Abstract
We propose a novel, graph-theoretic framework for distinguishing arteries from veins in a fundus image. We make use of the underlying vessel topology to better classify small and midsized vessels. We extend our previously proposed tree topology estimation framework by incorporating expert, domain-specific features to construct a simple, yet powerful global likelihood model. We efficiently maximize this model by iteratively exploring the space of possible solutions consistent with the projected vessels. We tested our method on four retinal datasets and achieved classification accuracies of 91.0%, 93.5%, 91.7%, and 90.9%, outperforming existing methods. Our results show the effectiveness of our approach, which is capable of analyzing the entire vasculature, including peripheral vessels, in wide field-of-view fundus photographs. This topology-based method is a potentially important tool for diagnosing diseases with retinal vascular manifestation.
Keywords: artery-vein classification, medical imaging, graph theory, tree topology, image analysis
I. Introduction
Various diseases affect blood circulation, thereby leading to either a thickening or a narrowing of arteries and veins [3], [17]. In particular, asymmetric changes in retinal arteriolar vs. venular diameter, as measured by the arteriolar-venular ratio (AVR) [4], have been correlated with a number of diseases including coronary heart disease and stroke [52], as well as atherosclerosis [18]. Additionally, a high AVR has been associated with higher cholesterol levels [18] and inflammatory markers, including high-sensitivity C-reactive protein, interleukin 6, and amyloid A levels [25]. Other conditions that can cause an abnormal AVR include high blood pressure and diseases of the pancreas [27]. Furthermore, a low AVR is a direct biomarker for diabetic retinopathy (DR), the leading cause of vision loss in working age individuals in developed countries [27]. This low AVR is caused by abnormal widening of the veins due to retinal hypoxia, which arises secondary to microvascular injury in the setting of chronic high blood sugar levels. These vascular changes often precede the onset of symptoms, and if detected, may allow preventive treatments that can reduce the risk of any vision loss.
These changes in the retinal vasculature can be captured using a variety of imaging methods, including fundus photography, fluorescein angiography, and optical coherence tomography. In particular, fundus photography is the preferred retinal imaging modality for both telemedicine and remote diagnostics due to its lower cost and ease of use. However, it is highly challenging to compute the AVR or any other measure of interest given a fundus image of a patient’s arteries and veins, even after accurately segmenting the vasculature from the image. Manually classifying arteries and veins requires expertise in retinal image interpretation and is a very labor-intensive process; in a high resolution image we can often detect over one hundred vessels or vessel segments, many of which are ambiguous and require careful viewing to classify. It is perhaps due to these time constraints that the classic AVR is only calculated using the six largest arteries and veins near the optic nerve [26]. We propose, however, that calculating a global AVR using the widths of smaller vessels might yield an even earlier biomarker of an underlying disease since smaller vessels are more vulnerable to changes in blood pressure.
However, it is time-prohibitive to manually calculate this global AVR due to the large number of vessels that need to be measured. Therefore, automatic or semi-automatic methods are needed to overcome the time constraints of manual classification. Traditionally, most computer-aided vessel analysis has focused on segmenting the vessels in the image [1], [8], [20], [21], [30], [44], [47], [54]. Artery-vein (AV) classification, on the other hand, goes one step further and seeks to classify the segmented vessels into either arteries or veins. However, even assuming a perfect segmentation, the aforementioned ambiguity of small and midsized vessels makes automatically classifying arteries and veins a very difficult computational task.
To address this problem, we propose a novel, graphtheoretic approach for classifying arteries and veins in a fundus image. Given a semi-automatically extracted planar graph representing the vasculature, we accurately estimate the label of each vessel. Two key features of our method are that it classifies the entire vasculature, not just the most prominent vessels, and it estimates the underlying topology—how the different vessels are anatomically connected to each other—not just the vessel types. A few methods based on graph theory have been proposed for this problem [5], [22], [29], [45]. However, these earlier approaches are limited because they determine the graph’s vessel labels either by relying on a set of initial hand-labeled edges [45], by using fixed rules for determining how the original vessels overlap each other [5], [22], or by constraining the vessels to a small region of interest (ROI) [29]. In contrast, we robustly estimate the most likely vessel labels by efficiently searching the space of valid labelings. Here, we extend our previous tree topology estimation work [7] in two ways: we incorporate global model features that are specifically tailored to arteries and veins and present a novel, heuristic optimization algorithm to better explore the space of possible vascular networks. Our features are based on expert, domain-specific knowledge about how arteries and veins perfuse the retina. Our proposed framework represents an important step towards developing a clinically viable, automatic AV classification method.
The main contributions of this work are as follows:
A global likelihood model that accurately captures the structural plausibility of a given set of vessel labels.
A novel best-first search algorithm that efficiently explores the space of possible vascular networks.
A non-metric, random optimization scheme that is able to avoid getting stuck in local minima.
The formalization of the artery-vein classification problem in terms of the underlying network topology.
A novel online dataset of wide-field retinal color images annotated for artery-vein classification.
The rest of our paper is organized as follows: in Section II, we first review prior work on segmenting and classifying arteries and veins. We provide an overview of our classification framework in Section III and present our likelihood model for labeled trees in Section IV. We then detail an efficient search method for exploring the space of possible solutions in Section V. Finally, we evaluate our algorithms on four retinal datasets in Section VI and conclude in Section VII.
II. Prior Work
The problem of classifying arteries and veins is relatively new. Compared to other vessel analysis tasks such as vessel segmentation [11], [14], [19], [50], [55] or vessel centerline extraction [10], [16], [23], [38], [48], there have been only a few automatic or semi-automatic AV classification methods proposed. We now review some of the most important existing approaches for this problem.
AV classification methods generally rely on some combination of vessel features, primarily pixel color, and connectivity constraints to assign one of the two labels to each vessel segment. Mirsharif et al. [32] used a three-step method: they first enhanced the input image, then they estimated different pixel color features and then corrected misclassifications at bifurcations. Konderman et al. [27] explored using both support vector machines and neural networks combined with principal component analysis (PCA) features obtained from small vessel image patches. Relan et al. [43] used a Gaussian mixture model on small vessel patches to classify the main vessels in each optic nerve-centered quadrant, while Vasquez et al. [51] employed a minimal path approach with which they connected a set of extracted concentric vessel segments. Zamperini et al. [57] focused on determining effective features for AV classification. They compared color, spatial, and size features and concluded that a mix of color and position features provided the best results. Other work has focused specifically on estimating the AVR using either the U.S. [4] or the Japanese [33] definition.
There has been some work on using graph theory to better classify arteries and veins. All these methods first establish a graph that represents the projected vascular network and then classify different parts of the graph as being either arteries or veins. Rothaus et al. [45], [46] used a semi-automatic method in which they propagate some initial manual edge labels throughout the graph by solving a constraint-satisfaction problem. Lau et al. [29] constructed their graph over a restricted ROI around the optic nerve and then assigned the vessel labels by approximating an optimal forest of subgraphs. Joshi et al. [22] first separated their vascular graph using Dijkstra’s shortest-path algorithm to find different subgraphs. They then labeled each subgraph as either artery or vein using a fuzzy classifier. Finally, Dashtbozorg et al. [5] also proposed a two-step graph estimation method for distinguishing arteries from veins. They first split the vascular graph into subgraphs based on the local angles between edges and then assigned a label to each subgraph using linear discriminant analysis.
Our proposed method differs from these earlier approaches in a number of ways. First, we do not rely on any manually labeled edges. Second, our vascular graph encompasses the entire visible retina instead of a narrow ROI. Finally, we do not assign edge labels based on fixed rules that define when two vessels cross and when a single vessel branches out. Instead, we robustly determine the most likely set of edge labels by efficiently searching through the space of possible labelings. Our likelihood model is robust and allows for arbitrary ways in which vessels could overlap each other. Finally, our algorithm outputs not only the edge labels, but also the undelying topology, which is also a clinically significant marker, particularly for hypertension [31] and diabetic retinopathy [34]. In the next section, we present an overview of our proposed classification framework.
III. Framework Overview
In this section, we provide an overview of our vessel classification framework, which relies on estimating the overall vascular topology in order to classify each individual vessel. Although arteries and veins are anatomically distinct, they overlap each other in a two-dimensional retinal image, as Figure 2 illustrates. In general, it is non-trivial to determine from a single image which vessels are arteries and which are veins. To tackle this problem, we build upon the graph-based, topology estimation approach of [7], which estimates the most likely topology of a rooted, directed, three-dimensional tree given a single two-dimensional image of it and a growth model for that type of tree. We solve this inverse problem through a combination of greedy approximation and heuristic search algorithms that explore the space of possible trees.
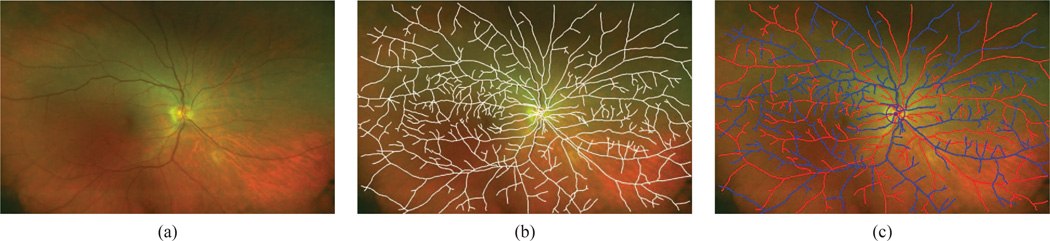
Fig. 2. Arteries vs. veins.
(This Figure is best viewed on-screen.) (a) Retinal arteries and veins overlap each other throughout a wide field-of-view color fundus image. (b) We construct a planar graph (overlaid in white) that captures the projected vascular topology. (c) Each edge in the graph is either an artery (in red) or a vein (in blue).
In our framework, we represent the original vascular network as a directed tree T with a set of artery-vein edge labels and the projected vascular network as an undirected planar graph G, respectively, such that the projection P(T) of T is G. Given G, we estimate the most likely topology and edge labels of T based on a parametric likelihood model that defines the functional and structural viability of every possible labeled tree.
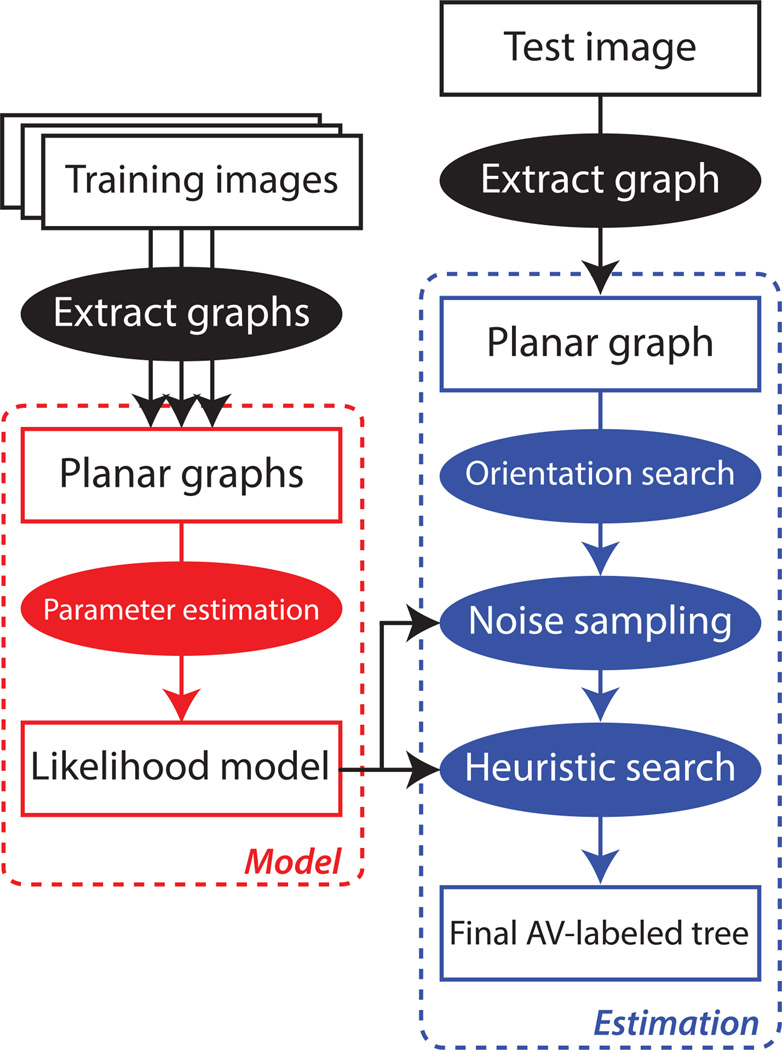
Figure 1 provides a visual summary of the pipeline that we use to estimate T. The left-hand side, in red, lists the steps needed to optimize the parameter values of our likelihood model. We describe this model in detail in Section IV and explain how to set its parameters by using a training set with ground-truth labels in Section VI-B4. The right-hand side in blue, on the other hand, describes the sequence of steps needed to estimate a labeled tree given an input image, once we have a trained model and semi-automatically estimated a planar graph. Our approach has three main steps:
We first estimate the most likely direction of flow for each vessel using the heuristic search algorithm detailed in [7].
