Abstract
Enterovirus A89 (EV-A89) is a novel member of the EV-A species. To date, only one full-length genome sequence (the prototype strain) has been published. Here, we report the molecular identification and genomic characterization of a Chinese EV-A89 strain, KSYPH-TRMH22F/XJ/CHN/2011, isolated in 2011 from a contact of an acute flaccid paralysis (AFP) patient during AFP case surveillance in Xinjiang China. This was the first report of EV-A89 in China. The VP1 coding sequence of this strain demonstrated 93.2% nucleotide and 99.3% amino acid identity with the EV-A89 prototype strain. In the P2 and P3 regions, the Chinese EV-A89 strain demonstrated markedly higher identity than the prototype strains of EV-A76, EV-A90, and EV-A91, indicating that one or more recombination events between EV-A89 and these EV-A types might have occurred. Long-term evolution of these EV types originated from the same ancestor provides the spatial and temporal circumstances for recombination to occur. An antibody sero-prevalence survey against EV-A89 in two Xinjiang prefectures demonstrated low positive rates and low titres of EV-A89 neutralization antibody, suggesting limited range of transmission and exposure to the population. This study provides a solid foundation for further studies on the biological and pathogenic properties of EV-A89.
Human enterovirus (EV) infections are usually asymptomatic or bring about only mild disease, such as the common cold or minor undifferentiated febrile illnesses. However, EVs are associated with outbreaks of more serious disease such as acute flaccid paralysis (AFP), acute haemorrhagic conjunctivitis, aseptic meningitis, encephalitis, myocarditis, and hand, foot, and mouth disease (HFMD)1,2,3,4, which result in considerable morbidity and occasionally in mortality.
EVs belong to the picornaviridae family and fall within the new order Picornavirales, which represents small non-enveloped RNA viruses with a single stranded positive-sense genome of approximately 7500 nucleotides5. The EV genome consists of a single open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs). The ORF is translated into a single, large polyprotein of 2200 amino acids (aa), which is subsequently cleaved by viral proteases into one capsid protein region (P1) and two non-structural regions (P2 and P3). The P1 region encodes four viral capsid proteins: viral protein 1–4 (VP1–VP4), and the P2 and P3 regions encode seven non-structural proteins 2A–2C and 3A–3D, respectively6. The 5′-UTR is about 740 nucleotides long and has an internal ribosome entry site (IRES) that is indispensable for translation initiation7,8. The approximately 100 nucleotide 3′-UTR, located between the ORF and the poly (A) stretch, forms highly conserved secondary and tertiary structures that are involved in RNA replication9.
Currently, more than 100 human EV serotypes have been described. They are currently classified into four species, EV-A, EV-B, EV-C, and EV-D, according to their genomic characteristics5,10,11. The classification of human EVs is based on sequence divergence in the VP1 coding region, which has been shown to completely correlate with the traditional classification made using antigenic properties12. Human EVs can be identified by comparison of the entire or partial VP1 sequence of an unknown EV to a database of prototype strain sequences. The unknown EV should be classified into the same serotype if they have more than 75% nucleotide identity (85% amino acid identity) in the VP1 coding region, or into different serotypes if they have less than 70% nucleotide identity (85% amino acid identity) in this region12,13. However, some isolates may occasionally demonstrate nucleotide identity between 70–75% in the VP1 coding region, which has been considered a “grey zone” of molecular typing of human EVs. Thus, the use of additional information such as complete P1 sequence identity for serotype identification may be beneficial for identifying the isolates14. The application of molecular typing methods to serologically “untypeable” EV strains has led to the discovery of a large number of new EV types within the four EV species15,16,17,18.
To date, species EV-A consists of 21 serotypes including Coxsackievirus 2–8, 10, 12, 14, 16, and EV-A71, as well as the new EV types EV-A76, EV-A89–A92, EV-A114, and EV-A119–A12119,20,21. EV-A89 is a newly identified serotype within the EV-A species. The prototype strain of EV-A89 (strain BAN00-10359/BAN/2000) was isolated from stool specimens of an AFP patient in Bangladesh in 200019. Subsequently, several other EV-A89 strains were isolated from AFP patients, acute gastroenteritis patients, or healthy individuals during disease surveillance activities (such as AFP case surveillance) in Bangladesh19,22, India23,24,25,26, and Egypt27. Currently, only one full-length genome sequence (the EV-A89 prototype strain) is available in the GenBank database. Besides the prototype strain, six entire VP1 sequences and several partial VP1 sequences of EV-A89 strains are available in the GenBank database. However, no EV-A89 sequences have been reported in China.
In this study, we report the molecular identification and genomic characterization of an EV-A89 strain (strain KSYPH-TRMH22F/XJ/CHN/2011, hereafter referred as strain KSYPH-TRMH22F) isolated in 2011 from a contact of an AFP patient during AFP case surveillance in Xinjiang Uygur Autonomous region of China. This was the first report of EV-A89 in China. Its genetic characteristics and phylogenetic relationship to other EV strains were also investigated.
Results
Isolation and molecular typing of the Chinese EV-A89 strain
The virus grew only in the RD cell line. Cell cultures were harvested after a complete EV-like cytopathic effect (CPE) was observed. We determined the partial VP1 coding region sequence of the KSYPH-TRMH22F strain, which was amplified by primer pairs 486 and 48828. Molecular typing of this strain based on the partial VP1 sequence was then performed using an online EV genotyping tool29. The results indicated that the KSYPH-TRMH22F strain belongs to the EV-A89 serotype.
