Abstract
Background and Objectives:
Methicillin resistance Staphylococcus aureus (MRSA) and coagulase negative staphylococci (MRCoNS) have recognized as the major cause of nosocomial infections that threat the burn patient's life. The aims of this study were to determine the frequency of MRSA and MRCoNS and their antibiotic resistance patterns among burn patients in a burn center in Ahvaz, Iran.
Material and Methods:
A total of 340 clinical specimens: (80%) wound and (20%) blood were obtained from patients in Taleghani burn hospital during February 2013–2014. Staphylococci species identification and antibiogram were performed by standard procedures using disk diffusion method. The Methicillin resistance strains were detected by Etest and PCR using mecA specific primers.
Results:
Out of 30.2% (103) isolates that were recognized as staphylococci, 82 % (84) and 18% (19) were identified as S. aureus and coagulase negative staphylococci (CoNS) respectively. Resistance to methicillin was detected in 60% and 63% of the S. aureus and CoNS isolates respectively. Seven different antimicrobial resistance patterns observed among methicillin resistant staphylococci. The MRSA and MRCoNS strains showed closed resistance phenotypes. All the methicillin resistant isolates showed a high rate resistance to the other studied antibiotics in comparison to methicilin sensitive isolates. Vancomycin and imipenem showed the greatest effect against methicillin resistant isolates. During 8 years in the studied burn hospital, no significant changes in the methicillin resistance staphylococci frequency were detected.
Conclusion:
The presence of multi resistant MRSA and MRCoNS strains is cause of concern in burn hospitals. Vancomycin remains as a drug of choice for methicillin resistance staphylococci infections.
Keywords: Methicillin Resistance, Staphylococci, Burn patients
INTRODUCTION
Staphylococcous aureus is common pathogen infecting the burn wound after Pseudomonas aeruginosa. Today methicillin-resistant staphylococci (MRS) have been recognized as a major cause of bacterial infection in hospitals and community worldwide. In addition methicillin resistant coagulase negative staphylococci (MRCoNS) as the normal inhabitants of skin have became a cause of concern in burn patients (1–3). MRSs are one of the major problems in hospitals and the rate of their incidence is rising considerably in recent years (4). MRSA is frequently associated with high rates of morbidity and mortality in the immonucompromized patients particularly burn patients. Burn patients due to damaged skin barrier, decreased bactericidal PMNs and immune deficiency are more vulnerable to colonization and infection with opportunist pathogens like staphylococci (5). Vancomycin is commonly used to treat infections caused by MRSA. Nowadays the options for treatment of MRSA infections are considerably limited and vancomycin remained as the last choice for MRSA treatment until recent years. Burn patients would be at risk if co-colonization of MRSA occurs with vancomycin resistant Enterococci (VRE) or Vancomycin Resistant staphylococci (VRSA) strains (6, 7). Taleghani burn hospital is the only referral burn center in Ahvaz city, Khuzestan provinces in southwest of Iran which cover many burn patients even from neighbor provinces in southwest of Iran. According to a few previous studies, high rate of MRSA has been reported (8–10). Thereby, considering the significance of following up and monitoring of antibiotic resistant pathogens among burn patients, the current study was focused on determine the prevalence of MRSA, MRCoNS, VRSA and antibiotic resistance patterns of isolates among burn patients and also we aimed to compare our results with findings of latest similar study in the same hospital in 2007 (9).
MATERIALS AND METHODS
Location and sampling procedure.
The sampling was carried out in Taleghani burn hospital. All the burn wound biopsies and blood cultures were tested during February 2013 to February 2014. Totally 340 samples from burn patients including 272 wound biopsy and 68 blood samples were cultured for staphylococci. All Staphylococci isolates were identified in the genus and species level by Gram staining, catalase and coagulase reaction, growth on manitol salt agar and other conventional microbiology and biochemistry tests (11).
Detection of methicillin and vancomycin resistance.
The Etest (Epsilometer test) gradient technology is based on a combination of the concepts of dilution and diffusion principles for susceptibility testing. In the present study the Etest oxacillin-vancomcin strips (Ezy MICTM, HiMedia, India) were used for simultaneous detection of resistant to methicillin and vancomycin. Etest strips were placed on plates on Muller –Hinton agar with added 2% NaCl for detection of MRSA. Minimum Inhibitory Concentration (MICs) for oxacillin and vancomycin were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines; MIC ≥4μg/ml and MIC ≥0.5μg/ml breakpoints for identification of oxacillin resistant S. aureus and coagulase - negative staphylococci (CoNS) isolates respectively, vancomycin resistant strains were detected with MIC≥16μg/ml and MIC ≥32μg/ml breakpoints for S. aureus and CoNS respectively (12).
