Abstract
SSRP1 is a subunit of the FACT complex, an important histone chaperone required for transcriptional regulation, DNA replication and damage repair. SSRP1 also plays important roles in transcriptional regulation independent of Spt16 and interacts with other proteins. Here, we report the crystal structure of the middle domain of SSRP1. It consists of tandem pleckstrin homology (PH) domains. These domains differ from the typical PH domain in that PH1 domain has an extra conserved βαβ topology. SSRP1 contains the well-characterized DNA-binding HMG-1 domain. Our studies revealed that SSRP1-M can also participate in DNA binding, and that this binding involves one positively charged patch on the surface of the structure. In addition, SSRP1-M did not bind to histones, which was assessed through pull-down assays. This aspect makes the protein different from other related proteins adopting the double PH domain structure. Our studies facilitate the understanding of SSRP1 and provide insights into the molecular mechanisms of interaction with DNA and histones of the FACT complex.
To overcome the inhibitory effects of nucleosomes in the accessibility of DNA during basic cellular processes, such as transcription, DNA replication and repair, many factors are required to alter the chromatin structure. The FACT (facilitates chromatin transcription) complex is an important histone chaperone that reorganizes nucleosomes without hydrolysing ATP1. The yeast FACT complex enhances DNA accessibility in nucleosomes without translocating histone octamers relative to the DNA2,3, whereas the human FACT complex is proposed to function as a histone chaperone that displaces a single H2A-H2B dimer from a nucleosome to promote DNA accessibility4,5,6. The FACT complex is a chromatin-specific factor that is required for both normal regulation of transcription1,7,8,9 and DNA replication9,10,11,12. It also plays important roles in DNA damage repair13,14,15,16. Consistent with its broad functional importance, the FACT complex is a highly conserved complex among all eukaryotes5,11,17,18. In S. cerevisiae, the FACT complex contains three proteins: ySpt16, Pob3 and Nhp61. In metazoans, the Pob3 and Nhp6 orthologs are fused to form a single polypeptide called SSRP1. Both SPT16 and SSRP1/Pob3 are essential genes for the viability of eukaryotes9,19. hSpt16 binds to H2A-H2B dimers and nucleosomes. In contrast, SSRP1 binds neither H2A-H2B dimers nor nucleosomes, but it does bind H3-H4 tetramers4.
The domain structure of the FACT complex is conserved throughout eukaryotes. Spt16 is composed of four domains, including an N-terminal domain (Spt16-N), a dimerization domain (Spt16-D), a middle domain (Spt16-M) and a C-terminal domain (Spt16-C)20,21. SSRP1 contains three well-defined domains: an N-terminal/dimerization domain (SSRP1-N), a middle domain (SSRP1-M) and a C-terminal high-mobility group (HMG)-1 domain (SSRP1-C)21. Pob3 includes two domains similar to SSRP1-N and SSRP1-M with a C-terminal acidic region. Nhp6 contains an HMG-l domain. Recently, the structures and the functions of the domains of the FACT complex were revealed12,18,20,22,23,24. Briefly, it was found that ySpt16-N, which is structurally similar to aminopeptidases, binds to histones H3-H418. ySpt16-M and Pob3-M both adopt tandem PH domain topologies20,22,23. ySpt16-M binds to H2A-H2B and H3-H4 histones22,23 whereas Pob3-M binds to only H3-H4 histones22,25. Notably, a related yeast histone H3-H4 chaperone Rtt106, which has sequence homology with Pob3 and SSRP1, has been identified26, and it contains a double PH domain25,27,28. Nhp6A adopts a typical L-shaped HMG fold and binds nonspecifically and stably to dsDNA24. However, no structural studies on the human FACT complex have been reported, and its ability to bind DNA and histone remains to be elucidated.
SSRP1 was initially identified as binding to specific DNA modified by the anticancer drug cisplatin29. The protein is expressed at high levels in proliferating cancerous tissues and at low levels in less-renewable and differentiated tissues30,31. Furthermore, SSRP1 can bind to cruciform DNA, linear duplex DNA and cisplatin-modified DNA32,33,34. SSRP1 has also been reported to act as an architectural transcription factor for many other genes33. Additionally, SSRP1 can function as a co-activator of several transcription factors, such as serum-response factor (SRF)35 and p6336. In addition, SSRP1 is a novel microtubule (MT)-binding protein that facilitates MT growth and bundling, and it is required for mitosis37. However, the mechanisms by which SSRP1 binds to DNA and interacts with other proteins and how each domain of SSRP1 contributes to the functions of SSRP1 are unclear. Among the three domains of SSRP1, the ability of the C-terminal HMG domain of SSRP1 to bind DNA has been studied most intensively16,34,38. However, the other domains of SSRP1 (SSRP1- (N + M)) are even more greathighly conserved than the HMG domain32,39, suggesting their importance for the functions of SSRP1. Consistently, SSRP1- (N + M) domains have been found to be critical for the interaction between SSRP1 and chromatin40. Although the N-terminal domain of SSRP1, (SSRP1-N), was reported to be required for the in vitro interaction of SSRP1 with Spt1641, little is known about the middle domain of SSRP1 (SSRP1-M), which is highly conserved across species (Fig. 1). In the present work, we determined the crystal structure of SSRP1-M at a resolution of 1.93 Å. The structure contains tandem pleckstrin homology (PH) domains and exhibits a positively charged region on each side. Furthermore, we investigated the dsDNA binding properties of this domain and tested its interaction with histones via biochemical assays.
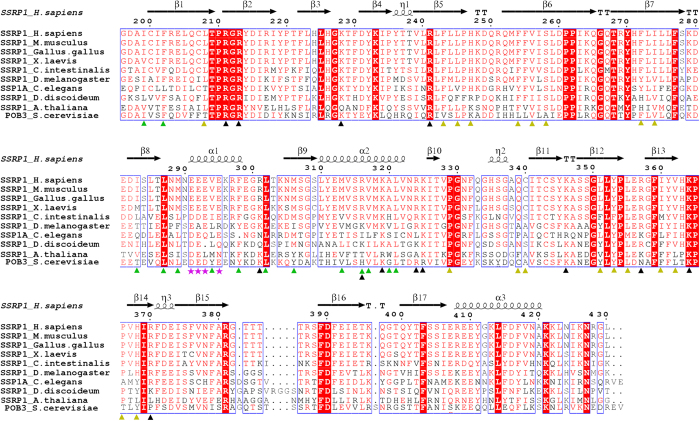
Figure 1. Sequence alignment of SSRP1-M with its homologues.