Complete genome sequence of the Chinese EV-A89 strain
The complete genome of the Chinese EV-A89 strain was determined. It was 7429 nucleotides in length, including a 5′-UTR of 747 nucleotides, a single ORF of 6588 nucleotides encoding a single polyprotein of 2195 amino acids, and a 3′-UTR of 94 nucleotides preceding the poly (A) stretch. Compared to the EV-A89 prototype strain (BAN00-10359/BAN/2000), no nucleotide insertions or deletions were observed in the whole genome. The overall base composition of the KSYPH-TRMH22F strain was 29.56% A, 22.43% C, 22.05% G, and 25.97% U.
A comprehensive comparison of the nucleotide sequence and deduced amino acid sequence of the Chinese EV-A89 strain with the EV-A89 prototype strain and other prototype strains belonging to EV-A is shown in Table 1. Overall, the complete genome sequence identity and the deduced amino acid sequence identity between the Chinese EV-A89 strain and the EV-A89 prototype strain were 91.7% and 98.3%, respectively. The VP1 coding sequence of this strain showed 93.2% nucleotide and 99.3% amino acid identity with the EV-A89 prototype strain, and 90.0–92.6% nucleotide and 98.6–98.9% amino acid identity with the other six reported EV-A89 strains available in the GenBank database. However, it had 55.7–68.7% nucleotide and 53.7–74.6% amino acid identity with the prototype strains of other EV-A serotypes, confirming that it belongs to the EV-A89 serotype, based on the molecular typing criteria30.
Table 1. The nucleotide sequence and deduced amino acid sequence identities of the Chinese EV-A89 strain KSYPH-TRMH22F with the EV-A89 prototype strain (BAN00-10359/BAN/2000) and other prototype strains belongs to EV-A types.
| Region | % nucleotide identity (%amino acid identity) |
|||
|---|---|---|---|---|
| Identity with BAN00-10359 (%) |
Identity with other EV-A(%) |
|||
| Nucleotide | Amino acid | Nucleotide | Amino acid | |
| 5′-UTR | 99.6 | 72.1–93.7 | ||
| VP4 | 93.7 | 100.0 | 61.8–84.0 | 65.2–100.0 |
| VP2 | 93.9 | 99.2 | 65.8–71.8 | 73.7–82.5 |
| VP3 | 93.8 | 99.5 | 66.3–71.1 | 71.7–84.3 |
| VP1 | 93.2 | 99.3 | 55.7–68.7 | 53.7–74.6 |
| 2A | 93.3 | 98.6 | 66.0–72.0 | 70.0–81.3 |
| 2B | 87.5 | 98.9 | 62.2–85.1 | 73.7–97.9 |
| 2C | 88.6 | 99.3 | 73.8–89.9 | 83.8–99.0 |
| 3A | 90.6 | 100.0 | 68.6–85.6 | 63.9–100.0 |
| 3B | 90.0 | 95.4 | 60.6–84.8 | 77.2–95.4 |
| 3C | 90.1 | 98.9 | 71.9–88.1 | 80.8–99.4 |
| 3D | 88.8 | 98.2 | 72.2–92.4 | 83.1–98.7 |
| 3′-UTR | 91.4 | 30.3–96.8 | ||
The 3′-UTR of both the Chinese EV-A89 strain and the EV-A89 prototype strain contain 94 nucleotides preceding the poly (A) stretch, which is the same length as EV-A76, EV-A90, and EV-A91. Further comparison based on this region revealed that the Chinese EV-A89 strain had 91.4% nucleotide identity with the EV-A89 prototype strain. Compared to the 3′-UTR sequences of the prototype strains of the EV-A76, EV-A90, and EV-A91, the Chinese strain had 88.2%, 94.6%, and 96.8% nucleotide identity, respectively. The Chinese strain demonstrates an obvious difference from the other EV-A types, which had only 30.3–59.4% nucleotide identity in the 3′-UTR.
Phylogenetic analysis of the Chinese EV-A89 strain and other EV-A prototype strains
Entire or partial VP1 nucleotide sequence analysis can be used to investigate the phylogenetic relationship among human EVs, however entire VP1 sequence analysis can provide more information12. Therefore, phylogenetic trees were generated from the 888nt (nucleotide 2452–3339) entire VP1 coding region of the Chinese EV-A89 strain and six other EV-A89 strains available in the GenBank database (Fig. 1). The Chinese EV-A89 strain segregates together with Bangladesh EV-A89 strains (including EV-A89 prototype strain) and India EV-A89 strains, and is monophyletic in comparison, with a longer genetic distance. This suggests a long evolution time and a high degree of genetic divergence among these EV-A89 strains.
Figure 1. Phylogenetic relationships based on partial VP1 genomic sequences of enterovirus A89 (EV-A89).
