Abstract
Nlrp9a, Nlrp9b and Nlrp9c are preferentially expressed in oocytes and early embryos in the mouse. Simultaneous genetic ablation of Nlrp9a and Nlrp9c does not affect early embryonic development, but the function of Nlrp9b in the process of oocyte maturation and embryonic development has not been elucidated. Here we show that both Nlrp9b mRNA and its protein are expressed in ovaries and the small intestine. Moreover, the NLRP9B protein was restricted to oocytes in the ovary and declined with oocyte aging. After ovulation and fertilization, NLRP9B protein was found in preimplantation embryos. Confocal microscopy demonstrated that it was mainly localized in the cytoplasm in the oocytes and blastomeres. Thus, this protein might play a role in oocyte maturation and early embryonic development. However, knockdown of Nlrp9b expression in GV-stage oocytes using RNA interference did not affect oocyte maturation or subsequent parthenogenetic development after Nlrp9b-deficient oocytes were activated. Furthermore, Nlrp9b knockdown zygotes could reach the blastocyst stage after being cultured for 3.5 days in vitro. These results provide the first evidence that the NLRP9B protein is dispensable for oocyte maturation and early embryonic development in the mouse.
Keywords: Early development, Mouse, NLRP9B protein, Oocyte maturation
There are 14 members of the NLRP gene family in primates, while 20 members have been identified in the mouse, with Nlrp1, Nlrp4 and Nlrp9 showing lineage-specific replication. However, Nlrp7, Nlrp8, Nlrp11 and Nlrp13 might have been lost during mouse evolution and have not yet been identified.
Phylogenetic analysis has shown that the Nlrp gene family has divided into two clades involving immunization- and reproduction-related functions in mice [1]. Initially, research on this gene family focused on apoptosis and inflammatory signaling pathways. Nevertheless, further studies have shown that some Nlrp genes also play a significant role in reproduction [2,3,4,5,6]. The reproduction-related genes are mainly expressed in the ovary and play vital roles in oogenesis and preimplantation embryo development [7,8,9,10,11,12,13,14,15]. Thus, investigation of the functions of reproduction-related genes is of great significance in understanding the mechanism of oogenesis, oocyte maturation and early embryonic development in the mouse.
In silico methods revealed that Nlrp2, Nlrp4a-Nlrp4g, Nlrp5/Mater, Nlrp9a–Nlrp9c and Nlrp14 are reproduction-related genes in the mouse [1]. Nlrp5/Mater was one of the first genes to be identified as having a maternal effect, as an Nlrp5 knockout experiment showed that Nlrp5-deficient embryos arrested at the 2-cell stage, causing female infertility, even though the formation of follicles, oocyte maturation and fertilization were all normal [2]. Further research showed that the NLRP5/MATER, FLOPED, TLE6 and FILIA proteins form a subcortical complex in the oocyte that plays an important role in the first zygotic cleavage [3, 4]. Knockdown of Nlrp2 or Nlrp14 expressions in the zygote led to most zygotes being arrested at the 1-cell to 8-cell stage [6, 9]. Besides Nlrp2, Nlrp5 and Nlrp14, Nlrp4a–Nlrp4g also show specificity or are mainly expressed in the ovary [14, 16]. The latest research has indicated that the Nlrp9a–Nlrp9c transcripts exist predominantly in oocytes and early embryos [15]. Furthermore, simultaneous genetic ablation of Nlrp9a and Nlrp9c does not affect early embryonic development in the mouse [17]. However, the function of Nlrp9b in the process of oocyte maturation and embryonic development in the mouse has not been elucidated.
In this study, RNA interference was employed to investigate the function of Nlrp9b in oocyte maturation and early embryonic development in the mouse. The results indicated that Nlrp9b-deficient oocytes can undergo normal maturation and that Nlrp9b knockdown zygotes could reach the blastocyst stage. These data provide the first evidence that Nlrp9b is dispensable for oocyte maturation and early embryonic development in the mouse.
Materials and Methods
Ethics statement
The experimental procedure was approved by the Animal Care Commission of the College of Animal Science, Fujian Agriculture and Forestry University. Adult male and female ICR strain mice were purchased from the Experimental Animal Center of Fujian Medical University (Fuzhou, P. R. China). They were provided with water and mouse chow ad libitum and maintained on a 14/10 h light/dark cycle in the Laboratory Animal Facility of the College of Animal Science, Fujian Agriculture and Forestry University.