We then sample multiple trees consistent with the chosen flow directions by randomly perturbing the feature values that we use to compute a tree’s likelihood.
Finally, we refine the best sampled tree by exploring the full space of possible labeled trees using best-first search.
We describe each of these steps in detail in Section V. In the following sections, however, we present the key concepts behind our approach: we first discuss how to assign anatomically valid labels to the edges of T and then formally describe the problem of estimating T from G.
Fig. 1. Estimation pipeline overview.
(This Figure is best viewed in color.) Given a planar graph semi-automatically extracted from a retinal image, we estimate the most likely AV-labeled tree. The red bubbles correspond to the steps for estimating the likelihood model’s parameters, while the blue bubbles indicate the steps needed to estimate an AV-labeled tree given a single planar graph. The ellipses correspond to processes, while the rectangles denote data structures. The model parameter estimation steps on the left are computed beforehand using a training set of graphs.
A. Trees and edge labelings
Let T = (VT, ET, rT) be a three-dimensional, rooted, directed tree. VT and ET are the vertices and edges of the tree, respectively, and rT ∈ VT is its root. In this work, T is a graph that represents the topology of the retinal vessels in a given eye. That is, the way the edges of T are connected to each other mirrors how the vessels branch out from the optic nerve. Thus, different retinal networks correspond to different directed trees. Anatomically, every edge in T corresponds to part of either an artery or a vein, but not both. We represent this relationship between edges and vessel types via a binary edge labeling that assigns one label to every artery edge (and assigns the opposite label to every vein edge).
Different edge labels correspond to different ways of classifying the retinal vessels. However, most binary edge labelings are anatomically implausible. In the retina, all arteries and veins emanate from the optic nerve (which corresponds to the root of T). Thus, all arteries are connected to each other, and so are all veins. Therefore, in order for a labeling to correspond to an anatomically plausible distribution of arteries and veins, all the artery-edges (and all the vein-edges, respectively) must be connected to each other and to the root. We refer to labelings that satisfy these adjacency constraints as AV-labelings.
More formally, given a binary edge-labeling ℓ of a graph T, let A, B ⊆ ET be the disjoint subsets of artery- and vein-edges, respectively and let TA and TB be their corresponding induced subgraphs of T. Also, let ET (rT) be the edges adjacent to the root in T. Then, ℓ is an AV-labeling if and only if:
TA (TB respectively) is either empty or has a single connected component.
If TA (TB respectively) is non-empty, then A ∪ E(rT) ≠ ∅. (B ∪ E(rT) ≠ ∅ respectively).
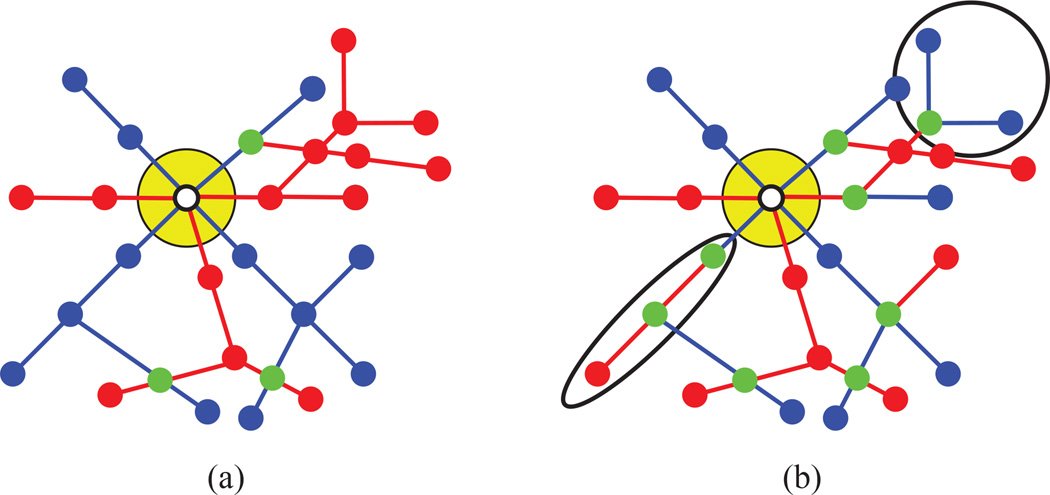
Figure 3 illustrates the difference between an AV- and a non-AV-labeling. The above definition is not limited to trees, but is applicable to general graphs. In order for a graph to admit AV-labelings that include both artery- and vein-edges, the degree of its root has to be greater than one, so in this work we assume that the roots of all graphs are adjacent to at least two edges.
Fig. 3. AV-labelings.
(This Figure is best viewed in color.) AV-labelings are a special subset of binary edge labelings in which the two subsets of edges induce connected subgraphs. (a) A projected graph assigned an AV-labeling. The white dot corresponds to the root and the yellow circle indicates the optic nerve. The arteries are marked in red and the veins in blue. Vertices that are connected to both vessels types are shown in green. Note that all the edges of the same label are connected to each other. (b) The same planar graph with a non-AV-labeling. In this labeling some arteries and veins are not connected to the root. Two of these disconnected sets of edges are circled in black.
We now show how to map a directed tree to a single AV-labeling consistent with it. The number of possible AV-labelings of a tree is given by:
where ℒ(T) is the set of AV-labelings of T and deg(rT) is the degree of the tree’s root [6]. The above equation follows directly from the second constraint: in an AV-labeling there must be at least one path from the root to every edge such that every edge along that path has the same label as the final edge. Since in a tree there is only one path from the root to each edge, every edge downstream of a root edge has to have the same label as its ancestor root edge.
In this work, we assume that deg(rT) is bounded, so we can list all the AV-labelings consistent with a given tree in constant time. In this case, given a likelihood function pM over AV-labeled trees, we can efficiently determine an AV-labeling ℓML that maximizes the tree’s likelihood:
| (1) |
where Tℓ indicates that T has been assigned the labeling ℓ. We detail our likelihood model in Section IV. Unless otherwise stated, whenever we refer to an AV-labeled tree, we assume that the tree has been assigned a labeling that maximizes its likelihood.
In summary, an AV-labeled tree captures how different retinal vessels are connected to each other and which vessels are arteries and which are veins. In the next subsection, we will detail how a retinal image obscures both the topology and the edge labels of the vascular network.
B. Tree projection
A retinal image is a projection of the retina onto two dimensions. In particular, a 2D image of the retinal vessels corresponds to a projection of T onto the plane. Thus, let G = (VG, EG, rG) be the undirected planar graph with root rG ∈ VG that results from projecting T, as detailed in [7]. Figure 2(b) shows a planar graph overlaid over a sample retinal image. In this work, we assume that G is a faithful representation of the projected vascular topology, so that any potential image noise is handled in the preprocessing stage which extracts a clean graph from the image. We leave more nuanced models of image formation for future work.
When we project a tree, different branches can overlap with each other, thereby obscuring both the original connectivity and the blood flow direction along each edge. Branches can even overlap each other over extended regions, such as when one vessel wraps around another or two vessels grow side-by-side. Therefore, in general many different trees are consistent with the same planar graph. However, under mild assumptions [7], projection preserves a one-to-one correspondence between the edges of the projected tree and the resulting planar graph, even in the presence of elongated overlaps. In this case, any AV-labeling of T maps to a unique AV-labeling of G, in which the corresponding edges have the same labels.
Now, let T* be the AV-labeled tree that best reflects the true retinal anatomy. To approximate T* given G, we construct a global likelihood model pM that predicts how likely it is for a given AV-labeled tree to be the true labeled tree. We then seek the most likely AV-labeled tree TML:
| (2) |
where 𝒯 (G) is the set of all directed trees consistent with G. If pM accurately models the topology and geometry of retinal vascular networks, then TML ≈ T*. In general, however, recovering the most likely tree given G is NP-hard [7]. To address this, in Section V, we present a novel, efficient search strategy for approximately optimizing (2). In Section VI, we show that this search strategy allows us to efficiently estimate the ground truth labeled trees for four different retinal datasets. In the following section, we present our likelihood model.
IV. Likelihood Model
In the previous section we defined our estimation problem, which consists of approximating the true AV-labeled tree T* given an input graph G. In this section we define a global likelihood model to estimate the anatomical plausibility of any possible AV-labeled tree. Our likelihood model extends our previous work [7], which defined the quality of each directed tree via a local likelihood model.
The purpose of retinal arteries and veins is to distribute oxygen and nutrients to the retinal tissues. As such, the vascular network evolved to best satisfy the fluid dynamic constraints involved in transporting blood. To satisfy these constraints, retinal arteries and veins grow subject to numerous, complex forces, including a genetic blueprint that directs when different parts of the vasculature will grow [13], as well as various feedback mechanisms that balance the growth of arteries and veins with other parts of the retina such as the nerve fiber layer or the plexiform layers [12]. The aforementioned forces and constrains lead most human retinas to display similar structural properties, such as a roughly 50–50 distribution of arteries and veins. While much is known about these anatomical processes, directly estimating these properties is generally not feasible. Due to image resolution limitations, we can never extract every artery and vein in the retina, so any flow calculations can only be approximate. Furthermore, angiogenesis models [39], [42] are computationally intensive and are not geared towards the problem of finding the most-likely AV classification in an existing vascular network.
Instead, in this work we rely on three features—local growth, overlap, and color—that capture some fluid dynamics and anatomical properties of arteries and veins, but are tractable to compute based on the extracted planar graph G. We developed our likelihood model based primarily on expert, domain-specific knowledge about how arteries and veins tend to perfuse the retina. Thus, we only make use of a small number of highly informative features to estimate the likelihood of each tree, whose efficacy is demonstrated by the experimental results in Section VI. We now describe each feature in turn.
A. Local growth model
We previously proposed a local generative prior model for the growth of various kinds of trees, including retinal vessels, plant roots, and lightning [7]. Our growth model defines the probability of a directed tree T as a function of its topology (the number of children per vertex) and geometry (the expected projected angles between the incoming edge of a parent and those of each of its children). More concretely, the local growth probability pg(T) is given by:
where vP is the projection of vertex υ onto the image plane and π(υ) and n(υ) are the parent and children of υ, respectively. The probability pa defines the likelihood of the angles between the incoming edge of υ and those of its children in the projection:
Here, fVM is a Von Mises distribution [24] with expected direction μi and concentration parameter κi; Vυ is the set of children of υ and eP (υ, wi) is the projection of the edge between υ and its child wi. The above set of Von Mises distributions determine what combinations of projected angles are most likely; in our experiments, we estimate their parameter values empirically.
The probability ps, on the other hand, defines the likelihood that υ has n(υ) children. In our experiments, it is given by a one-inflated Poisson distribution [28] that behaves as a Poisson distribution with rate λ at all values except 1:
where c = |n(υ)|. In retinal vessels, α >> Pois (1 ; λ) to account for the fact that a growing vessel is much more likely to continue growing, rather than stopping or splitting into two or more branches.
This model is local, in the sense that the expected number of children is the same for every vertex and their expected locations depend only on the current direction of growth and local environmental forces. Intuitively, our local growth model captures how likely it is, both topologically and geometrically, for blood to flow through the observed vessels in a given way.
B. Overlap
A given directed tree establishes which vertices of G are legitimate branch-points and which are crossings, i.e., points on the image where two different vessels overlap. As Figure 2 illustrates, though, arteries rarely cross other arteries and veins rarely cross other veins. Thus, we penalize crossings between vessels of the same type. Let m be the number of crossings of T and let ϕ and ψ be two indicator functions for pairs of crossings, such that:
Then, the overlap value is given by
Intuitively, po(T) is the percentage of crossings of T that are between an artery and a vein. Note that po(T) depends globally on the AV-labeled tree’s topology because an edge is constrained to have the same label as its ancestors. In other words, two AV-labeled trees that only differ in their topology at a single crossing may have vastly different percentages of artery-vein crossings, which makes overlap a global property of the tree’s topology.
C. Color
In color fundus images, veins generally appear darker than arteries because they carry deoxygenated blood back to the heart [56]. Human observers often make use of this color difference to differentiate the main arteries from the main veins when manually labeling an image. We likewise calculate the difference in color between the artery- and vein-edges, as follows. Let p be the vector that indexes all the pixels in the input image I. Now, let ω(e) be a vector of weights that define the overlap level between every pixel and the straight line segment between the two endpoints of edge e. We use Xiolin Wu’s line drawing algorithm [53] to determine each ω. Note that, in general, ω(e) is very sparse, i.e. most of the weights for each edge will be zero. Then, the red channel difference cr is given by:
where Ir(p) are the red channel pixel values for each pixel in the image and A and B are the sets of artery- and vein-edges respectively. We similarly calculate cg and cb for the green and blue channels, respectively. We then calculate the mean color value of the three channels:
Note that if the arteries are darker than the veins, then the color value is thresholded at 0. As with overlap, color is a global property of the AV-labeled tree’s topology because two trees that differ at only one crossing may have dramatically different color values.