The amino acid sequences of the 10 indicated species from human to yeast are aligned, with the secondary structural elements reported in this paper indicated above the sequences. The amino acids involved in the hydrophobic inter-domain interactions are designated with yellow triangles; residues involved in stabilizing the extra βαβ super-secondary structure are designated with green triangles; residues comprising the positively charged regions for dsDNA binding are designated with black triangles; and residues comprising the unique negatively charged regions are designated with magentas stars.
Materials and Methods
Protein expression and purification
cDNA fragments encoding SSRP1-N (residues 1–176), SSRP1-M (residues 196–430), and SSRP1-C (residues 513–709) were PCR-amplified from the full-length SSRP1 cDNA (Sanying). Each of the cDNAs was cloned into a modified pET-28a (+) plasmid lacking a thrombin cleavage site to produce target proteins as N-terminal 6 × His fusions. cDNA encoding the gene for Rtt106 was also cloned into the modified pET-28a (+) plasmid as reported27 for in vitro His-tag pull-down assays. cDNA encoding SSRP1-M or HIF was cloned into the pGEX-4T-2 vector to produce the GST-fused protein for in vitro GST pull-down assays. Mutations were generated using a QuikChange Site-directed Mutagenesis kit (Stratagene) according to the manufacturer’s protocol. All of the recombinant plasmids were confirmed by DNA sequencing (Invitrogen) and then transformed into Escherichia coli BL21 (DE3) (Novagen), for protein expression. E. coli cells were cultured in LB medium with 50 mg/l kanamycin or 100 mg/l ampicillin at 310 K until the OD600 reached 0.8–1.2. Then, the cells were induced by the addition of 0.25 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at 289 K for 20 h. The cells were then collected by centrifugation, resuspended in buffer A (20 mM Tris-HCl, 200 mM NaCl, 5 % glycerol, pH 7.0) and lysed via ultrasonication. The cell extracts were centrifuged at 14,000 × g for 30 min at 277 K. The supernatants were purified using Ni2+-nitrilotriacetate or GST affinity resin (GE Healthcare) pre-equilibrated with buffer A. The eluted proteins were concentrated via centrifugal ultrafiltration (Amicon Ultra-15, 10 kDa MWCO, Millipore). The concentrated poor was loaded onto a pre-equilibrated HiLoad 16/60 Superdex 200 column or a HiLoadTM 16/60 SuperdexTM 75 pg column (GE Healthcare) using an Äkta-System (GE Healthcare), and the protein was eluted at a flow rate of 1 ml/min with buffer B (20 mM Tris-HCl, 200 mM NaCl, pH 7.0) and collected in 1.2 ml fractions. The peak fractions were analysed using Tricine-SDS-PAGE (15 %, w/v) and stained with Coomassie brilliant (CB) blue R250. The purified fractions were pooled together and concentrated via centrifugal ultrafiltration. For crystallization, SSRP1-M was further purified by HiTrap SPFF (5 ml) ion exchange chromatography (GE Healthcare). The final protein pool in buffer C (5 mM Tris-HCl, 40 mM NaCl, pH 7.5) was concentrated to 40 mg/ml for crystallization trials.
Crystallisation and data collection
SSRP1-M was crystallized using the hanging drop vapour diffusion method by mixing together 1 μl protein solution and 1 μl reservoir solution at 287 K. The crystal with suitable X-ray diffraction was grown in a reservoir solution that contained 0.1 M Tris, and 15 % (w/v) polyethylene glycol 6,000, pH 8.5 (Hampton Research). Data collection was performed at 100 K with a cryoprotectant solution containing the reservoir solution with the addition of 20 % (v/v) glycerol. A set of diffraction data was collected using the beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF).
Structure determination and refinement
The diffraction data set was processed and scaled using the HKL2000 package42. The phases were determined using molecular replacement with the program Molrep43. The structure of Pob3-M (PDB code 2GCL) was used as the search model. Cycles of refinement and model building were carried out using the REFMAC544, Phenix45 and COOT46 software programs until the crystallography R-factor and free R-factor converged to 19.8 % and 21.7 %, respectively. The Ramachandran analysis showed that 92.7 % of the residues were in the most favoured region, and that an additional 7.3 % were within the allowed region. The quality of the structure was verified using the program MolProbity47. The details regarding the data collection and processing are summarized in Table 1. All the structural figures were prepared using PyMOL (DeLano Scientific).
Table 1. Data-Collection and Refinement Statistics for SSRP1-M.
| Data collection | |
|---|---|
| Space group | P3221 |
| Cell dimensions | a = b = 89.19 Å, c = 82.59 Å |
| Wavelength (Å) | 0.97954 Å |
| Resolution (Å) | 50.0 − 1.93 (1.96 − 1.93)a |
| Rmerge (%)c | 8.7 (49.6)a |
| I/σ (I) | 26.6 (4.9)a |
| CC (1/2) | 0.994 (0.970) |
| Completeness (%) | 99.9 (100)a |
| Redundancy | 10.7 (10.8) |
| Refinement | |
| Resolution (Å) | 36.4 − 1.93 |
| Number of reflections/test | 27578/1476 |
| Rwork/Rfree (%)c | 19.8/21.7 |
| Number of atoms Protein | 1883 |
| Number of waters | 97 |
| Rmsd bond (Å) | 0.009 |
| Rmsd angle (°) | 1.116 |
| B-value (Å2) | 39.8 |
| Ramachandran plot (%) | |
| most favoured | 92.7 |
| additional allowed | 7.3 |
aValues in parentheses are for the highest-resolution shell.
bRmerge = ∑|Ii − <I>|/∑|I|, where Ii is the intensity of an individual reflection and <I> is the average intensity of that reflection.
cRwork = ∑||Fo| − |Fc||/∑|Fo|, where Fo and Fc are the observed and calculated structure factors for reflections, respectively.
dRfree was calculated as Rwork using the 5% of reflections that were selected randomly and omitted from refinement.
Electrophoretic mobility shift assay (EMSA)
The protein-DNA interactions were evaluated using an EMSA. The DNA sequences used are as follows:
30-bp linker dsDNA, 5′-TCCAGTGCCGGTGTCGCTTGGGTCCCGAGG-3′;
AT-rich dsDNA, 5′-ATAATTTATATTTATTATTTTATTATAATT-3′;
GC-rich dsDNA, 5′-GCGGCCCGCGCCCGCCGCCCCGCCGCGGCC-3′;
The DNA segments were synthesized by Sangon (Shanghai). Two strands of complementary single-stranded DNA were annealed to form the dsDNA. Next, 8 μM dsDNA was mixed with increasing amounts of protein from 10–320 μM in buffer D (20 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 1 mM EDTA, pH 7.0) and then incubated on ice for 1 h to be resolved on a 1.0 % agarose gel at 60 V and 4 °C in 1 × TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 7.8), or an 8 % native polyacrylamide gel at 100 V and 4 °C in 0.5 × TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0). The gel was visualized using Gel-Red. The amount of DNA was estimated using ImageJ software48. The raw integrated intensity was plotted against the protein concentration.
