Abstract
Increasing evidence suggests that oxysterol-binding protein-related proteins (ORPs) localize at membrane contact sites, which are high-capacity platforms for inter-organelle exchange of small molecules and information. ORPs can simultaneously associate with the two apposed membranes and transfer lipids across the interbilayer gap. Oxysterol-binding protein moves cholesterol from the endoplasmic reticulum to trans-Golgi, driven by the retrograde transport of phosphatidylinositol-4-phosphate (PI4P). Analogously, yeast Osh6p mediates the transport of phosphatidylserine from the endoplasmic reticulum to the plasma membrane in exchange for PI4P, and ORP5 and -8 are suggested to execute similar functions in mammalian cells. ORPs may share the capacity to bind PI4P within their ligand-binding domain, prompting the hypothesis that bidirectional transport of a phosphoinositide and another lipid may be a common theme among the protein family. This model, however, needs more experimental support and does not exclude a function of ORPs in lipid signaling.
Keywords: lipid signaling, lipid transport, membrane contact site, oxysterol-binding protein (OSBP), OSBP-related protein, OSBPL gene
Introduction
The main site of lipid synthetic reactions in eukaryotic cells is the endoplasmic reticulum (ER). Although lipids lack intrinsic motifs specifying their distinct intracellular distributions, each organelle has its characteristic lipid composition that ensures optimal conditions for membrane-associated biochemical reactions and other organelle-specific functions such as ion transport and signal transduction. The compartmentalization of cells necessitates well-controlled lipid fluxes between organelles. Inter-organelle lipid transport occurs via three major mechanisms: (1) Movement as constituents of transport vesicles;1 (2) Transfer by lipid binding/transfer proteins (LTPs), which carry lipids within a hydrophobic pocket through the aqueous environment;2,3 and (3) Diffusion between closely apposed membrane leaflets. A wealth of evidence demonstrates that a substantial portion of the inter-organelle transport of lipids occurs via non-vesicular mechanisms.3 Of note, LTP-mediated transfer of lipid molecules over the narrow gap between closely apposed bilayers is envisioned to provide a markedly specific and efficient means of lipid transport.4–11
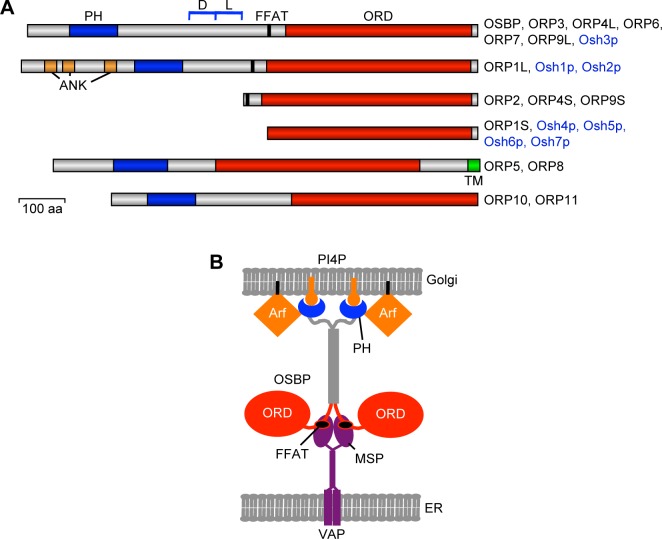
Oxysterol-binding protein (OSBP) is an LTP identified in the 1980s as a cytoplasmic high-affinity receptor for a variety of oxysterols.12 This ubiquitously expressed protein consists of a carboxy-terminal ligand-binding domain and an amino-terminal region that contains a pleckstrin homology (PH) domain interacting with phosphatidylinositol-4-phosphate (PI4P) in the trans-Golgi membranes.13–16 Between these domains, there is a dimerization motif, which mediates homodimer formation and heterodimerization with the closely related OSBP2/ORP4L,16,17 as well as a sequence motif (two phenylalanines in an acidic tract [FFAT]) that interacts with ER vesicle-associated membrane protein-associated proteins (VAPs) (Fig. 1).18 Families of proteins carrying an OSBP-related ligand-binding domain (ORD) have been identified in virtually all eukaryotic organisms for which sequence information is available: Mammals have 12 OSBPL genes encoding ORPs,19–21 the zebrafish Danio rerio has 14,22 Caenorhabditis elegans 4,23 Drosophila melanogaster 4,24 and the yeast Saccharomyces cerevisiae 725 OSBP homologue genes (Fig. 1), suggesting that the gene products serve fundamental and conserved functions in eukaryotic cell physiology.
Figure 1.
Structural organization of the ORPs. (A) Schematic diagram of the structure of mammalian and yeast ORPs. In the nomenclature of the proteins, L indicates “long” isoforms that contain a PH domain and S indicates “short” isoforms lacking this domain. Mammalian proteins are indicated with black, and yeast ones with blue print on the right. PH, pleckstrin homology domain; D, dimerization; L, leucine repeat domains delineated in OSBP and ORP4L; FFAT, “two phenylalanines in an acidic tract”; ORD, OSBP-related ligand-binding domain; ANK, ankyrin repeats; TM, trans-membrane segment. (B) A model of how OSBP is thought to bridge between ER and trans-Golgi membranes at a contact site (based on Ref. 32). Both OSBP and VAPs anchoring it to ER membranes are suggested to operate as dimers. PI4P, phosphatidylinositol 4-phosphate; Arf, the small GTPase ADP-ribosylation factor; MSP, major sperm protein domain; VAP, VAMP-associated protein.