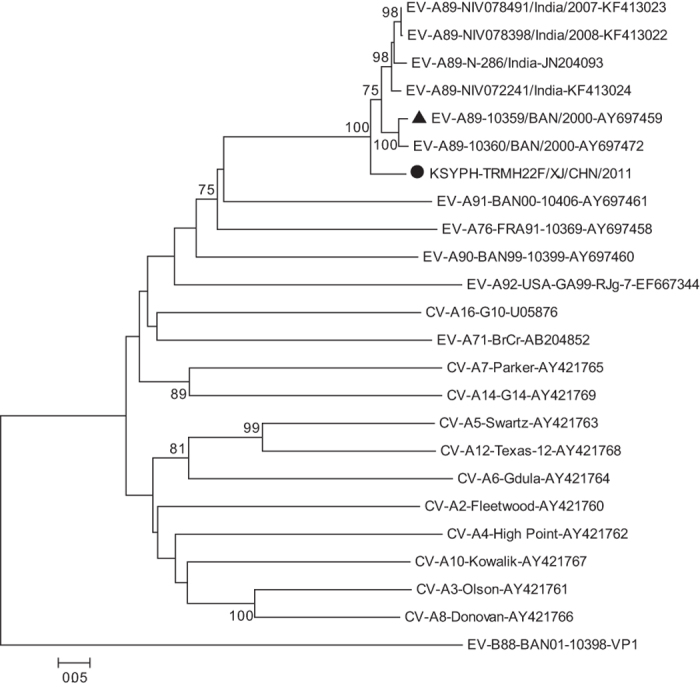
The Chinese EV-A89 strain isolated in this study (solid circles), other EV-A89 strains (available in the GenBank database), and other EV-A prototype strains were analysed based on the 888nt (nucleotide 2452–3339) entire VP1 coding region sequence. The triangle represents the EV-A89 prototype strain. The EV-B88 prototype strain served as an out-group.
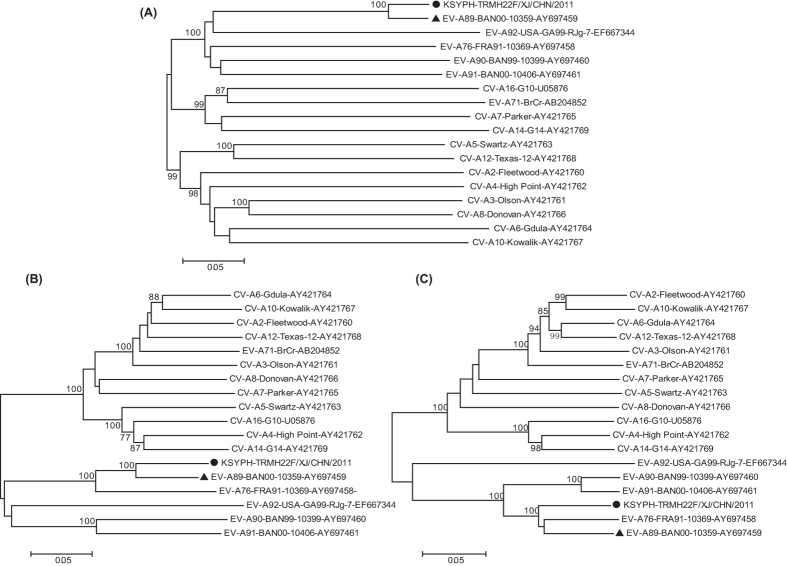
Moreover, we constructed phylogenetic trees based on the entire P1, P2, and P3 regions of the genome. In the P1 capsid coding regions, the Chinese EV-A89 strain clustered together with the EV-A89 prototype strain, with a bootstrap value of 100 (Fig. 2A), thus confirming the preliminary molecular typing results. The phylogenetic trees also showed considerable difference in the non-capsid coding regions. In the P2 and P3 coding regions, the Chinese EV-A89 strain shared a distinct high identity with the EV-A76, EV-A90, and EV-A91 prototype strains (Fig. 2B,C). The phylogenetic analysis indicated that one or more potential recombination events between EV-A89 and the three EV-A serotypes might have occurred.
Figure 2. Phylogenetic relationships based on the P1, P2, and P3 genome regions of enterovirus A (EV-A).
The Chinese EV-A89 strain (solid circles) and 17 EV-A prototype strains were analysed by nucleotide sequence alignment using the neighbour-joining algorithms implemented in the MEGA 5.0 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The solid triangle indicates the EV-A89 prototype strain. The scale bars represent the genetic distance. All panels have the same scale. (A) P1 coding sequences, (B) P2 coding sequences, and (C) P3 coding sequences.
Recombinant structure of the Chinese EV-A89 strain
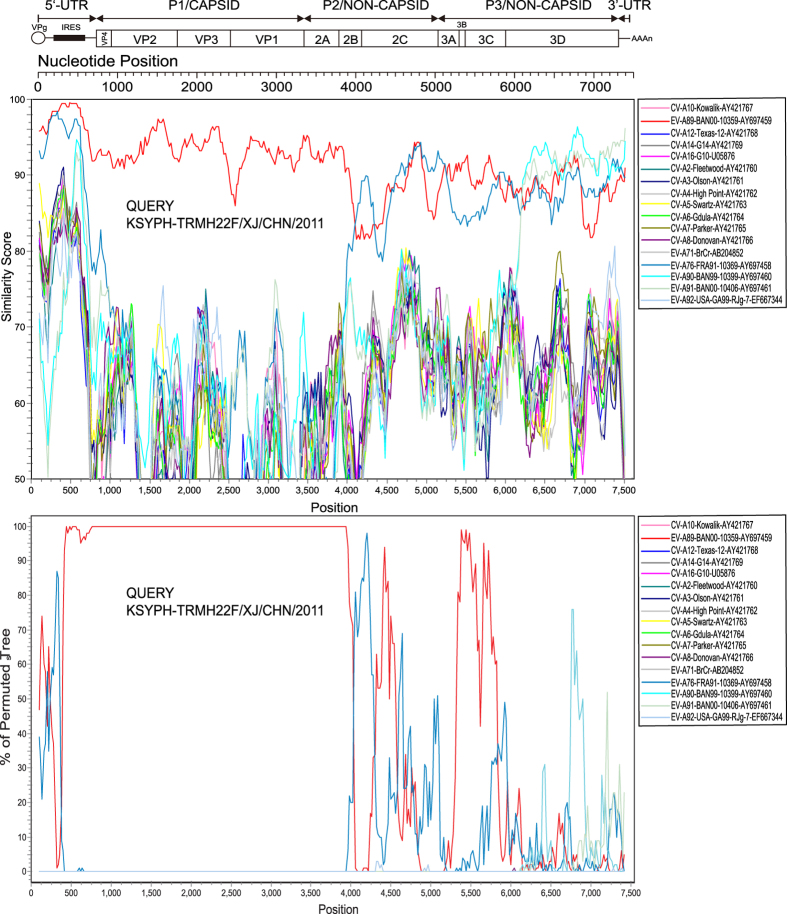
Similarity plot and bootscanning analyses were performed to confirm the existence of recombination events between the Chinese EV-A89 strain and the other EV-A prototype strains (Fig. 3). The Chinese EV-A89 strain was used as a query sequence and was compared to the EV-A89 prototype strain and other EV-A prototype strains. In the P1 and partial P2 (2A and 2B) coding regions, the Chinese EV-A89 strain demonstrated more than 80% identity with the EV-A89 prototype strain, reflecting a close relationship. However, in the 2C and partial P3 regions, the sequence similarities with the EV-A76, EV-A90, and EV-A91 prototype strains were relatively higher. Bootscanning analysis revealed the existence of recombination events between the Chinese EV-A89 strain and the EV-A76, EV-A90, and EV-A91 strains.