Antimicrobial susceptibility test.
Antimicrobial susceptibility testing was performed for MRSA and MRCoNS isolates against penicillin (10 μg), ampicillin (10 μg), ampicillin-sulbactam (10/10 μg), ciprofloxacin (5 μg), tetracycline (30 μg), erythromycin (30 μg), gentamicin (10 μg), tobramycin (10 μg), amikacin (30 μg), imipenem (10 μg), cefepime (30 μg), ceftazidime (30 μg) and ceftriaxone (30 μg) (Mast, UK), using disk diffusion method. The results were interpreted according to the CLSI criteria (12).
mecA gene detection.
DNAs of all isolates were extracted by using commercial DNA extraction Kit (Cinnagen, Iran). PCR assay were performed to detect mecA gene; encoding methicillin-resistance gene. For amplification of the mecA gene, primers mecA1 (5′-AAT CTT TGT CGG TAC ACG ATA TTC TTCACG-3′) and mecA2 (5′-CGT AAT GAG ATT TCA GTA GAT AAT ACA ACA-3′) were used to produce 174 bp fragment. S. aureus ATCC 43300 and S. aureus ATCC 25923 were used as positive and negative controls respectively (9, 13).
Statistical analysis.
Statistical analyses of the parameters were conducted using SPSS for Windows statistical software (version16; SPSS, Inc., Chicago, IL, USA). Categorical variables were analyzed using the Chi square test or Fisher exact test. A p-value<0.05 was considered as statistically significant.
RESULTS
Prevalence of methicillin resistant staphylococci.
Totally, staphylococci were isolated from 54.4% (103) of clinical samples. The frequency of staphylococci in wound and blood specimens were 65% (n= 67) and 35% (n=36) respectively. Among 103 staphylococci strains 82 % (n=84) and 18 % (n=19) were identified as S. aureus and CoNS respectively. Resistance to oxacillin and harboring the mecA gene were detected in 60% (n=50) of S. aureus and 63% (n= 12) of CoNS isolates. PCR of mecA gene confirmed all result of E test.
Antimicrobial susceptibility testing.
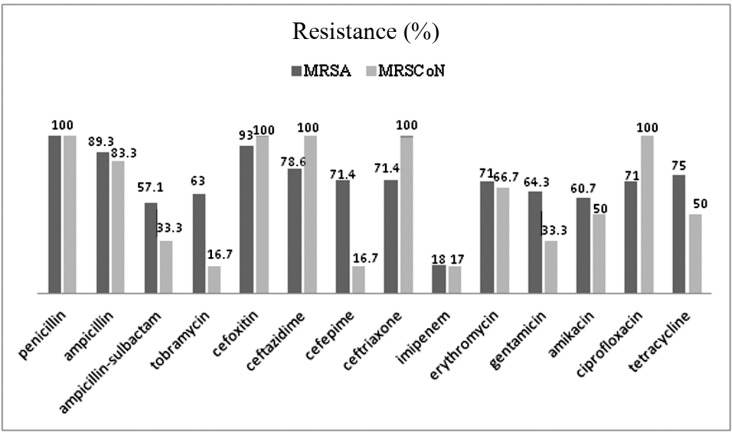
The antimicrobial susceptibility analysis showed high levels of resistance to penicillin (100%: 100%), cefoxitin (93%: 100%), ampicillin (89%: 83%), erythromycin (71%: 66%), tetracycline (75%: 50%), gentamicin (64%: 33%), ciprofloxacin (71%: 100%), ampicillin-sulbactam (57%: 33%), ceftazidime (79%: 100%), cefoxitin (93%: 100%) and ceftriaxone (71%: 100%) among MRSA and MRCoNS isolates respectively. Imipenem was the most effective antibiotic against both MRSA and MRCoNS isolates (18%: 17% respectively). It should be noted MRSA and MRCoNS strains showed significant variation (p<0.05) in resistance to tobramycin (63%: 16.7%) and cefepime (71.4%: 16.7%). The antibiotic resistance rates in MRSA and MRSCoNS is compared in Fig 1.
Fig. 1.