For a number of years, the nature of the function of ORPs remained enigmatic. The suggested functions included control of sterol homeostasis, intracellular sterol transport, and poorly understood roles in cell signaling and developmental processes. Clues of the physiologic roles of ORPs as we currently understand them were obtained from the seminal observations that (1) the PH domain of mammalian OSBP targets the Golgi complex;13,14 (2) a yeast ORP called Osh1p localizes at a membrane contact site (MCS), the nucleus–vacuole junction;26 and (3) most ORPs carry an FFAT motif that specifies interaction with the ER VAPs.18 These observations led us to propose that localization and function at MCSs, where ER membranes are closely apposed to other organelle membranes, might be a common denominator of the ORPs.27 MCSs are found ubiquitously and occur between the ER and virtually every other organelle type, including organelles not engaged in vesicle transport, mitochondria, peroxisomes, and cytoplasmic lipid droplets. Such membrane junctions execute a multitude of functions in lipid syntheses, inter-organelle lipid transport, Ca2+ regulation, signaling events, autophagy, mitochondrial fission, and organelle partitioning in yeast mitosis.6–11 In the past few years, a number of studies adding essential insight into the modes of ORP action have been published. In this review, I focus on selected key findings suggesting that the function of ORPs involves transport of specific lipid ligands over MCSs, lipid remodeling in trans over an MCS, or lipid signaling.
Localization of ORPs at Membrane Contact Sites
Membrane-targeting determinants on the ORPs
Eight of the mammalian ORPs (OSBP, ORP1, -2, -3, -4, -6, -7, and -9) and three yeast OSBP homologues (Osh1p, -2p, and -3p) possess upstream of their ORD a short motif designated FFAT.18 This motif interacts specifically with the major sperm protein (MSP) domain of VAPs, type 2 integral proteins of the ER.28 Moreover, mammalian ORP5 and -8 carry a carboxyterminal trans-membrane segment that targets the ER.29,30 On the other hand, most of the mammalian ORPs have a PH domain that is in several cases known to interact with phosphoinositides (PIPs) in distinct non-ER organelle membranes.31 In addition, ORPs interact with membrane-associated regulatory proteins, such as the small GTPase Arf1 in the case of OSBP15,32 and Rab7 in the case of ORP1L.33 The capacity of ORPs to target two distinct ER and non-ER organelle membranes is termed “dual targeting”.
Yeast Osh proteins at MCSs
Since the seminal observation on Osh1p (see Introduction), increasing evidence has accumulated for the localization of ORPs at MCSs both in yeast and in mammalian cells. Four of the seven yeast Osh proteins, namely Osh2p, Osh3p, Osh6p, and Osh7p, are enriched in the cortical ER (cER), where the ER forms extensive contacts with the plasma membrane (PM) (Table 1).34,35 Of note, the yeast VAP orthologues mediating the association of Osh2p and Osh3p with the ER, namely Scs2p and Scs22p, were found to be among the six genes the disruption of which caused disappearence of the cER and disturbed ER functions in general.36 While Osh2p and Osh3p (as well as Osh1p) carry an FFAT motif-mediating interaction with VAPs and a PH domain for targeting non-ER compartments, Osh6p and Osh7p lacked these motifs but were still found associated with the cER. Likewise, Osh4p, which lacks an FFAT motif and a PH domain, was shown to cluster ER- and PM-derived vesicles, apparently via two distinct membrane-binding surfaces delineated on the protein.34
Table 1.
Localization and suggested functions of human and yeast ORPs in lipid transport over MCSs.
| MAMMALIAN | MCS | LIPIDS TRANSPORTED | REFERENCE(S) |
|---|---|---|---|
| OSBP | ER1-trans-Golgi | Cholesterol, PI4P2 | 32 |
| ORP4/OSBP2 | ? | Cholesterol (B) | 17 |
| ORP1L | ER-LE3 | Cholesterol (B) | 23, 38, 65 |
| ORP2 | ER-LD4, ER-PM5? | Cholesterol | 39, 40, 83 |
| ORP3 | ER-PM | ? | 42 |
| ORP6 | ? | ? | |
| ORP7 | ? | ? | |
| ORP5 | ER-PM ER-LE? |
PS6, PI4P Cholesterol (in vitro) |
43 29 |
| ORP8 | ER-PM | PS, PI4P | 43 |
| ORP9 | ER-trans-Golgi? | Cholesterol (in vitro), PI4P (B) | 45, 84 |
| ORP10 | ? | PS (B) | 52 |
| ORP11 | trans-Golgi-LE7? | ? | 85 |
| S. cerevisiae | |||
| Osh1p | NVJ8 | Cholesterol (in vitro) | 26, 34 |
| Osh2p | ER-PM | Cholesterol (in vitro) | 34 |
| Osh3p | ER-PM | PI4P (B), cholesterol (in vitro) | 34, 51 |
| Osh4p | ER-PM? (Golgi, exocytotic vesicles, endosomes) | Sterols, PI4P, PIP29 | 34, 49, 50, 55, 69, 73, 75 |
| Osh5p | ? | Cholesterol (in vitro) | 34 |
| Osh6p | ER-PM | PS, PI4P | 35, 52, 53 |
| Osh7p | ER-PM | PS | 52 |
Notes:
Endoplasmic reticulum;
Phosphatidylinositol-4-phosphate;
Late endosome;
Lipid droplet;
Plasma membrane;
Phosphatidylserine;
Late endosome;
Nucleus-vacuole junction;
Phosphatidylinositol-4,5-bisphosphate; (B) There is evidence for binding of the lipid but whether it is transported by the protein in unclear.