In vitro pull-down assays
The plasmids encoding the Xenopus histones H2A, H2B, H3 and H4 were kind gifts from Dr. Guohong Li49. The fragments encoding the yeast histones H2A, H2B, H3 and H4 were amplified from yeast genomic cDNAs using PCR, and the plasmids were made as described previously50. The histones were bacterially expressed, and the histone octamers were reconstituted using an established protocol51. Whole calf thymus histones were purchased from Worthington Company. The in vitro pull-down assays were performed as described previously52. Briefly, 10 μg of GST or GST-fused SSRP1-M or GST-fused HIF was bound to 20 μl of glutathione-Sepharose beads (GE Healthcare). Then, the beads were incubated with reconstituted histone octamers or whole calf thymus histones in a binding buffer containing 50 mM Tris-HCl, 1 M NaCl, and 0.2 % Triton X-100 at pH 7.5 for 30 min at room temperature. The protein-bound glutathione resins were washed five times with the binding buffer, and the bound samples were subjected to SDS-PAGE. The gels were stained using Coomassie Blue. His-tag pull-down assays were carried out similarly, with the exceptions that His-tagged SSRP1 or His-tagged Rtt106 was used on Ni2+-nitrilotriacetate resins and reconstituted yeast histone octamers were input for Rtt106.
Results
Crystal structure of the middle domain of SSRP1
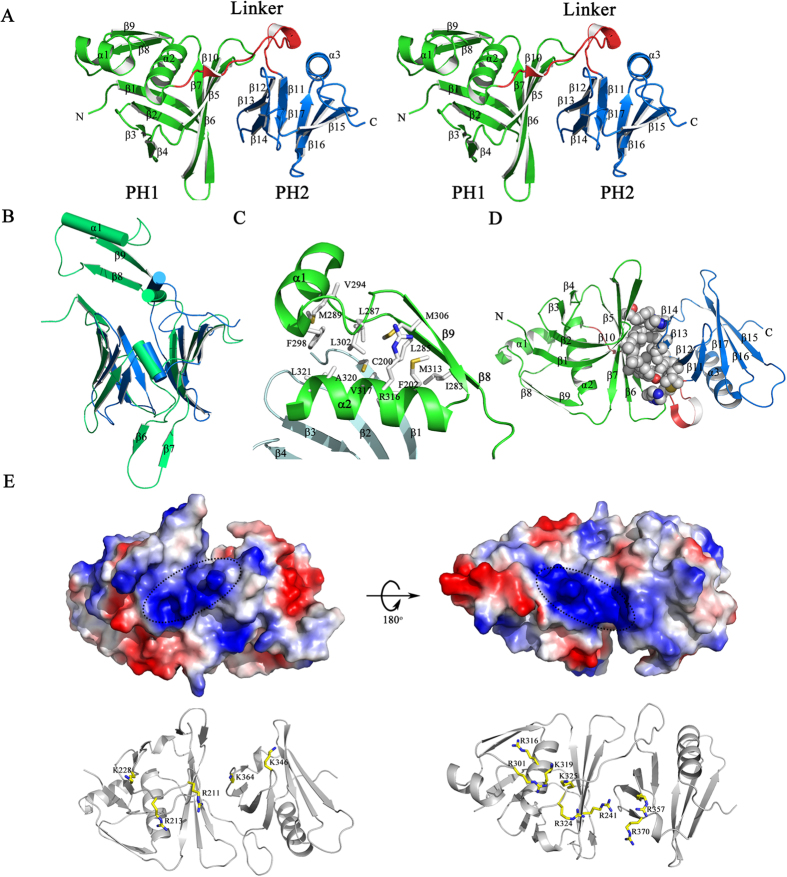
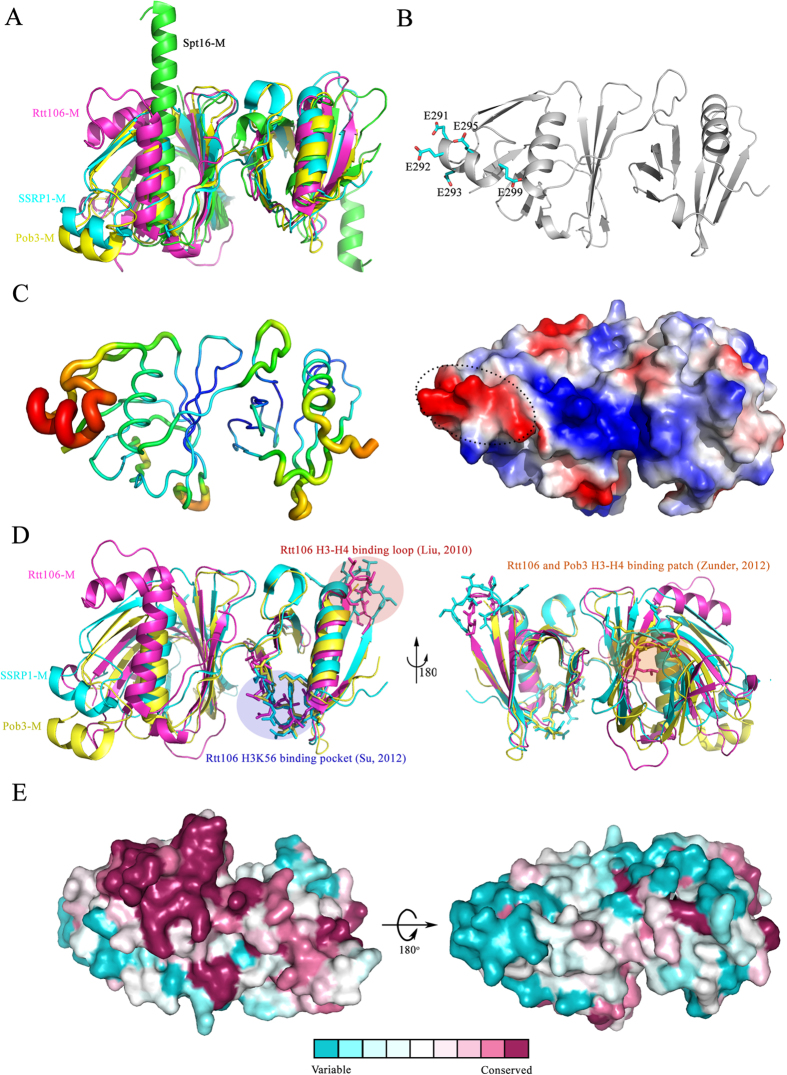
The crystal structure of the middle domain of SSRP1 (SSRP1-M) was determined at a resolution of 1.9 Å using molecular replacement; one molecular in the asymmetric unit is consistent with the observation that SSRP1-M is eluted as a monomer during the size exclusion chromatography of the purification process. The crystallographic statistics are summarized in Table 1. The structure contains tandem pleckstrin homology (PH) domains (Fig. 2A). In particular, the second PH domain (PH2) highly resembles the typical PH domain, which comprises of a seven-stranded antiparallel β-barrel capped by a C-terminal helix53. Superposition of the SSRP1-M PH1 domain (residues 197–328) with the PH2 domain (residues 329–427) revealed a root-mean-square (r.m.s.) deviation of 2.4 Å on 89 pairs of Cα atoms (Fig. 2B). In comparison with the second PH domain, the PH1 domain contains two extra antiparallel strands (β8 and β9) linked by a helix (α1), which are inserted between the last strand of the PH domain (β7) and the β-barrel-capping helix (α2). This, leads to a better-sealing cap. In addition, in the other corner of the β-barrel, the much longer β-strands (β6 and β7) in the PH1 domain form a tighter interaction composed of three inter-strand loops, which are usually potential binding sites. The extra secondary structure is stabilized by its hydrophobic interactions with residues that reside on the β1 and α2 of PH1, as shown in the enlarged view in Fig. 2C. The hydrophobic residues, which include I283, L285 and M306 on β8 and β9; M289, V294 and F298 on α1; L287 and L302 on the in-between linker; and M313, R316, V317, A320 and L321 on α2; and C200 and F202 on β1, form an extensive hydrophobic core. Notably, these residues are highly conserved among the homologues of SSRP1 (Fig. 1), suggesting that this extra topology is evolutionarily conserved and may be functionally significant. The two PH domains are associated with each other rather than