D. Global likelihood
We combine the three features outlined above—local growth, overlap, and color—to assign the final, global likelihood to each possible tree. For simplicity, in this work we model their joint likelihood as a linear combination of the three features:
| (3) |
where each λi is the weight, or importance, of its respective feature. In our experiments, we determined the value of each λi using a training set of graphs with known edge labels. Intuitively, we refrain from using more complicated ways of combining our features because their effects are largely independent. In other words, the value of one feature does not affect the preferred values of the other features. For instance, knowing the local growth likelihood pg(T) of a tree does not affect the fact that we wish to maximize the percentage of artery-vein crossings or have the arteries be brighter than the veins.
V. Search Algorithm
In the previous section, we presented a likelihood model to estimate the quality of any AV-labeled tree that is consistent with an input planar graph. As noted in Section III, however, optimizing this model is NP-hard in general. In this section we present an efficient three-step heuristic search algorithm for exploring the space of possible AV-labeled trees. In the next section, we show empirically that this algorithm efficiently approximates the ground truth trees for four retinal datasets.
Our algorithm has three steps:
We estimate the most likely edge orientations using the heuristic search algorithm detailed in our previous work [7].
We sample multiple trees consistent with the chosen orientation by randomly perturbing the local growth probabilities for each vertex.
We refine the best sampled tree by exploring the space of possible trees using best-first search.
The rationale for this sequence of steps is to first bring our search to a neighborhood of the correct solution with step 1, then trade off exploration and exploitation through a random search in step 2, and finally optimize locally the best randomly-found solution in step 3. We detail each step in turn below.
Steps 1 and 3 of our search algorithm iteratively explore spaces of possible solutions. They take advantage of the fact that both the space of possible edge orientations of the input graph G and the space of AV-labeled trees consistent with G are fully connected under a suitable neighborhood relationship. That is, given a current solution (orientation or tree) consistent with G, it is possible to define a simple, local transformation that will yield a different solution consistent with G. Furthermore, any two possible solutions can be transformed from one to the other by a series of such local transformations. We showed this property of edge orientations in [7] and prove that the same property holds for AV-labeled trees in the Appendix.
A. Orientation search
In order to be able to explore the space of possible AV-labeled trees, we need a good starting point. We first estimate the most likely direction of each edge in the input graph by using the heuristic search method defined in our previous work [7]. This method explores the space of possible orientations of the input graph, instead of the space of possible trees. As noted above, this space of orientations is fully connected if we explore it by iteratively making a local change (flipping the direction of one edge) to our current orientation.
Although multiple trees may be consistent with the same orientation, we can estimate the most likely tree–in terms of pg–in linear time for any orientation by selecting the most likely vertex partition at each of the orientation’s crossings. A vertex partition specifies which of the edges in the crossing are actually connected to each other in the original tree. Thus, it suffices to explore the space of orientations to optimize pg. Orientations are much fewer than trees, so we can explore the set of orientations more thoroughly. As suggested above, in this step we only use pg to estimate the quality of each potential orientation. This initial search ends up estimating the correct direction of most edges, so that the subsequent, full search over the space of trees will start much closer to the correct solution.
B. Noise sampling
In the previous step, we estimated a single, most likely topology for each set of possible edge orientations. However, although the local model pg generally estimates the correct vertex partition at most crossings, any errors, even at a single crossing, can lead to a tree that has very low overlap and color likelihoods. This is because the local model does not factor in these global, retina-specific properties of a given tree. Thus, even if we have correctly estimated most of the edge directions, the most likely tree, as defined by pg, may have a relatively low likelihood in terms of the global model pM.
Here, we use a random sampling scheme to approximate the most likely tree, in terms of pM, consistent with the edge directions obtained in the previous step. As noted above, there are many trees that are consistent with any fixed orientation. Randomly generating these trees, either uniformly or proportionally to their likelihood is nontrivial; instead, here we use a simple, non-metric method, which we refer to as noise sampling, for drawing weighted samples from a large combinatorial space. While noise sampling is statistically biased—that is, some configurations are slightly more likely to be selected than others—our experiments suggest that this bias is negligible for many graphs of interest. Furthermore, there is no need to reject samples; every sample is guaranteed to be part of the target space by construction.
Noise sampling uses a deterministic algorithm to optimize a target function, but perturbs the function values randomly, thereby yielding outputs that are generally different from what they would be if the original values were used. By repeating this optimization multiple times with different, independent realizations of noise, we can sample a large space of possible values non-metrically. Furthermore, each sampled tree is drawn independently, so successive samples may differ arbitrarily in their topologies.
Here, our deterministic optimization algorithm consists of optimizing pg given the chosen orientation. As noted above, we can find the most likely tree in terms of our local growth model in linear time by iteratively selecting the most likely vertex partition at each crossing. By randomly perturbing these local likelihoods, we can obtain different trees consistent with a given orientation.
More concretely, let w be a vector whose i-th entry is the likelihood of the i-th vertex partition. We can obtain a perturbed set of partition likelihoods by adding Gaussian noise:
where ν is a vector each of whose entries is drawn from a zero-mean Gaussian distribution 𝒩(0,σ). The most likely tree based on these perturbed values will likely differ from the original most likely tree and different noise values will yield different trees. If σ is small, then we draw trees that are very similar to the most likely tree, while if σ is large, then we draw trees more uniformly. In our experiments, we draw sample trees at different noise levels to both preferentially draw high-likelihood trees (high exploitation with small σ), as well as to cover the entire space of possible trees consistent with our initial orientation (high exploration with large σ). Finally, we retain the sampled tree that maximizes the global likelihood pM.
C. Best-first search
The random search in step 2 does not get stuck in local optima, but will generally return a solution that is itself not a local optimum. This solution is potentially improved by a final, deterministic search for a local optimum in the space of directed trees. We explore this space by iteratively expanding and contracting the input graph’s vertices.
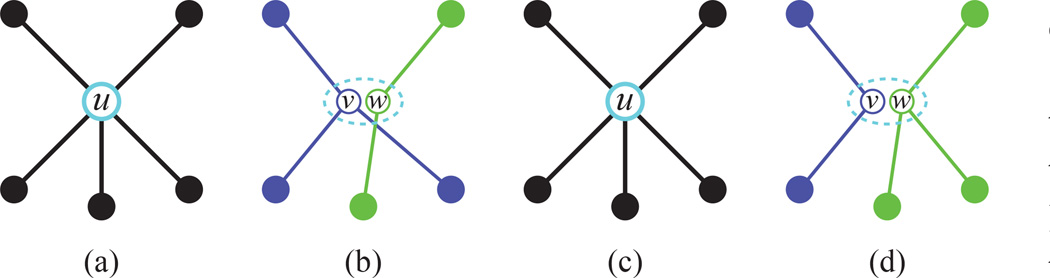
The contraction of vertices u and υ consists of first replacing the two vertices with a single vertex w and then adding an edge between w and every vertex that was adjacent to either u or υ. Vertex contraction is a key operation [40] in the theory of graph minors, and can be applied to any two vertices. In this paper, however, we only allow contractions between overlapping vertices, i.e. vertices with the same projection: P(u) = P(υ).
A vertex expansion is the reverse operation: First replace a vertex w with two new vertices u and υ and then assign every edge that is adjacent to w to either u or υ, such that:
where ET (w) is the set of edges that were adjacent to w and E′ is the edge set of the new graph the results from expanding w. We only allow a vertex expansion if the resulting graph is fully connected. Figure 4 illustrates how to expand and contract a given vertex. In general, we can obtain any tree consistent with a planar graph by expanding a subset of its vertices such that the resulting graph has a single path from the root to every other vertex.
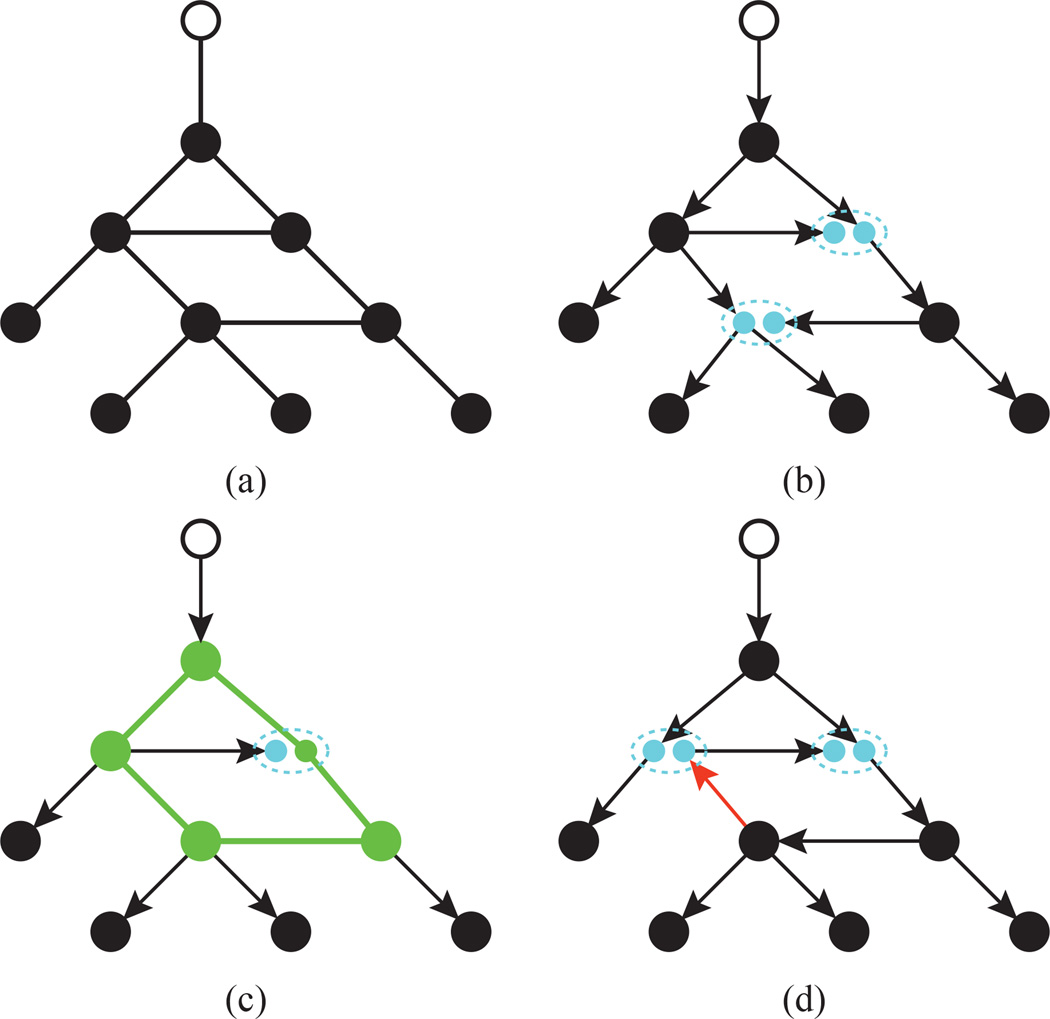
Fig. 4. Vertex contractions and expansions.
(This Figure is best viewed in color.) Expanding and contracting vertices allows us to change the topology of a graph. (a) A vertex u and its neighbors. In (b), u is expanded into two vertices υ (purple) and w (green). The dashed oval indicates that υ and w share the same location on the plane. (c) Contracting υ and w restores the original vertex u. (d) Vertex u can be expanded differently, so that υ and w are connected to different sets of vertices than in (b).
Furthermore, as we show in the Appendix, we can transform any directed tree for a graph G into any other directed tree for G by iteratively contracting and expanding individual vertices so that every intermediate graph is also a directed tree. This property defines a connected meta-graph over the set of directed trees 𝒯(G), where two trees are neighbors if and only if we can transform one into the other in a single contraction-expansion step.
In our final optimization step we look for a better solution by exploring this meta-graph using best-first search (BFS) [37]. We first add all of the neighbors of the sampled tree from the previous step to a priority queue. We then pop the queue to obtain the neighbor with the highest likelihood, in terms of the global model pM, and add all of its neighbors to the priority queue in turn.
We employ BFS, as opposed to either a simpler or a more complex search strategy, based on two assumptions. The first is that most of the edge directions in the noise-sampled tree are correct. Because of this, the sampled and optimal tree will generally only differ by a few partitions, so we do not need to stray too far from the current solution. Our second assumption, however, is that the search space is not convex, and a simple hill-climbing search will likely converge to a suboptimal solution. In the next section, we show that our three-step approach efficiently approximates the ground truth vascular networks for four different retinal datasets.
VI. Experiments
In order to validate the effectiveness of our proposed algorithms, we tested our method on four different retinal datasets with ground truth artery-vein labelings: a 30-image wide-field retinal dataset [7], two AV classification datasets [5], [41] based on the earlier 40-image DRIVE dataset [49], and a 40-image dataset which is used to validate algorithms that estimate the arteriolar-venular ratio (AVR). We have made the wide field-of-view images and the ground-truth labels for all four datasets freely available online. For each image, we constructed a planar graph and determined its ground truth labeled tree. We then quantified the distance between our algorithm’s final AV-labeled tree and the ground truth solution, as explained below.
