Abstract
Our study investigated whether microRNA-122 (miR-122) played important roles in the proliferation, invasion and apoptosis of human cholangiocarcinoma (CC) cells. QBC939 and RBE cells lines were chosen and divided into five groups: miR-122 mimic group, anti-miR-122 group, negative control (NC) group, mock group and blank group. MiR-122 expression was measured by qRT-PCR. Roles of miR-122 in cell proliferation, apoptosis and invasion were investigated using MTT assay, flow cytometer and Transwell invasion assay, respectively. MiR-122 expression was lower in CC tissues and QBC939 cell than that in normal bile duct tissues, HCCC-9810 and RBE cells. In both QBC939 and RBE cells lines, miR-122 expression was higher in miR-122 mimic group than that in NC group, mock group and blank group; opposite results were found in anti-miR-122 group. Cell proliferation and invasion were remarkably inhibited in miR-122 mimic group after 48 h/72 h transfection, while apoptotic cells numbers were much greater in miR-122 mimic group; the opposite results were obtained from anti-miR-122 group (all P < 0.05). MiR-122 expression was significantly weaker in CC tissues, and miR-122 overexpression might play pivotal roles in inhibiting proliferation, stimulating apoptosis and suppressing invasion of CC cells, suggesting a new target for CC diagnosis and treatment.
Cholangiocarcinoma (CC) is the second commonest primary liver malignancy after hepatocellular carcinoma (HCC) and is generally classified into three forms including intrahepatic cholangiocarcinoma (ICC), hilar cholangiocarcinoma and extrahepatic cholangiocarcinoma (ECC)1. CC is defined as a rare and fatal tumor, and the main challenge for treating CC is considered to be the accurate diagnosis at the early stage2. Incidence rate and mortality rate of ICC have steadily increased over the world in the past few decades while those of ECC decreased3. CC widely occurs in different regions with the highest in Southeast Asia and the lowest in Australia, and the rates are 96 per 100,000 in man and 38 per 100,000 in women4. The incidence rate is not controlled by improvements in diagnosis because the clinical characteristics of this cancer is not easy to identify5. Patients with respectable CC are usually recommended to take surgical resection and can achieve good prognosis, while the prognosis of advanced CC patients remains very poor6. Therefore, researchers are trying to develop novel therapeutic strategies to control CC progression7,8. MicroRNAs (miRNAs) have been found involved in several biological activities in some diseases, and regarded as potential targets for the diagnosis and treatment of CC9.
MiRNAs, the non-coding RNA molecules, act as post-transcription regulators of gene expression by binding to the complementary sequences in the 3′ untranslated region10. Each miRNA regulates hundreds of protein coding genes involved in cell proliferation, apoptosis and metastasis, and the overexpression of tumor suppressive miRNAs would help to block cell cycle, suppress tumor angiogenesis, and induce apoptosis11. MicroRNA-122 (miR-122) is the most abundantly expressed miRNA in human liver cells, and it plays an important role in suppressing cell proliferation and controlling cell viability by targeting antiapoptotic B-cell lymphoma 2 (Bcl-2) family protein Bcl-2 and activating caspase-3 (http://www.scirp.org/journal/PaperInformation.aspx?paperID=22969). The liver homoeostasis is considered to be closely associated with miR-122 expression level; low expression of miR-122 may cause cirrhosis and advanced HCC12. MiR-122 also plays a role in various types of cancer by regulating the expressions of the oncogenic proteins such as disintegrin, metalloproteinase domain 10, transcription factor and receptor tyrosine kinase, which are involved in cell adhesion, migration and invasion13. The function of miR-122 as tumor suppressor may inhibit the development of CC by binding to target genes related with tumor cell proliferation, apoptosis, invasion and angiogenesis14. In this study, we investigated the role of miR-122 in cell proliferation, apoptosis and invasion in CC.
Materials and Methods
Patients
Between January 2002 and January 2012, a total of 57 resected paraffin-embedded CC patients (30 males and 27 females; mean age: 61.04 ± 15.20 years; >65, 33 cases; ≤65, 24 cases; range: 28–79 years) were recruited from the Department of General Surgery, Hainan Provincial people’s Hospital, and 57 specimens were collected from surgical excision or biopsy. In addition, 10 normal bile duct tissues diagnosed under the endoscope from liver transplantation were collected to be the healthy control group. Each of the 57 patients was diagnosed by two experienced doctors. The diagnosis of CC was established by laboratory examination and imaging test, such as ultrasound and computed tomography (CT) scan15,16. The mean tumor diameter was 6.40 ± 4.01 cm (≤5 cm, n = 31; >5 cm, n = 26). Among all the CC patients, 22 cases had lymph nodes metastasis, and 35 cases didn’t. As for distant metastasis, 9 cases were diagnosed with distant metastasis (major metastasis sites were the liver, peritoneum and the greater retina), and 48 cases didn’t have distant metastasis. According to the degrees of differentiation, there were 33 cases in high-moderate differentiation and 24 cases in low differentiation. Tumor node metastasis (TNM) staging was based on the standards of the International Union Against Cancer/American Cancer Society guideline and the seventh edition of the TNM Classification of Malignant Tumors17 (stage I–II, n = 38; stage III–IV, n = 19). All the specimens were fixed with 10% neutral buffered formalin, followed by alcohol dehydration, xylene transparency, and wax dipping, and were finally embedded in serial 3–4 mm paraffin blocks and cut into 0.4 mm-thick slices. The clinical data of each patient were complete. Preoperative chemotherapy and biological treatment were not performed before surgery. This study was approved by the Hainan Provincial people’s Hospital, and carried out in accordance with the approved guidelines of the Hospital and the Declaration of Helsinki18. Written informed consents were obtained from all the participants and their families.