independent, burying a solvent-accessible surface area of approximately 680 Å2. They interact primarily through the hydrophobic contacts mediated by the residues located on β5 to β7 of the PH1 domain and on β2 to β4 of the PH2 domain (Fig. 2D). Interestingly, these residues are also well conserved across species (Fig. 1), suggesting that the compact topology is evolutionarily conserved. The analysis of the electrostatic potential surface of the SSRP1-M structure revealed that there is a positively charged region stretching across the PH domains on each side of SSRP1-M (Fig. 2E, upper panel). Several positively charged residues, including K228, R213, R211, K364 and K346 on one side and R316, R301, K319, K325, R324, R241, R357, and R370 on the other side, constitute the positively charged regions (Fig. 2E, lower panel). Most of these residues are highly conserved (Fig. 1) implying functional relevance.
Figure 2. Structural features of SSRP1-M.
(A) A stereoscopic representation of SSRP1-M. The secondary structural elements are labelled. The structure is coloured according to the domains, as follows: the PH1 domain (residues 197–323), green; the inter-domain linker (residues 324–340), red; and the PH2 domain (residues 341–427), blue. (B) The structural superposition of PH1 domain (green) and PH2 domain (blue) of SSRP1-M with the differences labelled. (C) The extra βαβ topology of the PH1 domain stabilized by hydrophobic interactions. (D) The hydrophobic contacts between the PH1 domain and PH2 domain. The involved residues are indicated as spheres coloured by atom-type (O, red; N, blue; C and H, grey). (E) Upper panel, the electronic potential surface of SSRP1-M. The two positively charged patches are highlighted. Lower panel, the basic residues comprising the positively charged patches.
SSRP1-M binds to DNA non-specifically
Because arginine and lysine residues are commonly involved in interactions with DNA24,27, the positively charged regions suggest the DNA-binding activity of SSRP1. It has been reported that SSRP1 binds to linker DNA among nucleosomes54, and it is generally attributed to the C-terminal HMG-1 domain, which enables the proteins to bind to DNA as it enters and exits a nucleosome55.
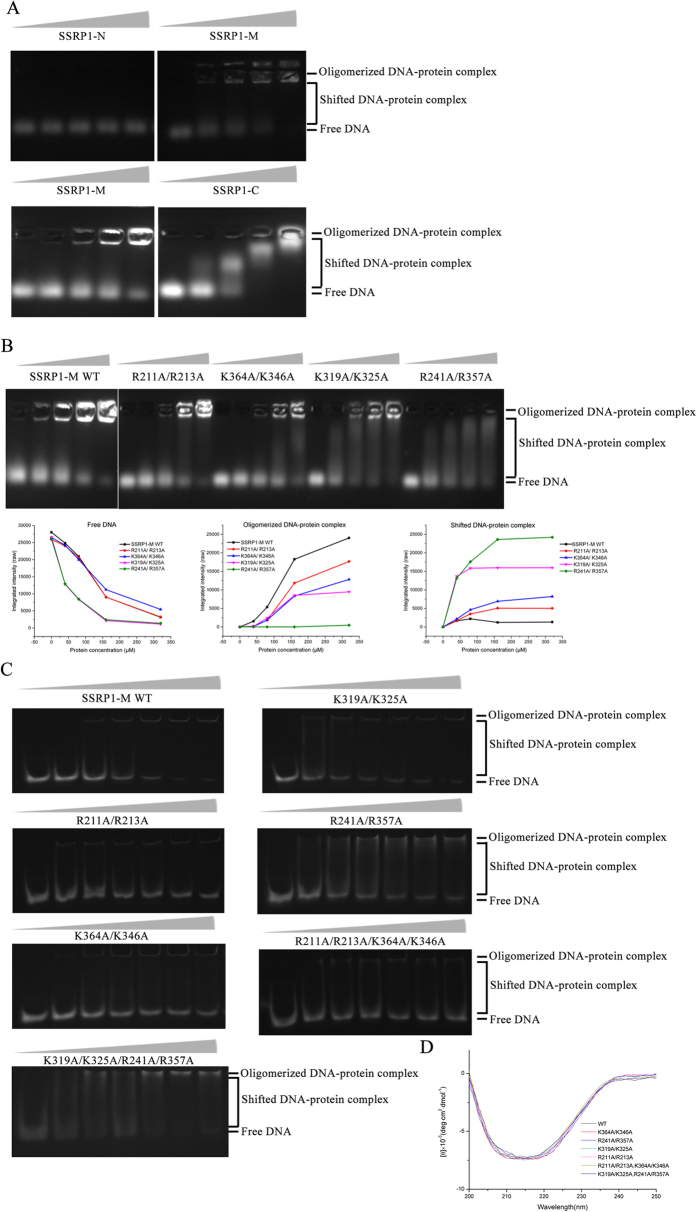
To investigate whether SSRP1-M contributes to the DNA binding of SSRP1, we tested the DNA-binding abilities of different constructs of SSRP1 via an agarose gel electrophoretic mobility shift assay (EMSA) using a 30-bp linker dsDNA, which has an identical sequence to the linker regions of the 207-bp nucleosomal DNA (601 sequence)56. As shown in the upper panel of Fig. 3A, no obvious change in the amount of free DNA in the gel was observed after the addition of SSRP1-N. For the addition of SSRP1-C, which contains the HMG domain and has been established as a DNA-binder, a shift from the free to the bound form in the gel was observed (Fig. 3A lower panel). Furthermore, the addition of increasing amounts of SSRP1-M caused the amount of free DNA in the gel to progressively decrease, and an increasing amount of DNA oligomerized in the wells (Fig. 3A upper and lower panels). As shown in our figures, the free DNA bands were labelled as “free DNA”; the shifting bands in the gel were labelled as “shifted DNA-protein complex”; and the bands in the well were labelled as “oligomerized DNA-protein complex”.