A. Materials
The WIDE dataset [7] consists of 30 high-resolution, wide-field, RGB images obtained using an Optos 200Tx ultra-wide-field device (Optos plc, Dunfermline, Scotland, UK). This dataset includes both healthy eyes, as well as eyes with age-related macular degeneration (AMD) which display geographic atrophy, drusen, or fibrotic scarring from neovascular AMD. All images were acquired at the Duke University Medical Center, Durham, NC, USA between either August 2010 to October 2012, or November 2013 to July 2014 under Duke IRB protocols Pro00015512 and Pro00056311. Each retinal image was taken from a different individual and was captured as an uncompressed TIFF file at the highest-resolution setting available for the Optos device (3900×3072 pixels). Each image was then manually cropped to remove the eyelashes and other non-retinal regions, and finally downsampled by a factor of 2.
The second dataset, which we refer to as the AV-DRIVE dataset, consists of 40 artery-vein labeled images (565 × 584 pixels) [41]. Here, three different human graders manually classified all the vessel pixels in each segmented image from the existing DRIVE dataset [49]. The three graders then jointly compared their classifications to arrive at a consensus labeling. The third dataset, which we denote as the CT-DRIVE dataset, consists of 20 artery-vein labeled images [41], corresponding to the test set of the DRIVE dataset only. For this dataset, a human expert manually classified the centerline pixels of all the vessels that were at least 3 pixels thick. Both the AV-DRIVE and the CT-DRIVE datasets provide ground-truth labels for the same 20 test images; however, as we show in Section VI-C, these labels differ somewhat from each other. These differences between the labels given by different human experts highlights the difficulty of this classification problem.
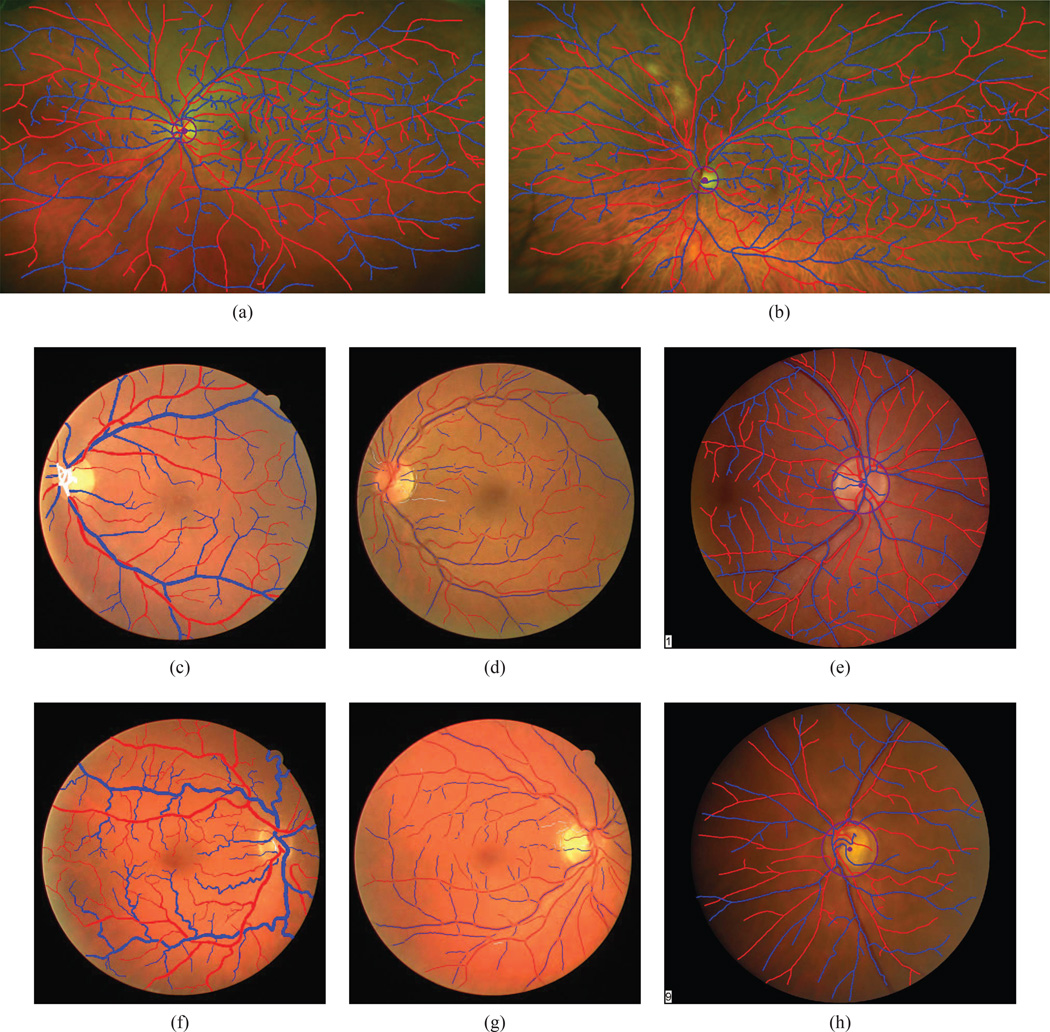
Finally, the fourth dataset (AV-INSPIRE) is based on the INSPIRE-AVR dataset [36], which was developed for validating methods that estimate the arteriolar-venular ratio, so every one if its images is centered on the optic nerve. It also consists of 40 images (2392×2048 pixels). Although each image has an associated ground-truth AVR, the original INSPIRE-AVR dataset did not include any ground-truth vessel labels, so we manually estimated these labels in a similar manner to the WIDE dataset, as detailed below. Figure 6 shows two sample images from each dataset.
Fig. 6. Artery-vein datasets.
(This Figure is best viewed on-screen.) We tested our algorithms on three different retinal datasets. (a,b) WIDE images with their corresponding ground truth labelings superimposed. (c,d) AV-DRIVE artery-vein pixel labelings superimposed on the original DRIVE images. (e,f) CT-DRIVE artery-vein pixel labelings superimposed on the original DRIVE images. The CT-DRIVE images have only the vessel centerline pixels of the main vessels classified as either artery of vein. (g,h) AV-INSPIRE images with their corresponding ground truth labels superimposed.
B. Methods
1) Error quantification
We quantified the error of an estimated AV-labeled tree relative to the ground truth tree in terms of three weighted scores: balanced accuracy, sensitivity, and specificity. The three terms are related as follows:
where
Here, tp, tn, fp, and fn are the true positives, true negatives, false positives, and false negatives, respectively, and ω is a vector of weights for each element. We interpret arteries as positives and veins as negatives. Intuitively, sensitivity defines how well a method can detect arteries, while specificity determines how well it can detect veins. Finally, the balanced accuracy quantifies the overall performance of the algorithm.
For the WIDE, AV-DRIVE, AND AV-INSPIRE datasets, we defined the errors in terms of edge labels; that is, we quantified what percentage of the edges have the same labels in the estimated and ground-truth trees. In this case, the weight ω(i) is the Euclidean distance between the two vertices adjacent to the i-th edge. For the CT-DRIVE, on the other hand, we quantified the error in terms of which pixels were correctly labeled because prior methods had reported their results on this dataset in terms of pixel classification. We obtained the pixel labels by assigning to each pixel the estimated label of the closest edge in the graph. Here, every ω(i) = 1 for this dataset.
2) Planar graph estimation
We obtained each planar graph semi-automatically. For the WIDE and AV-INSPIRE images, we first obtained a Gabor-enhanced image [9] and then extracted a noisy graph from it by building a set of tracks over the tree’s branches [7]. Our algorithm constructed each track by first setting a starting location based on the Gabor response (the higher the response, the more likely the pixel was to be part of a tubular structure such as a vessel). It then moved along the vessel by iteratively selecting the neighbor of the current position that was most likely to also be part of the vessel. Each track stopped when all neighbors fell below a salience threshold. Finally, it converted the set of tracks into a single graph by connecting overlapping tracks. We then manually edited this automatically-extracted graph using a graph editing software that we developed to correct any errors, such as missing or spurious edges due to low image quality or image artifacts. We obtained the planar graphs for the AVDRIVE images in a similar fashion. In this case, though, since we had access to the manual segmentations, we built the initial set of tracks using only the pixels that were part of a vessel. We used the same graphs for the CT-DRIVE and AV-DRIVE datasets.
3) Ground truth estimation
As noted above, the AV-DRIVE images included the ground truth labels for every pixel in the image. We automatically converted these pixel labels into edge labels by assigning to each edge the most common label of its corresponding pixels. For the WIDE and AV-INSPIRE images, authors one and two independently classified each image using a graph editing software that we developed for this task. Our software allows a user to classify each segment as either artery, vein, unsure, or non-vessel. Since even human experts differ on their assessment of vessel topology, we quantified the degree of inter-observer variability or uncertainty in our ground truth by comparing the labeled trees produced by our two human raters. We set the second rater’s trees as ground truth, due to his relevant clinical expertise as a fellowship-trained medical retina specialist. The balanced accuracy, sensitivity, and specificity of the first rater’s trees relative to the second rater’s, for the WIDE and AVINSPIRE datasets’ test sets, are listed in Tables I and IV.
TABLE I. WIDE dataset results.
Number of graphs n 15, Mean number of circuits μ(m): 101.8, Standard dev. σ(m): 23.15
| Method | Balanced accuracy | Sensitivity | Specificity | Time (sec) |
|---|---|---|---|---|
| Optimal AV-labeling | 0.999 (± 0.01) | 0.999 (± 0.01) | 1 (± 0.0) | – |
| Second human rater | 0.974 (± 0.02) | 0.970 (± 0.03) | 0.979 (± 0.02) | ~3000 |
| BFS search | 0.910 (± 0.06) | 0.910 (± 0.06) | 0.909 (± 0.06) | 777.35 (± 330.52) |
| Noise sampling | 0.888 (± 0.06) | 0.857 (± 0.11) | 0.919 (± 0.07) | 192.04 (± 77.95) |
| Orientation search | 0.776 (± 0.12) | 0.733 (± 0.17) | 0.819 (± 0.16) | 83.10 (± 39.48) |
TABLE IV. AV-INSPIRE dataset results.
Number of graphs n 20, Mean number of circuits μ(m): 44.0, Standard dev. σ(m): 10.3
| Method | Balanced accuracy | Sensitivity | Specificity | Time (sec) |
|---|---|---|---|---|
| Optimal AV-labeling | 1 (± 0.0) | 1 (± 0.0) | 1 (± 0.0) | – |
| Second human rater | 0.971 (± 0.03) | 0.972 (± 0.03) | 0.970 (± 0.02) | ~1800 |
| BFS search | 0.909 (± 0.1) | 0.915 (± 0.11) | 0.902 (± 0.1) | 117.68 (± 34.1) |
| Noise sampling | 0.883 (± 0.11) | 0.887 (± 0.13) | 0.879 (± 0.13) | 55.14 (± 10.33) |
| Orientation search | 0.793 (± 0.14) | 0.831 (± 0.16) | 0.754 (± 0.22) | 12.57 (± 3.49) |
For each dataset, human graders assigned one of four labels to each edge: {artery, vein, unsure, non-vessel}. The latter two categories capture both the inherent uncertainty in determining the anatomically correct labeling, as well as errors in the initial segmentation. Furthermore, graders were not constrained to use only AV-labelings; they were allowed to assign a disconnected label if they believed that the input graph was missing a segment that would have connected that edge to the rest of the edges with the same label.
Our method, however, used only two categories: artery or vein. We decided not to allow the algorithm any additional degrees of freedom because they represent a different, more complicated inference problem. Devising a method for not only distinguishing arteries from veins, but also determining if an edge is a valid vessel would require an augmented likelihood model, as well as exploring a different space of possible solutions. While these are interesting avenues for future work, they are beyond the scope of this paper.
4) Parameter estimation
For the WIDE dataset, we randomly chose 15 images as the testing set and kept the remaining 15 as the training set. We randomly split the images of the AV-INSPIRE dataset in a similar manner, with 20 images in the training set and 20 images in the test set. The AV-DRIVE dataset already had training and testing sets; the former had 20 images and the latter 19 images.1 Since the CT-DRIVE dataset did not include a training set, we used the parameter values that we estimated from the AV-DRIVE dataset for this dataset as well.
Our global likelihood model relies on three features: local growth, overlap, and color. The latter two are parameter-free; they use fixed formulas to determine the feature value of an estimated tree. Our local growth feature, on the other hand, required setting the parameters of two distributions, pa and pc: the former defines the probability of observing a given set of angles between a parent vertex’s and its children (local geometry), while the latter defines the probability that a vertex will have a given number of children (abstract topology). As noted in Section IV-A, pa is defined in terms of a set of von-Mises distributions. We estimated the most likely mean and variance parameters for this set using the angles of the ground-truth trees in the training set. We empirically determined pc, on the other hand, by tallying the numbers of children for each vertex of the ground-truth trees in the training set.