Chemicals
All chemicals and reagents were obtained from Sigma-Aldrich (St Louis, MO, USA) unless otherwise stated. Sterile plastic ware was purchased from Nunclon (Roskilde, Denmark).
Collection of oocytes and preimplantation embryos
In the experiments using germinal vesicle (GV)-stage oocytes, female ICR mice were injected with 10 IU pregnant mare serum gonadotropin (PMSG) by intraperitoneal injection 48 h before collection of oocytes. The ovaries were removed and placed in Hepes-buffered potassium simplex optimized medium (H-KSOM) containing 0.1 mM 3-isobutyl-1-methyl-xanthine to produce meiotic arrest, and oocytes were released by puncturing the edges of the ovaries with hypodermic needles. Metaphase II oocytes were obtained from the oviducts of female mice that had been given a second injection of 10 IU human chorionic gonadotropin 48 h after the PMSG injection. Cumulus masses were released into H-KSOM containing hyaluronidase (1 mg/ml). Parthenogenetic activation of oocytes and collection of preimplantation embryos were performed as described previously [6].
Reverse transcription polymerase chain reaction (RT–PCR) and quantitative real-time RT–PCR analysis
Total RNA was collected from 4-week-old mouse tissues (ovary, small intestine, testis, lung, heart, liver, brain, stomach, spleen and muscle) using RNeasy Mini Kits (Qiagen, Valencia, CA, USA). Complementary (c) DNA was synthesized using a PrimeScript II 1st strand cDNA Synthesis Kit (TaKaRa, Otsu, Japan). Oocytes (10 per group) were lysed, and first-strand cDNA was synthesized directly using a SuperScript® III CellsDirect cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The primer sequences used for RT–PCR and quantitative real-time RT–PCR were 5′–GACTTCACCAGTGACTGTTGTG–3′ (forward) and 5′–CCATACCAGACGAACACCC–3′ (reverse) for Nlrp9a, 5′– CACCAGTGACTGTTGTAAGGAC–3′ (forward) and 5′–TTCCTGCTGTTCCATACCAG–3′ (reverse) for Nlrp9b, 5′–TTCCAGTGAATCAACATCAGCT–3′ (forward) and 5′– GAACAGGGTAGCAAAGTACCA–3′ (reverse) for Nlrp9c and 5′– GCTTGCTGGTGAAAAGGACCTCTCGAAG–3′ (forward) and 5′– CCCTGAAGTACTCATTATAGTCAAGGGCAT–3′ (reverse) for Hprt1. The specificity of each primer pair and RT–PCR products were evaluated by electrophoresis and annealing temperature analysis to confirm the absence of primer dimers and to estimate fragment sizes. Quantitative real-time RT–PCR reactions were performed in triplicate for each individual sample in a final volume of 10 μl containing 2 × SYBR Premix ExTaq™ II, 100 nM each primer and 1 µl of cDNA on an ABI PRISM 7700 Sequence Detection System (Life Technologies, Carlsbad, CA, USA). The cycling conditions comprised 1 min preincubation at 95 C and 40 cycles at 95 C for 5 sec and 56–60 C for 30 sec. Expression of the target gene in each run was normalized to the housekeeping gene Hprt1. Relative expression was estimated using the 2−ΔΔCT method [18].
Western blotting
Protein samples from mouse tissues, oocytes, and preimplantation embryos were homogenized and solubilized in RIPA lysis buffer (P0013B, Beyotime Institute of Biotechnology, Jiangsu, P. R. China). The tissue lysates were centrifuged at 12,000 g for 20 min at 4 C. The supernatant fractions were collected, and the protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). Equal amounts of protein obtained from tissues or oocytes and preimplantation embryos were separated in 10% polyacrylamide gels containing 0.1% SDS and transferred to hydrophobic PVDF membranes (Millipore, Bedford, MA, USA). Membranes were blocked with Tris-buffered saline (TBS pH 7.4)/0.1% Tween 20 (TBST) containing 5% dry milk for 4 h at room temperature, incubated with antibodies against NLRP9B (NBP2-24661, Novus Biologicals, Littleton, CO, USA; 1:100) overnight at 4 C and then washed three times in TBST. A horseradish peroxidase-conjugated secondary antibody was added 2 h prior to processing using an ECL (enhanced chemiluminescence) Advance Western Blotting detection system (Pierce). β-Actin was used as a loading control.