Mammalian ORPs at organelle interfaces
ORP1L, which is highly expressed in the central nervous system and in macrophages,37 targets late endosome/lysosome (LE) limiting membranes via interaction with the small GTPase Rab7 and the ER via VAPs.33 We recently showed by employing bimolecular fluorescence complementation (BiFC) that ORP1L–VAP complexes in fact localize at ER–LE contacts (Table 1), which control the motility and positioning of the LE in a sterol-dependent manner.38,39 The closest homologue of ORP1, namely ORP2, possesses an FFAT motif but no PH domain. When expressed alone, this protein targets lipid droplet (LD) surfaces.40 However, complexes of ORP2 with VAP decorate ER structures with LD attached to them, suggesting that ORP2 has the capacity to mediate ER–LD interactions.39 Furthermore, we found evidence that the ORP2–VAP complexes have the capacity to facilitate the synthesis and to inhibit the degradation of cellular triglycerides in hepatoma cells, indicating that the ER–LD contacts mediated by these proteins execute an important function in the control of neutral lipid metabolism. A further family member, ORP3, which is expressed at highest levels in epithelia and lymphatic tissues/leukocytes,41 forms a physical complex with the small GTPase R-Ras. Our work in the embryonic kidney cell line HEK293 has demonstrated that ORP3 interacts with VAPs at the ER and ER–PM contact sites.42 Du et al29 presented evidence indicating that ORP5, which is anchored in the ER via a trans-membrane segment, interacts with the LE cholesterol transporter Niemann—Pick C1 protein (NPC1) and may reside at ER–LE contact zones; however, the latest report by De Camilli and coworkers suggests that ORP5, like its close homologue ORP8, rather localizes at ER–PM contacts.43
Importantly, the hallmark study of Mesmin et al32 has demonstrated that OSBP can annex together two types of giant unilamellar vesicles, one population decorated with VAP and the other containing PI4P, thus promoting the formation of an MCS in vitro. The authors also showed that double overexpression of VAP and OSBP constructs in HeLa cells induced extensive ER–Golgi MCSs. The localization of ORPs/Osh proteins at MCSs is associated with the observations that several ORPs are capable of accommodating not only one but two distinct types of lipid ligands within their ORD and new functional insight sprouting from these findings (see below).
ORPs are Not Exclusively Sterol-Binding Proteins
Structure of the ORD
The ORDs of a number of ORPs can accommodate a variety of oxysterols, cholesterol, or ergosterol.44–48 Im et al44 reported the high-resolution structure of the yeast ORP Osh4p complexed with five different sterols (cholesterol, ergosterol, 7-, 20-, and 25-hydroxycholesterol [25OHC]), revealing a β-barrel-like fold with a lipid-binding pocket, in which the bound sterol is oriented with its 3β-hydroxyl group facing the bottom of the cavity. The bound sterol stabilizes a closed conformation of a lid consisting of a two-stranded β-sheet and three α-helices. The lid-open Osh4p was suggested to expose basic amino acid residues near the mouth of the ligand cavity that interact with negatively charged head groups of membrane lipids, thus facilitating the extraction of sterol from the bilayer.
Accommodation of glycerophospholipids within the ORD
Raychaudhuri et al49 reported that Osh4p can also extract PIP2 from vesicles and mediate significant transfer of this lipid in vitro, suggesting that PIP2 could be fully or partially inserted into the ligand cavity of Osh4p. The first structural evidence for the binding of a PIP or, more generally, a glycerophospholipid within the ligand cavity of an ORP was provided by Saint-Jean et al,50 who determined the structure of Osh4p crystallized with PI4P in the ORD cavity. Furthermore, they demonstrated that a bound sterol, dehydroergosterol (DHE), was readily exchanged for PI4P. The authors suggested that the two lipids could be transported by Osh4p in opposite directions in cells, sterol from the ER to the trans-Golgi and PM and PI4P in the opposite direction. Intriguingly, Saint-Jean et al identified at the entrance of the ligand cavity a cleft that accommodated the phosphoinositol moiety of PI4P, and indicated that amino acid residues lining this cleft were highly conserved among all ORPs. Further proof for liganding of ORPs by glycerophospholipids was provided by Tong et al,51 who solved the structure of the Osh3p ORD with bound PI4P. This study suggested that the ligand cavity of Osh3p was too narrow to accommodate bulky sterol molecules. The above observations brought up the possibility that PI4P binding could be a unifying feature of many or even all ORPs, and that only a subset of these proteins could additionally accommodate sterol ligands.
Maeda et al52 demonstrated that yeast Osh6p and Osh7p specifically bind phosphatidylserine (PS) and can extract this phospholipid from membranes. The authors crystallized Osh6p and modeled PS into the ORD ligand cavity. The head group and the unsaturated sn-2 fatty acyl chain were oriented toward the entrance of the pocket, while a saturated sn-1 chain pointed toward the bottom of the cavity. The authors found Osh6p to be unable to extract ergosterol from membranes, suggesting that it, similar to Osh3p (see above), may be unable to bind sterol ligands. Of importance, the latest work by the group of Drin demonstrated that Osh6p can, in addition to PS, bind PI4P.53 Interestingly, sequence analyses by Maeda et al52 suggested that the mammalian ORP5, -8, -9, -10, and -11 belong to the same clade as Osh6p, and found that the ORDs of human ORP5 and -10 can in fact extract PS from liposomes. Consistently, Chung et al43 identified by mass spectrometry PS 36:1 and a PIP bound within the ORD of ORP8.
Among the mammalian ORPs, the archetype family member OSBP is the first one shown to accommodate either sterols or a glycerophospholipid (PI4P) within its ORD.32 This means that OSBP can interact with PI4P with both its PH domain and its ORD. The functional implications of this observation are addressed in detail in the following paragraph.
ORPs can Mediate Lipid Transfer Over MCSs
Intracellular sterol transport in yeast
It has been known for a long time that the transport of sterols from the yeast PM to the ER occurs largely via non-vesicular mechanisms.54 The study of Raychaudhuri et al49 initally suggested that the yeast Osh proteins played a role in this sterol transport process (Table 1). However, the first direct evidence that ORPs could mediate non-vesicular sterol transport over membrane contacts was provided by Schulz et al,34 who showed that sterol transport in vitro mediated by yeast Osh4p was markedly inhibited by a porous membrane separating the donor and acceptor vesicles, suggesting that the transport does not rely on diffusible carrier structures. In a recent study, von Filseck et al55 demonstrated, using real-time in vitro lipid transport assays, that yeast Osh4p can, as earlier suggested by Saint-Jean et al,50 indeed transport a sterol against its concentration gradient by dissipating the energy of a PI4P gradient. This transport is sustained by the PI4P-phosphatase Sac1p, which hydrolyzes the transported PI4P in the acceptor compartment.