Materials
Cholangiocarcinoma RBE, HCCC-9810, and QBC939 cell lines, were purchased from Cell Bank at the Chinese Academy of Sciences; fetal bovine serum (FBS) (Maverick, Beijing, China); streptomycin (Gibco, Grand Island, NY, USA); Trizol reagent (Molecular Science Research Center Inc., Cincinnati, Ohio); ABI 7500 real-time PCR system (Applied Biosystems, Warrington, United Kingdom); Roswell Park Memorial Institute (RPMI) 1640 medium (GIBCO. Uxbridge, UK); LipofectinTM2000 (Invitrogen, Carlsbad, USA); 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromid (MTT) and dimethyl sulphoxide (DMSO) (Sigma Chemical Co. St. Louis, USA); Matrigel (Corning Inc., Lowell, MA, USA); NCodeTM miRNA First-Strand complementary DNA synthesis Kit (Invitrogen, Carlsbad, USA); Fast SYBR® Green Master Mix (Roche Diagnostics GmbH, Mannheim, Germany); Transwell chamber (Corning Costar, Tewksbury, MA, USA) and miR-122 mimic and negative control (NC) (Shanghai GenePharma Co., Ltd, Shanghai, China).
Cell culture and transfection
RBE, HCCC-9810, and QBC939 cells were cultured in RPMI-1640 medium with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin, and were then stored in a 5% CO2 humidified incubator at 37 °C. After reaching 80% confluence, the cells were digested with 0.25% pancreatin-ethylene diamine tetraacetic acid diluted in the ratio of 1:5.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
CC specimens were gently crushed under liquid nitrogen and were then collected. Subsequently, cells were all harvested after 48 h of transfection, and the total RNA was extracted with the use of Trizol reagent. RNA purity and concentration were measured by UV spectroscopy. RNA integrity was evaluated using agarose gel electrophoresis. cDNA template was got by reverse transcription (RT) step using NCodeTM miRNA First-Strand complementary DNA synthesis kit, and then Fast SYBR Green Master Mix was used for real-time PCR with ABI 7500 real-time PCR system. Primer sequences for miR-122 were as follows. Forward primer: 5′-TTGAATTCTAACACCTTCGTGGCTACAGAG-′3; reverse primer: 5′-TTAGATCTCATTTATCGAGGGAAGGATTG-′3. U6 was used as internal control and the primer sequences were as follows. Forward primer 5′-CTCGCTTCGGCAGCACA-′3; reverse primer: 5′-AACGCTTCACGAATTTGCGT-′3. Real-time PCR protocol included an initial denaturation step (95 °C for 5 min) and 40 cycles of denaturation (95 °C for 10 s), annealing (60 °C for 20 s) and extension (72 °C for 10 s). The relative expression level of miR-122 was calculated using 2−ΔΔCt method.
Cells transfection and grouping
The miR-122 expression of different cell lines was measured through qRT-PCR, the result showed that the expression level of miR-122 in QBC93 cells was lower than that in HCCC-9810 and RBE cells, indicating that the QBC93 cell line was suitable for the following experiment. The reason might be that QBC93 cells were derived from liver metastasis foci of extrahepatic bile duct carcinoma, while HCCC-9810 and RBE cells were all from intrahepatic primary foci. Hence the malignant degree of QBC939 cells was comparatively high. One day before the transfection, RBE cells were seeded in 12-well plates at 0.5 × 105 cells/well. The cells were plated to 60% ~ 80% confluence within 24 hours, and 1 μL green fluorescent protein (GFP) (0.5 μg/μl) were then added to the transfected cells. After 72 h incubation, the expression of GFP was observed under the fluorescence microscope with a fluorescence intensity at 350–420 nm. Detailed grouping information were as follows: the miR-122 mimic group was transfected with miR-122 mimics; the anti-miR-122 group was transfected with miR-122 inhibitors; the negative control (NC) group was transfected with miR-122-NC; the Mock group was transfected with reagent by using LipofectinTM 2000; no transfection was performed on the Blank group. The sequences of transfected miR-122 mimics, miR-122 inhibitors and miR-122-NC (designed and synthesized by Shanghai GenePharma Co., Ltd) were 5′-UGGAGUGUGACAAUGGUGUUUG-′3, 5′-CAAACACCAUUGUCACACU-′3, and 5′-UUCUCCGAACGUGUCACGUTT-′3 respectively.
Cell proliferation assay
The proliferation of QBC939 and RBE cells were examined at the logarithmic growth phase. The cells were then recovered by digestion with 0.25% trypsin for 20 min, then a volume of RPMI-1640 containing 10% (v/v) FBS was used to terminate the digestion and to suspend the cells with a concentration of 2 × 105 cells/mL. After the incubation in a 24-well plate (500 μL/well), cells were grouped and transfected when these cell lines reached 80% confluency, and three parallel wells were measured in each group. After a 24 h transfection, 1 mL medium was used to culture the suspensions, and then the suspensions were transferred into the 96-well plates after a 1:5 dilution; six parallel wells were measured in each group. After 24 h, 48 h and 72 h of transfection, 10 μl MTT (5 mg/mL) was added to each well, and then the cells were incubated for 4 h in a 5% CO2 at 37 °C19. Subsequently, the crystallization (purple-red) was observed under the inverted microscope. After the supernatant was discarded, 100 μl DMSO was added to each well, and the mix was shaken for 15 min. The concentrations after 24 h, 48 h and 72 h were measured by absorbance of 570 nm wavelength with a reference wavelength of 630 nm using enzyme-linked immunosorbent assays. All experiments were repeated for at least three times. The cell growth curve was drawn to show cell proliferation rate.
Cell apoptosis assay
The transfected QBC939 and RBE cells were collected after 48 h (or 72 h) of transfection. Cells were fixed in 70% ethanol for 24 h and centrifuged once. Then DNA-binding dye propidium iodide (DAPI) (10 μg/ml) was added to the cell suspensions for staining, and the mixed solution was incubated at room temperature for 5 min in the dark. Then the samples were observed using the fluorescence microscope. Additionally, the transfected cells after 48 h were washed for three times by cold PBS and fixed in 70% ethanol. After fixation, the cell suspensions were stained in 500 μl precooled 1×binding buffer containing 5 μl annexin V/propidium iodide medium and 2.5 μl propidium iodide. Cell apoptosis was examined on FACScan flow cytometer (Becton-Dickinson, Sunny-vale, CA, USA)20.
