Abstract
Mouse models for Huntington’s Disease and HD patients demonstrate motor and behavioral dysfunctions, such as progressive loss of coordination and memory, and share similar transcriptional profiles and striatal neuron atrophy. Clear differences between the mouse and human diseases include almost complete striatal degeneration and rarity of intranuclear inclusions in HD, and the fact that mice expressing full length mutant huntingtin do not demonstrate a shortened lifespan characteristic of HD. While no clinical interventions tested in mouse models to date have delayed disease progression, the mouse models provide an invaluable tool for both investigating the underlying pathogenic processes and developing new effective therapies. Inherent differences between humans and mice must be considered in the search for efficacious treatments for HD, but the striking similarities between human HD and mouse models support the view that these models are a biologically relevant system to support the identification and testing of potential clinical therapies.
Keywords: Huntington’s Disease, neurodegeneration, mouse models, polyglutamine
Introduction
Huntington’s Disease [HD] is a progressive, fatal neurodegenerative disorder, characterized by motor, cognitive, behavioral, and psychological dysfunction. The cause of HD is an expansion within a poly(CAG) tract in exon 1 of the huntingtin (HTT) gene (HDCRG 1993). Age of onset is roughly inversely correlated with the length of the CAG tract, which causes disease when 39 or more CAG repeats are present (Norremolle et al., 1993). Affecting approximately 1 in 10,000 people worldwide (Myers et al., 1993), the most obvious pathology is progressive neurodegeneration, particularly within the striatum (caudate and putamen). The massive loss of neurons in this region, normally responsible (amongst many things) for facilitation of volitional movement, is believed to lead to the characteristic motor dysfunctions of HD, such as uncontrolled limb and trunk movements, difficulty maintaining gaze, and general lack of balance and coordination (Bates et al., 2002). Neuronal loss or dysfunction also leads to cognitive problems, behavioral abnormalities, and psychological dysfunction, some of which are reported before motor abnormalities are noticeable. Importantly, some patients present with a more rigid, Parkinsonian form of the disease, typical when age of onset is under 20 (so called juvenile onset cases). These children generally have large repeats, up to 120 for a 3 year old patient (Cannella et al., 2004). The distinctive features of juvenile HD cause many investigators to think of it as a discrete subform of HD which may involve distinctive pathological processes.
Expanded poly(CAG) HTT leads to production of huntingtin protein with an equally expanded polyglutamine (polyQ) stretch near the N-terminus. Despite a lack of consensus on the function of wild type huntingtin (wtHTT), it is well established through studies of human tissue, cellular models, and animal models that mutant polyQ huntingtin (mHTT) exerts a gain of toxic function through aberrant protein-protein interactions. Inclusions containing mHTT, wtHTT, ubiquitin, and many cellular proteins (Hoffner and Djian, 2002) are seen in patients and animal models. These aggregates are not necessarily toxic, but they are commonly observed wherever mHTT is expressed. That the same aggregates and cellular toxicity observed in humans are also seen in many models, with drastically different time scales (from days in tissue culture to decades in human HD), accentuates the importance of expression levels and protein context in cellular pathology. This is particularly evident in the wide variety of phenotypic progression seen in the many mouse models of HD, which is the subject of this review.
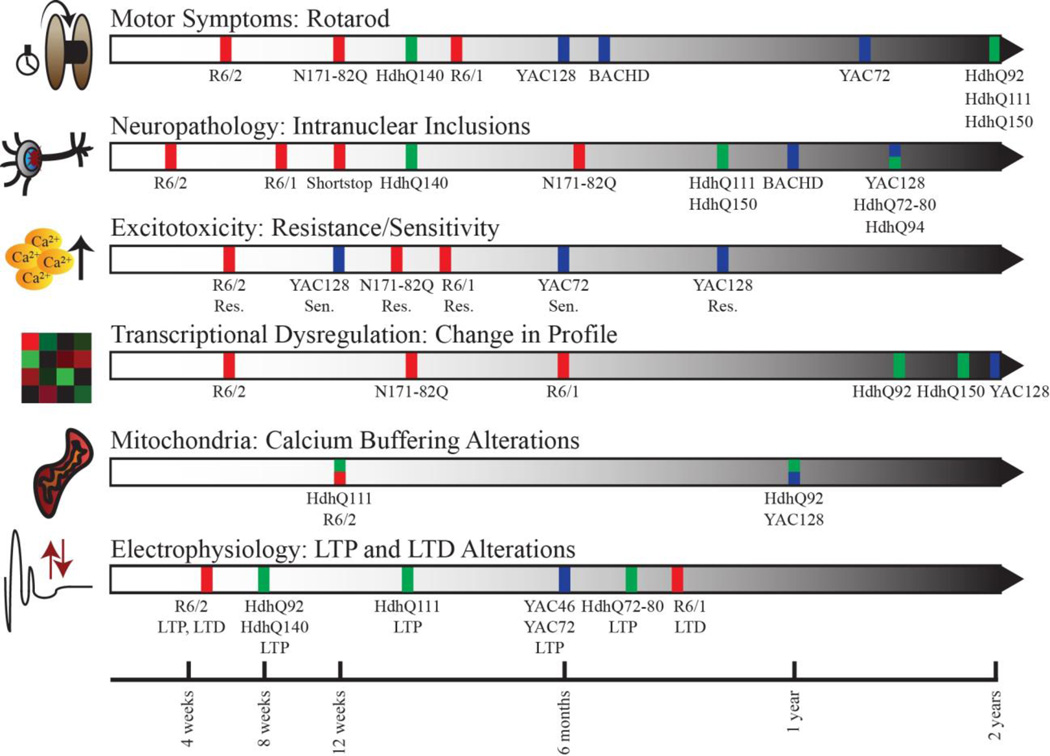
A mutant HD gene is present in the body of an individual from conception. The potential for beneficial therapeutic intervention is therefore present throughout the life of an affected individual. However, the physiological consequences of the presence of the HD mutation differ as life progresses. A key issue in utilizing a mouse model to test therapeutic intervention for HD is to assess which stage of disease a model corresponds to at any given point in time. Some strains display neuropathology from birth and early mortality, while others progress so slowly that visible phenotypes are not seen until the mice are very old, and do not present with morbidity. The age of onset of a number of frequently utilized behavioral and biological measures of pathology for HD mouse models are summarized in Figure 1.
Figure 1.
Timeline of behavioral and neuropathological symptoms in selected HD model mice. Strains are categorized by color: red (N-terminal transgenic), blue (full-length transgenic), and green (knock-in).
The first transgenic model of HD in mice was developed in 1996 (Mangiarini et al., 1996) by introducing a fragment of a juvenile HD patient’s HTT gene into the mouse genome. Although these strains (R6/2 and R6/1) were initially designed to study repeat expansion, these strains displayed motor and metabolic symptoms, including tremors, lack of coordination (rotarod balance difficulty), and excessive weight loss, leading to death at a very early age (~12–14 weeks in the R6/2 line). The rapid and reproducible progression of HD-like symptomology in R6/2 mice has made this line a mainstay of HD research. However, the limitations of R6/2, the absence of a full length mutant HTT protein and the extremely rapid progression of disease led to the development of quite a number of other animal models, each with their own unique genetic and phenotypic characteristics summarized in Table 1.
Table 1.
Commonly used mouse models of Huntington’s Disease.