Bidirectional transport of cholesterol and PI4P by mammalian OSBP
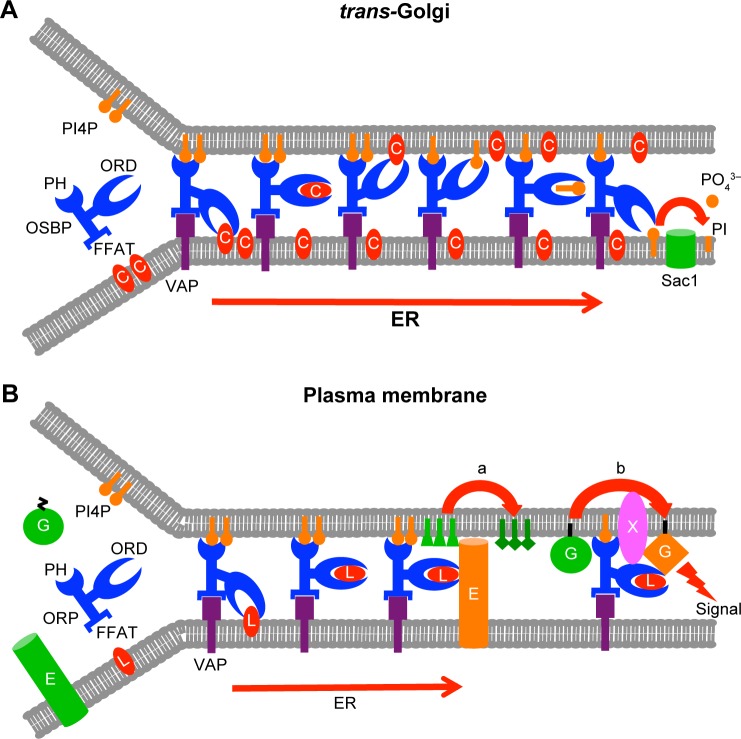
A similar mode of transport was reported for the mammalian OSBP by Mesmin et al,32 who provided elegant evidence that OSBP can tether ER and Golgi membranes and mediate the forward transport of cholesterol against its concentration gradient from the ER to trans-Golgi (Table 1; Fig. 2A). The transport is suggested to be driven by the retrograde transport of PI4P. The authors also showed that mutations inhibiting either the VAP or PI4P interaction of OSBP markedly impaired its lipid transport function. In the model arising from these observations, synthesis of PI4P in Golgi membranes and its hydrolysis in the ER by Sac1 sustain a gradient of membrane PI4P concentration driving the transport of cholesterol over MCSs mediated by OSBP. The fact that PI4P, which is retrogradely transported by the ORD of OSBP, also acts as a ligand for its PH domain in Golgi membranes provides an obvious mechanism of autoregulation of the OSBP transport function. The high-affinity oxysterol ligand of OSBP, namely 25OHC, was shown to inhibit the transport function, locking OSBP at Golgi membranes. Of note, the role of OSBP in lipid transport is not limited to the direct cholesterol/PI4P carrier function suggested above; OSBP was also reported to promote recruitment of the ceramide transporter CERT to Golgi sites,56 which most likely represent ER–Golgi MCSs.57 The two LTPs act in concert with Nir2, a phosphatidylinositol (PI) transfer protein that supplies Golgi membranes with PI for the generation of PI4P.58
Figure 2.
Models for the function of ORPs at membrane contact sites (MSCs). (A) OSBP transfers cholesterol (C) within its ligand-binding domain (ORD) over MCSs from the ER to trans-Golgi and PI4P in the opposite direction (based on Ref. 32). The phosphatidylinositol 4-phosphate (PI4P) retrogradely transported to the ER is hydrolyzed by the phosphatase Sac1 to phoshatidylinositol (PI) and inorganic phosphate. The PI4P gradient between Golgi and ER thus drives the forward transport of cholesterol against its concentration gradient. The fact that OSBP is targeted to the Golgi through binding of its pleckstrin homology (PH) domain to PI4P provides a mechanism of autoregulation of the process. FFAT, “two phenylalanines in an acidic tract,” motif mediating the interaction of OSBP with the VAMP-associated proteins (VAP) in the ER. (B) Models for ORP function as regulators of enzymatic or signaling processes at MCSs. An ORP binds a lipidous ligand (L), which regulates the conformation of the protein, allowing it to stimulate at an ER–plasma membrane contact (a) the activity of an enzyme (E) that is anchored in ER membranes and modifies in trans the structure of lipid substrates in the plasma membrane (based on Refs. 74, 75 on yeast Osh3p), or (b) the activation of a signaling component, eg, a small GTPase (G; based on Ref. 45 on ORP3). X, an activator of the GTPase. The inactive enzyme and GTPase are depicted in green and the active ones in orange. Regulation of the processes by the ligand (L) is in the cases of Osh3p and ORP3 hypothetical, and whether L is transported over the contact site is unknown.
Role of ORPs in the Generation of Viral Replication Organelles
An interesting extension of the above findings on a role of OSBP as a lipid transporter over MCSs is its essential role in the replication of enteroviruses and the hepatitis C virus (HCV). A recent study revealed that the antifungal agent itraconazole inhibits enterovirus and HCV replication by targeting OSBP and its close homologue ORP4.59 Consistently, OSBP knockdown and the small molecular antagonist of OSBP and ORP4, OSW-1, suppressed enterovirus replication. OSBP and ORP4 were also identified as targets of the minor enviroxime-like compounds, which inhibit enterovirus and HCV replication.60 The enteroviral RNA synthesis takes place in modified Golgi-derived membrane compartments designated “replication organelles”, which are enriched in PI4P. Certain virus-encoded proteins enhance phosphatidylinositol 4-kinase (PI4K) activity in the replication membranes. This results in the synthesis of PI4P to recruit OSBP, which transfers cholesterol to the membrane structures.61 Similarly, OSBP was shown to act as an effector of PI4K in HCV infection, contributing to cholesterol enrichment of the membranous web, which is the HCV replication compartment that arises as an extension of the ER.62 Moreover, replication of rhinoviruses causing common cold was shown to invariably require OSBP, and distinct rhinovirus subtypes also showed some dependence on other family members ORP1, -2, -5, -6, -7, -8, -9, -10, or -11.63 Even though the role of MCSs in the formation of the viral replication organelles has not been addressed in detail, the above results (1) suggest that lipid transport over MCSs by OSBP and possibly other ORPs may be required for generation and/or function of these subverted membrane structures, and (2) bring up the intriguing possibility of employing ORPs as targets for the development of new antiviral therapies.