In addition to these two distributions, we estimated the three λ weights in (3) based on how informative each of the three features was for the corresponding training dataset. We estimate these three values through a simple grid search. We did not use a least-squares fit because we required that all weights be non-negative. Our WIDE feature weights were λg = 0.43, λo = 0.29, and λc = 0.29, and our AV-INSPIRE weights were λg = 0.76, λo = 0.1, and λc = 0.14, while our AV-DRIVE and CT-DRIVE weights were λg = 0.3, λo = 0.3, and λc = 0.4.
5) Tree estimation
We applied our tree search algorithm to each planar graph and recorded the current solution after each of the three steps listed in Section V. The number of possible solutions for a planar graph is an exponential function of how many circuits it has. Thus, we ran both our initial orientation search until we explored 100m orientations, where m is the number of circuits in G. Then, in the noise sampling step, we sampled 1000 trees per graph. Finally, we ran our BFS search for another 100m steps to obtain our final estimated tree.
We recorded the cumulative mean running time in seconds for each of our method’s three steps on each dataset; that is, the running time for the noise sampling is the sum of the time needed to run this step plus the orientation search time. The best-first search time represents the total time needed to run all three steps. We ran our two algorithms on a Toshiba Satellite X870 laptop with a 2.4Ghz Intel I7 quad-core processor and 32GB of RAM.
C. Results
Our results are summarized in Tables I, II, III, and IV. In each table, n is the number of planar graphs in the test set and μ(m) and σ(m) are the mean and standard deviation of the number of circuits per graph, respectively. As noted above, we recorded the current solution at each of the three steps of our algorithm: Orientation search refers to the most likely tree, based only on our local growth model, found by exploring the space of possible orientations, while Noise sampling records the chosen sampled tree given the orientation found in the previous step. Finally, BFS search indicates our method’s final output, after refining the sampled tree using a local best-first search on the space of possible trees. The last column lists the cumulative mean running times for each step.
TABLE II. AV-DRIVE dataset results.
Number of graphs n 19, Mean number of circuits μ(m): 36.8, Standard dev. σ(m): 6.3
| Method | Balanced accuracy | Sensitivity | Specificity | Time (sec) |
|---|---|---|---|---|
| Optimal AV-labeling | 0.973 (± 0.03) | 0.967 (± 0.03) | 0.978 (± 0.04) | – |
| BFS search | 0.935 (± 0.05) | 0.93 (± 0.06) | 0.941 (± 0.07) | 131.32 (± 33.40) |
| Noise sampling | 0.901 (± 0.07) | 0.881 (± 0.12) | 0.922 (± 0.1) | 62.39 (± 14.37) |
| Orientation search | 0.537 (± 0.19) | 0.513 (± 0.26) | 0.56 (± 0.24) | 12.29 (± 4.12) |
TABLE III. CT-DRIVE dataset results.
Number of graphs n 19, Mean number of circuits μ(m): 36.8, Standard dev. σ(m): 6.3
| Method | Balanced accuracy | Sensitivity | Specificity | Time (sec) |
|---|---|---|---|---|
| AV-DRIVE ground truth | 0.962 (± 0.03) | 0.959 (± 0.06) | 0.965 (± 0.03) | – |
| BFS search | 0.917 (± 0.05) | 0.917 (± 0.07) | 0.917 (± 0.07) | 131.32 (± 33.40) |
| Noise sampling | 0.893 (± 0.08) | 0.883 (± 0.1) | 0.903 (± 0.09) | 62.39 (± 14.37) |
| Dashtbozorg et al. [5] | 0.874 | 0.9 | 0.84 | – |
| Niemeijer et al. [35] | N/A | 0.8 | 0.8 | – |
| Orientation search | 0.559 (± 0.2) | 0.58 (± 0.22) | 0.538 (± 0.22) | 12.29 (± 4.12) |
For the WIDE and AV-INSPIRE datasets, we have also included the results for the second human rater. Furthermore, for the WIDE, AV-DRIVE, and AV-INSPIRE datasets, we included the results for the optimal AV-labeling. Due to reduced field-of-view or ambiguity near the optic nerve, the ground truth labeling provided by the human experts might not exactly be an AV-labeling. Nevertheless, we restricted our algorithm to only consider AV-labelings, since they are anatomically correct. Intuitively, the optimal AV-labeling values are an upper-bound on how well an estimation algorithm can do on that dataset.
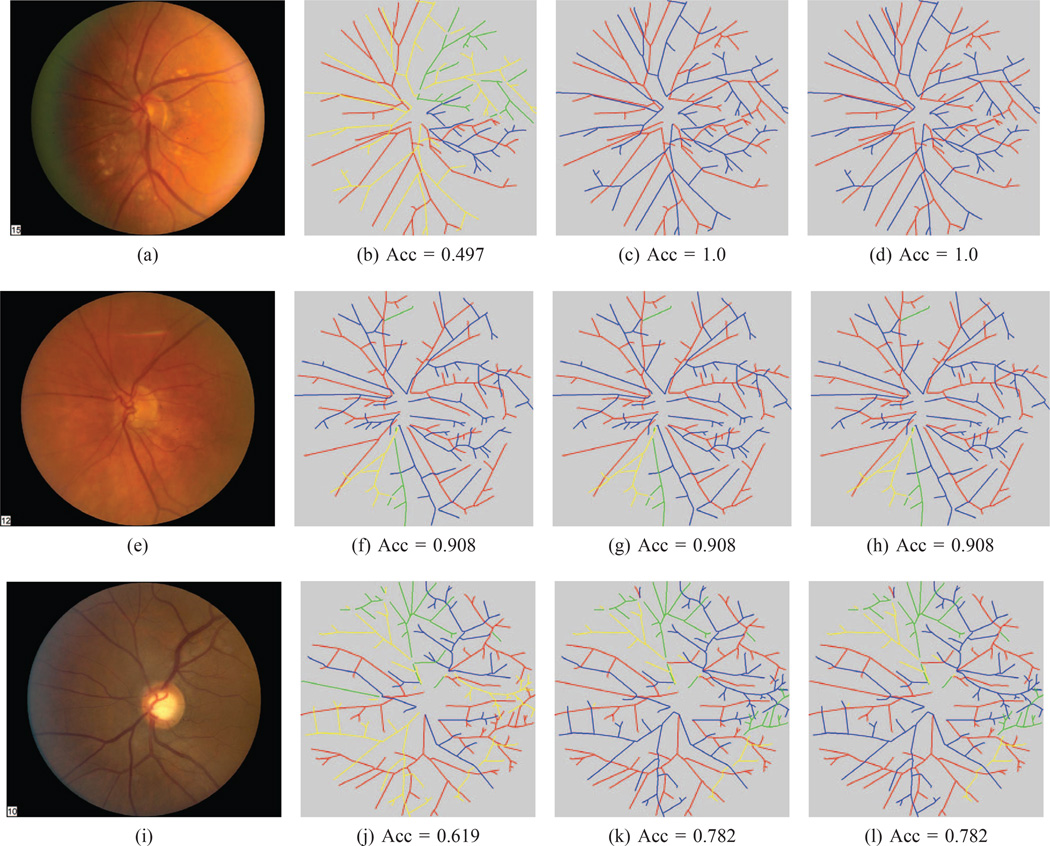
Figures 7, 8, 9, and 10 show three sample results from each dataset. In each of these figures, the top row is the graph on which our algorithm obtained the best result, the bottom row is the one on which it had the worst performance, and the middle row is the one whose accuracy is closest to the mean accuracy for that dataset.
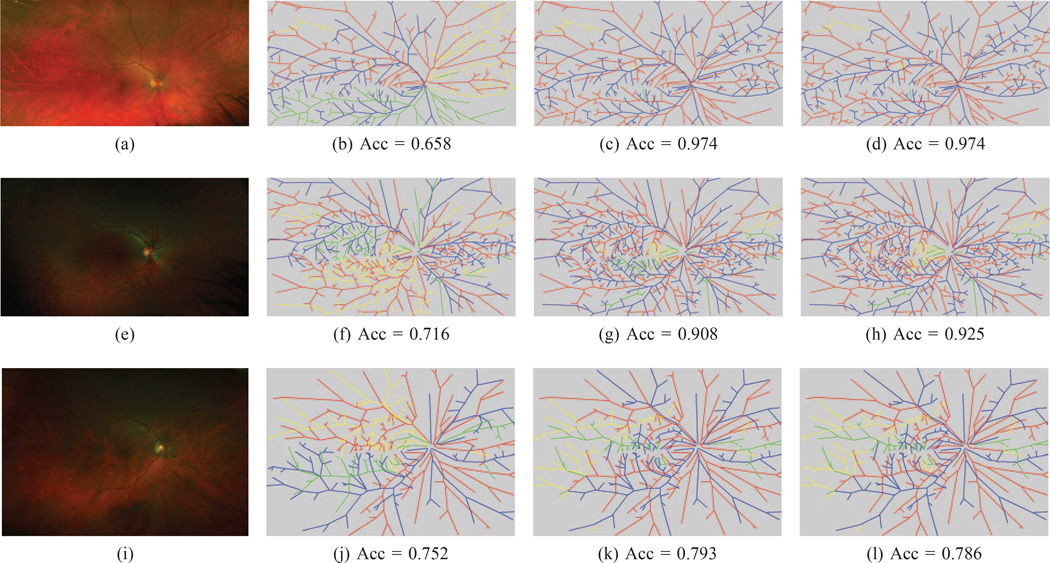
Fig. 7. WIDE classification results.
(This Figure is best viewed on-screen). Our method was able to accurately classify most arteries and veins in the test images of the three datasets. The top row is our method’s best result, the middle row is the result that is closest to the mean and the bottom row is the worst result for this dataset. In the first column is the original image. The second column is the best tree found after our orientation search. The third column is the best noise sampled tree, while the last column is the final tree after the BFS search. In each image, the correctly labeled arteries are marked in red and the correct veins in blue. Edges incorrectly labeled as arteries are in yellow, while incorrectly marked veins are shown in green. The corresponding balanced accuracy is listed below each image.
Fig. 8. AV-DRIVE classification results.
(This Figure is best viewed on-screen). The top row is our method’s best result on the AV-DRIVE dataset, the middle row is the result that is closest to the mean and the bottom row is the worst result for this dataset. In the first column is the original image. The second column is the best tree found after our orientation search. The second column is the best noise sampled tree, while the last column is the final tree after the BFS search. In each image, the correctly labeled arteries are marked in red and the correct veins in blue. Edges incorrectly labeled as arteries are in yellow, while incorrectly marked veins are shown in green. The corresponding balanced accuracy is listed below each image.
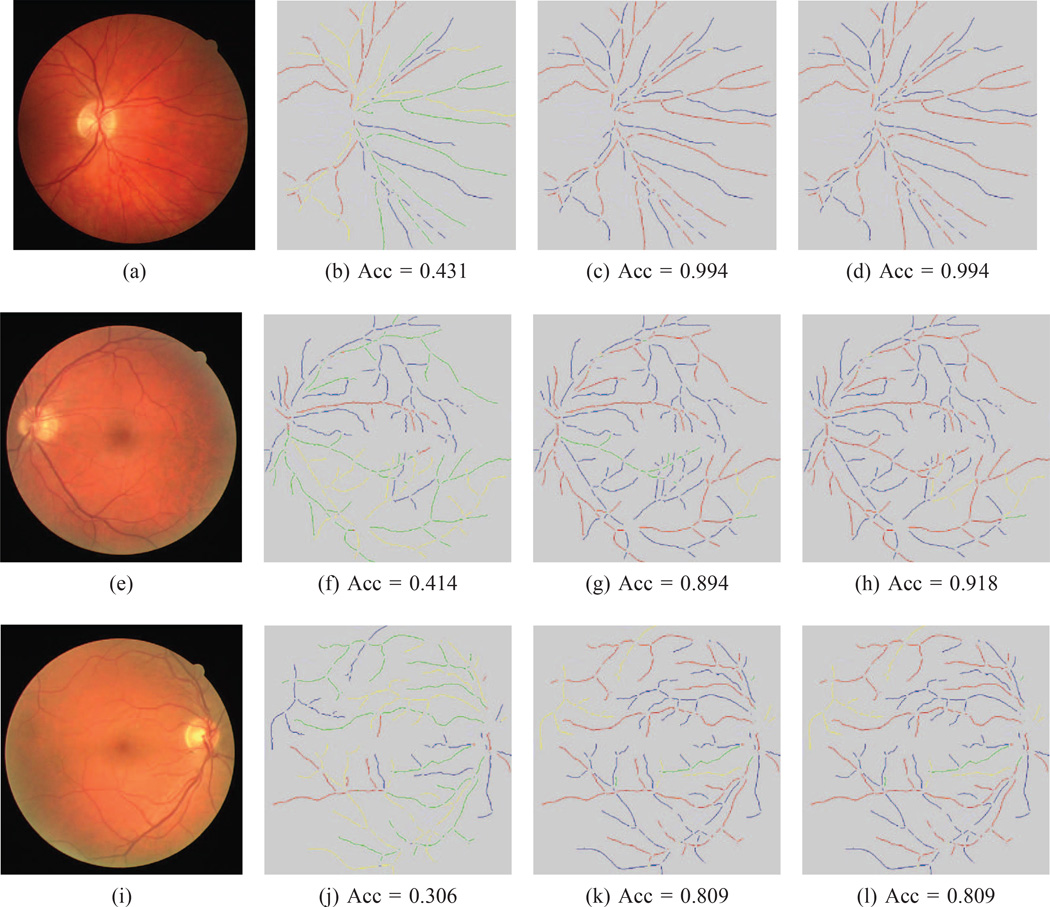
Fig. 9. CT-DRIVE classification results.