| Strain Name |
Transgenic or Knockin |
Gene Characteristics | Promoter | Repeat Length |
Motor Symptom Onset |
Lifespan | Background Strain(s) |
References |
|---|---|---|---|---|---|---|---|---|
| R6/2 | Transgenic fragment |
Exon 1 of human HTT gene |
1 kb of Human HTT |
~150 | 6 weeks | 10–13 wks | C57BL/6XCBA |
Mangiarini et al., 1996; Carter etal., 1999 |
| R6/1 | Transgenic fragment |
Exon 1 of human HTT gene |
1 kb of Human HTT |
116 | 18 weeks | 32–40 wks | C57BL/6xCBA or C57BL/6 |
Mangiarini et al., 1996; Hodges et al., 2008 |
| N171-82Q | Transgenic fragment |
First 171 AA of human HTT (exons 1, 2, part of 3) |
Prnp | 82 | 3 months | 16–22 wks | C57BL/6xC3H/He |
Schilling et al., 1999; Schilling et al., 2004 |
| Tg100 | Transgenic fragment |
First ~3 kb of human HTT cDNA |
Rat NSE | 100 | 3 months (nonspecific) |
Normal | C57BL/6xSJL | Laforet et al., 2001 |
| HD94 | Transgenic fragment |
Chimeric human/ mouse HTT exon 1 |
TetO + tTA | 94 | 4–8 weeks (clasping) |
Normal | C57BIV6xCBA | Yamamoto et al., 2000 |
| YAC72 | Transgenic full-length |
Full length human HTT gene |
Human HTT | 72 | 16 months | Normal | FVB/N |
Hodgson et al., 1999; Seo et al., 2008 |
| YAC128 | Transgenic full-length |
Full length human HTT gene |
Human HTT | 120 | 6 months | Normal | FVB/N | Slow et al., 2003 |
| BACHD | Transgenic full-length |
Full length human HTT gene (floxed exon 1) |
Human HTT | 97 (mixed) | 2 months | Normal | FVB/N | Gray et al., 2008 |
| HdhQ72, Q80 |
Knockin | Endogenous murine Htt gene, expanded CAG inserted |
Mouse Htt | 72,80 | 12 months | Normal | Mixed 129Sv, C57BIV6 |
Shelboume et al., 1999; Kennedy et al., 2003 |
| HdhQ111 | Knockin | Endogenous murine Htt gene, chimeric human/mouse exon 1 |
Mouse Htt | 109 | 24 months (gait) |
Normal | Mixed 129Sv, CD1 |
Wheeler et al., 1999; Wheeler et al., 2002 |
| HdhQ94 | Knockin | Endogenous murine Htt gene, chimeric human/mouse exon 1 |
Mouse Htt | 94 | 2 months (rearing) |
Normal | Mixed 129Sv, C57BL/6 |
Levine et al., 1999; Menalled et al., 2002 |
| HdhQ140 | Knockin | Endogenous murine Htt gene, chimeric human/mouse exon 1 |
Mouse Htt | 140 | 4 months | Normal | Mixed 129Sv, C57B176 |
Menalled et al., 2003; Hickey et al., 2008 |
| HdhQ150 | Knockin | Endogenous murine Htt gene, expanded CAG inserted |
Mouse Htt | 150 | 100 weeks | Normal | Mixed 12901a, C57BIV6 |
Lin et al., 2001; Heng et al., 2007 |
Mouse models of HD can be grouped into three categories, based on the genetic basis of their creation. N-terminal transgenic animals are those carrying a small 5’ portion of huntingtin, either human or chimeric human/mouse, at random in their genome. These animals tend to have the earliest onset of motor symptoms and diminished lifespan (Carter et al., 1999; Hodges et al., 2008; Mangiarini et al., 1996; Schilling et al., 1999; Schilling et al., 2004), thought to be because mHTT pathology is greatly enhanced by (though maybe not dependent on [Gray et al., 2008]) its proteolytic processing into N-terminal fragments (Graham et al., 2006; Li et al., 2000); these mouse models are probably a short cut to this particularly toxic state.
Transgenic models expressing full-length mHTT mice also exist, containing random insertions of the full-length human HTT gene with an expanded CAG repeat in the form of either YAC or BAC DNA (Gray et al., 2008; Hodgson et al., 1999; Seo et al., 2008; Slow et al., 2003). One interesting observation of the two most commonly used models in this category is the unexpected age of onset difference (~6 months in YAC128 mice and as early as 8 weeks in BACHD mice) despite the shorter repeat length of BACHD mice (97 vs. 128).
Several strains in which a pathological length CAG repeat is introduced into the mouse huntingtin (Htt) gene have also been created (so called knock-in strains) (Heng et al., 2007; Kennedy et al., 2003; Levine et al., 1999; Lin et al., 2001; Menalled et al., 2003; Menalled et al., 2002; Shelbourne et al., 1999; Wheeler et al., 1999; Wheeler et al., 2002). The longest repeat models (140 and 150 repeats) have motor symptom onset within 6 months, but the shorter models have little or no observable motor dysfunction for the first year of life, and no decrease in lifespan has been reported in any knock-in models. This may properly model the late adult onset of human HD but does not replicate the impaired quality of life and inevitable mortality.
As many models have brought into use significant differences among the models have emerged. It is important to note, however, that many cross-model studies underline significant pathological and molecular similarities in the different genetic models in spite of their inevitable differences (Bennett et al., 2007; Björkqvist et al., 2008; Ginés et al., 2006; Jenkins et al., 2005; Kuhn et al., 2007; Luthi-Carter et al., 2002; Menalled et al., 2000; Southwell et al., 2009; Strand et al., 2007; Walker et al., 2008; Woodman et al., 2007). In this review we have focusied on specific pathological aspects of HD to compare and contrast models.
Behavioral Symptoms
HD in patients is characterized by motor, cognitive and behavioral symptoms, and assays testing these broad categories are used to measure progression of pathology in HD mice. Motor phenotypes have been tested in a number of HD model mice, including limb clasping upon tail suspension, basal activity level, gait abnormalities, balance beam traversing time, swimming speed, suspended horizontal beam turning, and latency to remain on a fixed-speed or accelerating rotarod. The rotarod, in particular, has proven to be a robust quantitative measurement of balance and coordination deficits for which nearly every HD model mouse has demonstrated a deficiency. N-terminal transgenic mice consistently display an early onset of severe motor symptoms. R6/2 mice swim poorly by 5 weeks of age, and show beam walking and rotarod deficiencies by 6 weeks, both of which progressively worsen with age (Carter et al., 1999). R6/1 mice experience clear rotarod deficiency at 18 weeks (Hodges et al., 2008) with an earlier (13 week) onset of failure to turn around on a suspended horizontal rod (van Dellen et al., 2000), and N171-82Q mice display a subtle but progressive rotarod phenotype at 3 months (Schilling et al., 1999).
Full-length transgenic models display delayed motor symptoms compared to N-terminal transgenics; YAC72 mice do not display a significant rotarod phenotype until 16 months (Seo et al., 2008), while YAC128 mice decline starting at 6–7 months (Slow et al., 2003; Van Raamsdonk et al., 2005c). BACHD transgenics do show a significant reduction in rotarod latency as early as 4 weeks of age, but they do not precipitously decline in performance until 28 weeks; this is in contrast to R6/2 rotarod performance, which rapidly declines once a difference is measured (Menalled et al., 2009).
Knockin mice do not always display the characteristic motor phenotype seen in transgenic models, despite some strains carrying as many CAG repeats as R6/2 mice (~150) and having twice the gene dose as most transgenic strains (behavioral experiments carried out in knockin mice typically use homozygotes). This could reflect differences in chromosomal context, transgene expression, the chimeric nature of knockin Htt inserts, or strain background. HDHQ140 rotarod latency appears at 4 months at 30 rpm on a fixed-speed rotarod (Hickey et al., 2008), but another group reported no accelerating rotarod phenotype through 6 months (Dorner et al., 2007), while rotarod deficits are not seen in HdhQ92, HdhQ111, and HDHQ150 mice until about 2 years of age (Heng et al., 2007; Menalled et al., 2009; Trueman et al., 2009).
Cognitive phenotypes can again be measured in many ways, but tasks based on spatial learning and memory such as the Morris water maze or T maze (swimming or elevated) have been used to reveal deficits in initial task learning and re-learning upon parameter changes. 4–5 week old R6/2 mice learn the Morris water maze as well as wild types when the platform is visible, but display spatial memory deficits when the platform is hidden, and cannot re-learn upon platform movement as well as wild type mice. Two-choice swim testing revealed an earlier deficit in task reversal (6.5 weeks) than for initial visual learning of the task (10–11 weeks) (Lione et al., 1999). Initial visual learning deficiency of the two-choice swim test was also found in YAC128 mice (Van Raamsdonk et al., 2005c), but HDHQ150 knockins displayed no learning deficits on the Morris water maze (Heng et al., 2007).
Cognitive tests are challenging to standardize, as environmental conditions and spatial cues are difficult to replicate from lab-to-lab and can influence animals’ performance in behavioral tests. Despite these challenges, these consistent observations from many different labs demonstrating a clear effect on cognitive performance in HD model mice suggests that the cognitive decline commonly observed in HD patients is well represented by HD model mice.
Neuropathology
Human neuropathology is characterized by a severe loss of striatal volume (in particular the caudate nucleus). Medium spiny neurons but not interneurons are lost, and reactive gliosis is apparent (Sharp and Ross, 1996). Cortical degeneration is also prominent in late stages. HTT inclusions in patients are only found in a small fraction of cells (Gourfinkel-An et al., 1998), though they are visible in almost all HD patient brains with a clinical grade of at least 2 (Herndon et al., 2009). Within HD model mice, the progressive neuropathology is unique for each strain, but they share some commonalities.
N-terminal transgene strains display neuropathology at or prior to symptom onset. In contrast to patients, neuron loss is somewhat minimal, but R6/2 brains decrease in weight as much as 20% with enlargement of the lateral ventricles (Mangiarini et al., 1996). They demonstrate neuronal intranuclear inclusions (NIIs) as early as at birth (Stack et al., 2005), though NII’s are typically reported in this strain around 3–4.5 weeks (Davies et al., 1997; Meade et al., 2002; Morton et al., 2000), significantly prior to onset of easily observed symptoms. Inclusions were found in the cortex, striatum, cerebellum, spinal cord, and hippocampus, and progressively increase in prevalence and size (Meade et al., 2002). Despite this, chimera studies suggest that medium spiny neurons (MSNs) bearing large inclusions can survive for almost a year (Reiner et al., 2007) when surrounded by wild type cells. R6/2 MSN dendritic diameters and spine density also decrease with age (Klapstein et al., 2001).