ORPs in Phosphatidylserine Transport to the Plasma Membrane
The study by Maeda et al52 provided convincing data that yeast Osh6p and Osh7p, which localize at the cER, are instrumental for the transport of PS from the ER to the PM. The Osh6p/Osh7p-mediated transport also occurred at 4°C, consistent with a non-vesicular mechanism. Interestingly, the authors showed that it was possible to misdirect the flux of PS from the ER to the vacuole with a rapamycin-inducible Osh6p–Vph1p (a trans-membrane protein of the vacuole) complex, further supporting a specific function of Osh6p in the transport of PS and the notion that the process most likely involves MCSs. The latest report by van Filseck et al53 demonstrates that Osh6p can transport PS and PI4P in an exchange-type fashion and that the Osh6p-mediated PS transport in yeast cells depends on recognition of PI4P by the protein and on the presence of Sac1p, which hydrolyzes the counter-transported PI4P in the ER. In a concomitant study, Chung et al43 reported, based on experiments with HeLa cells and mouse embryonic fibroblasts, that ORP5 and ORP8 execute a similar PS/PI4P exchange function at ER–PM contacts in mammalian cells. These proteins are anchored by a trans-membrane span in ER membranes but localize in part at patches associated with the cell cortex. By employing the rapamycin-inducible FRB/FKBP heterodimerization approach, the authors showed that physical tethering of the ER to the PM by the ORPs was required for the PS/PI4P exchange to occur, demonstrating that the transport process occurs at MCSs.
Function of ORP1L at the ER–Late Endosome Interface—Lipid Sensor or Transporter
There is substantial evidence to suggest that liganding of ORP1L by sterols regulates distinct ER–LE contacts, the recruitment of dynein-dynactin motor complexes on the LE, and LE tethering mediated by the homotypic fusion and protein sorting (HOPS) complex. Depletion of cell cholesterol or expression of mutant ORP1L defective in sterol binding results in increased ER–LE contacts and a scattered subcellular distribution of LE, while liganding of ORP1L by cholesterol detaches LE from ER and facilitates recruitment of dynein–dynactin motor complexes on the LE. This leads to their microtubule (−)end-directed motility and clustering/fusion in the perinuclear region.38,64,65 However, whether the ER–LE contacts mediated by ORP1L and VAP play a role in inter-organelle lipid transport is thus far unclear. We showed that depletion of macrophage ORP1L by RNAi results in a defect in the efflux of endocytosed lipoprotein-derived cholesterol,38 and the data of Kobuna et al23 suggested the involvement of ORP1L in the formation of LE intralumenal vesicles. Interestingly, ORP1L was shown to interact with the adenoviral protein RIDa, which generates a virus-induced Niemann–Pick C1 (NPC1) independent pathway for cholesterol transport from late endocytic compartments to the ER for esterification.66,67 ORP1L is essential for this transport process, further implying that it either acts as a cholesterol carrier or generates distinct membane contacts over which the transport occurs. To conclude, there is extensive evidence that ORP1L functions as a sterol sensor that controls ER–LE MCSs and LE motility, positioning, and tethering. However, a lipid transporter function at ER–LE MCSs is also possible.
ORPs and Cell Signaling: Direct Function as Constituents of Signaling Complexes or Indirect Effects via Organelle Lipid Compositions
Role of OSBP in the control of extracellular-signal-regulated kinases
A number of reports have provided evidence for the roles of ORPs in cell signaling.42,48,68–70 In certain cases, they have been found to associate physically with signaling proteins, while on other occasions one can argue that ORP family members may affect signaling indirectly via effects on organelle lipid compositions. The best example of the former is the reported capacity of OSBP to scaffold two protein phophatases, namely the serine/threonine phosphatase PP2A and the tyrosine phosphatase HePTP, which control the activity of extracellular signal-regulated kinases.48 The cholesterol-bound OSBP was suggested to associate with the active phosphatases, while depletion of cholesterol or addition of 25OHC resulted in dissociation of the complex. These findings implied that OSBP forms a functional link between the cellular sterol status and mitogenic signaling cascades. The relationship between this signaling function and the role of OSBP in cholesterol transport from the ER to trans-Golgi, however, remains unclear. It seems plausible that, since 25OHC, which targets OSBP to Golgi membranes, dissociates the phosphatase complex, the active complex may not be associated with the Golgi or ER–Golgi MCSs but may rather occur in the cytosol. Although the above observations are highly interesting, it is puzzling that they have not been followed up by work from other research groups.
New evidence for ORP3 as a modulator of R-Ras signaling
We recently found that ORP3, which associates physically with the small GTPase R-Ras, is, in complex with its ER anchor VAPA, partially localized at ER–PM MCSs.42 Both the PM-targeting PH domain and the ER-targeting FFAT motif were demonstrated to be required for the capacity of ORP3 to activate R-Ras and its downstream signaling targets Akt and β1-integrins, suggesting that by targeting both membrane compartments, putatively at MCSs, ORP3 creates a platform for R-Ras activation (Fig. 2B). The lipid ligands of ORP3 are thus far unknown, but we find this protein unlikely to bind sterols.22 Thus, there is currently no information or hypothesis on its putative role as a lipid transporter at the ER–PM contacts.