(This Figure is best viewed on-screen). The top row is our method’s best result on the CT-DRIVE dataset, the middle row is the result that is closest to the mean and the bottom row is the worst result for this dataset. In the first column is the original image. The second column is the best tree found after our orientation search. The second column is the best noise sampled tree, while the last column is the final tree after the BFS search. In each image, the correctly labeled artery pixels are marked in red and the correct vein pixels in blue. Pixels incorrectly labeled as arteries are in yellow, while incorrectly marked vein pixels are shown in green. The corresponding balanced accuracy is listed below each image.
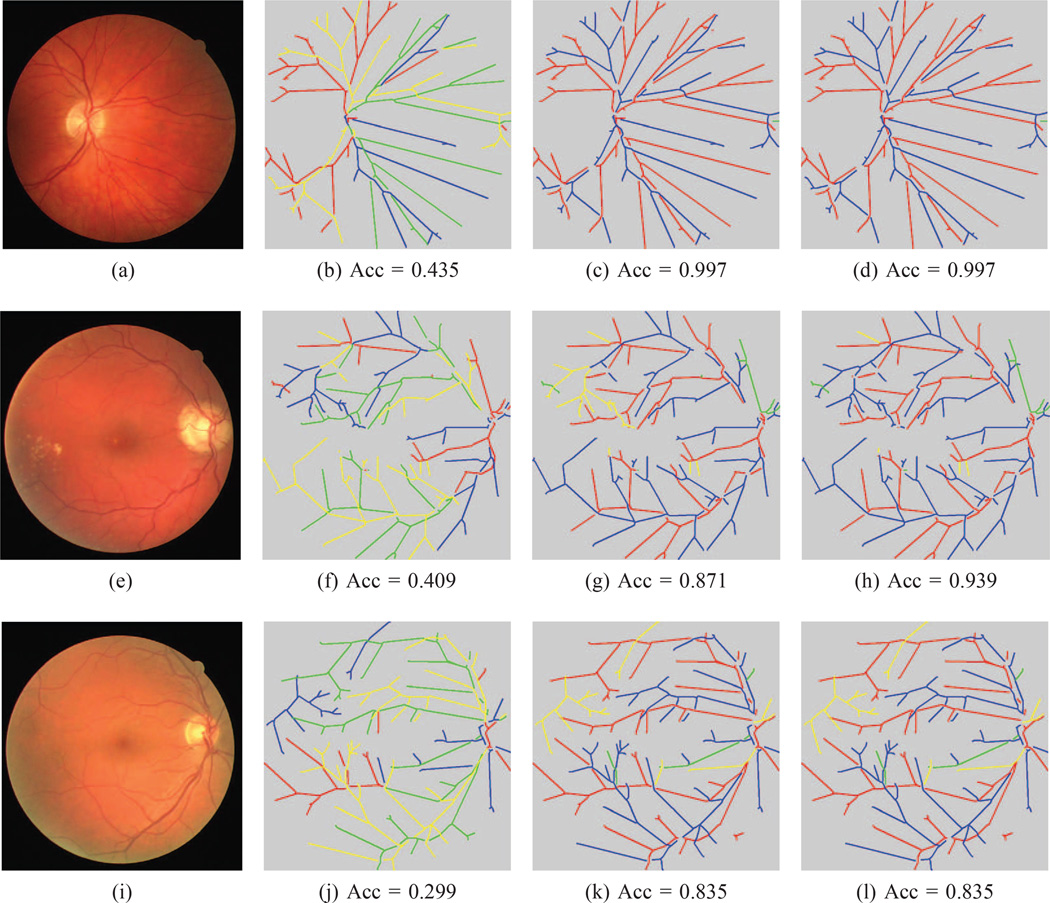
Fig. 10. AV-INSPIRE classification results.
(This Figure is best viewed on-screen). The top row is our method’s best result on the AV-INSPIRE dataset, the middle row is the result that is closest to the mean and the bottom row is the worst result for this dataset. In the first column is the original image. The second column is the best tree found after our orientation search. The second column is the best noise sampled tree, while the last column is the final tree after the BFS search. In each image, the correctly labeled arteries are marked in red and the correct veins in blue. Edges incorrectly labeled as arteries are in yellow, while incorrectly marked veins are shown in green. The corresponding balanced accuracy is listed below each image.
Overall, our proposed estimation framework correctly estimated over 90% of the vessel labels, including those for small vessels, on all four datasets. On the WIDE dataset, our algorithm obtained a classification accuracy on the test set of 91.0%, compared to 97.4% for the second human rater. Our algorithm performance on the AV-DRIVE and AV-INSPIRE datasets was 93.5% and 90.9%, respectively. Note that the results were computed in terms of edge labels for these three datasets.
We computed the accuracy in terms of pixel labels for the CT-DRIVE, in order to compare our results to prior methods, as reported in [5]. In this case, Niemeijer et al. [35] had achieved sensitivity and specificity values of around 0.8, while Dashtbozorg et al. [5] obtained values of 0.9 and 0.84, respectively, as well as an accuracy of 87.4%. In contrast, our algorithm obtained an accuracy of 91.7%, with identical sensitivity and specificity values of 91.7% each.
We now discuss each dataset in detail below, as well as analyze our algorithm’s performance in more depth.
D. Discussion
1) WIDE dataset
In this dataset, our algorithm achieved a performance of 91% on a set of challenging, high-resolution retinal images. On average, this dataset had over 100 circuits per graph, due to the wide field-of-view and high image resolution, so the set of possible trees consistent with each graph was extremely large (on the order of Ω(2100)). Nevertheless, our method was able to correctly approximate the vast majority of the edge labels by relying on the topological constraint that an edge must share its label with all its downstream edges.
2) AV-DRIVE dataset
We were also able to achieve very good results (93.5%) on this larger dataset, suggesting that our approach is applicable to a wide variety of retinal images. It is worth noting that the optimal accuracy that can be achieved by an AV-labeling on this dataset (97.3%) is considerably lower than that for the WIDE dataset (99.9%). This is due to the skewed and more narrow field-of-view of the DRIVE images. Nevertheless, since these images show a smaller region of the vascular network, their corresponding graphs have significantly fewer circuits than those of the WIDE images, so our method was able to achieve slightly better results on these graphs.
3) CT-DRIVE dataset
For the CT-DRIVE dataset, we were also able to achieve a high accuracy (91.7%), in spite of optimizing our parameters values using the training set of the AV-DRIVE dataset. It is worth noting that we already surpass prior methods in overall accuracy after noise sampling, which is only the second step of our algorithm.
We also note that if we compare the ground truth pixel values of the AV-DRIVE dataset to the CT-DRIVE dataset, they only agree ~96% of the time. Interestingly, this inter-observer agreement between these two sets of experts is close to the agreement between our two experts on both the WIDE and AV-INSPIRE datasets. While more data would be needed to draw firm conclusions, these results suggest that 96–97% is close to the best expected level of agreement that two independent observers, be it a human or an algorithm, can have with each other.
4) AV-INSPIRE dataset
Finally, we achieved a balanced accuracy of 90.9% on this fourth dataset, further validating the general applicability of our approach. This was the most challenging dataset for our algorithm, which we attribute primarily to the narrow field-of-view of these images. In this case, the vascular network has fewer branch-points–particularly far away from the optic nerve–so on average the label of one edge constrains fewer other edges downstream. Furthermore, most of the branch-points are clustered near the optic nerve, which is very difficult to classify, even for expert human graders. On the other hand, the graphs in this dataset are considerably smaller than the WIDE graphs (44 vs 102 average number of circuits), which allowed our algorithm to explore a larger portion of the search space for each graph and thus compensate for some of these ambiguities.
5) Analysis of each optimization step
Our three-step optimization scheme accurately approximated the most likely AV-labeling for a large number of graphs from different datasets. In Section V, we suggested how each of the three steps contributed to progressively yield a more accurate solution. Our experimental results empirically confirm this view, since each step markedly improves upon the previous one.
In more detail, the largest gain in accuracy occurred between the orientation search and noise sampling steps. On average, the balanced accuracy improved between 18% (WIDE) and 68% (AV-DRIVE) from the first to the second step. We believe that the main reason behind this drastic improvement—as we noted in Section V—is that the orientation search step estimates the vast majority of the edge directions correctly; however, relying solely on the local likelihood model may lead to local partitioning errors that may have a large impact on the resulting vessel labels. These large-impact errors arise because the local model is agnostic about vessel labels. The noise-sampling step, on the other hand, does make use of the global likelihood model; thus, it can more accurately predict the quality of each potential tree. The fact that the noise-sampled tree has a consistently high accuracy is given by the fact that the edge directions are mostly correct, so it only needs to determine the best way of partitioning the crossings defined by the chosen orientation.
The improvement between the noise sampling and best-first search steps is more modest, ranging from 2.5% (WIDE) to 3.8% (AV-DRIVE). However, it is important to note that for all datasets the BFS step consistently improved upon the best noise-sampled tree. This consistent performance strongly suggests that our global likelihood model accurately quantifies the plausibility of a given AV-labeling, especially considering that our search algorithm visited between 2500 and 14600 possible trees per graph, so it had to assess the quality a large number of solutions.
6) Current limitations
Although our AV classification pipeline generally yields good results, its performance is not optimal, since human graders are better able to estimate the vascular labels. Thus, we analyzed our algorithm’s results in detail in order to better understand its current limitations and outline ways of overcoming these weaknesses.
Currently, the main limitation of our optimization strategy is that the best-first step is myopic: it only estimates the value of exploring a possible tree based on the likelihood of the tree itself. Thus, this search strategy makes the tacit assumption that changes in likelihood are largely monotonic and independent. Unfortunately, this is not the case if the labels of a large region of the graph are switched (i.e. most arteries are labeled as veins and vice versa). Depending on the topology of the input graph it may take several contraction-expansion steps to switch this region to its correct labels. However, in this case correcting any one error will likely lead to a lower likelihood tree because the overlap likelihood will go down; it is only after multiple corrections that this likelihood will bounce back up. Since our search algorithm cannot forecast this likelihood increase down the road, it will tend to avoid changing large regions that are switched, unless its vessel labels can be reversed in a couple of steps.
It is worth noting that the orientation search step does make use of a heuristic—based on the fact that edges tend to grow away from the optic nerve—to better guide its search through the space of possible orientations. A similar heuristic for AV-labeled trees is less trivial to devise, however, and it represents an important avenue of future work.
Another minor current limitation is that our color likelihood is not context-dependent. Depending on the focus and lighting of the retinal image, vessels in one part of the image may appear consistently darker than in a different part of the image. Darker regions will bias the algorithm to label more of its vessels as veins (and conversely for arteries in light regions). We are currently exploring the use of local histogram equalization methods to minimize this bias.
VII. Conclusions
In this work, we developed a comprehensive, semiautomatic method for distinguishing arteries from veins in a retinal image. Our approach combines graph-theoretic methods with domain-specific knowledge to accurately estimate the correct vessel types on four different retinal datasets, outperforming existing methods. Moreover, this method is capable of analyzing vasculature in wide field-of-view fundus photographs, which is a potentially important tool for diagnosing diseases with peripheral retinal vascular manifestations including diabetic retinopathy, retinal vein occlusion, and sickle cell retinopathy.
Our future work will focus on using our classification method to better determine diagnostically relevant properties of arteries and veins, such as the arteriolar-venular ratio. We will also refine our automatic planar graph extraction step and extend our algorithm to handle noisy graphs that contain missing or spurious vessels in order to fully automate our framework, which we will then validate in a clinical setting. This robust algorithm will not only distinguish arteries from veins, but also determine if a given edge is a valid vessel, that is, it will also apply unsure and non-vessel categories. If available, we will take advantage of an existing vessel segmentation to constraint our extraction step. We also intend to focus on applying our vessel classification framework to other problems, including identifying the retinal layer in which a capillary lies [15] and distinguishing retinal from choroidal vessels.
Acknowledgments
This research was supported in part by NIH grant R01-EY022691 and NEI grant K12-EY016333-08. The authors would also like to thank Touseef Qureshi for providing the AV-DRIVE images and Behdad Dashtbozorg for providing the CT-DRIVE images.
Appendix
AV-Labeled Tree Space
Our heuristic tree search algorithm explores the space of possible AV-labeled trees consistent with a planar graph by iteratively expanding and contracting vertices. Here, we first prove that given a tree T consistent with an input graph G, we can obtain a different tree T′ by a single expansion-contraction step. We then use this result to prove that the space of possible trees 𝒯(G) consistent with G is connected in terms of vertex expansion-contraction operations. To the best of our knowledge, all the results and proofs are novel and may be of independent interest.