Figure 3. Recombination analyses of complete enterovirus A (EV-A) genomes.
(a) Similarity plot and (b) bootscanning analysis. A sliding window of 200 nucleotides was used, moving in 20-nucleotide steps. The Chinese EV-A89 strain KSYPH-TRMH22F/XJ/CHN/2011 was used as a query sequence.
Sero-prevalence of EV-A89 in Xinjiang of China
A total of 60 serum samples were collected from infants and children between 0 months and 4 years of age in the place where the virus was isolated in Xinjiang. Among these 60 serum samples surveyed, 30 were collected from the Kashgar region and the other 30 were collected from the Hotan region. A micro-neutralization assay against EV-A89 showed that 24 serum samples were positive for EV-A89 (>1:8) with a total positive rate of 40.0%. The geometric mean titre (GMT) was 1:43.97 among the positive sera samples. The composition ratios for the EV-A89 neutralization antibody titres of <1:8, 1:8–1:64, and >1:64 were 60.0%, 28.3%, and 11.7%, respectively (Table 2). However, compared to sero-epidemiology studies of other EVs in China, the positive rate of the EV-A89 neutralization antibody and GMT were relatively lower than that of other EVs such as EV-A71 and CV-A16 in the same age group (0–4 years old)31.
Table 2. The composition ratios for the EV-A89 neutralization antibody titers.
| Titers | Kashgar prefecture |
Hotan prefecture |
Total(%) | ||
|---|---|---|---|---|---|
| Number of samples | Ratio(%) | Number of samples | Ratio(%) | ||
| <1:8 | 14 | 46.7 | 22 | 73.3 | 36(60.0) |
| 1:8–1:64 | 9 | 30.0 | 8 | 26.7 | 17(28.3) |
| >1:64 | 7 | 23.3 | 0 | 0 | 7(11.7) |
| Total | 30 | 30 | 60 | ||
Although serum samples from the Kashgar and Hotan regions showed a lower positive rate of EV-A89 neutralization antibody and GMT, there were some differences between the two regions. In the Kashgar region, the positive rate of the EV-A89 neutralization antibody and GMTs were 53.3% and 1:66.8, respectively, while in the Hotan region, they were 26.7% and 1:19, respectively. The positive rate of the EV-A89 neutralization antibody and GMTs in the Kashgar region (in the place where the virus was isolated) were significantly higher than those in the Hotan prefecture (seroprevalence rate: p = 0.035 < 0.05, GMTs: p < 0.0001).
Discussion
In China, a high-sensitivity AFP case surveillance system to detect the presence of wild polioviruses in a population was established in 1994 as part of the Global Polio Eradication Initiative (PEI), and represents the gold standard surveillance system for PEI32. Although there is no special surveillance system to detect new EV types, a certain number of new EV types, known as non-polio EVs, have been isolated during the AFP case surveillance for polioviruses. These new EV types, such as EV-A76, EV-B81, EV-B85, EV-C96 and so on, which have been identified and analyzed using the molecular typing method, can provide valuable information on the molecular epidemiology of local novel EVs17,33,34,35. Here, we report the complete genome sequence of a novel EV type, EV-A89, which was identified for the first time in China, and was detected during AFP case surveillance.
The Chinese EV-A89 strain described in this study was isolated in 2011 in the Yopurga prefecture in the Kashgar region, which is located in the southern part of Xinjiang. Xinjiang is a frontier in China and is located in the center of Eurasia, adjacent to some Middle East and South Asian countries. All the EV-A89 strains deposited in the GenBank database to date are distributed in Middle East and South Asian countries, and almost all the EV-A89 strains were isolated from patients with AFP except for a strain from Egypt27. Therefore, we speculate that EV-A89 infection may correlate with AFP, although there is not enough evidence to demonstrate this relationship since only a few EV-A89 strains have been discovered worldwide. Therefore, more information and data are needed to reveal the biological and pathological properties of EV-A89.