Cell invasion assay
At 48 h after the cell transfection, cells were added to 1.5 ml EP tube and centrifuged at 2000 rpm for 5 min. Then the supernatant was discarded and the cells were re-suspended in 200 μl serum-free Dulbecco’s modified Eagle’s medium. The prepared 1 × 105cell was added to the upper chamber coated with Matrigel (8 μm pore size) and polycarbonate membrane. RPMI1640 medium with 10% FBS was placed in the lower chamber21. After 24 h at 37 °C, the filters with the mixture of methanol and acetic acid (3:1) were fixed for 30 min, and the cells were stained for 15 min with crystal violet solution. Invasive cells (five high-power fields) were counted using a light microscope.
Statistical analysis
All the experiments were repeated for three times independently. Enumeration data comparisons were conducted with I2 test. Numerical data were presented as means ± standard deviation. The comparisons between groups were performed using t test; comparisons among multiple groups were conducted using One-way analysis of variance (ANOVA). Robust tests of homogeneity of variances were conducted before the analysis. Interactions among multiple groups were tested by Fishers-LSD t-tests. SPSS 19.0 software (SPSS, Chicago, IL) was applied for statistical analyses, and P < 0.05 was considered to be statistically significant.
Results
Expression of miR-122 in CC tissues and cell lines
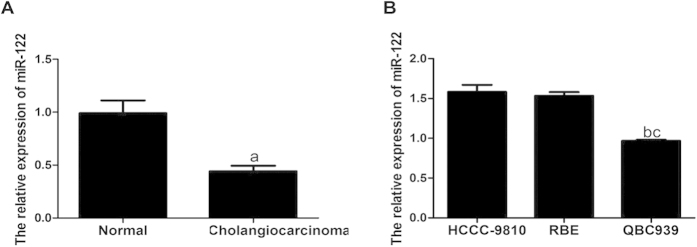
As shown in Fig. 1A, qRT-PCR results indicated that the expression of miR-122 was significantly lower in CC tissues than that in the normal bile duct tissues, and the relative expression levels (2−ΔΔCt) were (0.442 ± 0.051) and (0.990 ± 0.121), respectively (t = 14.10, P < 0.001). The relative expressions of miR-122 in HCCC-9810, RBE and QBC939 were (1.582 ± 0.088), (1.534 ± 0.047), and (0.968 ± 0.012), respectively. Furthermore, the expression level of miR-122 in QBC939 was significantly lower than those of the first two cell lines (both P < 0.05), while there was no significant difference in the expression levels of miR-122 between HCCC-9810 and RBE (P > 0.05) (Fig. 1B).
Figure 1. Quantitative real-time polymerase chain reaction (qRT-PCR) detected the relative expression of microRNA-122 (miR-122) in cholangiocarcinoma (CC) tissues and cell lines.
(A) Comparison of the relative expression of miR-122 between CC tissues and normal bile duct tissues; (B) Comparison of the relative expression of miR-122 among three difference cell lines, HCCC-9810, RBE and QBC939. acompared to the normal bile duct tissues, P < 0.05; bcompared to HCCC-9810 cell line, P < 0.05; ccompared to RBE cell line, P < 0.05.
Association of miR-122 expression with the clinicopathological features of CC patients
The associations between miR-122 expression and the clinicopathological features of the CC patients were presented in Table 1. The results showed that there was no significant association between miR-122 expression and gender, age, tumor size or differentiation (all P > 0.05). However, the expression of miR-122 was found significantly associated with lymph node metastasis and distant metastasis (both P < 0.05).
Table 1. Association of the relative expression of microRNA-122 (miR-122) and clinicopathological features in cholangiocarcinoma (CC) patients.
| Indexes | Cases | Relative expressionof miR-122(2−ΔΔCt) | t | P |
|---|---|---|---|---|
| Gender | ||||
| male | 30 | 0.452 ± 0.051 | ||
| female | 27 | 0.430 ± 0.049 | 1.657 | 0.103 |
| Age (years) | ||||
| ≤65 | 24 | 0.432 ± 0.044 | ||
| >65 | 33 | 0.449 ± 0.055 | 1.250 | 0.217 |
| Tumor size (cm) | ||||
| ≤5 | 31 | 0.448 ± 0.048 | ||
| >5 | 26 | 0.433 ± 0.048 | 1.175 | 0.245 |
| Histological grade | ||||
| well-moderate | 33 | 0.452 ± 0.052 | ||
| low | 24 | 0.429 ± 0.048 | 1.702 | 0.094 |
| Lymph node metastasis | ||||
| with | 22 | 0.392 ± 0.023 | ||
| without | 35 | 0.473 ± 0.037 | 9.195 | <0.001 |
| Distant metastasis | ||||
| with | 9 | 0.369 ± 0.017 | ||
| without | 48 | 0.456 ± 0.043 | 5.947 | <0.001 |
| TNM stage | ||||
| I–II | 38 | 0.446 ± 0.050 | ||
| III–IV | 19 | 0.434 ± 0.053 | 0.837 | 0.406 |
Note: TNM, tumor node metastasis.