Immunohistochemistry
Mouse ovaries were fixed in 4% paraformaldehyde overnight, embedded in paraffin wax and sectioned at 6 μm. These sections were then deparaffinized, rehydrated and heated in 10 mM sodium citrate buffer (pH 6.0) with microwaves for antigen retrieval. Immunohistochemistry of ovarian tissues using UltraSensitive™ SAP IHC kits (Fuzhou Maixin Biotech, P. R. China) was performed as described previously [6]. In brief, sections were incubated with 10% nonimmune goat serum and then with an anti-NLRP9B polyclonal antibody (1:100) overnight at 4 C, followed by reaction with a biotinylated goat anti-rabbit secondary antibody. After several washes, streptavidin–peroxidase and a diaminobenzidine substrate solution were applied to the sections. Negative controls were performed via processing sections as above with the absence of the primary antibody.
Immunofluorescence
Oocytes and embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature, washed and permeabilized with 0.2% Triton X-100 in PBS for 20 min. Samples were first incubated in PBS containing 3% BSA for 1 h to block nonspecific interactions. Immunofluorescence staining was performed by incubating the samples with an antibody against NLRP9B (1:200) overnight at 4 C, followed by an incubation in Alexa Fluor 488-conjugated goat anti-rabbit (Beyotime, A0428) diluted 1:500 for 2 h. DNA was stained with DAPI (C1005, Beyotime Institute of Biotechnology) for 5 min. Fluorescence was detected using a Zeiss LSM 510 confocal microscope equipped with differential interference contrast optics (Carl Zeiss, Thornwood, NY, USA).
Electroporation of customized short interfering (si)RNA
A mixture of three target-specific 19–25 nt siRNA that specifically target the Nlrp9b transcripts was purchased from Santa Cruz Biotechnology (sc-149819, Dallas, TX, USA). Electroporation was employed to deliver siRNA into GV-stage oocytes and zygotes as described previously [6]. Briefly, GV-stage oocytes and zygotes were incubated in prewarmed acid Tyrode’s solution for 12 sec to weaken the zona pellucida. The siPORT Amine Transfection Agent (AM4502, Ambion, Austin, TX, USA) and the negative control siRNA (Silencer® negative control No. 1, AM4611, Ambion) or the custom Nlrp9b siRNA were diluted in Opti-MEM® I (Invitrogen) medium according to the manufacturers’ protocols, respectively. After 10 min, the diluted siRNA and the siPORT Amine Transfection Agent were combined and incubated for 10 min at room temperature to form transfection complexes. The transfection complexes and oocytes or zygotes were added to a flat electrode chamber and arranged linearly between two electrodes. The electroporation parameters were DC 20 volts/1 msec pulse length, 3 pulses and 0 repeats. The electroporation control (EP control) was performed as mentioned above in the absence of siRNA. A negative control siRNA was used as the negative control.
After electroporation, GV-stage oocytes were cultured in MEM supplemented with 3 mg/ml bovine serum albumin (MEM-BSA) and 0.1 mM 3-isobutyl-1-methyl-xanthine for 24 h, washed three times and incubated in MEM-BSA for another 12 h, followed by calculation of the maturation rates of the oocytes. Zygotes were incubated in KSOMaa medium containing 4 mg/ml bovine serum albumin (KSOMaa-BSA) in a humidified atmosphere of 5% CO2/95% air at 37 C.
Statistical analysis
All experiments were repeated at least three times. The results are presented as the mean ± SEM. Data were analyzed by one-way ANOVA and LSD tests using the SPSS version 13.0 software (SPSS, Chicago, IL, USA). Differences were considered significant when P < 0.05.
Results
Tissue distribution of Nlrp9b
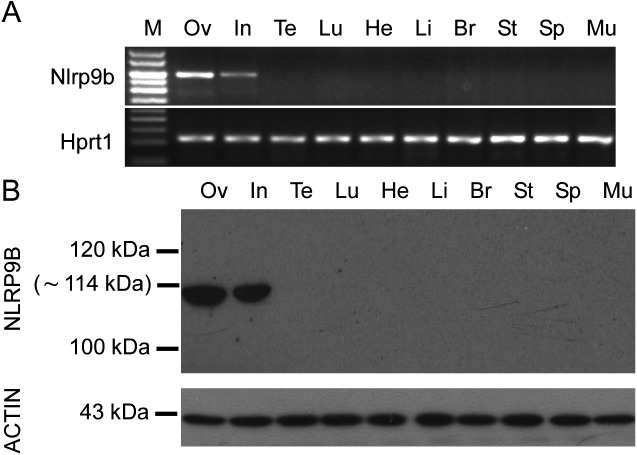
RT–PCR analysis detected transcripts of Nlrp9b in ovaries and the small intestine (Fig. 1A). The expression pattern of NLRP9B protein, as determined by western blotting, was similar to that determined by RT–PCR assay (Fig. 1B).