The phylogenetic trees based on the P2 and P3 coding region sequences showed that the Chinese EV-A89 strain is closely related to EV-A76, EV-A90, and EV-A91, suggesting the existence of recombination events in these regions. These four EVs (EV-A76, EV-A89, EV-A90 and EV-A91) may have evolved from the same ancestor due to their similar genomic structures and high nucleotide and amino acid similarities. Particularly, there is high nucleotide identity in the P2 region, P3 region, and 3′-UTR between EV-A76 and EV-A89, which implies an evolutionary relationship between these two novel EV types. A recombination analysis of the Chinese EV-A89 strain and other EV-A prototype strains in all regions of the genome demonstrated that, regardless of regions (P1, P2 or P3), the Chinese EV-A89 strain is clustered to a distinct monophyletic subgroup with the EV-A76, EV-A90, and EV-A91 prototype strains, and is distinct from the other EV-A members, indicating that the recombination barrier may exist within EV-A species19. Based on the findings of the similarity comparison and recombination analysis, it is reasonable to conclude that long-term evolution of these viruses originated from the same ancestor, which provides the spatial and temporal circumstances for recombination to occur.
With the usage of EV molecular typing method, more and more new EVs types were identified, such as EV-B7336, EV-A7633, EV-B8534, and EV-B10637, lots of new EV types are proved to be novel recombination variant, indicated that recombination is a frequent event in EV evolution that usually occurs between viruses of the same species and is correlated with the appearance of new EV lineages or EV types. In this respect, the Chinese EV-A89 strain is no exception.
Previous considerable researches indicated the recombination and mutation among enteroviruses have been generally recognized as the frequent mechanisms and features in the EV evolution, based on a large number of epidemiological investigations38,39. Due to the absence of proof reading mechanism, the mispairing rate of the viral polymerase of EVs is unusual high, under these circumstances, recombination plays a crucial and high effective role in viral evolution by repairing deleterious mutations in EV genomes, thus frequent recombination contributes to their genetic diversity and provides the ability for large evolutionary transitions, by producing in a single step significantly divergent genomes, better response and adapt to new environmental challenges40,41. For instance, EV-A71 is the most heavily studied EV-A type due to numerous large HFMD outbreaks with high morbidity and mortality caused by EV-A71 have occurred in Asian countries and regions in recent years, a lot of epidemiological and etiological studies indicated that recombination is a frequent phenomenon and is the main force for the evolution of EV-A7142,43,44,45.
Recombination with closely related non–poliovirus EV-C species frequently occurs during wild poliovirus circulation and is an indication of person-to-person transmission, and appears to facilitate the emergence of circulating vaccine-derived polioviruses (cVDPV) by replacement of attenuating sites in a single event. However, recombination with EV-C may not be obligatory for cVDPV emergence especially type 1 VDPV, due to type 1 cVDPV circulating locally in China46 and the United States47 had non-recombinant genomes. Although the possible role of recombination in the phenotypic drift of vaccine strains to a wild strain is not very clear, we propose that recombination with species EV-C may not be essential and is not correlated with the phenotypic drift to higher neurovirulence and higher transmissibility, but it may be an epidemiology indicator of the duration of viral circulation in the human community.
EV-A89 is a newly identified serotype within the EV-A species. Currently, only one full-length genome sequence (the EV-A89 prototype strain) and 12 other EV-A89 sequences are available in the GenBank database, and this is the first and the only report of EV-A89 in China. Due to the limited number of EV-A89 isolates in China and the world, the EV-A89 epidemiological data is very short, so the biological and pathogenic properties of EV-A89 are currently difficult to study in detail. However, we conducted a small range of sero-epidemiology studies, and our research team is currently using reverse genetic methods to explore the advantages of virus recombination found in the Chinese EV-A89 strain.
To investigate the sero-epidemiology of EV-A89 in Xinjiang China, a sero-epidemiology survey was conducted in the places where the virus was isolated. The results showed a prominent difference in the positive rates of neutralizing antibody and GMTs of EV-A89 between the Kashgar and Hotan regions in Xinjiang, indicating a geographical difference in EV-A89 infection. The seropositive rate of EV-A89 neutralization antibody and GMT were relatively higher in the Kashgar region than in the Hotan region (p < 0.05). This suggests that the EV-A89 strain, KSYPH-TRMH22F, isolated in 2011 in the Yopurga prefecture of the Kashgar region may have circulated in the Kashgar region more widely than in the Hotan region. However, the seropositive rate and GMT of EV-A89 neutralization antibody in the two regions were generally lower than other EVs prevalent in China, such as EV-A71 and CV-A1631, suggesting limited range of transmission and exposure of the novel EV type EV-A89 to the population. This is consistent with the low viral isolation rate of EV-A89 strains in China (only one EV-A89 strain has been isolated since 1988), and further research will be required to corroborate these conclusions.
In conclusion, this is the first report of the complete genome sequence of a recently described novel EV type, EV-A89, in China. The extremely rare isolation rate suggests that it has not been a prevalent EV serotype in China or even in the world until now. The Chinese EV-A89 strain is closely related to EV-A76, EV-A90, and EV-A91, suggesting the existence of recombination events in non-structure regions. Long-term evolution of EV-A76, EV-A89, EV-A90, and EV-A91 originated from the same ancestor, in order to provide the spatial and temporal circumstances for recombination to occur. Although the biological and pathogenic properties of EV-A89 are currently difficult to study in detail due to the limited number of EV-A89 isolates, we believe that this study provides a solid foundation for further studies on EV-A89.