Comparison of Antibiotic Resistance (%) in MRSA and MRSCoN strains
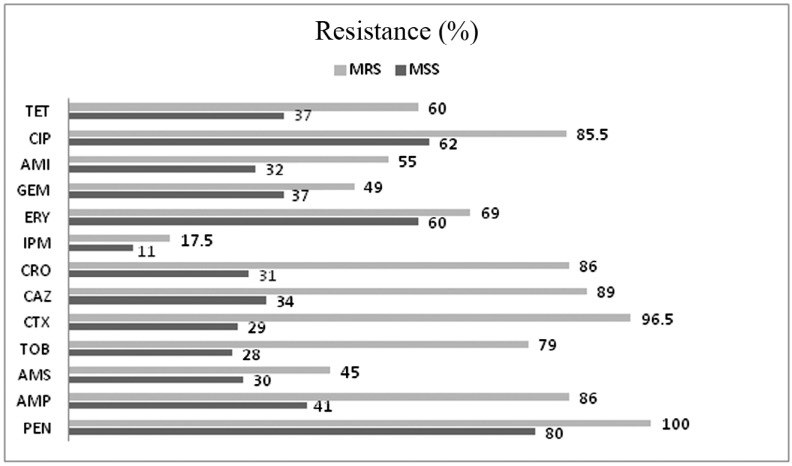
We also evaluated the susceptibility pattern between methicillin resistance and methicillin susceptible Staphylococci using different antibiotic disks. All the methicillin resistant staphylococci (MRS) showed a high rates of resistance to the other antibiotics in comparison to methicillin susceptible staphylococci (MSS). However high resistance against ciprofloxacin (62%), penicillin (80%) and erythromycin (60%) was observed in MSS strains. It should be noted that MSS strains also showed low level resistant to imipenem (17%) like MRS strains (Fig. 2). Comparison of antibiotic resistance rates between MSS and MSS strains is demonstrated in Fig. 2.
Fig. 2.
Comparison of antibiotics resistance (%) in methicillin resistance staphylococci (MRS) and methicillin susceptible staphylococci (MSS). PEN; penicillin, AMP; ampicillin, AMS; ampicillin-sulbactam, TOB; tobramycin, CTX; cefoxitin, CAZ; ceftaziime, CRO; ceftriaxone, IPM; imipenem, ERY; erythromycin, GEM; gentamicin, AMI; amikacin, CIP; ciprofloxacine, TET; tetracycline
MIC breakpoint for oxacillin in all of MRSA and MRSCoNS isolates were detected in MIC ≥8μg/ml range. The MICs of vancomycin for all strains were in sensitive ranges.
Seven different patterns of antibiotic resistance were recognized in staphylococci isolates which is shown in Table 1. In addition approximately 50% of MRSA, 47% of MRSCoNS and 15% of methicillin susceptible staphylococci (MSS) isolates were resistant to at least three antibiotics from different antibiotic families and were considered as multidrug-resistant (MDR) isolates. The most common observed pattern was resistance to penicillin and ceftazidime (45%).
Table 1.
Antibiotic resistance patterns of methicillin- resistant staphylococci (MRS) and methicillin susceptible staphylococci (MSS) isolated from burn patients
| Antibiotic resistance patterns | MRS | MSS |
|---|---|---|
| PEN | 62 (100%) | 33 (80%) |
| PEN+CAZ | 31 (50%) | 23 (57%) |
| PEN + CAZ + IPM | 6 (9.6%) | 4 (9.7%) |
| PEN+CAZ+ AM/S+GEM | 8 (12.9%) | 7 (17%) |
| PEN+CAZ+ AM/S+ GEM+ ERY | 8(12.9%) | 6 (14.6%) |
| PEN+CAZ+ +AM/S+GEM+ERY+ IPM | 4(6.4%) | 2 (4.8%) |
| PEN+CAZ+IPM+AM/S+GEM+ERY+TET | 4(6.4%) | 0 (0%) |
| PEN+CAZ+IPM+AM/S+GEM+ERY+TET+CIP | 1(1.6%) | 0 (0%) |
DISCUSSION
MRSA is one of the most common causes of bacterial infection in burn patients. Our study revealed that 54.4% of specimens from burn patients were infected with staphylococci and among them about 60% and 63% were identified as MRSA and MRCoNS. In consistent with our finding, Ohadian Moghadam et al. have been reported that frequency of MRSA in burn patients was 61.5% (14). The results of this study was not different from previous work done in 2007 as the prevalence of MRSA and MRCoNS in staphylococci isolates was 61% and 60% at that time (9). Although the total number of VRSA has been reported worldwide is not significant, however, these reports are very important particularly in immunocompromised patients such as burn patients (15–17).