Fig. 1.
Expression of Nlrp9b transcripts and proteins in different mouse tissues. (A) Reverse-transcription polymerase chain reaction (RT-PCR) was applied to total RNA extracted from the 4-week-old mouse ovary (Ov), intestines (In), testis (Te), lung (Lu), heart (He), liver (Li), brain (Br), stomach (St), spleen (Sp), and muscle (Mu). Hprt1 was used as a control. (B) Western blotting analyses with total proteins extracted from the 4-week-old mouse ovary (Ov), intestines (In), testis (Te), lung (Lu), heart (He), liver (Li), brain (Br), stomach (St), spleen (Sp) and muscle (Mu). Molecular masses (kDa) are indicated on the left. ACTIN was used as an internal control.
NLRP9B expression was restricted to oocytes and declined with oocyte aging in the ovary
NLRP9B protein was detected in oocytes by immunohistochemistry (Fig. 2A). Furthermore, its expression declined with oocyte aging (Fig. 2B).
Fig. 2.
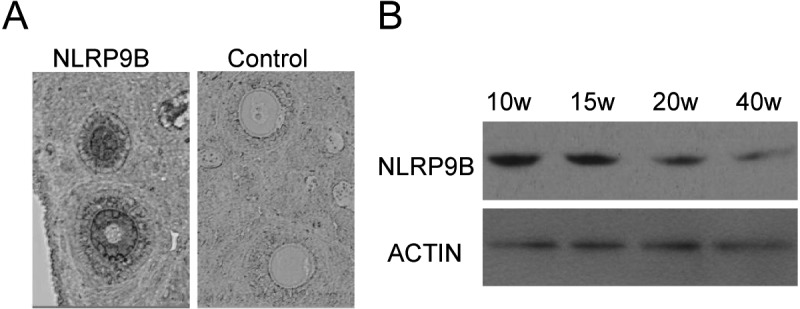
Expression of NLRP9B proteins in the ovary and GV-stage oocytes. (A) Immunohistochemistry of ovarian sections using the anti-NLRP9B antibody. The original magnification was × 100. (B) Detection of NLRP9B proteins in GV-stage oocytes from mouse ovaries at 10, 15, 20 and 40 weeks of age using western blotting. ACTIN was used as an internal control.
Expression and localization of NLRP9B in preimplantation embryos
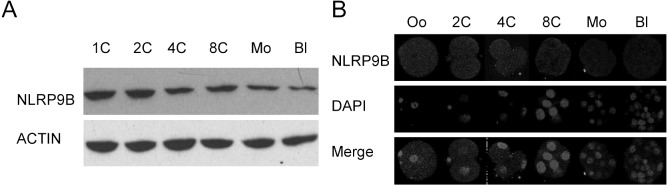
Western blotting confirmed that the NLRP9B protein was detected in all preimplantation embryos, although its expression declined with further development (Fig. 3A). Confocal microscopy demonstrated a mainly cytoplasmic localization of the NLRP9B protein in mature oocytes and preimplantation embryos (Fig. 3B).
Fig. 3.
Expression and localization of NLRP9B proteins in preimplantation embryos. (A) Western blotting of lysates isolated from preimplantation embryos. Oo, mature oocytes; 1C, 1-cell stage; 2C, 2-cell stage; 4C, 4-cell stage; 8C, 8-cell stage; Mo, morula; Bl, blastocyst. ACTIN was used as an internal control. (B) Confocal microscopic images of oocytes and preimplantation embryos. Each sample was counterstained with DAPI to visualize DNA (blue). The original magnification was × 200.