Yeast Osh3p in the control of enzymes at ER–plasma membrane junctions
Yeast Osh3p, which localizes in the cER, has been assigned functions as a regulator of enzymes active at the ER–PM contact sites. Stefan et al71 provided evidence that Osh3p recruits the ER integral PI4-phosphatase Sac1p to ER–PM MCSs, where it can access in trans its substrate PI4P at the PM. In this way Osh3p would contribute to regulation of the PI4P content of the PM or its specific domains, with impacts on PM PIP signaling and vesicle transport. Analogously, Tavassoli et al72 reported that Osh3p has the capacity to recruit another ER enzyme Opi3p, a phosphatidylethanolamine (PE) N-methyltransferase, at ER–PM contacts, where it converts plasma membrane PE in trans to phosphatidylcholine (PC). These articles on Osh3p provide examples of ORP function as modulators of organelle lipid composition via mechanisms not involving inter-organelle lipid transfer (Fig. 2B). However, the data do not exclude the possibility that Osh3p could also act as a lipid transporter over the ER–PM MCSs—the ORD of this protein binds PI4P,51 which could plausibly be a retrogradely transported ligand, the other putative lipid ligand(s) being thus far unknown. Of note, the findings of Stefan et al71 could also reflect retrograde transport of PI4P by Osh3p and hydrolysis of the PIP in cis by Sac1p in the ER.
Yeast Osh4p: Lipid Transporter or Signal Transducer
Quite distinct from the suggested role in the bidirectional sterol/PI4P transport function suggested by Saint-Jean et al50 and von Filseck et al,55 the group of Bankaitis has reported results implying a signaling function of yeast Osh4p.69 They found that a sterol-binding deficient point mutant Osh4Y97F, originally characterized by Alfaro et al,73 interferes with yeast proliferation due to its enhanced association with the trans-Golgi network (TGN) and endosomes. These data suggested that sterol binding within the ORD of Osh4p acts as a negative regulatory signal that detaches the protein from the TGN/endosomal membranes. Whether Osh4p could act as both a sterol-dependent signal mediator and a transporter of the bound sterol is thus far unclear, but this possibility cannot be excluded. The Osh4Y97F-mediated growth arrest observed by Mousley et al69 was associated with amino acid deficiencies caused by defects in the trafficking of amino acid permeases, suppression of gene expression driven by GCN4, a transcriptional activator of the general amino acid control (GAAC) regulon, and disturbed target of the rapamycin complex 1 (TORC1) signaling. The precise nature of the signal generated on TGN/endosomal membranes by Osh4p remains enigmatic, and the associated context of interaction partners mediating the input from Osh4p to the downstream signaling is unknown. The authors suggested that the signal might involve enrichment of sphingolipids in these membranes, an idea supported by the study of LeBlanc et al,74 who observed sphingolipid defects in Δosh4 cells. In addition to the above function, Osh4p was shown to reside on yeast secretory vesicles and to regulate their PI4P content, with impacts on polarized exocytosis.73,75 Moreover, work by the group of Menon indicated that the Osh proteins may rather control the distribution of sterols between lateral membrane domains than their inter-organelle transport.76 The above investigations do not support a simple function of Osh4p as a sterol/PI4P transporter, but provide implications of a more complex role of this protein in TGN/endosomal PI4P/sphingolipid signaling or sterol organization in membranes. However, one cannot exclude the possibility that the protein may execute more or less interconnected transport and signaling functions.
Future Pespectives
Membrane contact sites are currently moving into the focus of cell biological research. It is quite obvious that the functional interplay of distinct organelles is orchestrated via direct physical contacts to a much larger extent than previously appreciated. Functions of such membrane junctions in the inter-organelle flow of small molecules and information, as well as their molecular constituents, are being characterized at a rapid pace. The concept that MCSs act as high-capacity and high-specificity platforms of inter-organelle lipid transfer is gaining ground, and there is substantial evidence that certain ORPs (mammalian OSBP, ORP5, and ORP8; yeast Osh4p, -6p, and 7p) play important roles as sterol, PI4P, or PS transporters over MCSs. An attractive hypothesis is that ORPs could use the energy deposited in the intracellular gradient of PI4P (or possibly other PIPs) to drive the transport of lipids against their concentration gradients. Substantial evidence is accumulating in support of this “bidirectional lipid transporter” hypothesis. However, further evidence is required, and one needs to elucidate how widely this functional hypothesis is applicable among the large eukaryotic ORP families. An urgent key task is the identification of the endogenous cellular lipid ligands of the remaining ORP family members from different organisms. This information will be instrumental in further testing of the bidirectional lipid transporter hypothesis. Another crucial line of study will be the identification and characterization of the proteomes and functional protein complexes located at distinct MCSs and their relationships with the inter-organelle bridges formed by ORPs. To place all this new molecular information into its physiologic context, intense research efforts aimed at understanding the function of ORPs in developmental processes, cell fate determination, metabolism, and signaling in vivo are constantly warranted. Of note, no ORP has thus far been associated with ER–mitochondrion contacts, the biogenesis of mitochondria, or mitochondrial partition in mitosis. Furthermore, the role of this machinery in disease-associated processes such as ER stress responses or autophagy has not been addressed. The first reports connecting genetic defects of ORPs (OSBPL genes) to human disease are beginning to emerge: Mutations in OSBPL1A encoding ORP1L were found to be associated with extremely low serum high-density lipoprotein (HDL) cholesterol levels,77 and a frameshift mutation in OSBPL2 encoding ORP2 was suggested to cause a form of autosomal dominant hearing loss.78,79 Moreover, studies on knock-out mice have suggested roles of ORP8 in HDL regulation and of the macrophage protein in atherogenesis,80,81 as well as of ORP4 in the postmeiotic differentiation of male germ cells.82 Future work on the molecular functions of ORPs in specific cell types is required to uncover their roles in human disease and their potential as novel therapy targets.
Footnotes
ACADEMIC EDITOR: Tim Levine, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 565 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by the Academy of Finland (grant 285223), the Sigrid Juselius Foundation, the Liv och Hälsa Foundation, the Finnish Foundation for Cardiovascular Research, and the Magnus Ehrnrooth Foundation. The author confirms that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: The author discloses no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Developed the structure and arguments for the paper: VMO. Wrote the manuscript: VMO. The author reviewed and approved of the final manuscript.