Figure 3. SSRP1-M is able to bind to DNA.
(A) Agarose EMSA results of different constructs of SSRP1 with 30-bp linker dsDNA. In each lane, 8μM dsDNA was incubated with increasing amounts of protein (0, 40, 80, 160, and 320 μM for SSRP1-N and SSRP1-M, and 0, 10, 20, 40 and 80 μM for SSRP1-C). Upper panel, the gels were stained for 10mins; lower panel, the gels were stained for 15mins. The relevant bands were labelled. (B) Upper panel: agarose EMSA results of SSRP1-M and its four mutants, R211A/R213A, K364A/K346A, K319A/K325A and R241A/R357A. The concentration of dsDNA in each lane was 8 μM. The concentrations of protein were the same as SSRP1-M in (A). Lower panel: raw quantified curves for the amount of free DNA, oligomerized DNA-protein complex and shifted DNA-protein complex as labeled in each gel against the protein concentration. (C) Native polyacrylamide EMSA results of SSRP1-M and its mutants, R211A/R213A, K364A/K346A, K319A/K325A, R241A/R357A, R211A/R213A/K364A/K346A and K319A/K325A/R241A/R357A, with 30-bp linker dsDNA. The concentration of dsDNA in each lane was 8 μM. The concentrations of proteins in each gel were 0, 10, 20, 40, 80, 160, and 320 μM, respectively. (D) CD spectra of SSRP1-M and its six mutants as indicated.
To investigate why DNA is oligomerized in the well, we mutated some basic residues on each positively charged surface of SSRP1-M to alanine according to its structure and observed their behaviors in gel shift assays. In agarose EMSA (Fig. 3B), all the four mutants caused less oligomerized DNA-protein complex but more shifted DNA-protein complex than the wild-type protein, as judged from both gel graphs and raw quantified curves. While the amount of free DNA at the end of the run is similar for the wild type and R211A/R213A across the protein concentrations tested, the graphs and curves show that K364A/K346A had more free DNA than the wild-type, which indicates that this mutant has reduced DNA-binding ability. K319A/K325 and especially R241A/ R357A, caused the amounts of free DNA and oligomerized DNA-protein complex greatly reduced and correspondingly, the amount of shifted DNA highly increased by comparing with the wild-type protein. The result shows that the two mutants relieved the highly oligomerized DNA-protein complexes by shifting them into smaller complexes, which are able to run in the gel. This finding suggests that the four residues, R319, K325, especially R241 and R357, play key roles in the oligomerization of DNA-protein complexes. For the native polyacrylamide EMSAs (Fig. 3C), the mutants R211A/R213A and K364A/K346A caused more free DNA than the wild-type protein. While a little gel shifting appeared in some lanes with the K319A/K325A mutant, much more shifted DNA-complex was observed in each lane with the mutant R241A/R357A than the wild-type protein. Our results showed that SSRP1-M is able to bind to DNA and form DNA-protein complexes. After binding DNA, these complexes oligomerize to form high molecular oligomers that are retarded in the well region of the gel. Residues R211, R213, especially K364 and K346 are important for the interaction between SSRP1-M and DNA to form a complex, whereas the residues K319, K325, especially R241 and R357, contribute to the high oligomerization of DNA-protein complexes. Consistently, the results of the native polyacrylamide EMSA assay of the four-site mutants showed that the R211A/R213A/K364A/K346A mutant obviously decreased the DNA binding affinity of SSRP1-M, while the R241A/R357A/K319A/K325A mutant caused increasing gel shifting compared with the wild-type protein (Fig. 3C). All of the mutants exhibited similar CD spectra to that of wild-type SSRP1-M (WT), indicating that the mutation did not affect the folding of the proteins (Fig. 3D).
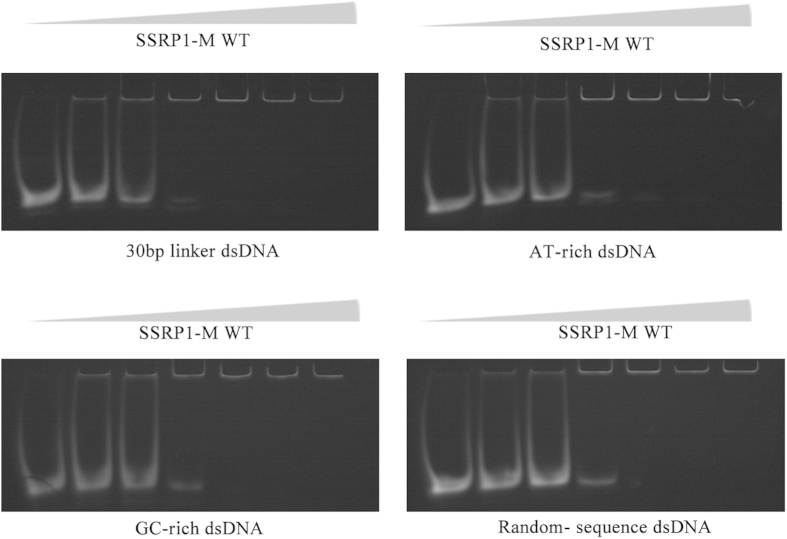
To investigate whether SSRP1-M has a DNA sequence preference, we performed binding studies of SSRP1-M with a 30-bp linker dsDNA, AT- or GC-rich sequences or a random-sequence DNA using EMSA assays. As shown in Fig. 4, no obvious differences in the DNA-binding affinity of SSRP1-M were observed for the different types of dsDNA, indicating that SSRP1-M binds to DNA with no sequence preference.