Transfection efficiency of miR-122 in CC QBC939 and RBE cell lines
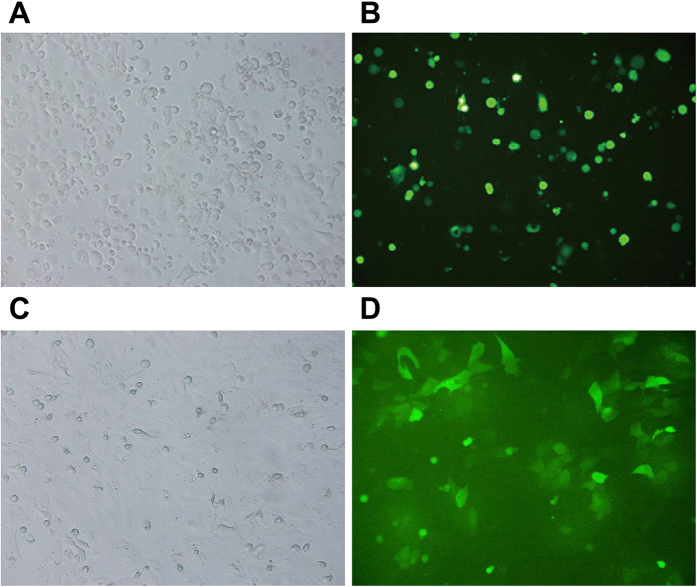
The transfection efficiency of GFP was observed by fluorescence microscope for many times for this kind of transient transfection assay, and results indicated that the transfection efficiency of liposome miR-122mimics/inhibitor and Lip2000 in QBC939 and RBE cells were both better than those in the other cells, with the transfection efficiency reached to 70%–80% (Fig. 2).
Figure 2. The transfection efficiency of microRNA-122 (miR-122) observation in cholangiocarcinoma under fluorescence microscope.
(A) QBC939 cells before transfection under fluorescence microscope; (B) QBC939 cells after transfection under fluorescence microscope; (C) RBE cells before transfection under fluorescence microscope; (D) RBE cells after transfection under fluorescence microscope.
MiR-122 expression level after transfection
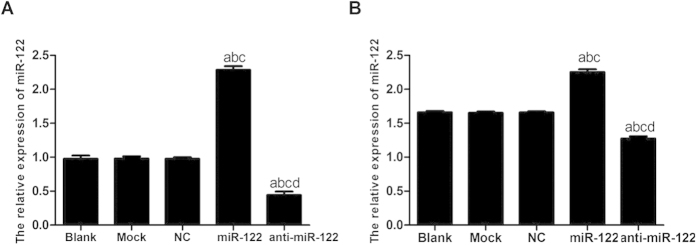
The amplification curve of target gene miR-122 was in accordance with the smooth curve with four characteristic stages (baseline period, exponential phase, exponential phase, platform stage). As shown in Fig. 3, the dissolution curve had only one peak and no impurity peak, suggesting that the gene was effectively amplified with a single amplification product, which met the experiment requirement. After the transfection with miR-122 mimics, the miR-122 expression levels in QBC939 and RBE cells of the NC group, the Mock group and the Blank group were significantly lower than that of miR-122 mimic group (all P < 0.05). However, the expression levels of miR-122 in QBC939 and RBE cells of the NC group, the Mock group and the Blank group were significantly higher than that of anti-miR-122 group (all P < 0.05). No significant difference in miR-122 level was found among the NC group, the Mock group and the Bland group (all P > 0.05) (Fig. 4 and Table 2).
Figure 3.
(A) The amplification curve of target gene miR-122; (B) The dissolution curve of target gene miR-122.
Figure 4.
Quantitative real-time polymerase chain reaction (qRT-PCR) testing microRNA-122 (miR-122) expression level after transfection in different groups, (A) QBC939 cells; (B) RBE cells. NC: negative control; acompared to the blank group, P < 0.05; bcompared to the mock group, P < 0.05; ccompared to the NC group, P < 0.05; dcompared to the miR-122 mimic group, P < 0.05.
Table 2. Quantitative real-time polymerase chain reaction (qRT-PCR) testing microRNA-122 (miR-122) expression level after transfection in different groups.
| Group | Relative expression of miR-122 (2−ΔΔCt) |
|
|---|---|---|
| QBC939 | RBE | |
| Blank group | 0.977 ± 0.048 | 1.661 ± 0.020 |
| Mock group | 0.978 ± 0.036 | 1.654 ± 0.017 |
| NC group | 0.977 ± 0.022 | 1.659 ± 0.018 |
| miR-122 mimic group | 2.284 ± 0.057a,b,c | 2.254 ± 0.041a,b,c |
| anti-miR-122 group | 0.442 ± 0.053a,b,c,d | 1.475 ± 0.033a,b,c,d |
| F | 693.300 | 488.600 |
| P | <0.001 | <0.001 |
Note: NC: negative control.
acompared to the blank group, P < 0.05.
bcompared to the mock group, P < 0.05.
ccompared to the NC group, P < 0.05.
dcompared to the miR-122 group, P < 0.05.
The role of miR-122 in inhibiting CC cell proliferation
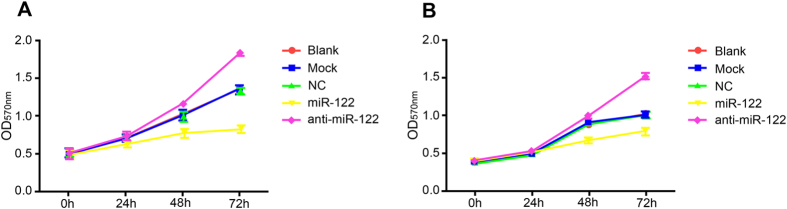
At 24 h after co-culture, QBC939 and RBE cell proliferation had no significant difference among the groups (all P > 0.05). However, compared to the NC group, Mock group and Blank group, QBC939 and RBE cell proliferations were both significantly inhibited in the miR-122 mimic group at the time points of 48 h and 72 h after the transfection (P < 0.05), while the cell proliferation was remarkably promoted in the anti-miR-122 group (Tables 3-4 and Fig. 5).