A. Switching between directed trees
Here we show how to obtain different tree T′ given a current tree T using vertex expansion and contraction. Let H be the graph that results from contracting two overlapping vertices of T. We will show that H contains exactly one simple circuit; that is, H is a pseudo-tree or unicyclic graph. We first note that the circuit rank ρ of a graph G is given by:
where |NG| is the number of connected components of G [2]. The circuit rank is the minimum number of edges that we need to remove from G to remove all its circuits. The circuit rank of any connected tree is 0, since |VT| = |ET | + 1 and |NT | = 1. However, a vertex contraction reduces the number of vertices by one, keeps the number of edges the same, and, in the case of trees in 𝒯(G), also keeps the number of connected components the same. Thus, ρ(H) = 1, which implies that H has exactly one circuit.
We will now show how to obtain a new directed tree T′ from H, such that T ≠ T′. An example of this process is shown in Figure 11. First, let CH ⊂ VH \ {rH} be the set of vertices, excluding the root, that are part of the sole circuit of H. By definition, |CH| ≥ 2.2 Also, w ∈ CH, since w is adjacent to all the vertices that were adjacent to either u or υ. Thus, we can reach w from rH by either the path that used to connect υ and rT or the one that used to connect u and rT. Furthermore, each vertex x ∈ CH has exactly two adjacent edges which are part of the circuit. To obtain a directed tree T′ from H, we have to expand one x ∈ CH such that each of two circuit edges of x is assigned to a new vertex. We can see this by noting that we can reach any such x from the root by two different paths: one that uses the first edge that is part of the circuit and another which uses the second circuit edge. Since the two edges are reachable from the root through different paths, then expanding x such that the two circuit edges are assigned to different vertices will not affect the reachability of any other vertex in H. Thus, the resulting graph T′ is a tree. On the other hand, if we expand x such that the two circuit edges are assigned to the same new vertex, then the resulting graph will not be a tree because the new graph still has a circuit. Finally, we note that T′ will be different from T, if either x ≠ w or if we partition the edges incident to w differently than how they were partitioned in T.
Fig. 11. Switching between directed trees.
(This Figure is best-viewed in color.) A directed tree can be converted to different directed tree by contracting two overlapping vertices and then expanding one of the vertices in the resulting circuit. (a) A small planar graph. The root is marked in white. (b) A directed tree consistent with the planar graph. Its overlapping vertices are shown in small light blue circles. The dotted oval around them indicates that the pairs of vertices overlap each other when projected. (c) The intermediate graph that results from contracting the lower two overlapping vertices into one. Note how the contraction creates an undirected circuit (shown in green). Every vertex in this circuit can be potentially expanded to obtain a new tree. (d) A new directed tree obtained by expanding the left-most green vertex. The overlapping vertices are again shown in light blue. Note how this second tree defines a different orientation for the edge highlighed in orange.
B. Directed tree meta-graph
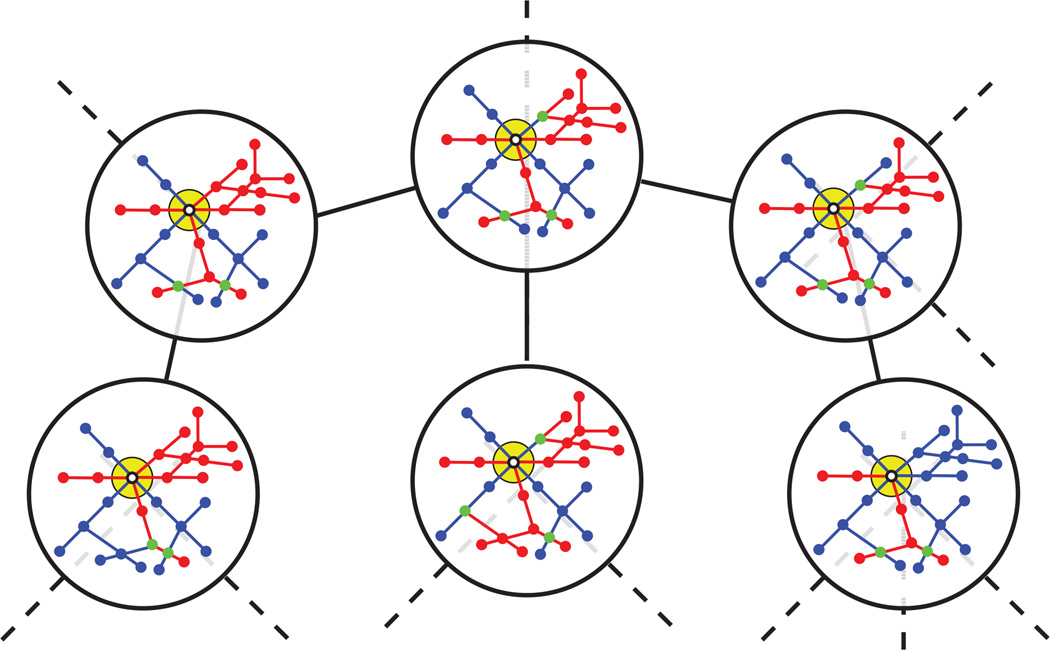
Here, we prove that all directed trees consistent with a given input graph are connected by vertex expansions and contractions. A pair of contraction-expansion steps allow us to convert a directed tree T into a different tree T′ that is also consistent with the same planar graph. We refer to pairs of directed trees that differ by a single contraction-expansion pair as neighbors of each other. Thus, the pairs of vertex contraction and expansion operations induce a meta-graph over 𝒯(G) in which every tree is a node in the meta-graph and there is an edge between two trees if and only if they are neighbors. Figure 5 illustrates a portion of the meta-graph for a small graph. We will now show that this meta-graph is fully connected. That is, we can convert any directed tree T into any other directed tree T′ such that each intermediate tree is also a directed tree.
Fig. 5. The directed tree meta-graph.
(This Figure is best viewed in color.) The meta-graph of AV-labeled trees consistent with a small graph G. Neighboring trees differ by a single expansion-contraction step. All pairs of directed trees consistent with a given planar graph can be converted into each other by one or more expansion-contraction steps, so the meta-graph, shown in black, is fully connected. In each tree, arteries are shown in red and veins in blue; pairs of vertices of different types which overlap are shown in purple.
We prove this statement by constructing an algorithm that, given starting and target trees T and T′, converts the former into the latter by traversing the meta-graph; that is, it iteratively eliminates any differences between the two trees.
More concretely, there are three ways in which two trees in the meta-graph can differ. The first possible difference is that the sets of crossings that they induce on the planar graph may be different. In this case, there is at least one vertex in G that is a crossing with respect to one tree, but not the other. For instance, note how the vertices that we have to partition to obtain the trees in Figures 11(b) and 11(d) are different.
The second way in which two trees can differ is in their edge orientations. As Figures 11(b) and 11(d) illustrate, the edge highlighted in orange is assigned a different orientation in the two trees, such that in 11(b) an upper vertex is the parent of a lower vertex, while in 11(d) these roles are reversed. It is important to note that even if two trees induce identical crossings on G, their edges may be oriented differently.
The third way in which two trees can differ is by defining a different parent for one or more of their vertices. As Figures 4(b) and 4(d) illustrate, there is often more than one way to partition the same vertex and different ways of partitioning a vertex give rise to different trees. Finally, we note that two trees which induce identical crossings and edge orientations may still differ in their vertex partitions.
Our algorithm for converting the starting tree T into the target tree T′ systematically eliminates these three kinds of differences. It thus consists of three steps:
First, we shift the location of every crossing to match the target.
Then, we flip any induced edge orientations to match the target.
Finally, we ensure that each edge has the same parent as in the target.
The first step forces all the crossings to be the same, while the second step ensures that every pair of corresponding edges is oriented in the same direction. Finally, the last step adjusts the partition at each crossing to match the target tree.
We will first detail the initial step: shifting the locations of the crossings to match T′. Let u ∈ VG be a vertex which is a crossing with respect to T, but not T′. Since u is not a crossing for T′, we first contract two of the vertices of T that project down to u to obtain a pseudo-tree H. T′ does not have any circuits, in spite of not having a partition at u; therefore, there exists some other vertex υ ∈ VH which is part of the sole circuit of H and whose projection is a crossing with respect to T′ (but not T). Thus, let T″ be any tree that results from applying a valid expansion at υ. By construction, T″ has one more crossing in common with T′ than T did. Furthermore, note that the above operation did not modify any vertices other than u and υ. Thus, we can iteratively apply the above procedure to each subsequent tree to converge to a tree T̂ that induces the same set of crossings as T′.
We will now detail the second step: adjusting the induced edge directions so that they match T′. Although T̂ has the same set of crossings as T′, some of the edges may be oriented differently in the two trees. In order to modify the edge directions without changing the set of crossings, we have to flip the orientations of entire paths between crossings en masse. More concretely, let EP ∈ EG be a path between two crossings u and υ that is oriented differently in the two trees and which is not incident to any other crossings. Note that, by construction, EP has only two possible orientations. Without loss of generality, assume that EP is oriented from u to υ with respect to T̂. Since both u and υ are crossings regardless of the orientation of EP, then each vertex has at least two other incident edges and there are at least three vertices in T̂ that project down to υ. To change the orientation of EP, we first contract the vertex in T̂ which projects down to υ and is incident to EP with one of the other vertices that project down to υ. This creates a pseudo-tree H, which we then convert to a tree by expanding the vertex in H that projects down to u and is incident to EP. Clearly, both u and υ remain crossings after this contraction-expansion step, but now EP is oriented in the opposite direction. Since this operation only affects u, υ, and EP, we can iteratively flip the edge orientations of all the edges on which T̂ and T′ differ.
We will now detail the final step: adjusting the parent of each edge to match T′. Let T̃ be the tree that results from adjusting all the edge orientations of T̂ to match T′. In other words, T̃ and our target T′ have the same crossings and edge orientations, but may differ in how their crossings are partitioned. Thus, let u be a crossing which is partitioned differently for the two trees. To correct this partition, first contract two of the vertices of T̃ which project down to u and then expand them again, such that their incident edges are partitioned as defined by T′. Figures 4(b) – 4(d) illustrate this procedure. Since each contraction-expansion step only modifies the two target vertices, we can iteratively repeat the above procedure to ensure all the partitions match T′.
In summary, we can convert any two directed trees consistent with G into each other by iteratively contracting and expanding their vertices such that every intermediate graph is itself a directed tree. Thus, the contraction-expansion operation induces a connected meta-graph over the space of directed trees 𝒯(G).
Footnotes
We excluded test image 11 from our experiments because its field-of-view was too narrow, so a significant proportion of the vessels were disconnected from the optic nerve.
Contributor Information
Rolando Estrada, Email: rolando.estrada@duke.edu, Department of Ophthalmology, Duke University, Durham, NC 27708 USA.
Michael J. Allingham, Email: mike.allingham@duke.edu, Department of Ophthalmology, Duke University, Durham, NC 27708 USA.
Priyatham S. Mettu, Email: priyatham.mettu@duke.edu, Department of Ophthalmology, Duke University, Durham, NC 27708 USA.
Scott W. Cousins, Email: scott.cousins@duke.edu, Department of Ophthalmology, Duke University, Durham, NC 27708 USA.
Carlo Tomasi, Email: tomasi@cs.duke.edu, Department of Computer Science, Duke University, Durham, NC 27708 USA.
Sina Farsiu, Email: sina.farsiu@duke.edu, Departments of Biomedical Engineering, Ophthalmology, Electrical and Computer Engineering, and Computer Science, Duke University, Durham, NC, 27708 USA.