R6/1 mice share most of the R6/2 pathology, but at a later age. NII’s appear by 9 weeks (Naver et al., 2003) and also show minimal gliosis (Yu et al., 2003) and similar dendritic spine atrophy by 8 months (Spires et al., 2004). Apoptotic and necrotic cells are rarely seen in the striatum of R6/2 and R6/1 mice, despite significant atrophy and ventricular enlargement; instead, electron micrographs contain so called dark neurons, displaying condensation of the cytoplasm and nucleus without the chromatin fragmentation and nuclear blebbing characteristic of apoptosis (Yu et al., 2003). In contrast, 3 month old N171-82Q mice do demonsrate cortical and striatal apoptotic neurons, with reactive gliosis by 4 months. Note that in old (22–30 week) R6/2 chimeras, gliosis is apparent in regions densely populated in transgenic neurons (Reiner et al., 2007), and particularly old R6/2 animals (17 weeks) show astrocytes with processes enveloping degenerating neurons (Turmaine et al., 2000). Therefore, the signals necessary to develop gliosis in R6/2 mice may be present, but the mice may die before glial recruitment and activation. N171-82Q mice also presented with striatal degeneration and ventricular enlargement by 17 weeks (Gardian et al., 2005) and NII’s in many brain regions (cortex, hippocampus, cerebellum, and striatum among others) by late endstage of 6.5 months.
NII’s are not seen until far after symptom onset in full-length transgenic HD lines. YAC128 mice display behavioral symptoms at 12 months, and striatal neuron loss of ~15% is seen by this time (Slow et al., 2003) along with increased intranuclear HTT staining of certain brain structures (Van Raamsdonk et al., 2005a). However, NII’s did not show up until 18 months of age, and only populated ~30% of striatal neurons and ~5% of cortical neurons. NII’s were absent in the YAC128 hippocampus, a site of NII staining in endstage R6/2’s (Morton et al., 2000). In the other distinct full-length transgenic strain, BACHD mice also display atrophy of the cortex and striatum by as much as 30% at 12 months (Gray et al., 2008), with 14% of striatal neurons with the aforementioned “dark” morphology. Interestingly and as opposed to R6 mice, inclusions (over 90%) were extranuclear and were more common in the cortex than striatum, a feature reminiscent of adult onset HD.
R6/2 chimaeras suggest that inclusions themselves may be neither toxic nor markers of cells about to die, and a strain arising with a spontaneous mutation in the YAC128 transgene (termed Shortstop or Ss for its early termination) provides further evidence to this end (Slow et al., 2005). The mutation truncated the transgene after exon 2, providing a product with 128 glutamines and an expected and observed protein size similar to that encoded by the R6/2 transgene. NII’s are particularly common (90% of striatal, cortical, and hippocampal cells) and appear earlier in Ss mice versus YAC128s; however, Ss mice had no obvious phenotype at all ages examined. Why this strain is free from the early onset behavioral symptoms one would expect in what is essentially an N-terminal transgene HD model is still under investigation.
Knockin mouse neuropathology, as with their symptoms, usually occurs very late in life and is minor in comparison to transgenic strains. HdhQ72–80 mice demonstrate loss of brain weight by 16 months (Shelbourne et al., 1999), while NII’s are primarily seen in striatal MSNs. Knockins with 94 repeats demonstrate striatal NII’s by 18 months (Menalled et al., 2002) while HdhQ111 mice show NII appearance at an earlier age (10 months), and mHtt nuclear accumulation is evident at a very early (6 weeks) age (Wheeler et al., 2000). HDHQ150 knockins lose as much as 40% of striatal volume and neurons by 23 months, but significant gliosis and NII’s appear by 10–14 months (Heng et al., 2007; Lin et al., 2001; Tallaksen-Greene et al., 2005; Yu et al., 2003). Degenerating neurons are not apoptotic in this strain, though occasional dark neurons are encountered. Knockin mice with 140 CAG repeats display relatively early onset striatal NII’s and neuropil aggregates (4 months) becoming progressively stronger in other brain regions as well by 6 months (Menalled et al., 2003). These data do not imply that knockin brains only present with abnormality in old age, although HdhQ111 embryos demonstrate impaired neurogenesis as early as embryonic day 13.5 (Molero et al., 2009).
Protein context clearly influences neuropathology and symptoms in HD, as other polyglutamine disorders such as SBMA, DRPLA and the various spinocerebellar ataxias (reviewed in Yamada et al., 2008) produce distinct vulnerable neuronal populations and motor symptoms. Post-translational modifications influence progression, as ablation of caspase-6 cleavage sites within the YAC transgene yields a mouse which demonstrates no behavioral symptoms or striatal atrophy, along with a delay in mHTT translocation to the nucleus (Graham et al., 2006). Also, while only 17 residues lie N-terminal to the polyQ repeat in human HTT, these play a role in neuropathology; phosphomimetic mutations of serines 13 and 16 prevent aggregation and symptom onset in parallel BACHD lines (Gu et al. 2009).
While inclusions are a historic histological hallmark of HD, evidence continues to mount that their presence does not correlate with toxicity, as seen in chimaeric R6/2 or in Ss mice. The theory that inclusions represent a sequestration site for mHTT, while smaller oligomers mediate toxicity, is consistent with neuropathological data in mice but is hard to prove with causative data. It represents an intriguing therapeutic option though, that perhaps altering aggregation kinetics by accelerating macro-aggregation or reducing oligomerization would slow toxicity.
Cell autonomous vs. non-autonomous pathology
Neurodegeneration in HD affects multiple brain regions, but striatal degeneration has been the focus of much of the research field, for two reasons: the massive loss of neurons suggests a particular vulnerability of striatal MSN’s, and more specifically, the characteristic motor phenotype is classically attributed to the early loss of one of two nearly identical arms of the corticostriatal loop, the indirect (striatopalladial) pathway. Nevertheless, recent research suggests that multiple cell types in the brain contribute to pathology. Driving an expanded poly(CAG) HTT fragment in glial (GFAP+) cells also induces many features in common with other mouse models of HD (clasping, failure to keep on weight, rotarod phenotype, and premature death), albiet at a later time than is common for models expressing N-terminal transgenes in neurons (Bradford et al., 2009). This is interesting when one considers the stark phenotype of the N171-82Q mice, whose N-terminal transgene is driven primarily in neurons by the prion promoter (Schilling et al. 1999). However, a conditional model of HD suggests that expression of mutant HTT in multiple cell types is required for motor symptoms. A lox-STOP-lox poly(CAG) HTT exon 1 strain mated to Nestin-Cre mice (pan-neuronal expression) induced a behavioral phenotype at 6 months of age, but mating it to Emx1-Cre (cortical pyramidal cell expression) (Gu et al., 2005) or Dlx5/6-Cre mice (striatal MSN expression) produced EM48+ aggregates in the expected brain regions but no observed motor phenotype; the animal’s short lifespan may limit phenotypic progression in these models. Taken as a whole, we can see that mutant HTT can cause neuropathology (aggregate formation at the least) in nearly every neuronal or glial cell in which it is expressed, and while MSN expression plays a large role, cells other than MSNs can contribute to manifest disease in mice. This has particular importance from a therapeutic perspective, as it suggests that drugs which by default cannot affect neurons (e.g. the target enzyme is not expressed in neurons) should not a priori be set aside.
Excitotoxicity
An important and unanswered question in the HD field is what mediates the specific vulnerability of striatal MSNs, leaving striatal interneurons, glia, and other brain regions less damaged. The observation that kainic acid (KA), a structural analog of the excitatory neurotransmitter glutamate, produced striatal degeneration reminiscent of HD (Coyle and Schwarcz, 1976) while sparing dopaminergic projections suggested overactivation of postsynaptic glutamate receptors damages MSNs. Another glutamate analog, quinolinic acid (QA), was later tested (Beal et al., 1991; Beal et al., 1986) and produced a similar lesion as KA, but spared cholinergic interneurons, making it a particularly similar animal model for HD. These experiments brought forward the excitotoxicity hypothesis, that MSNs in HD are sensitive to overactivation of glutamate receptors (specifically, NMDA receptors) resulting in excessive Ca2+ and other ionic influx and selective death.
Excitotoxicity was later assayed in genetic HD mouse models. R6/1 and R6/2 mice injected presymptomatically (18 weeks and 6 weeks, respectively) displayed marked resistance to excitotoxic lesions compared to wild type littermates (Hansson et al., 1999); this resistance was progressive with age. N171-82Q mice displayed resistance to intrastriatal QA administered at 15 weeks (Jarabek et al., 2004), and asymptomatic shortstop mice are also QA resistant (Slow et al., 2005), but this phenotype is not ubiquitous among the N-terminal transgene strains. TgHD100 mice, which express the N-terminal 1/3 of HTT with 100 CAGs at about 30% endogenous levels, display no alteration of QA lesion size (Petersén et al., 2002). Older R6 mice have five-fold higher basal levels of Ca2+, suggesting that resistance might be the result of compensatory mechanisms (Hansson et al., 2001). Modest protection from mHTT is observed upon decortication or administration of glutamate release inhibitors, glutamate transporter upregulators, mGluR5 antagonists, and mGluR2/3 agonists (Miller et al., 2008; Schiefer et al., 2002; Schiefer et al., 2004; Stack et al., 2007). YAC mice display early QA sensitivity but a progressive loss of sensitivity, becoming resistance to QA in 10 month YAC128 mice (Graham et al., 2009).