Table 3. 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromid (MTT) assay detected the role of microRNA-122 (miR-122) in inhibiting QBC939 cell proliferation.
| Group | 0 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| Blank group | 0.494 ± 0.053 | 0.711 ± 0.047 | 1.029 ± 0.054 | 1.360 ± 0.059 |
| Mock group | 0.508 ± 0.055 | 0.705 ± 0.051 | 1.015 ± 0.075 | 1.355 ± 0.063 |
| NC group | 0.514 ± 0.051 | 0.690 ± 0.037 | 0.995 ± 0.067 | 1.340 ± 0.045 |
| miR-122 mimic group | 0.502 ± 0.055 | 0.637 ± 0.041 | 0.776 ± 0.059a,b,c | 0.829 ± 0.050a,b,c |
| anti-miR-122 group | 0.514 ± 0.066 | 0.752 ± 0.059 | 1.175 ± 0.026a,b,c,d | 1.855 ± 0.039a,b,c,d |
| F | 0.069 | 2.286 | 17.860 | 146.300 |
| P | 0.990 | 0.132 | <0.001 | <0.001 |
Note: NC: negative control.
acompared to the blank group, P < 0.05.
bcompared to the mock group, P < 0.05.
ccompared to the NC group, P < 0.05.
dcompared to the miR-122 group, P < 0.05.
Table 4. 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromid (MTT) assay detected the role of microRNA-122 (miR-122) in inhibiting RBE cell proliferation.
| Group | 0h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| Blank group | 0.371 ± 0.021 | 0.514 ± 0.029 | 0.871 ± 0.034 | 1.017 ± 0.037 |
| Mock group | 0.408 ± 0.030 | 0.505 ± 0.020 | 0.915 ± 0.031 | 1.015 ± 0.041 |
| NC group | 0.394 ± 0.025 | 0.497 ± 0.017 | 0.905 ± 0.037 | 1.021 ± 0.045 |
| miR-122 mimic group | 0.402 ± 0.035 | 0.521 ± 0.031 | 0.669 ± 0.041a,b,c | 0.791 ± 0.050a,b,c |
| anti-miR-122 group | 0.404 ± 0.029 | 0.534 ± 0.024 | 1.007 ± 0.039a,b,c,d | 1.527 ± 0.044a,b,c,d |
| F | 0.812 | 1.001 | 34.970 | 116.200 |
| P | 0.546 | 0.451 | <0.001 | <0.001 |
Note: NC: negative control.
acompared to the blank group, P < 0.05.
bcompared to the mock group, P < 0.05.
ccompared to the NC group, P < 0.05.
dcompared to the miR-122 group, P < 0.05.
Figure 5.
3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromid (MTT) assay detected the role of microRNA-122 (miR-122) in inhibiting QBC939 and RBE cell proliferation, (A) QBC939 cells; (B) RBE cells. NC: negative control; OD, optical density.
The role of miR-122 in stimulating CC cell apoptosis
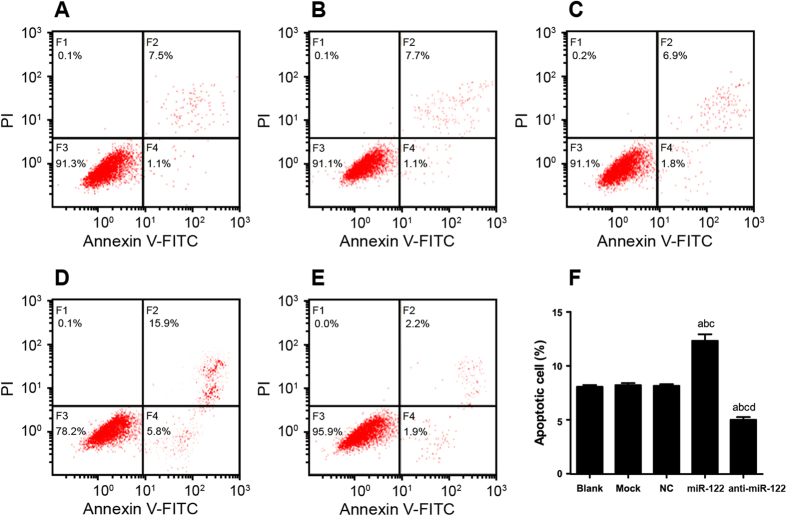
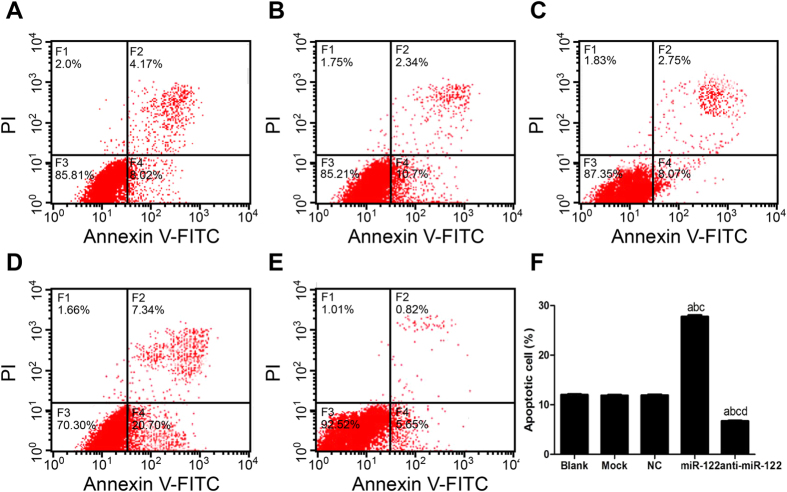
As shown in Fig. 6, the rates of QBC939 cell apoptosis in the NC group, the Mock group and the Blank group were (8.76 ± 0.15) %, (8.72 ± 0.18) % and (8.57 ± 0.16) % respectively, with no significant difference between any two (all P > 0.05). Similar results were also observed with respect to the rate of RBE cell apoptosis in the NC group (11.91 ± 0.15) %, the Mock group (11.87 ± 0.11) % and the Blank group (12.01 ± 0.13) %, showing no apparent statistically difference (all P > 0.05) (Fig. 7). Besides, in the miR-122 mimic group, the rate of QBC939 cell apoptosis was (21.63 ± 0.60) %, and in the anti-miR-122 group, the rate of QBC939 cell apoptosis was (4.11 ± 0.25) %, suggesting that rates of apoptosis in the NC group, the Mock group and the Blank group were significantly different from those in the miR-122 and the anti-miR-122 group (all P < 0.05). Similarly, the rate of RBE cell apoptosis in the miR-122 mimics group and the anti-miR-122 group was (27.76 ± 0.31) % and (6.71 ± 0.10) %, respectively, which were significantly different from those in the NC group, the Mock group and the Blank group (all P < 0.05).