REFERENCES
- 1.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Angelo G, Vicinanza M, De Matteis MA. Lipid-transfer proteins in biosynthetic pathways. Curr Opin Cell Biol. 2008;20(4):360–370. doi: 10.1016/j.ceb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11(10):739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 4.Alpy F, Tomasetto C. START ships lipids across interorganelle space. Biochimie. 2014;96:85–95. doi: 10.1016/j.biochi.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Gatta AT, Wong LH, Sere YY, et al. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife. 2015;4:e07253. doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833(11):2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 7.Henne WM, Liou J, Emr SD. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol. 2015;35:123–130. doi: 10.1016/j.ceb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18(4):371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36(11):616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Klecker T, Bockler S, Westermann B. Making connections: interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 2014;24(9):537–545. doi: 10.1016/j.tcb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Toulmay A, Prinz WA. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr Opin Cell Biol. 2011;23(4):458–463. doi: 10.1016/j.ceb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor FR, Saucier SE, Shown EP, Parish EJ, Kandutsch AA. Correlation between oxysterol binding to a cytosolic binding protein and potency in the repression of hydroxymethylglutaryl coenzyme A reductase. J Biol Chem. 1984;259(20):12382–12387. [PubMed] [Google Scholar]
- 13.Lagace TA, Byers DM, Cook HW, Ridgway ND. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem J. 1997;326(pt 1):205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol. 1998;8(13):729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 15.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12(9):695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 16.Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116(2):307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyles JP, Perry RJ, Ridgway ND. Characterization of the sterol-binding domain of oxysterol-binding protein (OSBP)-related protein 4 reveals a novel role in vimentin organization. Exp Cell Res. 2007;313(7):1426–1437. doi: 10.1016/j.yexcr.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in opi1p binds VAP. EMBO J. 2003;22(9):2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anniss AM, Apostolopoulos J, Dworkin S, Purton LE, Sparrow RL. An oxysterol-binding protein family identified in the mouse. DNA Cell Biol. 2002;21(8):571–580. doi: 10.1089/104454902320308942. [DOI] [PubMed] [Google Scholar]
- 20.Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR. A family of 12 human genes containing oxysterol-binding domains. Genomics. 2001;78(3):185–196. doi: 10.1006/geno.2001.6663. [DOI] [PubMed] [Google Scholar]
- 21.Lehto M, Laitinen S, Chinetti G, et al. The OSBP-related protein family in humans. J Lipid Res. 2001;42(8):1203–1213. [PubMed] [Google Scholar]
- 22.Zhou Y, Wohlfahrt G, Paavola J, Olkkonen VM. A vertebrate model for the study of lipid binding/transfer protein function: conservation of OSBP-related proteins between zebrafish and human. Biochem Biophys Res Commun. 2014;446:675–680. doi: 10.1016/j.bbrc.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Kobuna H, Inoue T, Shibata M, et al. Multivesicular body formation requires OSBP-related proteins and cholesterol. PLoS Genet. 2010;6(8):e1001055. doi: 10.1371/journal.pgen.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, Liu Z, Huang X. OSBP- and FAN-mediated sterol requirement for spermatogenesis in Drosophila. Development. 2010;137(22):3775–3784. doi: 10.1242/dev.049312. [DOI] [PubMed] [Google Scholar]
- 25.Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157(3):1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine TP, Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol Biol Cell. 2001;12(6):1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olkkonen VM, Levine TP. Oxysterol binding proteins: in more than one place at one time? Biochem Cell Biol. 2004;82(1):87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13(7):1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Du X, Kumar J, Ferguson C, et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192(1):121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan D, Mäyränpää MI, Wong J, et al. OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J Biol Chem. 2008;283(1):332–340. doi: 10.1074/jbc.M705313200. [DOI] [PubMed] [Google Scholar]
- 31.Olkkonen VM, Li S. Oxysterol-binding proteins: sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Prog Lipid Res. 2013;52(4):529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155(4):830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 33.Johansson M, Lehto M, Tanhuanpää K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16(12):5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz TA, Choi MG, Raychaudhuri S, et al. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187(6):889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Duan W, Munn AL, Yang H. Molecular characterization of Osh6p, an oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae. FEBS J. 2005;272(18):4703–4715. doi: 10.1111/j.1742-4658.2005.04886.x. [DOI] [PubMed] [Google Scholar]
- 36.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23(6):1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Johansson M, Bocher V, Lehto M, et al. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol Biol Cell. 2003;14(3):903–915. doi: 10.1091/mbc.E02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vihervaara T, Uronen RL, Wohlfahrt G, Björkhem I, Ikonen E, Olkkonen VM. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell Mol Life Sci. 2011;68(3):537–551. doi: 10.1007/s00018-010-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber-Boyvat M, Kentala H, Peränen J, Olkkonen VM. Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci. 2015;72(10):1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J Lipid Res. 2009;50(7):1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehto M, Tienari J, Lehtonen S, Lehtonen E, Olkkonen VM. Subfamily III of mammalian oxysterol-binding protein (OSBP) homologues: the expression and intracellular localization of ORP3, ORP6, and ORP7. Cell Tissue Res. 2004;315(1):39–57. doi: 10.1007/s00441-003-0817-y. [DOI] [PubMed] [Google Scholar]