In at least four HD mouse models, there is consistent resistance to excitotoxic stress, either pre-symptomatic (R6/1, R6/2, and N171-82Q) or after symptom onset (YAC128). The nature of the resistance phenotype is still under investigation, but may be mediated by adjustments to higher basal Ca2+ levels (Hansson et al., 2001) combined with decreases in dendritic spine density and length (Klapstein et al., 2001; Spires et al., 2004). All told, we see that MSNs are particularly vulnerable to excessive Ca2+ influx, but that over time, the neurons compensate for this to a certain extent. However, even the loss of normal glutamatergic afferents increases neuronal survival, suggesting that despite tolerance to acute excitotoxic insult, corticostriatal glutamate signaling still contributes to neuropathology in HD.
Mitochondria and Energy Imbalance
Neurons, requiring very high metabolic ATP synthesis for maintenance of membrane polarization, are sensitive to perturbations of mitochondrial activity. Rodent medium spiny neurons seem particularly sensitive. Chronic systemic administration of a low dose of succinate dehydrogenase inhibitor 3-nitropropionate (3-NP) in rats induced a massive loss of MSNs but relative sparing of interneurons and dopaminergic afferents (Beal et al., 1993). The toxicity of 3-NP in rats is significantly ameliorated by dietary creatine supplements (Matthews et al., 1998), a compound that also improved survival, rotarod latency, weight, and neuronal atrophy in R6/2 (Ferrante et al., 2000) and N171-82Q mice (Andreassen et al., 2001). R6/2, HdhQ92 and HdhQ111 striatal mitochondria become progressively desensitized to Ca2+ depolarization over time by 3, 12, and 3 months of age, respectively (Brustovetsky et al., 2005). Total forebrain mitochondria also show increase in Ca2+ buffering capacity in 12 week R6/2 and 12 month YAC128 mice prior to permeabilization, though no difference was seen in 16 week HDHQ150 mice (Oliveira et al., 2007), at an age when subtle gait abnormalities are visible (Lin et al., 2001). These observations may be indicative of progressive compensation to heightened baseline Ca2+ levels in aged HD mouse striatal neurons, perhaps also mediating QA resistance. Mitochondrial biogenesis deficiencies may be a contributing factor, as PGC-1α, a positive regulator of mitochondrial biogenesis, is reduced in both human HD samples and 12 month HDHQ140 striatal MSNs (but is elevated in interneurons) (Cui et al., 2006). Disturbances of mitochondria in HD are perhaps not surprising, given neuronal ATP requirements, and the data demonstrate that mHTT can perturb mitochondrial calcium sensitivity.
The hypersensitivity of MSNs to a mitochondrial poison, combined with the mouse mitochondrial perturbations and a general metabolic deficit in HD patients, strongly suggest that both CNS and peripheral symptoms of HD are influenced by altered mitochondrial function.
Electrophysiology
Many motor and behavioral symptoms in HD arise from the massive loss of MSNs, and the motor symptoms that acute 3-NP and QA toxicity produce are reminiscent of advanced Huntington’s Disease. However, many mouse models of HD demonstrate almost no neuronal death. That neurons can be intact but still clearly malfunctioning, combined with the cognitive and memory deficits seen in most patients, suggest that synaptic abnormalities may be significant in HD pathology.
Disturbances in long term potentiation (LTP) and long term depression (LDT) are presented as evidence of a synaptic plasticity dysfunction, and such abnormal responses to LTP and LTD are seen in almost all mouse HD models. Asymptomatic heterozygous HdhQ72–80 mice displayed impaired hippocampal LTP between 8 and 14 months of age (Usdin et al., 1999), as did YAC46 and YAC72 by 6 months (Hodgson et al., 1999), R6/2 averaged from 5 weeks to endstage (Murphy et al., 2000), where as HdhQ92 and HdhQ111 displayed impairment at 2 months and 4–6 months old, respectively, and HDHQ140 mice at only 8 weeks old. (Simmons et al., 2009). LTD was also abnormal in R6/2s (Murphy et al., 2000) and R6/1s (Cummings et al., 2007; Cummings et al., 2006). LTP or LTD deficits have not been reported in BACHD mice, but reduction in high amplitude mEPSCs of MSNs at 6 months (Gray et al., 2008), as well as cortical synaptic alterations at the same age (Spampanato et al., 2008) demonstrate some corticostriatal circuitry impairment in this strain as well. As impaired performance at cognitive tasks such as the Morris water maze or T maze is seen in R6/2s (Lione et al., 1999) and YAC128 (Van Raamsdonk et al., 2005c) animals, as well as somatosensory associative memory problems in R6/1s (Cybulska-Klosowicz et al., 2004), the LTP and LTD impairments likely represent behaviorally relevant plasticity deficits. Because these phenotypes exist in mice which recapitulate many HD patholgic features but without massive neuronal loss, these studies suggest that in spite of the massive loss of neurons in HD, it is likely that defects within existing neuronal circuits contribute to early behavioral symptoms.
CAG Expansion
The CAG repeats within human HD and mouse HD models are prone to mutation, both in the germline and in somatic tissue. Germline expansions are more common in males (Wheeler et al., 2007), correlating with baseline mutant repeat length, and are thought to occur during mitosis, based on the very high percentage of sperm found with mutated alleles (averaging over 80%) (Leeflang et al., 1999).
R6/2 mice are notoriously prone to intergenerational CAG repeat expansion (Morton et al., 2009). This has prompted many labs studying this strain to adopt a selective breeding strategy using only breeders with the desired number of repeats. R6/1 mice are almost as prone to expansions as R6/2s (Mangiarini et al., 1997), but contractions are also seen, notably an R6/1 substrain with 89 CAG repeats that demonstrates a later onset of neuropathology and motor symptoms than standard R6/1s (Vatsavayai et al., 2007). Interestingly, in spite of the fact that CAG repeat length is the strongest correlate for age of onset in HD, R6/2 substrains carrying anywhere from 150 to over 400 repeats have demonstrated that in this transgene and background, higher CAG lengths strongly correlate with a later age of onset (Morton et al., 2009), perhaps due to changes in mHTT subcellular localization. Knockin mice also demonstrate intergenerational CAG repeat length instability, with more mutations seen in mice with higher repeat lengths (HdhQ92, HdhQ111) and higher rates in males (Ishiguro et al., 2001; Shelbourne et al., 1999; Wheeler et al., 1999). We are not aware of germline instability in YAC HD model mice, but BACHD mice do not expand due to the alternating CAA-CAG repeats of the transgene (Gray et al., 2008).
Somatic poly(CAG) instability is also observed in most HD model mice; that BACHD mice display symptoms despite the absence of CAG instability demonstrates that somatic expansions are not required for neuropathology. However, knockins (HdhQ111) lacking DNA mismatch repair enzyme Msh2 had delayed intranuclear mHtt accumulation with absence of somatic CAG repeat expansion (Wheeler et al., 2003). Msh2 knockout R6/1 mice also lacked somatic expansion (Manley et al., 1999). HdhQ72–80 knockins also display prominent striatal, cortical and cerebellar expansions, and HDHQ150 animals show somatic expansions as early as at 4 months of age. (Kennedy et al., 2003; Kennedy and Shelbourne, 2000).
The phenotype of BACHD mice clearly demonstrates that somatic CAG expansion is unlikely to be a major driving force in early disease onset. A possible propensity to cancer that could arise from reducing the activity of mismatch repair proteins also demands caution in exploring this specific pathway for HD therapy. Nevertheless, the mouse Msh2 ablation studies and correlation of expansions to patient samples demonstrate that somatic expansion may contribute to HD.
Transcriptional Dysregulation
Abnormal interactions between mHTT and transcription factors may play a prominent role in neuropathology, and as they are expected to be quite pleiotropic, it suggests both an intriguing explanation for the wide-ranging systems disrupted in HD neurons as well as a promising target for therapy. The reduction of neurotransmitter receptors in the HD striatum (Glass et al., 2000; Pavese et al., 2003; Weeks et al., 1996) is one of the earliest observed symptoms, and mHTT is known to interact with or sequester numerous transcription factors (Boutell et al., 1999; Dunah et al., 2002; Huang et al., 1998; Nucifora et al., 2001; Steffan et al., 2000). The advent of more advanced transcriptional profiling in the last 10 years along with a bevy of mouse models of HD have provided ample opportunity for assaying this dysregulation and attempting therapies.