Materials and Methods
Sample collection
This study did not involve human participants or experimentation. The only human material used was a stool sample collected from a close contact of an AFP patient at the instigation of the Ministry of Health P. R. of China for public health purposes. Written informed consent for the use of clinical samples was obtained from all patients involved in this study. This study was approved by the second session of the Ethics Review Committee of the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, and the methods were carried out in accordance with the approved guidelines.
The EV-A89 strain (strain KSYPH-TRMH22F/XJ/CHN/2011) was isolated from a stool sample of an asymptomatic contact residing in the Yopurga prefecture of the Kashgar region, in the southern part of the Xinjiang of China. The sample was collected in 2011 during the course of poliovirus surveillance activities in support of the global PEI.
Sixty healthy children ≤ 5 years of age were surveyed for a seroprevalence study of EV-A89 antibodies. Serum samples were collected randomly in 2013, with informed parental consent, by the Xinjiang Center for Disease Control and Prevention: 30 samples were collected in the Kashgar region in the place where the virus was isolated and 30 were collected in the Hotan prefecture (neighboring Kashgar). None of the participants demonstrated any signs of disease at the time of sample collection.
Viral isolation
The stool sample from the healthy contact was collected and processed according to the World Health Organization (WHO) laboratory manual48. The processed sample was used to inoculate three cell lines for viral isolation: a genetically engineered mouse cell line expressing the human poliovirus receptor (L20B cell), human rhabdomyosarcoma (RD), and human laryngeal epidermoid carcinoma (HEp-2). These cell lines were obtained from the WHO Global Poliovirus Specialized Laboratory USA, but were originally purchased from the American Type Culture Collection.
Molecular typing
The viral RNA was extracted from the viral isolates using a QIAamp Viral RNA Mini Kit (Qiagen, USA) according to the manufacturer’s instructions, and stored at −80 °C.Primer pairs 486 and 488 were used for amplifying the partial VP1 coding region28. RT-PCR (reverse transcription–polymerase chain reaction) was performed using the PrimeScript One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) as previously described17. Amplicons were purified using the QIAquick PCR purification kit (Qiagen, Germany) and sequenced using an ABI 3130 Genetic Analyser (Applied Biosystems, Hitachi, Japan). Every nucleotide position of each strand was sequenced at least once. The obtained VP1 sequences were compared with sequences available in the GenBank database for molecular typing using the EV Genotyping Tool29.
Whole genomic sequencing
The 5′ end sequence of the genome was obtained using a 5′ rapid amplification of cDNA ends (RACE) core set (Takara Biomedicals) according to the manufacturer’s instructions. The 3′ end sequence of the genome was obtained by amplification using an oligo-dT primer (primer 7500A)49 as the downstream primer. The rest of the genome was determined using the primer walking method. The primers used for PCR amplification and sequencing are listed in Table 3.
Table 3. PCR and sequencing primer.
| Primer | Nucleotide position (nt) | Primer sequence (5′-3′) | Orientation | Reference |
|---|---|---|---|---|
| 0001S48 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTAAAACAGCTCTGGGGTT | Forward | 49 | |
| EV89-874A | 854-874 | TAGTGGCCGAAGCTGCGTATG | Reverse | This study |
| EVP4 | 541-560 | CTACTTTGGGTGTCCGTGTT | Forward | 52 |
| 0L68-1 | 1178-1197 | GGTAAYTTCCACCACCANCC | Reverse | 52 |
| EV89-982S | 982-1000 | AGTGACAGAGTGGCGCAAC | Forward | This study |
| EV89-2470A | 2451-2470 | TGTCTTCCATGGGGTCACCT | Reverse | This study |
| E486 | 2297–2322 | TGGTAICARACIAAITWYGTIGTNCC | Forward | 28 |
| E488 | 3063–3038 | GTIGGRTAICCITCITARAACCAYTG | Reverse | 28 |
| EV89-3521A | 3502-3521 | CCGGTTGTGCACTGACATCT | Reverse | This study |
| EUG3a | 3002-3021 | TGGCAAACTTCCWCCAACCC | Forward | 53 |
| EUC2b | 4469-4488 | GGTTCAATACGGTGTTTGCT | Reverse | 53 |
| EV89-3827S | 3827-3846 | TTGGCACAG GAT TCA CAGAC | Forward | This study |
| EV89-4873A | 4854-4873 | CATCCAATCTTCCAGACTCC | Reverse | This study |
| EV89-4716S | 4716-4735 | TGTCTCCTTCACCTCTAAGT | Forward | This study |
| EV89-5897A | 5878-5897 | CTGCCATTCCCTCCAATGTG | Reverse | This study |
| EV89-5412S | 5412-5431 | CTTTGCCCTTTCCCTACTCA | Forward | This study |
| EV89-6256A | 6237-6256 | TTCCATAACAAGCATCCTCC | Reverse | This study |
| EV89-6001S | 6001-6020 | GGCCCAACCAAGACTAAACT | Forward | This study |
| EV89-6962A | 6943-6962 | ATAGGGAAGGGGTAACTAGC | Reverse | This study |
| EV89-6789S | 6789-6808 | CTGTGTAATGGGCGGAATGC | Forward | This study |
| 7500A | GGGGACCACTTTGTACAAGAAAGCTGGG(T)24 | Reverse | 49 |
Phylogenetic trees and bioinformatics analyses
The nucleotide and deduced amino acid sequences of the Chinese EV-A89 strain were compared with those of the prototype EV-A strains by pairwise alignment using the MEGA program (version 5.03)50. Phylogenetic trees were constructed by the neighbour-joining method, implemented in the MEGA program, using the Kimura 2-parameter model. Bootstrapping was performed with 1000 bootstrap replicates and bootstrap values greater than 80% were considered statistically significant for grouping. Similarity plots and bootscanning analyses were performed using the SimPlot 3.5.1 program. A 200-nucleotide window was moved in 20-nucleotide steps and bootscanning analyses were operated using the neighbour-joining method51.