Effect of Nlrp9b knockdown on oocyte maturation
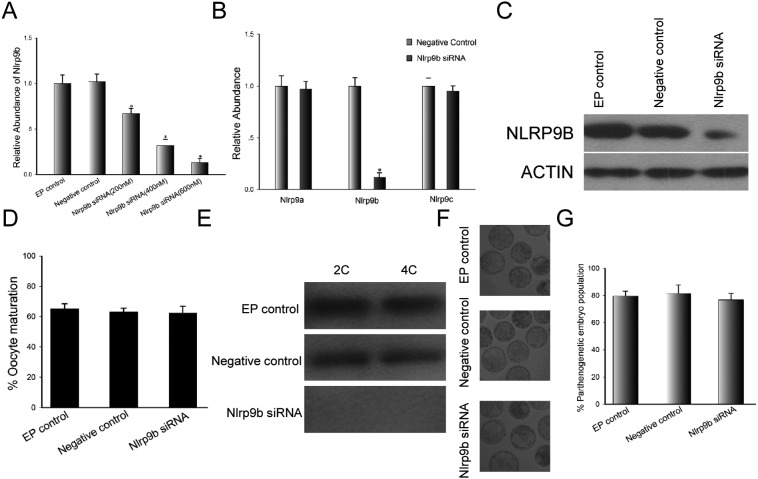
As shown in Fig. 4A, the expression of Nlrp9b was downregulated in a dose-dependent manner, and a concentration of 600 nM siRNA was suitable in terms of efficiency. The Nlrp9b knockdown effect was specific, because the expressions of Nlrp9a and Nlrp9c were not affected by the delivery of Nlrp9b siRNA (Fig. 4B). Furthermore, NLRP9B protein levels declined sharply in mature oocytes 24 h after electroporation with Nlrp9b siRNA compared with control groups (Fig. 4C). However, the decline of Nlrp9b transcripts and protein in oocytes did not affect oocyte maturation (Fig. 4D). Mature oocytes obtained from oocytes electroporated at the GV stage were subjected to parthenogenetic activation and then cultured in KSOMaa-BSA medium. The 2-cell and 4-cell stage parthenogenetic embryos obtained from mature knockdown oocytes were collected and subjected to western blot analysis. We found that NLRP9B protein was not detected in the knockdown group, while the protein was detected in control groups (Fig. 4E). The embryo development stages and morphological appearances of the embryos after they were cultured for 3.5 days are presented in Fig. 4F. Parthenogenetic embryos obtained from the control and knockdown groups reached the blastocyst stage, and there was no significant difference between the three groups with regard to the rate of blastocyst formation (Fig. 4G).
Fig. 4.
GV-stage oocyte maturation after electroporation with Nlrp9b siRNA. (A) Relative abundance of Nlrp9b transcripts following electroporation of GV oocytes with custom siRNA at different concentrations (200 nM, 400 nM and 600 nM) and culture for 48 h. The data were normalized to the electroporation control (EP control). Statistical comparisons were made using ANOVA and LSD tests (* P < 0.05). (B) Nlrp9a, Nlrp9b and Nlrp9c gene expression by qRT-PCR in mouse oocytes at 24 h after electroporation with Nlrp9b siRNA (600 nM). Results were normalized to the negative control (600 nM). * P < 0.05. (C) Western blot analysis of mouse oocytes derived from the EP control, negative control (600 nM) and Nlrp9b siRNA group (600 nM) after 24 h culture. ACTIN was used as an internal control. (D) Oocyte maturation rate following electroporation of GV-stage oocytes in the EP control, negative control (600 nM) and Nlrp9b siRNA group (600 nM). (E) Western blot analysis of mouse parthenogenetic embryos derived from the EP control, negative control (600 nM) and Nlrp9b siRNA group (600 nM). 2C, 2-cell stage; 4C, 4-cell stage. (F) Morphological appearance of parthenogenetic embryos obtained from the control and Nlrp9b siRNA groups after culture for 3.5 days. The original magnification was × 100. (G) Percentage of parthenogenetic embryos derived from the control and Nlrp9b siRNA groups after culture for 3.5 days.
Nlrp9b is dispensable for early embryonic development
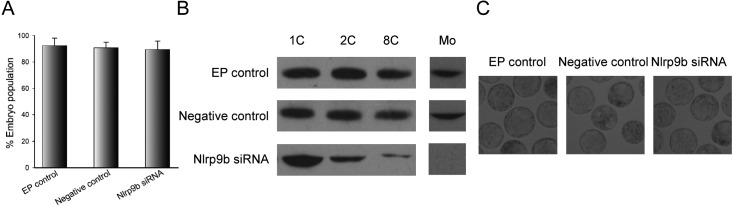
To further test the function of Nlrp9b in preimplantation development, the zygotes were electroporated with Nlrp9b siRNA and cultured in KSOMaa-BSA medium for 3.5 days. As shown in Fig. 5A and 5B, the rates of blastocyst formation showed no obvious differences between the two control groups and the Nlrp9b knockdown group, although the introduction of Nlrp9b siRNA led to a notable reduction in the NLRP9B protein level. Thus, embryos obtained from all three groups reached the blastocyst stage successfully (Fig. 5C).