There are some studies considering the presence of VRSA and MRSA strains in hospitals in Iran (18–20). VRSA status in the recent and previous studies in Taleghani hospital suggesting vancomycin is still an effective antibiotic to treatment of the infections due to MRSA and MRCoNS strains in burn patients. However, emergence of VRE and VRSA in hospitals is a critical concern; hence vancomycin must be prescribed with cautions.
Following vancomycin, imipenem was detected as the most effective antibiotic against MRSA, MRCoNS and also MSS strains. According to our findings and previous reports in 2007, no increasing trend was found in methicillin resistance rates in the studied hospital (9). We did not detect any VRSA which can be considered as remarkable finding. This success may be achieved by sticking to infection control guideline by hospital's infection controlling team to manage the infection due to MRSA and MRCoNS strains. However, their actions were not efficient enough to reduce MRS rates. Another hypothesis is circulation of same clones or clones with similar antibiotic susceptibility in the hospital environment as well as patients in this hospital during 8 years. Thereby molecular epidemiology studies would be valuable and useful to detect the exact clonal relatedness of strains in various courses.
Resistance to most available antibiotics is increasing among staphylococci (14–21, 22). Our finding showed that about half of MRSA and MRCoNS and also 15% of MSS showed MDR phenotypes with different resistant phenotypes. This diversity should be considered as an important point to manage and control the MDR staphyoccooci and “prescribe the right antibiotic rather than empirical treatment”. In consistent with our finding Ohadian Moghadam et al. has been reported high rates of resistance to amikacin, ceftriaxone, ciprofloxacin, erythromycin, gentamicin, tetracycline, and tobramycin among MRSA strains isolated from Motahari burn care center in Tehran (14). Rahimi et al. reported the lower frequency (30%) of MRSA but high rates of MDR strains: 93% and 61% of MRSA and MSSA isolates were MDR respectively in Tehran hospitals during 2007–2011 (21). We should also consider the multi drug resistant MRCoNS as the one of the important nosocomial pathogen beside the MRSA in hospitals especially in burn patients because of their immune deficiency status. CoNS can adhere to medical devices and surfaces and may easily colonize and spread within hospital environment (23, 24). Accordant to our results, Kokasal et al. from Turkey reported high rate of methicillin resistance (67.5%) and extremely high resistant to ceftriaxone, erythromycin, tetracycline and gentamicin and any vancomycin resistance among MRCoNS. They also found that 14.5% of staphylococci belonged to CoNS in emergency burn care unit (25).
In conclusion, the current study results showed the prevalence of MRSA and MRCoNS in burn hospital remain high after 8 years. Also multi drug resistant MRSA and MRCoNS strains which are resistant to many available antibiotics, are cause of concern and limitation for treatment options of burn patients. The follow-up and continuous monitoring of antibiotic resistance profiles of hospital pathogens specially staphylococci and also profound studies on hospital resident clones are suggested.
ACKNOWLEDGMENTS
This research was funded partially by a grant (No. U-92004) from infection and tropical disease research center, Jundishapur University of Medical Sciences, Ahvaz, Iran. We would like to thank all members of microbiology laboratory of Taleghani hospital in Ahvaz.