Figure 6.
The apoptosis of QBC939 cells stimulated by microRNA-122 (miR-122) detected by flow cytometry in (A) Blank group, (B) Mock group, (C) NC group, (D) miR-122 mimic group, (E) anti-miR-122 group; (F) Histogram results of the apoptotic QBC939 cells of different groups. NC: negative control; acompared to the blank group, P < 0.05; bcompared to the mock group, P < 0.05; ccompared to the NC group, P < 0.05; dcompared to the miR-122 mimic group, P < 0.05.
Figure 7.
The apoptosis of RBE cells stimulated by microRNA-122 (miR-122) detected by flow cytometry in (A) Blank group, (B) Mock group, (C) NC group, (D) miR-122 mimic group, (E) anti-miR-122 group; (F) Histogram results of the apoptotic RBE cells of different groups. NC: negative control; acompared to the blank group, P < 0.05; bcompared to the mock group, P < 0.05; ccompared to the NC group, P < 0.05; dcompared to the miR-122 mimic group, P < 0.05.
The role of miR-122 in suppressing CC cell invasion
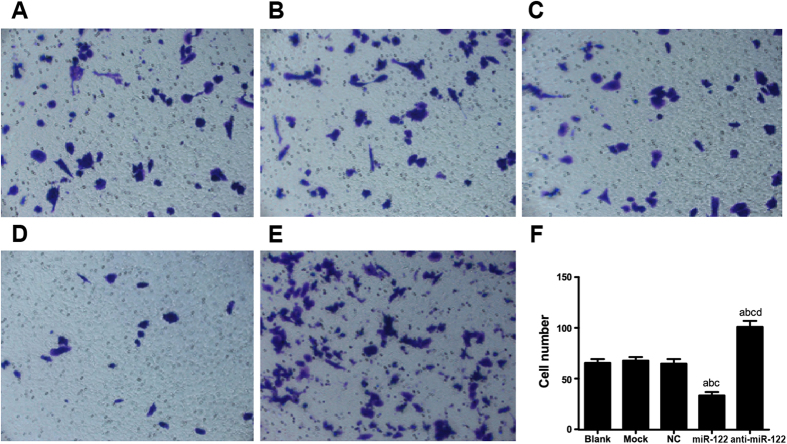
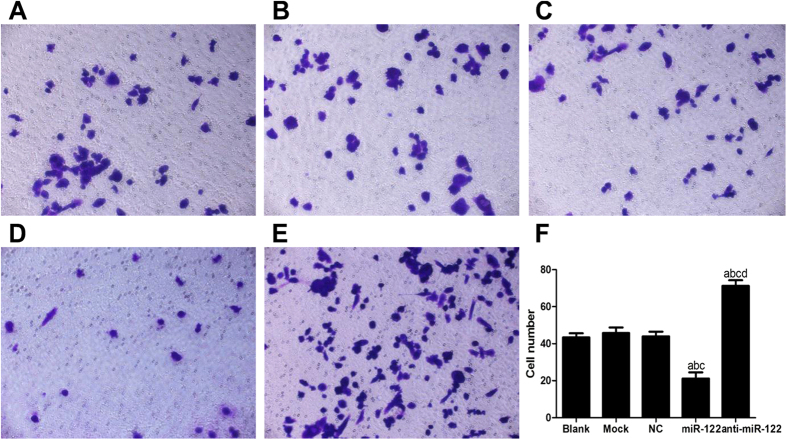
The cell invasion of QBC939 were measured after 24 h using Transwell invasion assay and Matrigel. The number of QBC939 cells migrating into Matrigel in the miR-122 mimic group (33.47 ± 3.292) was significantly smaller than that in the NC group (64.87 ± 4.42), the Mock group (67.87 ± 3.502) and the Blank group (65.53 ± 3.796) (all P < 0.05). However, the number of invasive QBC939 cells into Matrigel in anti-miR-122 group was (101.02 ± 5.92), significantly greater than those in the NC group, the Mock group and the Blank group (all P < 0.05). The number of QBC939 cells migrating into Matrigel was not significantly different among the NC group, the Mock group and the Blank group (all P > 0.05) (Fig. 8). On the other side, the numbers of RBE cells migrating into Matrigel in the miR-122 mimic group and the anti-miR-122 group were (21.19 ± 3.292) and (71.21 ± 3.091), respectively. In contrast, the number of migrating cells in the miR-122 mimic group was significantly greater than that in the NC group (43.96 ± 2.567), the Mock group (45.76 ± 3.011) and the Blank group (43.42 ± 2.201), while the number in the anti-miR-122 group was significantly smaller than the latter three groups (all P < 0.05) (Fig. 9).
Figure 8.
Transwell assay investigated the role of miR-122 in suppressing QBC939 cell invasion in (A) Blank group, (B) Mock group, (C) NC group, (D) miR-122 mimic group, (E) anti-miR-122 group; (F) Histogram results of QBC939 cell invasion detection within different groups. NC: negative control; acompared to the blank group, P < 0.05; bcompared to the mock group, P < 0.05; ccompared to the NC group, P < 0.05; dcompared to the miR-122 mimic group, P < 0.05.
Figure 9.
Transwell assay investigated the role of miR-122 in suppressing RBE cell invasion in (A) Blank group, (B) Mock group, (C) NC group, (D) miR-122 mimic group, (E) anti-miR-122 group; (F) Histogram results of RBE cell invasion detection within different groups. NC: negative control; acompared to the blank group, P < 0.05; bcompared to the mock group, P < 0.05; ccompared to the NC group, P < 0.05; dcompared to the miR-122 mimic group, P < 0.05.