Fig. 5.
Development of zygotes after electroporation with Nlrp9b siRNA. (A) The blastocyst formation rate of zygotes obtained from the control and Nlrp9b siRNA groups after culture for 3.5 days. (B) Western blotting of mouse embryos at 4 h (1 cell), 28 h (2 cells), 52 h (8 cells) and 64 h (morula) after electroporation. (C) Morphology of Nlrp9b knockdown embryos after culture for 3.5 days. The original magnification was × 100.
Discussion
Molecular evidence has shown that maternal factors are important for oogenesis and the development of preimplantation embryo in mammals [19,20,21,22,23,24,25,26]. Some of the NLRP proteins transcribed and translated by the Nlrp gene family are maternal factors such as NLRP9A, NLRP9B and NLRP9C in the mouse. Surprisingly, female mice obtained from a Nlrp9a and Nlrp9c simultaneous knockout model could give birth to offspring [17]. However, the developmental potential of Nlrp9b-deficient GV-stage oocytes and zygotes is uncertain.
Both Nlrp9b transcripts and proteins were expressed in ovaries and the small intestine, unlike Nlrp2, which is detected only in mouse ovaries [6]. This pattern differs from NLRP9, which is expressed in bovine ovaries as well as testes [27]. We found here that the NLRP9B protein was restricted to oocytes and declined with oocyte aging in the ovary, suggesting that this protein might play an essential role in oogenesis. The expression pattern of Nlrp9b in mouse ovaries was similar to that reported for Nlrp4a, Nlrp4f, Nlrp4g and Nlrp14 [9, 13].
After ovulation and fertilization, the NLRP9B protein was present throughout early embryogenesis, while the protein level diminished gradually with embryo development. This expression pattern was similar to those of NLRP2, NLRP4G and NLRP5 during preimplantation embryo development [6, 8, 16]. Confocal microscopy revealed that the NLRP9B protein was predominantly localized in the cytoplasm of blastomeres. This distribution of NLRP9B protein is similar to those of NLRP2 and NLRP4G [6, 16] and differs from that of NLRP5, which is mainly located in the subcortical regions of oocytes and preimplantation embryos [3, 4].
The expression and accumulation of Nlrp9b transcripts and proteins in immature oocytes might reflect a physiological role in the process of oocyte maturation. To test this hypothesis, custom siRNAs targeting Nlrp9b transcripts were delivered into GV-stage oocytes. However, these oocytes progressed through the metaphase of meiosis I and emitted the first polar body, suggesting that the NLRP9B protein is dispensable for oocyte maturation. The function of Nlrp9b is unlike the known maternal-effect genes Formin-2 and basonuclin, which are required for oocyte maturation [20, 21]. Of course, there is a possibility that a tiny amount of NLRP9B protein was present in knockdown oocytes and enabled the GV-stage oocytes to emit the first polar body. The Nlrp9b-deficient embryos obtained from Nlrp9b knockdown oocytes or zygotes underwent normal development in the terms of the rate of blastocyst formation, implying that this maternal factor is not required for early embryo development in the mouse. The function of Nlrp9b differs from those of Nlrp2, Nlrp5 and Nlrp14, which are essential for early embryogenesis, suggesting that different Nlrp genes have different functions even though they are mainly expressed in oocytes and early embryos. Moreover, Nlrp9b might play a key role in intestinal development or intestinal mucosal immunity, as its transcripts and proteins were also detected in the small intestine. Knockout models for this gene or other targeted inhibition experiments should be carried out to uncover the function of Nlrp9b in intestinal development or intestinal mucosal immunity.
In summary, the Nlrp9b gene was expressed in ovaries and the small intestine in the mouse. In the ovary, the NLRP9B protein was restricted to oocytes and declined with oocyte aging. After ovulation and fertilization, NLRP9B protein was present throughout early embryogenesis and located in the cytoplasm of blastomeres. Furthermore, targeted gene knockdown experiments indicated that the NLRP9B protein was not required for oocyte maturation or preimplantation embryo development in the mouse, implying that Nlrp9b might be an immunization-related gene involved in intestinal mucosal immunity in mice.