Neutralization tests against EV-A89
In this study, neutralizing antibodies against EV-A89 were detected using the micro-neutralization assay using the human RD cell line as previously described, with some modifications31. Serum samples were inactivated at 56 °C for 30 min before use, and were serially diluted from 1:4 to 1:1024.Virus samples (50 μL) with a tissue culture infective dose (TCID50) of 100 were then mixed with the appropriate serum dilution (50 μL), added to a 96-well microplate, and incubated at 37 °C for 2 hours. RD cells (2 × 105 cells/ml) were then added, and the mixture was further incubated at 36 °C in a CO2 incubator for 7 days. Virus back titration was simultaneously performed to verify whether the micro-neutralization assay was effective (virus back titration values between 32 and 320 TCID50/well indicated that the test was effective). The highest dilution of serum that protected 50% of the cultures, based on the observation of EV-like CPE, was recorded. The serum sample was considered positive if the neutralization antibody titre was 1:8. The GMT was then calculated for positive samples.
Statistical analysis
Statistical analyses were carried out using the SPSS version 13.0 software (SPSS Inc., Chicago, IL, USA). A chi-square test was used to compare the sero-prevalence rates of EV-A89 between the Kashgar and Hotan prefectures, and a Mann-Whitney test was used to determine the significance of EV-A89 GMTs. Titres below 1:8 were assumed to be 1:4 for calculation. An error value of p < 0.05 indicated statistical significance.
Nucleotide sequence accession numbers
The complete genomic sequence of the EV-A89 strains (KSYPH-TRMH22F) described in this study was deposited in the GenBank database under the accession number KT277550.
Additional Information
How to cite this article: Fan, Q. et al. A Novel Recombinant Enterovirus Type EV-A89 with Low Epidemic Strength in Xinjiang, China. Sci. Rep. 5, 18558; doi: 10.1038/srep18558 (2015).
Acknowledgments
We would like to acknowledge the staff of the national polio eradication program in the Xinjiang Uygur Autonomous Region Center for Disease Control and Prevention for investigating AFP cases and collecting stool specimens for use in this study. The study was supported by the National Key Technology R&D Program of China (project nos. 2013ZX10004-101 and 2013ZX10004-202) and the National Natural Science Foundation of China (project nos. 81101303 and 81373049).
Footnotes
Author Contributions Y.Z. and W.X. conceived and designed the experiments. Q.F., L.H., Q.S., H.C., D.Y., H.S., H.T., D.W., Z.Z. and S.Z. performed the experiments. Q.F., L.H. and Y.Z. analysed the data. Q.F. and Y.Z. wrote the main manuscript and Q.F. prepared all the figures. All authors reviewed the manuscript.
References
- Zhang Y. et al. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J Clin Virol 44, 262–7 (2009). [DOI] [PubMed] [Google Scholar]
- Tang J. et al. Molecular epidemiology and recombination of human enteroviruses from AFP surveillance in Yunnan, China from 2006 to 2010. Sci Rep 4, 6058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D. et al. Outbreak of acute hemorrhagic conjunctivitis in Yunnan, People’s Republic of China, 2007. Virol J 7, 138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. & Xu W. B. Molecular epidemiology of enteroviruses associated with hand, foot, and mouth disease in the mainland of China. Biomed Environ Sci 26, 875–6 (2013). [DOI] [PubMed] [Google Scholar]
- Oberste M. S., Penaranda S., Maher K. & Pallansch M. A. Complete genome sequences of all members of the species Human enterovirus A. J Gen Virol 85, 1597–607 (2004). [DOI] [PubMed] [Google Scholar]
- Picornaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (Elsevier, San Diego, 2011).