Discussion
MiR-122 serves as functional miRNA involved in regulating lipid metabolism, cell differentiation, hepatic metabolism and hepatitis C virus replication22. The design of the experiment constituted a knockout mouse model modeling the loss of miRNA in hepatocytes, and finally liver disruptions including steatosis, inflammation and hepatocyte apoptosis were identified, suggesting that miRNAs played a key role in the liver function and that miR-122 may be associated with hepatocarcinogenesis by influencing hepatocyte survival and tumor progression23,24. MiR-122 deficiency in CC patients may contribute to the dysregulation of mitochondrial functions related with liver function, so its loss of expression may lead to increased morbidity and mortality and may predict poor prognosis of CC patients25. Our study confirmed that the expression of miR-122 was significantly lower in CC tissues than that in normal bile duct tissues, indicating that the decreased expression of miR-122 might be closely related to the development of CC. Therefore, we suggested that regulating the level of miR-122 could be used in controlling the progression of CC, which was consistent with the previous study findings revealing that the modulation of miR-122 level may be one of the targets for the prognosis prediction and the design of effective therapies for CC patients26. Furthermore, miR-122 participates in the tumor cell survival, proliferation, differentiation and migration as a tumor suppressor27. Previous evidence have shown that compared with adjacent benign liver, miR-122 appears to be down regulated in deficient liver, suggesting the potential of miR-122 as a novel biomarker for liver injury28. Further, the magnitude of decrease of the miR-122 level was previously proved to be helpful for the measurement the increase in the grade of hepatocellular injury29. Our study also confirmed that the decreased expression of miR-122 was also associated with lymph node metastasis and distant metastasis. Similarly, it was also previously reported that serum miR-122 level was an important indicator in the diagnosis of liver injury and could be used as a biomarker and therapeutic target for diagnosis and treatment of CC-30. In addition, we identified that the expression level of miR-122 in QBC939 was significantly lower than those of HCCC-9810 and RBE cells, while there was no significant difference between HCCC-9810 and RBE. These results supported that QBC939 cells were promising for further in vitro experiment; the reason might be that QBC939 cells widely exist in metastatic foci in liver of extrahepatic bile duct carcinoma, while HCCC-9810 and RBE cells mainly exist in the primary foci in the liver, so QBC939 had a higher degree of malignancy31.
In our study, we found that the overexpression of miR-122 played pivotal roles in inhibiting proliferation, stimulating apoptosis and suppressing invasion of QBC939 and RBE cells. As the most abundant miRNA in the liver, miR-122 is well known for its biologic function in maintaining liver homeostasis, as well as its role in regulating cell growth, differentiation, apoptosis and metabolism in the carcinogenesis in the liver, which is detrimental to normal liver function32,33. The over expression of insulin-like growth factor 1 receptor (Igf1R) in the development of tumors stimulates cell growth, survival and proliferation and regulates the initiation of cancer cell metastasis; the level of Igf1R is negatively associated with the level of miR-122 expression, implying that the overexpression of miR-122 can inhibit tumor cell growth and proliferation by suppressing Igf1R expression34. MiR-122 also functions as a key modulator of cyclin G1 expression and there is also a negative association between the levels of miR-122 and cyclin G1 (CCNG1)35. Decreased level of miR-122 was detected in the CC patients, leading to the increased level of CCNG1, which is associated with accumulation of tumor cells via affecting cell cycle (http://wenku.baidu.com/link? url=qineu6YlskaIZAh01hq9dV1Uw9rC6aU_JUHsgmy_NaJyMuVaIYFt4BErfqFSganqod6GceBfAMuN5rOiL1NZAZ1yikvVs_mgqjoeFy232we). The imbalance between miR-122 and CCNG1 may help to inhibit the tumor cell proliferation of CC through triggering p53 tumor suppressor gene36.
A previous study demonstrated that the abnormal expression of miR-122 was responsible for hepatocarcinogenesis; the loss of miR-122 led to the down-regulation of tumor cell apoptosis37. MiR-122 expression in tumor cells is suppressed in the early phase of CC, resulting in severe metastasis of tumor cells, and the restoration of its expression may help to control tumor progression of CC patients38. As a vital apoptosis regulator, and the mechanism of miR-122 in CC cells involves suppressing Bcl-W mRNA and the protein level, consequently leading to large reduction of cell motility39. Bcl-W activity can inhibit cancer cell apoptosis, and the overexpression of miR-122 can inhibit the expression of Bcl-W and CCNG1 to induce cell apoptosis and cell cycle arrest40. Thus, down-regulated miR-122 is potential to be an independent predictor of the development and progression of CC characterized by the loss of anti-apoptotic effect41. Our study also found that the role of miR-122 in antitumor activity is manifested in suppressing tumor cell invasion. Based on previous study results, miR-122 down-regulation was identified to be associated with hepatic cell invasion, intrahepatic metastasis and reduction of tumor cell sensitivity to drug agent resulting in tumor aggressiveness42. As a tumor suppressor, miR-122 can inhibit intrahepatic invasion and migration of CC cells by suppressing angiogenesis through regulating the disintegrin and metalloprotease 17 activity43.
In summary, we found that miR-122 expression significantly decreased in CC tissues, and the overexpression of miR-122 played a pivotal role in inhibiting proliferation, stimulating apoptosis and suppressing invasion of CC cells. Finally, our study suggested that miR-122 could be a promising biomarker and target used for the diagnosis and treatment of CC.
Additional Information
How to cite this article: Liu, N. et al. The Roles of MicroRNA-122 Overexpression in Inhibiting Proliferation and Invasion and Stimulating Apoptosis of Human Cholangiocarcinoma Cells. Sci. Rep. 5, 16566; doi: 10.1038/srep16566 (2015).