Figure 4. SSRP1-M binds to DNA without a sequence preference.
EMSA results of SSRP1-M with four different types of dsDNA, including 30-bp linker dsDNA, an AT- rich sequence, a GC-rich sequence and a random-sequence DNA. In each lane, 8 μM dsDNA was incubated with increasing amounts of SSRP1-M (0, 20, 40, 80, 160, and 320 μM, respectively).
In summary, our results indicated that SSRP1-M is able to bind to DNA nonspecifically and the positively charged patch, including residues R211, R213, K364 and K346, is important for the DNA-binding ability of SSRP1-M, while the other positively charged patch on the opposite side takes part in the oligomerization of DNA-protein complexes. This is the first time that the DNA-binding ability of SSRP1 has been demonstrated outside the SSRP1-C domain.
SSRP1-M was unable to bind to histones
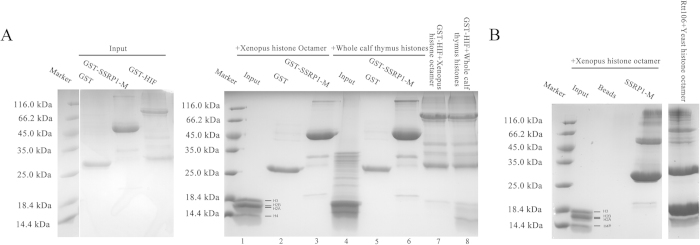
The FACT complex, which is composed of Spt16 and SSRP1, possesses intrinsic histone chaperone activity4, and different components of the yFACT complex, including Spt16-N, Spt16-M and Pob3-M, have been reported to bind to histones18,22,23,25. However, no study has reported about the histone-binding ability of the individual domains of SSRP1. Therefore, it is of interest to determine whether SSRP1-M binds to histones. First, we performed GST pull-down assays using the recombinant GST fusion proteins. We reconstituted Xenopus histone octamers from recombinant histones and tested the interaction between SSRP1-M and the octamers. By comparison with the proteins on their own, it was found that SSRP1-M does not bind to histones, whereas the HIF protein, which has been reported to selectively interact with H3 and H4 histones57, binds to the H3 histone (Fig. 5A, lanes 1–3 and 7). Furthermore, we performed pull-down assays with whole calf thymus histones, and similarly, there was no obvious binding detected for SSRP1-M, in contrast to the HIF protein (Fig. 5A, lanes 4–6 and 8). In addition, we performed His-tag pull-down assays, and the same results were observed. There was no obvious binding detected between SSRP1-M and the Xenopus histone octamers. In contrast, Rtt106, which is reported to bind to histone H3-H427, substantially bound to yeast histone octamers (Fig. 5B) under the same experimental conditions. In summary, our pull-down assay results showed that SSRP1-M binds weakly, if at all, to histones compared with HIF and Rtt106.
Figure 5. SSRP1-M cannot bind to histones, as determined by in vitro pull-down assays.
(A) The input proteins alone are shown on the left. The results of the GST pull-down assays of SSRP1-M with reconstituted Xenopus histone octamers and whole calf thymus histones are shown on the right, using GST-HIF as a positive control. The bands of the histones are indicated. (B) The His-tag pull-down assays of SSRP1-M with reconstituted Xenopus histone octamers, using His-Rtt106 bound to yeast histone octamers as a positive control. The bands of the histones are indicated.
Discussion
Our studies revealed that the middle domain of SSRP1 adopts a compacted structure with tandem PH domain, similar to that of its yeast homologue Pob3 and the related proteins Rtt106-M27 and Spt16-M22,23. By superimposing the structures of these proteins with that of SSRP1-M, we found that the PH2 domains are quite similar, with the exception of an additional C-terminal α-helix in Spt16-M, and that the primary differences within the PH1 domain reside in the region between β7 and the last α-helix (Fig. 6A). Rtt106-M contains an inserted α-helix. In Spt16-M, there is no insertion, but the last α-helix is extended, forming a much longer helix. However, SSRP1-M and Pob3-M contain two extra antiparallel strands linked by a helix. As mentioned above, this extra topology is evolutionarily conserved and may be functionally significant (Fig. 1). In particular, five acidic residues located in the linking helix (E291, E292, E293, E295 and E299) make up a negatively charged region on the electrostatic potential surface of SSRP1-M (Fig. 6B). Most of these residues are highly conserved (Fig. 1), suggesting that they are associated with various biological roles, such as the participation in the reported interaction between SSRP1-M and p63γ36 and the interaction among SSRP1-M and α-, β-, and γ-tubulins37. Consistently, the linking helix displays a higher temperature factor within the structure, implying flexibility in its function (Fig. 6C). In addition, the structure of the FACT heterodimerization domain was reported to possess a single PH domain in Spt16-D and a tandem PH domain in Pob3-N22. The purpose of the duplication of the PH domain within the FACT complex remains to be addressed.
Figure 6. A structural comparison of SSRP1-M with other proteins adopting double PH domains and surface conservation analysis of SSRP1-M.
(A) The structural superposition of the tandem PH domains of SSRP1-M (cyan, PDB ID: 4IFS), Pob3-M (yellow, PDB ID: 2GCL; average r.m.s.d. of 1.52 Å on 225 Cα-atoms), Rtt106-M (magentas, PDB ID: 3GYP; average r.m.s.d. of 2.44 Å on 174 Cα-atoms), and Spt16-M (green, PDB ID: 4IOY; average r.m.s.d. of 2.55 Å on 178 Cα-atoms). (B) Upper panel: the acidic residues comprising the negatively charged region; Lower panel: the electronic potential surface of SSRP1-M with the unique negatively charged region labelled. (C) The B-factor distribution of SSRP1-M. The wider and redder tubing indicates higher B-factor. (D) The structural superposition of SSRP1-M (PDB ID: 4IFS), Pob3-M (PDB ID: 2GCL) and Rtt106-M (PDB ID: 3GYP). The reported histone binding surfaces of Pob3 and Rtt106 are indicated. (E) The surface conservation analysis of SSRP1-M. The orientations are kept the same as Fig. 2E.