- 42.Weber-Boyvat M, Kentala H, Lilja J, et al. OSBP-related protein 3 (ORP3) coupling with VAMP-associated protein A regulates R-Ras activity. Exp Cell Res. 2015;331(2):278–291. doi: 10.1016/j.yexcr.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Chung J, Torta F, Masai K, et al. INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349(6246):428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437(7055):154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngo M, Ridgway ND. Oxysterol binding protein-related protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20(5):1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suchanek M, Hynynen R, Wohlfahrt G, et al. The mammalian OSBP-related proteins (ORP) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, JeBailey L, Ridgway ND. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem J. 2002;361(pt 3):461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307(5714):1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 49.Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol. 2006;173(1):107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Saint-Jean M, Delfosse V, Douguet D, et al. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195(6):965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong J, Yang H, Eom SH, Im YJ. Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 2013;21(7):1203–1213. doi: 10.1016/j.str.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Maeda K, Anand K, Chiapparino A, et al. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501(7466):257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- 53.Moser von Filseck J, Čopič A, Delfosse V, et al., editors. INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349(6246):432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Prinz WA. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J Biol Chem. 2004;279(43):45226–45234. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 55.Moser von Filseck J, Vanni S, Mesmin B, Antonny B, Drin G. A phosphati-dylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- 56.Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17(6):2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanada K, Kumagai K, Tomishige N, Yamaji T. CERT-mediated trafficking of ceramide. Biochim Biophys Acta. 2009;1791(7):684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19(9):3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strating JR, van der Linden L, Albulescu L, et al. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015;10(4):600–615. doi: 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arita M, Kojima H, Nagano T, Okabe T, Wakita T, Shimizu H. Oxysterol-binding protein family I is the target of minor enviroxime-like compounds. J Virol. 2013;87(8):4252–4260. doi: 10.1128/JVI.03546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arita M. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol Immunol. 2014;58(4):239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146(5):1373–85.e1. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roulin PS, Lötzerich M, Torta F, et al. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16(5):677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 64.van der Kant R, Fish A, Janssen L, et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013;126(pt 15):3462–3474. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- 65.Rocha N, Kuijl C, van der Kant R, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185(7):1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cianciola NL, Greene DJ, Morton RE, Carlin CR. Adenovirus RIDalpha uncovers a novel pathway requiring ORP1L for lipid droplet formation independent of NPC1. Mol Biol Cell. 2013;24(21):3309–3325. doi: 10.1091/mbc.E12-10-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shah AH, Cianciola NL, Mills JL, Sonnichsen FD, Carlin C. Adenovirus RIDalpha regulates endosome maturation by mimicking GTP-Rab7. J Cell Biol. 2007;179(5):965–980. doi: 10.1083/jcb.200702187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lessmann E, Ngo M, Leitges M, Minguet S, Ridgway ND, Huber M. Oxysterol-binding protein-related protein (ORP) 9 is a PDK-2 substrate and regulates Akt phosphorylation. Cell Signal. 2007;19(2):384–392. doi: 10.1016/j.cellsig.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 69.Mousley CJ, Yuan P, Gaur NA, et al. A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2012;148(4):702–715. doi: 10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugawara K, Morita K, Ueno N, Shibuya H. BIP, a BRAM-interacting protein involved in TGF-beta signalling, regulates body length in Caenorhabditis elegans. Genes Cells. 2001;6(7):599–606. doi: 10.1046/j.1365-2443.2001.00444.x. [DOI] [PubMed] [Google Scholar]
- 71.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144(3):389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 72.Tavassoli S, Chao JT, Young BP, et al. Plasma membrane—endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013;14(5):434–440. doi: 10.1038/embor.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alfaro G, Johansen J, Dighe SA, Duamel G, Kozminski KG, Beh CT. The sterol-binding protein Kes1/Osh4p is a regulator of polarized exocytosis. Traffic. 2011;12(11):1521–1536. doi: 10.1111/j.1600-0854.2011.01265.x. [DOI] [PubMed] [Google Scholar]
- 74.LeBlanc MA, Fairn GD, Russo SB, et al. The yeast oxysterol binding protein kes1 maintains sphingolipid levels. PLoS One. 2013;8(4):e60485. doi: 10.1371/journal.pone.0060485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ling Y, Hayano S, Novick P. Osh4p is needed to reduce the level of phosphatidylinositol-4-phosphate on secretory vesicles as they mature. Mol Biol Cell. 2014;25(21):3389–3400. doi: 10.1091/mbc.E14-06-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic. 2011;12(10):1341–1355. doi: 10.1111/j.1600-0854.2011.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Motazacker MM, Peter J, Treskes M, Shoulders CC, Kuivenhoven JA, Hovingh GK. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler Thromb Vasc Biol. 2013;33(7):1521–1528. doi: 10.1161/ATVBAHA.113.301505. [DOI] [PubMed] [Google Scholar]
- 78.Thoenes M, Zimmermann U, Ebermann I, et al. OSBPL2 encodes a protein of inner and outer hair cell stereocilia and is mutated in autosomal dominant hearing loss (DFNA67) Orphanet J Rare Dis. 2015;10:15. doi: 10.1186/s13023-015-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xing G, Yao J, Wu B, et al. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Genet Med. 2015;17(3):210–218. doi: 10.1038/gim.2014.90. [DOI] [PubMed] [Google Scholar]
- 80.Béaslas O, Metso J, Nissilä E, et al. Osbpl8 deficiency in mouse causes an elevation of high-density lipoproteins and gender-specific alterations of lipid metabolism. PLoS One. 2013;8(3):e58856. doi: 10.1371/journal.pone.0058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Kampen E, Beaslas O, Hildebrand RB, et al. Orp8 deficiency in bone marrow-derived cells reduces atherosclerotic lesion progression in LDL receptor knockout mice. PLoS One. 2014;9(10):e109024. doi: 10.1371/journal.pone.0109024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Udagawa O, Ito C, Ogonuki N, et al. Oligo-astheno-teratozoospermia in mice lacking ORP4, a sterol-binding protein in the OSBP-related protein family. Genes Cells. 2014;19(1):13–27. doi: 10.1111/gtc.12105. [DOI] [PubMed] [Google Scholar]
- 83.Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12(2):218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 84.Liu X, Ridgway ND. Characterization of the sterol and phosphatidylinositol 4-phosphate binding properties of Golgi-associated OSBP-related protein 9 (ORP9) PLoS One. 2014;9(9):e108368. doi: 10.1371/journal.pone.0108368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Y, Li S, Mäyränpää MI, et al. OSBP-related protein 11 (ORP11) dimerizes with ORP9 and localizes at the Golgi-late endosome interface. Exp Cell Res. 2010;316(19):3304–3316. doi: 10.1016/j.yexcr.2010.06.008. [DOI] [PubMed] [Google Scholar]