Microarray transcriptional profiles were compiled for R6/2 mice both before (6 weeks) and after (12 weeks) onset of overt motor symptoms. ~1.5% of transcripts displayed altered levels at each age, with a majority (75%) displaying decreased expression (Luthi-Carter et al., 2000). Many of these transcriptional changes were verified in N171-82Q mice, though they were not shared by YAC72 mice (Chan et al., 2002). Further analysis from this group demonstrated that 12 week old R6/2, 16 week old N171-82Q, and 12 month old animals modeling DRPLA (a disorder resulting from polyglutamine expansion in the Atrophin-1 gene) all show significant overlap of cerebellar profiles (Luthi-Carter et al., 2002). That cerebellar tissue and also laser-capture microdissected interneurons (Zucker et al., 2005) of R6/2 mice demonstrate transcriptional dysregulation suggest that this phenomenon is not unique to the cells most vulnerable to degeneration, nor are inclusion-bearing cells more prone to transcriptionally-altered neurotransmitter receptor levels (Sadri-Vakili et al., 2006). What has been particularly striking is the significant similarities in transcriptional profiles of most genetic HD mouse models tested, both among each other and with human HD. Simultaneous profiling of R6/1, R6/2, HDHQ150, HdhQ92, and YAC128 mice demonstrated that every model correlated significantly with every other model and with human HD, with the caveat that the strains had to be aged appropriately (Kuhn et al., 2007). Other studies have reached similar conclusions (Hodges et al., 2008; Strand et al., 2007).
Given that the global transcriptional changes are more commonly downregulations than upregulations in HD model mice (Luthi-Carter et al., 2000), and that there are altered chromatin dynamics associated with repressed transcription (increased methylation, decreased acetylation) (Stack et al., 2007), it has been investigated whether general modifications to histone dynamics in the form of histone deacetylase (HDAC) modulation could be therapeutic. Suberoylanilide hydroxamic acid (SAHA) and sodium butyrate, two HDAC inhibitors, both caused a delay in motor symptom onset in R6/2 animals, though SAHA was toxic. (Ferrante et al., 2003; Hockly et al., 2003). HDAC inhibitors 4b and valproate alleviated locomotor deficits in R6/2 and N171-82Q mice, respectively (Thomas et al., 2008; Zádori et al., 2009). N171-82Q mice demonstrated marked improvement in lifespan, striatal atrophy, and histone methylation:acetylation ratio upon administration of HDAC inhibitor phenylbutyrate after symptom onset, an important result for a disease in which not every carrier chooses to know their gene status and may only initiate treatment after overt symptoms are detected (Gardian et al., 2005). In order to limit the off target effects of a general HDAC inhibitor, drugs with tighter specificity are needed as well as more focused targets. Recently, R6/2 mice have been bred to strains carrying either homozygous (if not lethal) or heterozygous deletions in HDACs to parse out which HDACs are the best modifiers of pathology. So far HDAC7 has been discounted as a potential modifier (Benn et al., 2009), but HDAC4 does show some promise (Gillian Bates, personal communication).
The similarity in transcriptional profiles between many aged HD model mice and patient samples suggests a fundamental consequence of mHTT on basal levels of transcription, either by direct interaction with transcription factors, attempts at compensatory changes, or both. That this phenotype is directly quantifiable in mice and correlates so strongly with patient samples supports its utility as a biological measure for pathology.
From Preclinical to Clinical Treatment of HD
As we have discussed above, mouse models give insight into many aspects of pathology observed in HD patients. That said, the central question in front of us is how can mouse models best be used to develop therapeutic intervention for HD.
Candidate based approaches and success in mouse models have resulted in many drug trials progressing from preclinical mouse work to patients. The authors direct the readers to three excellent reviews: Li et al. (2005) and Gil and Rego (2009) enumerate the trials conducted in the R6 lines, and Mestre et al. (2009) provide a detailed summary and discussion of published HD clinical trials. Of those treatments tested in rodents, many have made it to clinical trials; nine trials passing the Mestre et al. criteria (randomized, placebo-controlled, symptomatic therapy with at least 10 participants), plus the combination remacemide / Coenzyme Q10 trials, are listed in Table 2. The majority of these selected clinical trials were aimed at safety and tolerability, rather than efficacy, so it comes as little surprise that no improvement in clinical outcome was seen for most. To date, only tetrabenazine (TBZ) has demonstrated a reduction of motor symptoms in both mice and patients (Frank, 2009; Group, 2006; Tang et al., 2007; Wang et al., 2010), and though the mouse studies showed a reduction in striatal atrophy and motor symptoms, reduced neurodegeneration has not been documented in human patients on TBZ.
Table 2.
Selection of drugs tested in both HD model mice and in clinical trials with HD patients.
| Drug | Animal Model | Animal Dosage | Animal Duration | Animal Effect | Patient Dosage | Patient Duration | Patient Effect | References |
|---|---|---|---|---|---|---|---|---|
| Cannabidiol | 3-NP Rat | 5 mg/kg daily | From 12 weeks for 5 days |
a | 10 mg/kg daily | 6 weeks drug 15 weeks obs. |
n.s. |
Sagredo et al., 2007; Consroe et al., 1991 |
| Creatine | R6/2 | 2% in chow ad lib |
From 21 days | 18% improved survival |
8 g daily | 16 weeks drug 24 weeks obs. |
n.s. |
Fenante et al., 2000; Hersch et al., 2006 |
| Ethyl-EPA | YAC128 | 1 % in chow ad lib |
From 7 months | 44% improved rotarod |
2 g daily | 6 or 12 months drug |
n.s. |
Van Raamsdonk et al., 2005b; Huntington Study Group TREND-HD Investigators, 2008 |
| Fluoxetine | R6/1 | 20 mg/kg daily | 10 to 20 weeks | b | 20 mg daily | 4 months drug | n.s. |
Grate et al., 2005; Como et al., 1997 |
| L-carnitine | N171-82Q | 250 mg/kg 5x/week |
From 6 weeks | 15% improved survival |
45 mg/kg daily | 1 week drug 4 weeks obs. |
n.s. |
Vamos et al., 2010; Qoetz et al., 1990 |
| Minocycline | R6/2 | 5 mg/kg daily | From 6 weeks | 13% improved survival |
200 mg daily | 8 weeks drug | n.s. |
Chen et al., 2000; The Huntington Study Group 2004 |
| Remacemide | R6/2 | 0.007% in chow ad lib |
From 21 days | 16% improved survival |
200 or 600 mg daily |
6 weeks drug | n.s. |
Fenante et al., 2002; Kieburtz et al., 1996 |
| Remacemide + CoQ10 | R6/2 | 0.007% Rem 0.2% CoQ10 in chow ad lib |
From 21 days | 32% improved survival |
600 mg each daily |
30 months drug | n.s. |
Ferrante et al., 2002; The Huntington Study Group, 2001 |
| Riluzole | R6/2 | 10 mg/kg daily | From 21 days | 10% improved survival |
200 mg daily | 8 weeks drug | n.s. |
Schiefer et al., 2002; The Huntington Study Group, 2003 |
| Tetrabenazine | YAC128 | 5 mg/kg 3x/week |
From 2 months | ~60% improved rotarod |
Up to 100 mg daily |
12 weeks drug | 3.5 UHDRS units improvement |
Wang et al., 2010; The Huntington Study Group 2006 |
No behavior tested; reduced striatal lesion size and some transcript reduction.
n.s. on rotarod; protected against degeneration in dentate gyrus.
A current challenge to therapeutic development in HD is the identification of validated targets for HD therapy. Currently, there is only one such target: huntingtin itself. Reduction in levels of expression of HTT should be beneficial to HD patients if they can be achieved. Mouse models strongly support this contention. Early work in conditional, reversible models of HD (Yamamoto et al., 2000) demonstrated that silencing of the mutant locus, even relatively late in pathology, results in not only halting of disease progression but reversal of some pathologic sequelae. More recently, two studies have shown that reduction of mutant HTT levels in the brain of model mice, either by reducing translational output of HTT via viral siRNA delivery (Boudreau et al., 2009) or increasing protein clearance of HTT by intrabody (intracellular antibody) expression (Southwell et al., 2009), have a beneficial effect on behavior and neuropathology in HD model mice. The demonstration of a therapeutic benefit of these approaches in mouse models suggests that these approaches could benefit patients as well. Perhaps equally importantly, these studies give confidence that if new validated targets are identified mouse models will be valuable in assessing how effective therapeutic intervention against these targets might be.
However, refinements in the measurements of pathology are needed to make the most out of mouse model studies. In the last few years, clinical studies (volumetric MRI and functional) have begun to provide useful measures to characterize HD progression prior to the point in disease formally designated by functional decline as onset. The modeling of this period (pre-manifest HD) requires the development and validation of a set of measures in the mouse which clearly correspond appropriately to the progression of HD during this period in the human; for example, imaging modalities such as MRI are being minaturized for use in HD model mice (Sawiak et al., 2009; Zhang et al., 2009), and show promise. We don’t yet have this correspondence well established in the mouse for several reasons. First, and perhaps foremost, many of the findings on premanifest HD are quite recent. Second, assay strategies, particularly at the biological level, may require deeper insight into the mechanisms of molecular pathology in premanifest HD including more powerful transcriptional and translational profiling; for example, modern transcriptional profiling by RNAseq will provide additional insight as it allows linearity over a greater range of transcript levels than arrays provide.