All three proteins, Spt16-M, Pob3-M and Rtt106-M, have been reported to readily bind to the H3-H4 histones22,25,28. Although no structures of the complexes formed by these proteins with the H3-H4 histones have been reported, various regions of the proteins have been identified as important for histone binding. These regions include the basic patch in the PH1 domains of Rtt106-M and Pob3-M25, and a loop, and a hydrophobic area within the PH2 domain of Rtt106-M25,27,28. Spt16-M lacks the basic patch in its PH1 domain, but a prominent acidic patch in PH2 may account for the binding23. SSRP1-M has the highest sequence identity (38%) with Pob3-M, and the structure of SSRP1-M highly resembles that of Pob3-M, with an r.m.s. deviation of 1.52 Å on the 225 Cα-atoms (Fig. 6D). Similar to Pob3-M, SSRP1-M contains all the identified histone-binding regions of Rtt106-M, except the loop. The corresponding sequences of the loop are similar in SSRP1-M and Pob3-M (the amino acids between β15 and β16 in Fig. 1), although a loop was not observed in the structure of Pob3. However, SSRP-M showed little ability to bind histones in our pull-down assays, in contrast to all the three other proteins. It suggests that although the structures and sequences among SSRP1-M, Pob3-M and Rtt106 are quite similar, the histone binding ability is different. It may stem from the slight difference in human histones and yeast histones leading to the different binding models. Just as shown in Fig. 6D, even for the same protein Rtt106, the histone-binding sites can be different. SSRP1-M may lack the required site not shown here to bind human histones. By comparing the domain structures of SSRP1 and Pob3 plus Nhp6, SSRP1 has one unique intrinsically disordered (IDD) domain just on the C terminal of SSRP1-M21,58. Considering the flexibility of IDD domain, it may either bind histone alone or assist SSRP1-M to bind histone. The mechanisms of histone binding remain unclear. Therefore, further assays are required to uncover the detailed mechanisms.
The SSRP1 protein is one of many HMG domain-containing proteins, which usually either contain multiple HMG domains or a single HMG domain without any other DNA-binding region59,60. However, our study showed that SSRP1-M also binds to dsDNA non-specifically. One of the positively charged patches on the surface of SSRP1-M, the R211A/R213A side, participates in the binding of dsDNA binding. The opposite positively charged patch, which includes residues K319, K325, R241 and R357, participates in the high oligomerization of the DNA-protein complexes. Considering that SSRP1-M exists as a monomer as determined by gel filtration, the oligomerization of the DNA-protein complexes is promoted by the addition of DNA. The binding of DNA through the R211A/R213A side may cause the confirmation change of SSRP1-M, which makes the opposite side more exposed to the solvent, inducing the oligomerization among DNA-protein complexes. However, the oligomerization should be only observed for our construct, which makes the R241A/R357A side of SSRP1-M exposed to the solvent. For the in vivo full-length SSRP1 protein, such a positively charged patch may bind to other parts of SSRP1 to form a compact protein, avoiding the oligomerization. Furthermore, there is an acidic residues-rich region between the SSRP1-M and SSRP1-C domains61, which may interact with the R241A/R357A side of SSRP1-M. When comparing the electronic potential surfaces of Rtt106-M and SSRP1-M, the positively charged patch on the R211A/R213A side of SSRP1-M corresponds to the only positively charged patch on the surface of Rtt106, which plays important roles in the binding of DNA and is essential for its functions27. Consistently, the positively charged patch on R211A/R213A side of SSRP1-M is much more conserved than the opposite side according to the surface conservation analysis (Fig. 6E). This suggests that such a positively charged patch on the structure of SSRP1-M might be crucial for the binding of DNA in vivo In addition, Pob3-M has also been found to bind to dsDNA27, suggesting its conserved role in DNA binding. It has been reported that SSRP1 requires linker DNA to interact stably with the nucleosome and that the presence of linker DNA positively influences the occupancy of the FACT complex54. According to our studies, SSRP1-M also directly contributes to the binding of linker DNA, except for the high-affinity DNA binding domain HMG-1. Therefore, SSRP1-M may act as an important fragment that helps SSRP1 and the FACT complex bind to DNA or chromatin. It was reported that during apoptosis, SSRP1-C is released from chromatin, while SSRP1- (N + M) remains bound to Spt16 in a tight association with chromatin40. The nonspecific DNA-binding ability of SSRP1-M may contribute to the binding of the truncated FACT complex to chromatin at this stage. Given the multi-domains of SSRP1, the domains may interact with each other, thus intermolecularly masking the C-terminal HMG-1 domain. When Spt16 is present to bind to SSRP1-N13,21, a conformational change occurs that unveils the HMG domain of SSRP1, which is consistent with the report that the presence of Spt16 notably increases the nucleosomal DNA binding of SSRP1 and even the FACT complex16,54. The DNA-binding ability of SSRP1-M may also contribute to other DNA-binding functions of SSRP1 independent of Spt16, such as transcriptional regulation32.
In summary, in this study, we present the crystal structure of SSRP1-M. We uncovered its dsDNA-binding ability, which is in addition to the well-recognized ability of the HMG-1 domain to bind DNA, for the first time. Although SSRP1 is known to bind H3-H4 histones4, we found that the SSRP1-M domain does not substantially bind to histones. Our structural and biochemical studies of SSRP1-M shed light on the functions of the multiple domains in the SSRP1 protein and facilitate the understanding of the molecular mechanisms of interaction with DNA and histones of the FACT complex.
Additional Information
Accession codes: Atomic coordinates and structure factors for human SSRP1 middle domain have been deposited in the Protein Data Bank under the accession codes 4IFS.
How to cite this article: Zhang, W. et al. Crystal Structure of Human SSRP1 Middle Domain Reveals a Role in DNA Binding. Sci. Rep. 5, 18688; doi: 10.1038/srep18688 (2015).
Acknowledgments
The authors thank the staff of BL17U at SSRF for assistance with synchrotron diffraction data collection. Financial support for this project was provided by the Chinese National Natural Science Foundation (Grant No. 31130018), the Chinese Ministry of Science and Technology (Grant No. 2012CB917200), the Chinese National Natural Science Foundation (Grant Nos. 31370732, 31270014 and U1432107), the Scientific Research Grant of Hefei Science Center of CAS (Grant No. 2015SRG-HSC042) and the Science and Technological Fund of Anhui Province for Outstanding Youth (Grant No. 1308085JGD08).
Footnotes
Author Contributions X.L., W.Z. and M.T. designed the research and wrote the manuscript; W.Z., F.Z., C.S., S.L. and H.L. performed the experiments; Y.L., Y.S. and L.N. discussed the project and provided reagents; all authors reviewed the paper.