REFERENCES
- 1. Boyce JM, Landry M, Deetz TR, Dupont HL. Epidemiologic studies of an outbreak of nosocomial methicillin- resistant Staphylococcus aureus infections. Infect Control 1981; 2: 110– 116. [DOI] [PubMed] [Google Scholar]
- 2. Reardon CM, Brown TP, Stephenson AJ, Freedlander E. Methicillin-resistant Staphylococcus aureus in burns patients-why all the fuss? Burns 1998; 24 : 393– 397. [DOI] [PubMed] [Google Scholar]
- 3. Roth RR, James WD. Treatment of infections due to coagulase-negative staphylococci. Annu Rev Microbiol 1988; 42: 441. [DOI] [PubMed] [Google Scholar]
- 4. Palavecino E. Clinical, epidemiological and laboratory aspects of methicillin-resistant Staphylococcus aureus (MRSA) infections. Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols. Humana Press, 2007: 1– 19 . [DOI] [PubMed] [Google Scholar]
- 5. Barlow Y. T lymphocytes and immunosuppression in the burned patient: a review. Burns 1994; 20: 487– 490. [DOI] [PubMed] [Google Scholar]
- 6. Ruef C. Epidemiology and clinical impact of glycopeptide resistance in Staphylococcus aureus. Infection 2004; 32: 315– 27. [DOI] [PubMed] [Google Scholar]
- 7. Furuno JP, Perencevich EN, Johnson JA, Wright MO, McGregor JC, Morris JR, et al. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci co-colonization. Emerg Infect Dis 2005; 11: 539– 1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ekrami A, Kalantar E. Bacterial infections in burn patients at a burn hospital in Iran. Indian Med Res 2007; 12: 541. [PubMed] [Google Scholar]
- 9. Ekrami A, Samarbafzadeh A, Alavi M, Kalantar E, Hamzeloi F. Prevalence of methicillin resistant Staphylococcus species isolated from burn patients in a burn center, Ahvaz, Iran. Jundishapur J Microbiol 32 ( 2011): 84– 91. [Google Scholar]
- 10. Abbasi Montazeri E, Khosravi A D, Feizabadi M M, Goodarzi H, Khoramrooz S, Mirzaii S, et al. The prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolates with high-level mupirocin resistance from patients and personnel in a burn center. Burns 2013; 39:, 650– 654. [DOI] [PubMed] [Google Scholar]
- 11. Baron J, Fingold S. Methods for identification of ethologic agent of infection disease. Baily & Scotts diagnostic microbiology. 10 th ed St. Louis, USA: Mosby, Inc; 1996: 327– 529 . [Google Scholar]
- 12. CLSI Performance Standards for Antimicrobial Susceptibility Testing; 21th ed Wayne, PA: Clinical and Laboratory Standards Institute; 2011; 31: 84– 87. [Google Scholar]
- 13. Unal S, Hoskins J, Flokowitsh J, Wu C, Preston D, Scatrud P. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol 1992; 30: 1685– 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moghadam Ohadian S, Pourmand MR, Aminharati F. “Biofilm formation and antimicrobial resistance in methicillin-resistant Staphylococcus aureus isolated from burn patients, Iran. J Infect Dev Countr 2014; 8: 1511– 1517. [DOI] [PubMed] [Google Scholar]
- 15. Thati V, Shivannavar C T, Gaddad SM. Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J Med Res 2011; 134: 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover F C, Downes FP, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. New England J Med 2003; 348: 1342– 1347. [DOI] [PubMed] [Google Scholar]
- 17. Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. Vancomycin-resistant Staphylococcus aureus in the United States, 2002– 2006. Clin infect 2008; 46: 668– 674. [DOI] [PubMed] [Google Scholar]
- 18. Dezfulian A, Aslani MM, Oskoui M, Farrokh P, Azimirad M, Dabiri H, et al. Identification and characterization of a high vancomycin-resistant Staphylococcus aureus harboring vanA gene cluster isolated from diabetic foot ulcer. Iran J Basic Med Sci 2012; 15: 803. [PMC free article] [PubMed] [Google Scholar]
- 19. Askari E, Zarifian A, Pormand MR, Naderi-Nasab M. High-Level Vancomycin-Resistant Staphylococcus aureus (VRSA) in Iran: A Systematic Review. J Med Bacteriol 2012; 1: 53– 61. [Google Scholar]
- 20. Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Mirab Samiee S, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolated from respiratory tract of a hospitalized patient in a university hospital in north east of Iran. J Clin Microbiol 2012; 50: 3581– 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahimi F, Bouzari M, Katouli M, Pourshafie MR. (2013). Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbiol 2013; 6: 144– 149. [Google Scholar]
- 22. Mayhall CG. Hospital epidemiology and infection control, 3rd ed Philadelphia: Lippincott Wiliamand Wilkins; 2004. p. 495– 510. [Google Scholar]
- 23. Huang SY, Tang RN, Chen SY. Coagulase-negative staphylococcal bacteremia in critically ill children: risk factors and antimicrobial susceptibility. J Microbiol Immunol Infect 2003; 36: 51– 5. [PubMed] [Google Scholar]
- 24. Knauer A, Fladerer P, Strempfl C. Effect of hospitalization and antimicrobial therapy on antimicrobial resistance of colonizing Staphylococcus epidermidis. Wien Klin Wochenschr 2004; 116: 489– 94. [DOI] [PubMed] [Google Scholar]
- 25. Koksal F, Yasar H, Samasti M. Antibiotic resistance patterns of coagulase-negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiol Res 2009; 164: 404– 410. [DOI] [PubMed] [Google Scholar]