The mouse models of HD demonstrate a clear pathology, and while some of the phenotypes (rotarod latency for example) have limited direct relation to measurable patient symptoms, many others (transcriptional profile changes) bear striking resemblance to patients. What the field now has is a collection of mouse models suited to study one or more of the different stages of human disease, a collection of measurable phenomena in these mice (many of which are directly relatable to patient symptoms), and demonstration that targeting the protein at the root of the pathology will effectively reduce or delay the symptoms. Continued studies of the mice give us a tremendous opportunity to use a mammalian nervous system under similar stresses to HD patients, identify therapeutic candidates relevant to a specific disease stage, and test therapies with the knowledge that it is possible to at least partially rescue the cells from the toxic insult of mHTT. It is hopefully only a matter of time before such studies yeild one or more therapeutics that effectively reduce neuropathology in patients.
Selected Reading
- Andreassen OA, Dedeoglu A, Ferrante RJ, Jenkins BG, Ferrante KL, Thomas M, Friedlich A, Browne SE, Schilling G, Borchelt DR, et al. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington’s disease. Neurobiology of Disease. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- Bates G, Harper P, Jones L. Huntington’s Disease. 3rd edn. USA: Oxford University Press; 2002. [Google Scholar]
- Beal MF, Brouillet E, Jenkins BG, Ferrante RJ, Kowall NW, Miller JM, Storey E, Srivastava R, Rosen BR, Hyman BT. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Ferrante RJ, Swartz KJ, Kowall NW. Chronic quinolinic acid lesions in rats closely resemble Huntington’s disease. J Neurosci. 1991;11:1649–1659. doi: 10.1523/JNEUROSCI.11-06-01649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Kowall NW, Ellison DW, Mazurek MF, Swartz KJ, Martin JB. Replication of the neurochemical characteristics of Huntington’s disease by quinolinic acid. Nature. 1986;321:168–171. doi: 10.1038/321168a0. [DOI] [PubMed] [Google Scholar]
- Benn CL, Butler R, Mariner L, Nixon J, Moffitt H, Mielcarek M, Woodman B, Bates GP. Genetic knock-down of HDAC7 does not ameliorate disease pathogenesis in the R6/2 mouse model of Huntington’s disease. PLoS ONE. 2009;4:e5747. doi: 10.1371/journal.pone.0005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu K-Y, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Björkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL. Aberrant interactions of transcriptional repressor proteins with the Huntington’s disease gene product, huntingtin. Hum Mol Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- Bradford J, Shin J-Y, Roberts M, Wang C-E, Li X-J, Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci USA. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustovetsky N, LaFrance R, Purl KJ, Brustovetsky T, Keene CD, Low WC, Dubinsky JM. Age-dependent changes in the calcium sensitivity of striatal mitochondria in mouse models of Huntington’s Disease. J Neurochem. 2005;93:1361–1370. doi: 10.1111/j.1471-4159.2005.03036.x. [DOI] [PubMed] [Google Scholar]
- Cannella M, Gellera C, Maglione V, Giallonardo P, Cislaghi G, Muglia M, Quattrone A, Pierelli F, Di Donato S, Squitieri F. The gender effect in juvenile Huntington disease patients of Italian origin. Am J Med Genet B Neuropsychiatr Genet. 2004;125B:92–98. doi: 10.1002/ajmg.b.20110. [DOI] [PubMed] [Google Scholar]
- Carter RJ, Lione LA, Humby T, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EYW, Luthi-Carter R, Strand A, Solano SM, Hanson SA, DeJohn MM, Kooperberg C, Chase KO, Difiglia M, Young AB, et al. Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington’s disease. Human Molecular Genetics. 2002;11:1939–1951. doi: 10.1093/hmg/11.17.1939. [DOI] [PubMed] [Google Scholar]
- Chen M, Ona VO, Li M, Ferrante RJ, Fink KB, Zhu S, Bian J, Guo L, Farrell LA, Hersch SM, et al. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- Como PG, Rubin AJ, O’Brien CF, Lawler K, Hickey C, Rubin AE, Henderson R, McDermott MP, McDermott M, Steinberg K, Shoulson I. A controlled trial of fluoxetine in nondepressed patients with Huntington’s disease. Mov Disord. 1997;12:397–401. doi: 10.1002/mds.870120319. [DOI] [PubMed] [Google Scholar]
- Consroe P, Laguna J, Allender J, Snider S, Stern L, Sandyk R, Kennedy K, Schram K. Controlled clinical trial of cannabidiol in Huntington’s disease. Pharmacol Biochem Behav. 1991;40:701–708. doi: 10.1016/0091-3057(91)90386-g. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Schwarcz R. Lesion of striatal neurones with kainic acid provides a model for Huntington’s chorea. Nature. 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Milnerwood AJ, Dallérac GM, Vatsavayai SC, Hirst MC, Murphy KPSJ. Abnormal cortical synaptic plasticity in a mouse model of Huntington’s disease. Brain Res Bull. 2007;72:103–107. doi: 10.1016/j.brainresbull.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Milnerwood AJ, Dallérac GM, Waights V, Brown JY, Vatsavayai SC, Hirst MC, Murphy KPSJ. Aberrant cortical synaptic plasticity and dopaminergic dysfunction in a mouse model of Huntington’s disease. Human Molecular Genetics. 2006;15:2856–2868. doi: 10.1093/hmg/ddl224. [DOI] [PubMed] [Google Scholar]
- Cybulska-Klosowicz A, Mazarakis NK, van Dellen A, Blakemore C, Hannan AJ, Kossut M. Impaired learning-dependent cortical plasticity in Huntington’s disease transgenic mice. Neurobiology of Disease. 2004;17:427–434. doi: 10.1016/j.nbd.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Dorner JL, Miller BR, Barton SJ, Brock TJ, Rebec GV. Sex differences in behavior and striatal ascorbate release in the 140 CAG knock-in mouse model of Huntington’s disease. Behav Brain Res. 2007;178:90–97. doi: 10.1016/j.bbr.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim Y-M, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Jenkins BG, Dedeoglu A, Kuemmerle S, Kubilus JK, Kaddurah-Daouk R, Hersch SM, Beal MF. Neuroprotective effects of creatine in a transgenic mouse model of Huntington’s disease. J Neurosci. 2000;20:4389–4397. doi: 10.1523/JNEUROSCI.20-12-04389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington’s disease. J Neurosci. 2002;22:1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, Hersch SM. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S. Tetrabenazine as anti-chorea therapy in Huntington disease: an open-label continuation study. Huntington Study Group/TETRA-HD Investigators. BMC Neurol. 2009;9:62. doi: 10.1186/1471-2377-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardian G, Browne SE, Choi D-K, Klivenyi P, Gregorio J, Kubilus JK, Ryu H, Langley B, Ratan RR, Ferrante RJ, Beal MF. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Gil JM, Rego AC. The R6 lines of transgenic mice: a model for screening new therapies for Huntington’s disease. Brain research reviews. 2009;59:410–431. doi: 10.1016/j.brainresrev.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Ginés S, Bosch M, Marco S, Gavaldà N, Díaz-Hernández M, Lucas JJ, Canals JM, Alberch J. Reduced expression of the TrkB receptor in Huntington’s disease mouse models and in human brain. Eur J Neurosci. 2006;23:649–658. doi: 10.1111/j.1460-9568.2006.04590.x. [DOI] [PubMed] [Google Scholar]
- Glass M, Dragunow M, Faull RL. The pattern of neurodegeneration in Huntington’s disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington’s disease. NSC. 2000;97:505–519. doi: 10.1016/s0306-4522(00)00008-7. [DOI] [PubMed] [Google Scholar]
- Goetz C, Tanner CM, Cohen JA, Thelen JA, Carroll VS, Klawans HL, Fariello RG. L-acetyl-carnitine in Huntington’s disease: double-blind placebo controlled crossover study of drug effects on movement disorder and dementia. Mov Disord. 1990;5:263–265. doi: 10.1002/mds.870050317. [DOI] [PubMed] [Google Scholar]
- Gourfinkel-An I, Cancel G, Duyckaerts C, Faucheux B, Hauw JJ, Trottier Y, Brice A, Agid Y, Hirsch EC. Neuronal distribution of intranuclear inclusions in Huntington’s disease with adult onset. NeuroReport. 1998;9:1823–1826. doi: 10.1097/00001756-199806010-00028. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Graham RK, Pouladi MA, Joshi P, Lu G, Deng Y, Wu N-P, Figueroa BE, Metzler M, André VM, Slow EJ, et al. Differential susceptibility to excitotoxic stress in YAC128 mouse models of Huntington disease between initiation and progression of disease. J Neurosci. 2009;29:2193–2204. doi: 10.1523/JNEUROSCI.5473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu X-H, Tao J, Yamazaki I, Li S-H, Sun YE, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group HS. Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology. 2006;66:366–372. doi: 10.1212/01.wnl.0000198586.85250.13. [DOI] [PubMed] [Google Scholar]