References
- Formosa T. et al. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 20, 3506–3517 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades A. R., Ruone S. & Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol Cell Biol 24, 3907–3917 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H. et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell 35, 365–376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M. & Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 (1999). [DOI] [PubMed] [Google Scholar]
- Reinberg D. & Sims R. J. 3rd. de FACTo nucleosome dynamics. J Biol Chem 281, 23297–23301 (2006). [DOI] [PubMed] [Google Scholar]
- Biswas D., Yu Y., Prall M., Formosa T. & Stillman D. J. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol 25, 5812–5822 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster N. K., Johnston G. C. & Singer R. A. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol Cell Biol 21, 3491–3502 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger M. B. & Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155, 1593–1606 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162, 1557–1571 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K. et al. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol 9, 341–350 (1999). [DOI] [PubMed] [Google Scholar]
- VanDemark A. P. et al. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J Biol Chem 283, 5058–5068 (2008). [DOI] [PubMed] [Google Scholar]
- Keller D. M. & Lu H. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem 277, 50206–50213 (2002). [DOI] [PubMed] [Google Scholar]
- Keller D. M. et al. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 7, 283–292 (2001). [DOI] [PubMed] [Google Scholar]
- Sand-Dejmek J. et al. Concordant and opposite roles of DNA-PK and the “facilitator of chromatin transcription” (FACT) in DNA repair, apoptosis and necrosis after cisplatin. Mol Cancer 10, 74, doi: 10.1186/1476-4598-10-74. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell A. T., Oh S., Reinberg D. & Lippard S. J. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J Biol Chem 276, 25736–25741 (2001). [DOI] [PubMed] [Google Scholar]
- Brewster N. K., Johnston G. C. & Singer R. A. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem 273, 21972–21979 (1998). [DOI] [PubMed] [Google Scholar]
- Stuwe T. et al. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci USA 105, 8884–8889 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S. et al. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol 23, 5301–5307 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark A. P. et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell 22, 363–374 (2006). [DOI] [PubMed] [Google Scholar]
- Winkler D. D. & Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286, 18369–18374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M. et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013). [DOI] [PubMed] [Google Scholar]
- Kemble D. J. et al. Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J Biol Chem 288, 10188–10194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse J. E. et al. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol 323, 263–284 (2002). [DOI] [PubMed] [Google Scholar]
- Zunder R. M., Antczak A. J., Berger J. M. & Rine J. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc Natl Acad Sci USA 109, E144–153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res 30, 3643–3652 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J Biol Chem 285, 4251–4262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D. et al. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature 483, 104–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., King D. L., LaBonne C. & Kafatos F. C. A Drosophila single-strand DNA/RNA-binding factor contains a high-mobility-group box and is enriched in the nucleolus. Proc Natl Acad Sci USA 90, 6488–6492 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel L. et al. The HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp Cell Res 250, 313–328 (1999). [DOI] [PubMed] [Google Scholar]
- Xiang Y. Y. et al. Expression of structure-specific recognition protein mRNA in fetal kidney and Fe-nitrilotriacetate-induced renal carcinoma in the rat. Cancer Lett 106, 271–278 (1996). [DOI] [PubMed] [Google Scholar]
- Dyer M. A., Hayes P. J. & Baron M. H. The HMG domain protein SSRP1/PREIIBF is involved in activation of the human embryonic beta-like globin gene. Mol Cell Biol 18, 2617–2628 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zeng S. X., Landais I. & Lu H. Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J Biol Chem 282, 6936–6945 (2007). [DOI] [PubMed] [Google Scholar]
- Shirakata M. et al. HMG1-related DNA-binding protein isolated with V-(D)-J recombination signal probes. Mol Cell Biol 11, 4528–4536 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J. A., Baron M. H. & Olson E. N. Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J Biol Chem 274, 15686–15693 (1999). [DOI] [PubMed] [Google Scholar]
- Zeng S. X., Dai M. S., Keller D. M. & Lu H. SSRP1 functions as a co-activator of the transcriptional activator p63. The EMBO journal 21, 5487–5497 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S. X. et al. Structure-specific recognition protein 1 facilitates microtubule growth and bundling required for mitosis. Mol Cell Biol 30, 935–947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn S. L., Pil P. M., Essigmann J. M., Housman D. E. & Lippard S. J. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc Natl Acad Sci USA 89, 2307–2311 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn S. L., Housman D. E. & Lippard S. J. Isolation and characterization of cDNA clones encoding the Drosophila homolog of the HMG-box SSRP family that recognizes specific DNA structures. Nucleic Acids Res 21, 1643–1646 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais I., Lee H. & Lu H. Coupling caspase cleavage and ubiquitin-proteasome-dependent degradation of SSRP1 during apoptosis. Cell Death Differ 13, 1866–1878 (2006). [DOI] [PubMed] [Google Scholar]
- Keller Dm, L. H. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem 277, 50206–50211 (2002). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Vagin A. & Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Davis I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat Meth 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. P. et al. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 19, 1287–1292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones. Biochem J 462, 465–473 (2014). [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T. J. & Richmond T. J. Preparation of nucleosome core particle from recombinant histones. Method Enzymol 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- He C., Li F., Zhang J., Wu J. & Shi Y. The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem 288, 4692–4703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A. Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32, 707–711 (2004). [DOI] [PubMed] [Google Scholar]
- Winkler D. D., Muthurajan U. M., Hieb A. R. & Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem 286, 41883–41892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. & Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 54, 35–100 (1996). [DOI] [PubMed] [Google Scholar]
- Lowary P. T. & Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- Ai X. & Parthun M. R. The Nuclear Hat1p/Hat2p Complex: A Molecular Link between Type B Histone Acetyltransferases and Chromatin Assembly. Molecular cell 14, 195–205 (2004). [DOI] [PubMed] [Google Scholar]
- Tsunaka Y. et al. Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J Biol Chem 284, 24610–24621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Giese K. & Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10, 94–100 (1994). [DOI] [PubMed] [Google Scholar]
- Landsman D. & Bustin M. A signature for the HMG-1 box DNA-binding proteins. Bioessays 15, 539–546 (1993). [DOI] [PubMed] [Google Scholar]
- Bruhn S. L., Pil P. M., Essigmann J. M., Housman D. E. & Lippard S. J. Isolation and Characterization of Human Cdna Clones Encoding a High Mobility Group Box Protein That Recognizes Structural Distortions to DNA Caused by Binding of the Anticancer Agent Cisplatin. P Natl Acad Sci USA 89, 2307–2311 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]