 10.1038/nrmicro2283 [DOI] [PubMed] [Google Scholar]
- Srikhanta Y. N., Gorrell R. J., Steen J. A., Gawthorne J. A., Kwok T., Grimmond S. M., et al. (2011). Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS ONE 6:e27569 10.1371/journal.pone.0027569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikhanta Y. N., Maguire T. L., Stacey K. J., Grimmond S. M., Jennings M. P. (2005). The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. U.S.A. 102 5547–5551. 10.1073/pnas.0501169102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D. C., Gunn J. S., Radlinska M., Piekarowicz A. (1995). Restriction and modification systems of Neisseria gonorrhoeae. Gene 157 19–22. 10.1016/0378-1119(94)00649-D [DOI] [PubMed] [Google Scholar]
- Stepanovic S., Vukovic D., Dakic I., Savic B., Svabic-Vlahovic M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40 175–179. 10.1016/S0167-7012(00)00122-6 [DOI] [PubMed] [Google Scholar]
- Supuran C. T. (2011). Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med. Chem. 3 1165–1180. 10.4155/fmc.11.69 [DOI] [PubMed] [Google Scholar]
- Swanson J. (1978). Studies on gonococcus infection. XII. Colony color and opacity variants of gonococci. Infect. Immun. 19 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28 33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Nicholas R. A. (2012). Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 7 1401–1422. 10.2217/fmb.12.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77 53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2012). Global Action Plan to Control the Spread and Impact of Antimicrobial Resistance in Neisseria gonorrhoeae. Geneva: World Health Organization Press. [Google Scholar]
- Williams R. J. (2003). Restriction endonucleases: classification, properties, and applications. Mol. Biotechnol. 23 225–243. 10.1385/MB:23:3:225 [DOI] [PubMed] [Google Scholar]
- Wion D., Casadesús J. (2006). N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4 183–192. 10.1038/nrmicro1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.