Acknowledgments
The authors thank Prof Jilong Chen and Dr Song Wang for their generous technical assistance. This work was supported by FAFU Program for Distinguished Young Scholars (grant number XJQ201509), the Foundation of Fujian Educational Committee (grant number JK2014013), Fujian Provincial Natural Science Foundation (grant number 2014J01071) and the National Natural Science Foundation of China (grant number 31402079).
References
- 1.Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol 2009; 9: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 2000; 26: 267–268. [DOI] [PubMed] [Google Scholar]
- 3.Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development 2008; 135: 259–269. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 2008; 15: 416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X. Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta). Hum Reprod 2009; 24: 415–424. [DOI] [PubMed] [Google Scholar]
- 6.Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, Wang Y, Liu J, Zhang B, Quan F, Guo Z, Zhang Y. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS ONE 2012; 7: e30344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong ZB, Bondy CA, Zhou J, Nelson LM. A human homologue of mouse Mater, a maternal effect gene essential for early embryonic development. Hum Reprod 2002; 17: 903–911. [DOI] [PubMed] [Google Scholar]
- 8.Tong ZB, Gold L, De Pol A, Vanevski K, Dorward H, Sena P, Palumbo C, Bondy CA, Nelson LM. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology 2004; 145: 1427–1434. [DOI] [PubMed] [Google Scholar]
- 9.Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet 2004; 13: 2263–2278. [DOI] [PubMed] [Google Scholar]
- 10.Dadé S, Callebaut I, Paillisson A, Bontoux M, Dalbiès-Tran R, Monget P. In silico identification and structural features of six new genes similar to MATER specifically expressed in the oocyte. Biochem Biophys Res Commun 2004; 324: 547–553. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P, Dixon M, Zucchelli M, Hambiliki F, Levkov L, Hovatta O, Kere J. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLoS ONE 2008; 3: e2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDaniel P, Wu X. Identification of oocyte-selective NLRP genes in rhesus macaque monkeys (Macaca mulatta). Mol Reprod Dev 2009; 76: 151–159. [DOI] [PubMed] [Google Scholar]
- 13.Peng H, Zhang W, Xiao T, Zhang Y. Nlrp4g is an oocyte-specific gene but is not required for oocyte maturation in the mouse. Reprod Fertil Dev 2014; 26: 758–768. [DOI] [PubMed] [Google Scholar]
- 14.Peng H, Zhuang Y, Wu X, Li H, Hong Z, Zhang X, Lin X, Zhang W. Expression analysis of Nlrp4a-Nlrp4f during mouse development. Journal of Animal and Veterinary Advances 2013; 12: 754–759. [Google Scholar]
- 15.Peng H, Zhang W, Xiao T, Zhang Y. Expression patterns of Nlrp9a, Nlrp9b and Nlrp9c during mouse development. Biologia 2014; 69: 107–112. [Google Scholar]
- 16.Peng H, Lin X, Li W, Zhang W. Expression and localization of Nlrp4g in mouse preimplantation embryo. Zygote 2014; 11: 1–6. [DOI] [PubMed] [Google Scholar]
- 17.Wei Y, Li L, Zhou X, Zhang QY, Dunbar A, Liu F, Kluetzman K, Yang W, Ding X. Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model. Drug Metab Dispos 2013; 41: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 19.Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature 2000; 407: 693–694. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Zeng F, Schultz RM, Tseng H. Basonuclin: a novel mammalian maternal-effect gene. Development 2006; 133: 2053–2062. [DOI] [PubMed] [Google Scholar]
- 21.Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, Maas R, Leder P. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol 2002; 4: 921–928. [DOI] [PubMed] [Google Scholar]
- 22.Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 2003; 13: 2110–2117. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet 2003; 33: 187–191. [DOI] [PubMed] [Google Scholar]
- 24.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 2003; 300: 633–636. [DOI] [PubMed] [Google Scholar]
- 25.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 2006; 20: 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci USA 2009; 106: 7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalbiès-Tran R, Papillier P, Pennetier S, Uzbekova S, Monget P. Bovine mater-like NALP9 is an oocyte marker gene. Mol Reprod Dev 2005; 71: 414–421. [DOI] [PubMed] [Google Scholar]