- Andreev D. E. et al. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res 40, 5602–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Jang S. K., Paul A. V., Reuer Q. & Wimmer E. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature 356, 255–7 (1992). [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Maslova S. V., Sinyakov A. N. & Agol V. I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res 20, 1739–45 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B., Oberste M. S., Maher K. & Pallansch M. A. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J Virol 77, 8973–84 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K. & Pallansch M. A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J Virol 78, 855–67 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R. & Pallansch M. A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73, 1941–8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S. et al. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol 38, 1170–4 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. A. et al. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J Gen Virol 90, 1713–23 (2009). [DOI] [PubMed] [Google Scholar]
- Oberste M. S. et al. Molecular identification of 13 new enterovirus types, EV79-88, EV97, and EV100-101, members of the species Human Enterovirus B. Virus Res 128, 34–42 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Molecular typing and characterization of a new serotype of human enterovirus (EV-B111) identified in China. Virus Res 183, 75–80 (2014). [DOI] [PubMed] [Google Scholar]
- Hu L. et al. Phylogenetic evidence for multiple intertypic recombinations in enterovirus B81 strains isolated in Tibet, China. Sci Rep 4, 6035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M., Schnurr D., Maher K., al-Busaidy S. & Pallansch M. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J Gen Virol 82, 409–16 (2001). [DOI] [PubMed] [Google Scholar]
- Oberste M. S. et al. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species Human enterovirus A. J Gen Virol 86, 445–51 (2005). [DOI] [PubMed] [Google Scholar]
- Ayukekbong J. et al. Shift of Enterovirus species among children in Cameroon - Identification of a new enterovirus, EV-A119. J Clin Virol 58, 227–32 (2013). [DOI] [PubMed] [Google Scholar]
- Razafindratsimandresy R., Joffret M. L., Delpeyroux F. & Heraud J. M. First full genome sequence of a human enterovirus a120, isolated in madagascar. Genome Announc 2 e00568–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S. et al. Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J Virol 87, 558–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmivandana R., Yergolkar P., Gopalkrishna V. & Chitambar S.D. Characterization of the non-polio enterovirus infections associated with acute flaccid paralysis in South-Western India. PLoS One 8, e61650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitambar S. et al. Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western India. Int J Environ Res Public Health 9, 895–915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C. D., Yergolkar P. & Shankarappa K. S. Antigenic diversity of enteroviruses associated with nonpolio acute flaccid paralysis, India, 2007–2009. Emerg Infect Dis 18, 1833–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil P. R., Chitambar S. D. & Gopalkrishna V. Molecular surveillance of non-polio enterovirus infections in patients with acute gastroenteritis in Western India: 2004–2009. J Med Virol 87, 154–61 (2015). [DOI] [PubMed] [Google Scholar]
- Blomqvist S., Paananen A., Savolainen-Kopra C., Hovi T. & Roivainen M. Eight years of experience with molecular identification of human enteroviruses. J Clin Microbiol 46, 2410–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S. et al. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol 87, 119–28 (2006). [DOI] [PubMed] [Google Scholar]
- Kroneman A. et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol 51, 121–5 (2011). [DOI] [PubMed] [Google Scholar]
- Oberste M. S. et al. Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 37, 1288–93 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. et al. Retrospective seroepidemiology indicated that human enterovirus 71 and coxsackievirus A16 circulated wildly in central and southern China before large-scale outbreaks from 2008. Virol J 7, 300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Type 2 vaccine-derived poliovirus from patients with acute flaccid paralysis in china: current immunization strategy effectively prevented its sustained transmission. J Infect Dis 202, 1780–8 (2010). [DOI] [PubMed] [Google Scholar]
- Xu A. et al. The complete genome sequence of an enterovirus 76 isolate in China reveals a recombination event. Archives of virology 156, 1685–9 (2011). [DOI] [PubMed] [Google Scholar]
- Sun Q. et al. Transmission of human enterovirus 85 recombinants containing new unknown serotype HEV-B donor sequences in Xinjiang Uighur autonomous region, China. PLoS One 8, e55480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. Whole genomic sequence and replication kinetics of a new enterovirus C96 isolated from Guangdong, China with a different cell tropism. PLoS One 9, e86877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H., Bjerregaard L. & Magnius L. O. Open reading frame sequence of an Asian enterovirus 73 strain reveals that the prototype from California is recombinant. J Gen Virol 83, 1721–8 (2002). [DOI] [PubMed] [Google Scholar]
- Tang J. et al. Complete genome characterization of a novel enterovirus type EV-B106 isolated in China, 2012. Sci Rep 4, 4255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santti J., Hyypia T., Kinnunen L. & Salminen M. Evidence of recombination among enteroviruses. J Virol 73, 8741–9 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmblat B. et al. Nonhomologous recombination between defective poliovirus and coxsackievirus genomes suggests a new model of genetic plasticity for picornaviruses. MBio 5, e01119–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou Z., Pliaka V., Amoutzias G. D. & Markoulatos P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes 50, 177–88 (2015). [DOI] [PubMed] [Google Scholar]
- Lukashev A. N. Role of recombination in evolution of enteroviruses. Rev Med Virol 15, 157–67 (2005). [DOI] [PubMed] [Google Scholar]
- McWilliam Leitch E. C. et al. The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol 86, 2676–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sanden S. et al. Detection of recombination breakpoints in the genomes of human enterovirus 71 strains isolated in the Netherlands in epidemic and non-epidemic years, 1963–2010. Infect Genet Evol 11, 886–94 (2011). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Analysis of recombination and natural selection in human enterovirus 71. Virology 398, 251–61 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Emergence and transmission pathways of rapidly evolving evolutionary branch c4a strains of human enterovirus 71 in the central plain of china. PLoS One 6, e27895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. et al. An outbreak of poliomyelitis caused by type 1 vaccine-derived poliovirus in China. J Infect Dis 194, 545–51 (2006). [DOI] [PubMed] [Google Scholar]
- Alexander J. P. et al. Transmission of imported vaccine-derived poliovirus in an undervaccinated community in Minnesota. J Infect Dis 199, 391–7 (2009). [DOI] [PubMed] [Google Scholar]
- Isolation and identification of polioviruses. WHO Polio laboratory manual, 4th edn. (World Health Organization, Geneva, 2004).
- Yang C. F. et al. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J Virol 77, 8366–77 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M. O., Carr J. K., Burke D. S. & McCutchan F. E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11, 1423–5 (1995). [DOI] [PubMed] [Google Scholar]
- Ishiko H. et al. Molecular diagnosis of human enteroviruses by phylogeny-based classification by use of the VP4 sequence. J Infect Dis 185, 744–54 (2002). [DOI] [PubMed] [Google Scholar]
- Caro V., Guillot S., Delpeyroux F. & Crainic R. Molecular strategy for ‘serotyping’ of human enteroviruses. J Gen Virol 82, 79–91 (2001). [DOI] [PubMed] [Google Scholar]