Footnotes
Author Contributions Conceived and designed the experiments: N.L., F.J. Performed the experiments: J.K.Z., J.Z., T.L.H. and G.X.J. Analyzed the data: L.P.C. and P.C.K. Wrote the paper: X.Y.Z. and C.W. Revised and approved the final version of the paper: T.Y.L., Y.F.C. and C.W.
References
- Tyson G. L. et al. Secular trends in the incidence of cholangiocarcinoma in the USA and the impact of misclassification. Dig Dis Sci 59, 3103–3110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth C. A. et al. Genetic factors in the pathogenesis of cholangiocarcinoma. Dig Dis 29, 93–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A. et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 61, 1657–1669 (2012). [DOI] [PubMed] [Google Scholar]
- Charbel H. & Al-Kawas F. H. Cholangiocarcinoma: epidemiology, risk factors, pathogenesis, and diagnosis. Curr Gastroenterol Rep 13, 182–187 (2011). [DOI] [PubMed] [Google Scholar]
- Tyson G. L. & El-Serag H. B. Risk factors for cholangiocarcinoma. Hepatology 54, 173–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marubashi S. et al. Prediction of the postoperative prognosis of intrahepatic cholangiocarcinoma (ICC): importance of preoperatively- determined anatomic invasion level and number of tumors. Dig Dis Sci 59, 201–213 (2014). [DOI] [PubMed] [Google Scholar]
- Deng G. et al. Downregulation of ROS-FIG inhibits cell proliferation, colonyformation, cell cycle progression, migration and invasion, while inducing apoptosis in intrahepatic cholangiocarcinoma cells. Int J Mol Med 34, 661–668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li W. et al. Mycoepoxydiene induces apoptosis and inhibits TPA-induced invasion in human cholangiocarcinoma cells via blocking NF-kappaB pathway. Biochimie 101, 183–191 (2014). [DOI] [PubMed] [Google Scholar]
- Patel T. Extracellular vesicle noncoding RNA: new players in the diagnosis and pathogenesis of cholangiocarcinoma. Hepatology 60, 782–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P. & Peter M. E. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene 33, 269–278 (2014). [DOI] [PubMed] [Google Scholar]
- Khare S., Zhang Q. & Ibdah J. A. Epigenetics of hepatocellular carcinoma: role of microRNA. World J Gastroenterol 19, 5439–5445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer A. J. et al. Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122. J Viral Hepat 20, 158–166 (2013). [DOI] [PubMed] [Google Scholar]
- Henry J. C., Azevedo-Pouly A. C. & Schmittgen T. D. MicroRNA replacement therapy for cancer. Pharm Res 28, 3030–3042 (2011). [DOI] [PubMed] [Google Scholar]
- Nakao K., Miyaaki H. & Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol 49, 589–593 (2014). [DOI] [PubMed] [Google Scholar]
- Charatcharoenwitthaya P. et al. Utility of serum tumor markers, imaging, and biliary cytology for detecting cholangiocarcinoma in primary sclerosing cholangitis. Hepatology. 48, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- Gores G. J. Cholangiocarcinoma: current concepts and insights. Hepatology. 37, 961–969 (2003). [DOI] [PubMed] [Google Scholar]
- Novak J. & Fabian P. Comments on the TNM classification of malignant tumours—7th edition. Klin Onkol. 24, 149–150 (2011). [PubMed] [Google Scholar]
- Krleza-Jeric K. & Lemmens T. 7th revision of the Declaration of Helsinki: good news for the transparency of clinical trials. Croat Med J. 50, 105–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z. et al. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med Oncol 29, 886–892 (2012). [DOI] [PubMed] [Google Scholar]
- Liu Q. et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36, 5391–5404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer 46, 2828–2836 (2010). [DOI] [PubMed] [Google Scholar]
- Tsai W. C. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122, 2884–2897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand N. J., Master Z. R., Le Lay J. & Friedman J. R. Hepatic function is preserved in the absence of mature microRNAs. Hepatology 49, 618–626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S. et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304–2315 e2301-2304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard J. et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol 6, 402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakatsanis A. et al. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 52, 297–303 (2013). [DOI] [PubMed] [Google Scholar]
- Takaki Y. et al. Silencing of microRNA-122 is an early event during hepatocarcinogenesis from non-alcoholic steatohepatitis. Cancer Sci 105, 1254–1260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 50, 136–142 (2011). [DOI] [PubMed] [Google Scholar]
- Starckx S. et al. Evaluation of miR-122 and other biomarkers in distinct acute liver injury in rats. Toxicol Pathol 41, 795–804 (2013). [DOI] [PubMed] [Google Scholar]
- Shifeng H. et al. Circulating liver-specific miR-122 as a novel potential biomarker for diagnosis of cholestatic liver injury. PLoS One 8, e73133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. H. et al. The growth-inhibition effect of tamoxifen in the combination chemotherapeutics on the human cholangiocarcinoma cell line QBC939. Mol Biol Rep. 37, 2693–2701 (2010). [DOI] [PubMed] [Google Scholar]
- Coulouarn C. et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 28, 3526–3536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay H. et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem 99, 671–678 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 284, 32015–32027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L. et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 67, 6092–6099 (2007). [DOI] [PubMed] [Google Scholar]
- Fornari F. et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 69, 5761–5767 (2009). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. miR-122 affects the viability and apoptosis of hepatocellular carcinoma cells. Scand J Gastroenterol 44, 1332–1339 (2009). [DOI] [PubMed] [Google Scholar]
- Cho W. C. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 42, 1273–1281 (2010). [DOI] [PubMed] [Google Scholar]
- Lima R. T. et al. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer 47, 163–174 (2011). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biol Ther 9, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- Lin C. J. et al. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 375, 315–320 (2008). [DOI] [PubMed] [Google Scholar]
- Yang F. et al. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia 13, 590–600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. C. et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49, 1571–1582 (2009). [DOI] [PubMed] [Google Scholar]