References
- 1.Benmansour F, Cohen L. Tubular structure segmentation based on minimal path method and anisotropic enhancement. International Journal of Computer Vision. 2011:1–19. [Google Scholar]
- 2.Berge C. The Theory of Graphs. Courier Dover Publications; 2001. [Google Scholar]
- 3.Cheung CY-l, Ikram MK, Sabanayagam C, Wong TY. Retinal microvasculature as a model to study the manifestations of hypertension. Hypertension. 2012;60(5):1094–1103. doi: 10.1161/HYPERTENSIONAHA.111.189142. [DOI] [PubMed] [Google Scholar]
- 4.Dashtbozorg B, Mendonca A, Campilho A. An automatic method for the estimation of arteriolar-to-venular ratio in retinal images; Computer-Based Medical Systems (CBMS), 2013 IEEE 26th International Symposium on; 2013. Jun, pp. 512–513. [Google Scholar]
- 5.Dashtbozorg B, Mendonca A, Campilho A. An automatic graphbased approach for artery/vein classification in retinal images. Image Processing, IEEE Transactions on. 2014 Mar;23(3):1073–1083. doi: 10.1109/TIP.2013.2263809. [DOI] [PubMed] [Google Scholar]
- 6.Diestel R. Graph Theory. 4 edition. Heiderberg: Springer-Verlag; 2010. [Google Scholar]
- 7.Estrada R, Schmidler SC, Tomasi C, Farsiu S. Tree Topology Estimation. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2015 doi: 10.1109/TPAMI.2014.2382116. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrada R, Tomasi C, Cabrera M, Wallace D, Freedman S, Farsiu S. Exploratory Dijkstra forest based automatic vessel segmentation: applications in video indirect ophthalmoscopy (VIO) Biomedical Optics Express. 2012;3:327–339. doi: 10.1364/BOE.3.000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada R, Tomasi C, Trager M, Wallace D, Freedman S, Farsiu S. Enhanced video indirect ophthalmoscopy (VIO) via robust mosaicing. Biomedical Optics Express. 2011;2:2871–2887. doi: 10.1364/BOE.2.002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraz M, Barman S, Remagnino P, Hoppe A, Basit A, Uyyanonvara B, Rudnicka A, Owen C. An approach to localize the retinal blood vessels using bit planes and centerline detection. Computer Methods and Programs in Biomedicine. 2012;108(2):600–616. doi: 10.1016/j.cmpb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Fraz M, Remagnino P, Hoppe A, Uyyanonvara B, Rudnicka A, Owen C, Barman S. Blood vessel segmentation methodologies in retinal images - a survey. Computer Methods and Programs in Biomedicine. 2012;108(1):407–433. doi: 10.1016/j.cmpb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Gariano RF, Gardner TW. Retinal angiogenesis in development and disease. Nature. 2004;438(7070):960–966. doi: 10.1038/nature04482. [DOI] [PubMed] [Google Scholar]
- 13.Geudens I, Gerhardt H. Coordinating cell behaviour during blood vessel formation. Development. 2011;138(21):4569–4583. doi: 10.1242/dev.062323. [DOI] [PubMed] [Google Scholar]
- 14.González G, Türetken E, Fleuret F, Fua P. Delineating trees in noisy 2D images and 3D image-stacks; Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on; 2010. pp. 2799–2806. [Google Scholar]
- 15.Hendargo HC, Estrada R, Chiu SJ, Tomasi C, Farsiu S, Izatt JA. Automated non-rigid registration and mosaicing for robust imaging of distinct retinal capillary beds using speckle variance optical coherence tomography. Biomed. Opt. Express. 2013 Jun;4(6):803–821. doi: 10.1364/BOE.4.000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Zhang J, Huang Y. An automated computational framework for retinal vascular network labeling and branching order analysis. Microvascular Research. 2012;84(2):169–177. doi: 10.1016/j.mvr.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Ikram MK, Cheung CY, Lorenzi M, Klein R, Jones TL, Wong TY and for the NIH/JDRF Workshop on Retinal Biomarker for Diabetes Group. Retinal vascular caliber as a biomarker for diabetes microvascular complications. Diabetes Care. 2013;36(3):750–759. doi: 10.2337/dc12-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikram MK, de Jong FJ, Vingerling JR, Witteman JCM, Hofman A, Breteler MMB, de Jong PTVM. Are retinal arteriolar or venular diameters associated with markers for cardiovascular disorders? The Rotterdam study. Investigative Ophthalmology & Visual Science. 2004;45(7):2129–2134. doi: 10.1167/iovs.03-1390. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa H, Geiger D, Cole R. Finding tree structures by grouping symmetries. Computer Vision, IEEE International Conference on. 2005;2:1132–1139. [Google Scholar]
- 20.Jan J, Odstrcilik J, Gazarek J, Kolar R. Retinal image analysis aimed at blood vessel tree segmentation and early detection of neurallayer deterioration. Computerized Medical Imaging and Graphics. 2012;36(6):431–441. doi: 10.1016/j.compmedimag.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, DeBuc DC, Rundek T, Lam BL, Wright CB, Shen M, Tao A, Wang J. Automated segmentation and fractal analysis of high-resolution non-invasive capillary perfusion maps of the human retina. Microvascular Research. 2013;89(0):172–175. doi: 10.1016/j.mvr.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi VS, Reinhardt JM, Garvin MK, Abramoff MD. Automated method for identification and artery-venous classification of vessel trees in retinal vessel networks. PloS one. 2014;9(2):e88061. doi: 10.1371/journal.pone.0088061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaleel HH, Rahmat RO, Zamrin D, Mahmod R, Mustapha N. Vessel centerline extraction using new center of gravity equations. IAENG International Journal of Computer Science. 2013;40(1) [Google Scholar]
- 24.Khatri C, Mardia K. The von Mises-Fisher matrix distribution in orientation statistics. Journal of the Royal Statistical Society. Series B (Methodological) 1977;39(1):95–106. [Google Scholar]
- 25.Klein R, Klein B, Knudtson M, Wong T, Tsai M. Are inflammatory factors related to retinal vessel caliber? The Beaver Dam eye study. Archives of Ophthalmology. 2006;124(1):87–94. doi: 10.1001/archopht.124.1.87. [DOI] [PubMed] [Google Scholar]
- 26.Knudtson MD, Lee KE, Hubbard LD, Wong TY, Klein R, Klein BE. Revised formulas for summarizing retinal vessel diameters. Current eye research. 2003;27(3):143–149. doi: 10.1076/ceyr.27.3.143.16049. [DOI] [PubMed] [Google Scholar]
- 27.Kondermann C, Kondermann D, Yan M. Blood vessel classification into arteries and veins in retinal images. Proc. SPIE. 2007;6512:651247–651247–9. [Google Scholar]
- 28.Lambert D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics. 1992;34(1):1–14. [Google Scholar]
- 29.Lau Q, Lee ML, Hsu W, Wong TY. Simultaneously identifying all true vessels from segmented retinal images. Biomedical Engineering, IEEE Transactions on. 2013 Jul;60(7):1851–1858. doi: 10.1109/TBME.2013.2243447. [DOI] [PubMed] [Google Scholar]
- 30.Lee N, Laine A, Smith R. A hybrid segmentation approach for geographic atrophy in fundus auto-fluorescence images for diagnosis of age-related macular degeneration; Engineering in Medicine and Biology Society, 2007. EMBS 2007. 29th Annual International Conference of the IEEE; 2007. Aug, pp. 4965–4968. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Perez M, Hughes AD, Stanton AV, Thom SA, Chapman N, Bharath AA, Parker KH. P-289: Changes of retinal blood vessel tree topology in hypertension. American Journal of Hypertension. 2001;14(S1):125A. [Google Scholar]
- 32.Mirsharif Q, Tajeripour F, Pourreza H. Automated characterization of blood vessels as arteries and veins in retinal images. Computerized Medical Imaging and Graphics. 2013;37(7–8):607–617. doi: 10.1016/j.compmedimag.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Muramatsu C, Hatanaka Y, Iwase T, Hara T, Fujita H. Automated selection of major arteries and veins for measurement of arteriolar-tovenular diameter ratio on retinal fundus images. Computerized Medical Imaging and Graphics. 2011;35(6):472–480. doi: 10.1016/j.compmedimag.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen T, Wong T. Retinal vascular changes and diabetic retinopathy. Current Diabetes Reports. 2009;9(4):277–283. doi: 10.1007/s11892-009-0043-4. [DOI] [PubMed] [Google Scholar]
- 35.Niemeijer M, van Ginneken B, Abràmoff MD. Automatic classification of retinal vessels into arteries and veins. SPIE Conference on Medical Imaging. 2009;7260(1F):1–8. [Google Scholar]
- 36.Niemeijer M, Xu X, Dumitrescu A, Gupta P, van Ginneken B, Folk J, Abramoff M. Automated measurement of the arteriolar-to-venular width ratio in digital color fundus photographs. Medical Imaging, IEEE Transactions on. 2011 Nov;30(11):1941–1950. doi: 10.1109/TMI.2011.2159619. [DOI] [PubMed] [Google Scholar]
- 37.Pearl J. Heuristics: Intelligent Search Strategies for Computer Problem Solving. Boston, MA, USA: Addison-Wesley Longman Publishing Co., Inc.; 1984. [Google Scholar]
- 38.Pechaud M, Keriven R, Peyre G. Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on. IEEE; 2009. Extraction of tubular structures over an orientation domain; pp. 336–342. [Google Scholar]
- 39.Peirce SM. Computational and mathematical modeling of angiogenesis. Microcirculation. 2008;15(8):739–751. doi: 10.1080/10739680802220331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pemmaraju SV, Skiena SS. Computational Discrete Mathematics: Combinatorics and Graph Theory with Mathematica. Cambridge University Press; 2003. [Google Scholar]
- 41.Qureshi T, Habib M, Hunter A, Al-Diri B. A manually-labeled, artery/vein classified benchmark for the drive dataset; Computer-Based Medical Systems (CBMS), 2013 IEEE 26th International Symposium on; 2013. Jun, pp. 485–488. [Google Scholar]
- 42.Qutub A, Mac Gabhann F, Karagiannis E, Vempati P, Popel A. Multiscale models of angiogenesis. Engineering in Medicine and Biology Magazine, IEEE. 2009 Mar;28(2):14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Relan D, MacGillivray T, Ballerini L, Trucco E. Retinal vessel classification: Sorting arteries and veins; Engineering in Medicine and Biology Society (EMBC) 2013 35th Annual International Conference of the IEEE; 2013. Jul, pp. 7396–7399. [DOI] [PubMed] [Google Scholar]
- 44.Rivest-Henault D, Cheriet M. 3-d curvilinear structure detection filter via structure-ball analysis. Image Processing, IEEE Transactions on. 2013 Jul;22(7):2849–2863. doi: 10.1109/TIP.2013.2240005. [DOI] [PubMed] [Google Scholar]
- 45.Rothaus K, Jiang X, Rhiem P. Separation of the retinal vascular graph in arteries and veins based upon structural knowledge. Image and Vision Computing. 2009;27(7):864–875. 7th IAPR-TC15 Workshop on Graph-based Representations (GbR 2007). [Google Scholar]
- 46.Rothaus K, Rhiem P, Jiang X. Separation of the retinal vascular graph in arteries and veins. In: Escolano F, Vento M, editors. Graph-Based Representations in Pattern Recognition, volume 4538 of Lecture Notes in Computer Science. Springer Berlin Heidelberg; 2007. pp. 251–262. [Google Scholar]
- 47.Soares J, Leandro J, Cesar R, Jr, Jelinek H, Cree M. Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Transactions on Medical Imaging. 2006;25(9):1214–1222. doi: 10.1109/tmi.2006.879967. [DOI] [PubMed] [Google Scholar]
- 48.Sofka M, Stewart C. Retinal vessel centerline extraction using multiscale matched filters, confidence and edge measures. Medical Imaging, IEEE Transactions on. 2006 Dec;25(12):1531–1546. doi: 10.1109/tmi.2006.884190. [DOI] [PubMed] [Google Scholar]
- 49.Staal J, Abramoff M, Niemeijer M, Viergever M, van Ginneken B. Ridge-based vessel segmentation in color images of the retina. Medical Imaging, IEEE Transactions on. 2004 Apr;23(4):501–509. doi: 10.1109/TMI.2004.825627. [DOI] [PubMed] [Google Scholar]
- 50.Türetken E, González G, Blum C, Fua P. Automated reconstruction of dendritic and axonal trees by global optimization with geometric priors. Neuroinformatics. 2011;9(2–3):279–302. doi: 10.1007/s12021-011-9122-1. [DOI] [PubMed] [Google Scholar]
- 51.Vázquez S, Cancela B, Barreira N, Penedo M, Rodríguez-Blanco M, Pena Seijo M, Tuero G, Barceló M, Saez M. Improving retinal artery and vein classification by means of a minimal path approach. Machine Vision and Applications. 2013;24(5):919–930. [Google Scholar]
- 52.Wang JJ, Liew G, Klein R, Rochtchina E, Knudtson MD, Klein BE, Wong TY, Burlutsky G, Mitchell P. Retinal vessel diameter and cardiovascular mortality: pooled data analysis from two older populations. European Heart Journal. 2007;28(16):1984–1992. doi: 10.1093/eurheartj/ehm221. [DOI] [PubMed] [Google Scholar]
- 53.Wu X. An efficient antialiasing technique. SIGGRAPH Comput. Graph. 1991 Jul;25(4):143–152. [Google Scholar]
- 54.Xiao C, Staring M, Wang Y, Shamonin D, Stoel B. Multiscale bi-Gaussian filter for adjacent curvilinear structures detection with application to vasculature images. Image Processing, IEEE Transactions on. 2013;22(1):174–188. doi: 10.1109/TIP.2012.2216277. [DOI] [PubMed] [Google Scholar]
- 55.Yedidya T, Hartley R. Tracking of blood vessels in retinal images using Kalman filter. Digital Image Computing: Techniques and Applications (DICTA), 2008. 2008:52–58. [Google Scholar]
- 56.Yu H, Barriga S, Agurto C, Nemeth S, Bauman W, Soliz P. Automated retinal vessel type classification in color fundus images. Proc. SPIE. 2013;8670:86700P–86700P–8. [Google Scholar]
- 57.Zamperini A, Giachetti A, Trucco E, Chin KS. Effective features for artery-vein classification in digital fundus images; Computer-Based Medical Systems (CBMS), 2012 25th International Symposium on; 2012. Jun, pp. 1–6. [Google Scholar]