- Grote HE, Bull ND, Howard ML, van Dellen A, Blakemore C, Bartlett PF, Hannan AJ. Cognitive disorders and neurogenesis deficits in Huntington’s disease mice are rescued by fluoxetine. Eur J Neurosci. 2005;22:2081–2088. doi: 10.1111/j.1460-9568.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson O, Guatteo E, Mercuri NB, Bernardi G, Li XJ, Castilho RF, Brundin P. Resistance to NMDA toxicity correlates with appearance of nuclear inclusions, behavioural deficits and changes in calcium homeostasis in mice transgenic for exon 1 of the huntington gene. Eur J Neurosci. 2001;14:1492–1504. doi: 10.1046/j.0953-816x.2001.01767.x. [DOI] [PubMed] [Google Scholar]
- Hansson O, Petersén A, Leist M, Nicotera P, Castilho RF, Brundin P. Transgenic mice expressing a Huntington’s disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc Natl Acad Sci USA. 1999;96:8727–8732. doi: 10.1073/pnas.96.15.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng MY, Tallaksen-Greene SJ, Detloff PJ, Albin RL. Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington’s disease. J Neurosci. 2007;27:8989–8998. doi: 10.1523/JNEUROSCI.1830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon ES, Hladik CL, Shang P, Burns DK, Raisanen J, White CL. Neuroanatomic profile of polyglutamine immunoreactivity in Huntington disease brains. J Neuropathol Exp Neurol. 2009;68:250–261. doi: 10.1097/NEN.0b013e318198d320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2’dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- Hickey MA, Kosmalska A, Enayati J, Cohen R, Zeitlin S, Levine MS, Chesselet M-F. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington’s disease mice. NSC. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PAS, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Hughes G, Brooks S, Elliston L, Holmans P, Dunnett SB, Jones L. Brain gene expression correlates with changes in behavior in the R6/1 mouse model of Huntington’s disease. Genes Brain Behav. 2008;7:288–299. doi: 10.1111/j.1601-183X.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Hoffner G, Djian P. Protein aggregation in Huntington’s disease. Biochimie. 2002;84:273–278. doi: 10.1016/s0300-9084(02)01398-6. [DOI] [PubMed] [Google Scholar]
- Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group TREND-HD Investigators [TREND-HD] Randomized controlled trial of ethyl-eicosapentaenoic acid in Huntington disease: the TREND-HD study. Arch Neurol. 2008;65:1582–1589. doi: 10.1001/archneur.65.12.1582. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Yamada K, Sawada H, Nishii K, Ichino N, Sawada M, Kurosawa Y, Matsushita N, Kobayashi K, Goto J, et al. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington’s disease gene. J Neurosci Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- Jarabek BR, Yasuda RP, Wolfe BB. Regulation of proteins affecting NMDA receptor-induced excitotoxicity in a Huntington’s mouse model. Brain. 2004;127:505–516. doi: 10.1093/brain/awh058. [DOI] [PubMed] [Google Scholar]
- Jenkins BG, Andreassen OA, Dedeoglu A, Leavitt B, Hayden M, Borchelt D, Ross CA, Ferrante RJ, Beal MF. Effects of CAG repeat length, HTT protein length and protein context on cerebral metabolism measured using magnetic resonance spectroscopy in transgenic mouse models of Huntington’s disease. J Neurochem. 2005;95:553–562. doi: 10.1111/j.1471-4159.2005.03411.x. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Evans E, Chen C-M, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Human Molecular Genetics. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Human Molecular Genetics. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- Kieburtz K, Feigin A, McDermott M, Como P, Abwender D, Zimmerman C, Hickey C, Orme C, Claude K, Sotack J, et al. A controlled trial of remacemide hydrochloride in Huntington’s disease. Mov Disord. 1996;11:273–277. doi: 10.1002/mds.870110310. [DOI] [PubMed] [Google Scholar]
- Klapstein GJ, Fisher RS, Zanjani H, Cepeda C, Jokel ES, Chesselet MF, Levine MS. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington’s disease transgenic mice. J Neurophysiol. 2001;86:2667–2677. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha J-HJ, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Human Molecular Genetics. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, et al. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington’s disease. Journal of Neuroscience. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang EP, Tavaré S, Marjoram P, Neal CO, Srinidhi J, MacFarlane H, MacDonald ME, Gusella JF, de Young M, Wexler NS, Arnheim N. Analysis of germline mutation spectra at the Huntington’s disease locus supports a mitotic mutation mechanism. Human Molecular Genetics. 1999;8:173–183. doi: 10.1093/hmg/8.2.173. [DOI] [PubMed] [Google Scholar]
- Levine MS, Klapstein GJ, Koppel A, Gruen E, Cepeda C, Vargas ME, Jokel ES, Carpenter EM, Zanjani H, Hurst RS, et al. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knockin mouse models of Huntington’s disease. J Neurosci Res. 1999;58:515–532. [PubMed] [Google Scholar]
- Li H, Li SH, Johnston H, Shelbourne PF, Li XJ. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat Genet. 2000;25:385–389. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- Li JY, Popovic N, Brundin P. The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx : the journal of the American Society for Experimental NeuroTherapeutics. 2005;2:447–464. doi: 10.1602/neurorx.2.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Human Molecular Genetics. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Lione LA, Carter RJ, Hunt MJ, Bates GP, Morton AJ, Dunnett SB. Selective discrimination learning impairments in mice expressing the human Huntington’s disease mutation. J Neurosci. 1999;19:10428–10437. doi: 10.1523/JNEUROSCI.19-23-10428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Human Molecular Genetics. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand AD, Hanson SA, Kooperberg C, Schilling G, La Spada AR, Merry DE, Young AB, Ross CA, Borchelt DR, Olson JM. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington’s disease mouse models reveal context-independent effects. Human Molecular Genetics. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- Matthews RT, Yang L, Jenkins BG, Ferrante RJ, Rosen BR, Kaddurah-Daouk R, Beal MF. Neuroprotective effects of creatine and cyclocreatine in animal models of Huntington’s disease. J Neurosci. 1998;18:156–163. doi: 10.1523/JNEUROSCI.18-01-00156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade CA, Deng Y-P, Fusco FR, Del Mar N, Hersch S, Goldowitz D, Reiner A. Cellular localization and development of neuronal intranuclear inclusions in striatal and cortical neurons in R6/2 transgenic mice. J. Comp Neurol. 2002;449:241–269. doi: 10.1002/cne.10295. [DOI] [PubMed] [Google Scholar]
- Menalled L, El-Khodor BF, Patry M, Suárez-Fariñas M, Orenstein SJ, Zahasky B, Leahy C, Wheeler V, Yang XW, MacDonald M, et al. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiology of Disease. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled L, Zanjani H, MacKenzie L, Koppel A, Carpenter E, Zeitlin S, Chesselet MF. Decrease in striatal enkephalin mRNA in mouse models of Huntington’s disease. Experimental Neurology. 2000;162:328–342. doi: 10.1006/exnr.1999.7327. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet M-F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Wu Y, Olivieri M, Li X-J, Li H, Zeitlin S, Chesselet M-F. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J Neurosci. 2002;22:8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre T, Ferreira J, Coelho MM, Rosa M, Sampaio C. Therapeutic interventions for symptomatic treatment in Huntington’s disease. Cochrane Database Syst Rev. 2009:CD006456. doi: 10.1002/14651858.CD006456.pub2. [DOI] [PubMed] [Google Scholar]
- Miller BR, Dorner JL, Shou M, Sari Y, Barton SJ, Sengelaub DR, Kennedy RT, Rebec GV. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero AE, Gokhan S, Gonzalez S, Feig JL, Alexandre LC, Mehler MF. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington’s disease. Proc Natl Acad Sci USA. 2009;106:21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Glynn D, Leavens W, Zheng Z, Faull RLM, Skepper JN, Wight JM. Paradoxical delay in the onset of disease caused by super-long CAG repeat expansions in R6/2 mice. Neurobiology of Disease. 2009;33:331–341. doi: 10.1016/j.nbd.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Morton AJ, Lagan MA, Skepper JN, Dunnett SB. Progressive formation of inclusions in the striatum and hippocampus of mice transgenic for the human Huntington’s disease mutation. J Neurocytol. 2000;29:679–702. doi: 10.1023/a:1010887421592. [DOI] [PubMed] [Google Scholar]
- Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH, MacDonald ME, Koroshetz WJ, Duyao MP, Ambrose CM, Taylor SA, Barnes G, Srinidhi J, Lin CS, Whaley WL, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Nat Genet. 1993;5:168–173. doi: 10.1038/ng1093-168. [DOI] [PubMed] [Google Scholar]
- Naver B, Stub C, Møller M, Fenger K, Hansen AK, Hasholt L, Sørensen SA. Molecular and behavioral analysis of the R6/1 Huntington’s disease transgenic mouse. NSC. 2003;122:1049–1057. doi: 10.1016/j.neuroscience.2003.08.053. [DOI] [PubMed] [Google Scholar]
- Nørremølle A, Riess O, Epplen JT, Fenger K, Hasholt L, Sørensen SA. Trinucleotide repeat elongation in the Huntingtin gene in Huntington disease patients from 71 Danish families. Human Molecular Genetics. 1993;2:1475–1476. doi: 10.1093/hmg/2.9.1475. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Oliveira JMA, Jekabsons MB, Chen S, Lin A, Rego AC, Gonçalves J, Ellerby LM, Nicholls DG. Mitochondrial dysfunction in Huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J Neurochem. 2007;101:241–249. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- Pavese N, Andrews TC, Brooks DJ, Ho AK, Rosser AE, Barker RA, Robbins TW, Sahakian BJ, Dunnett SB, Piccini P. Progressive striatal and cortical dopamine receptor dysfunction in Huntington’s disease: a PET study. Brain. 2003;126:1127–1135. doi: 10.1093/brain/awg119. [DOI] [PubMed] [Google Scholar]
- Petersén A, Chase K, Puschban Z, DiFiglia M, Brundin P, Aronin N. Maintenance of susceptibility to neurodegeneration following intrastriatal injections of quinolinic acid in a new transgenic mouse model of Huntington’s disease. Experimental Neurology. 2002;175:297–300. doi: 10.1006/exnr.2002.7885. [DOI] [PubMed] [Google Scholar]
- Reiner A, Del Mar N, Deng Y-P, Meade CA, Sun Z, Goldowitz D. R6/2 neurons with intranuclear inclusions survive for prolonged periods in the brains of chimeric mice. J. Comp. Neurol. 2007;505:603–629. doi: 10.1002/cne.21515. [DOI] [PubMed] [Google Scholar]
- Sadri-Vakili G, Menon AS, Farrell LA, Keller-McGandy CE, Cantuti-Castelvetri I, Standaert DG, Augood SJ, Yohrling GJ, Cha J-HJ. Huntingtin inclusions do not down-regulate specific genes in the R6/2 Huntington’s disease mouse. Eur J Neurosci. 2006;23:3171–3175. doi: 10.1111/j.1460-9568.2006.04871.x. [DOI] [PubMed] [Google Scholar]
- Sagredo O, Ramos JA, Decio A, Mechoulam R, Fernández-Ruiz J. Cannabidiol reduced the striatal atrophy caused 3-nitropropionic acid in vivo by mechanisms independent of the activation of cannabinoid, vanilloid TRPV1 and adenosine A2A receptors. Eur J Neurosci. 2007;26:843–851. doi: 10.1111/j.1460-9568.2007.05717.x. [DOI] [PubMed] [Google Scholar]
- Sawiak SJ, Wood NI, Williams GB, Morton AJ, Carpenter TA. Use of magnetic resonance imaging for anatomical phenotyping of the R6/2 mouse model of Huntington’s disease. Neurobiology of Disease. 2009;33:12–19. doi: 10.1016/j.nbd.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Schiefer J, Landwehrmeyer GB, Lüesse H-G, Sprünken A, Puls C, Milkereit A, Milkereit E, Kosinski CM. Riluzole prolongs survival time and alters nuclear inclusion formation in a transgenic mouse model of Huntington’s disease. Mov Disord. 2002;17:748–757. doi: 10.1002/mds.10229. [DOI] [PubMed] [Google Scholar]
- Schiefer J, Sprünken A, Puls C, Lüesse H-G, Milkereit A, Milkereit E, Johann V, Kosinski CM. The metabotropic glutamate receptor 5 antagonist MPEP and the mGluR2 agonist LY379268 modify disease progression in a transgenic mouse model of Huntington’s disease. Brain Res. 2004;1019:246–254. doi: 10.1016/j.brainres.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Human Molecular Genetics. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Schilling G, Savonenko AV, Coonfield ML, Morton JL, Vorovich E, Gale A, Neslon C, Chan N, Eaton M, Fromholt D, et al. Environmental, pharmacological, and genetic modulation of the HD phenotype in transgenic mice. Experimental Neurology. 2004;187:137–149. doi: 10.1016/j.expneurol.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Seo H, Kim W, Isacson O. Compensatory changes in the ubiquitin-proteasome system, brain-derived neurotrophic factor and mitochondrial complex II/III in YAC72 and R6/2 transgenic mice partially model Huntington’s disease patients. Human Molecular Genetics. 2008;17:3144–3153. doi: 10.1093/hmg/ddn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AH, Ross CA. Neurobiology of Huntington’s disease. Neurobiology of Disease. 1996;3:3–15. doi: 10.1006/nbdi.1996.0002. [DOI] [PubMed] [Google Scholar]
- Shelbourne PF, Killeen N, Hevner RF, Johnston HM, Tecott L, Lewandoski M, Ennis M, Ramirez L, Li Z, Iannicola C, et al. A Huntington’s disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Human Molecular Genetics. 1999;8:763–774. doi: 10.1093/hmg/8.5.763. [DOI] [PubMed] [Google Scholar]
- Simmons DA, Rex CS, Palmer L, Pandyarajan V, Fedulov V, Gall CM, Lynch G. Up-regulating BDNF with an ampakine rescues synaptic plasticity and memory in Huntington’s disease knockin mice. Proc Natl Acad Sci USA. 2009;106:4906–4911. doi: 10.1073/pnas.0811228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, et al. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang Y-Z, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Human Molecular Genetics. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- Southwell AL, Ko J, Patterson PH. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J Neurosci. 2009;29:13589–13602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Gu X, Yang XW, Mody I. Progressive synaptic pathology of motor cortical neurons in a BAC transgenic mouse model of Huntington’s disease. NSC. 2008;157:606–620. doi: 10.1016/j.neuroscience.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires TL, Grote HE, Garry S, Cordery PM, van Dellen A, Blakemore C, Hannan AJ. Dendritic spine pathology and deficits in experience-dependent dendritic plasticity in R6/1 Huntington’s disease transgenic mice. Eur J Neurosci. 2004;19:2799–2807. doi: 10.1111/j.0953-816X.2004.03374.x. [DOI] [PubMed] [Google Scholar]
- Stack EC, Dedeoglu A, Smith KM, Cormier K, Kubilus JK, Bogdanov M, Matson WR, Yang L, Jenkins BG, Luthi-Carter R, et al. Neuroprotective effects of synaptic modulation in Huntington’s disease R6/2 mice. J Neurosci. 2007;27:12908–12915. doi: 10.1523/JNEUROSCI.4318-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack EC, Kubilus JK, Smith K, Cormier K, Del Signore SJ, Guelin E, Ryu H, Hersch SM, Ferrante RJ. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington’s disease transgenic mice. J. Comp. Neurol. 2005;490:354–370. doi: 10.1002/cne.20680. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, Cleren C, Beal MF, Jones L, Kooperberg C, Olson JM, Jones KR. Expression profiling of Huntington’s disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallaksen-Greene SJ, Crouse AB, Hunter JM, Detloff PJ, Albin RL. Neuronal intranuclear inclusions and neuropil aggregates in HdhCAG(150) knockin mice. NSC. 2005;131:843–852. doi: 10.1016/j.neuroscience.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Tang T-S, Chen X, Liu J, Bezprozvanny I. Dopaminergic Signaling and Striatal Neurodegeneration in Huntington’s Disease. Journal of Neuroscience. 2007;27:7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group [HDCRG] A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- The Huntington Study Group. A randomized, placebo-controlled trial of coenzyme Q10 and remacemide in Huntington’s disease. Neurology. 2001;57:397–404. doi: 10.1212/wnl.57.3.397. [DOI] [PubMed] [Google Scholar]
- The Huntington Study Group. Dosage effects of riluzole in Huntington’s disease: a multicenter placebo-controlled study. Neurology. 2003;61:1551–1556. doi: 10.1212/01.wnl.0000096019.71649.2b. [DOI] [PubMed] [Google Scholar]
- The Huntington Study Group. Minocycline safety and tolerability in Huntington disease. Neurology. 2004;63:547–549. doi: 10.1212/01.wnl.0000133403.30559.ff. [DOI] [PubMed] [Google Scholar]
- The Huntington Study Group. Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology. 2006;66:366–372. doi: 10.1212/01.wnl.0000198586.85250.13. [DOI] [PubMed] [Google Scholar]
- Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci USA. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman RC, Brooks SP, Jones L, Dunnett SB. Rule learning, visuospatial function and motor performance in the Hdh(Q92) knock-in mouse model of Huntington’s disease. Behav Brain Res. 2009;203:215–222. doi: 10.1016/j.bbr.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Turmaine M, Raza A, Mahal A, Mangiarini L, Bates GP, Davies SW. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2000;97:8093–8097. doi: 10.1073/pnas.110078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin MT, Shelbourne PF, Myers RM, Madison DV. Impaired synaptic plasticity in mice carrying the Huntington’s disease mutation. Human Molecular Genetics. 1999;8:839–846. doi: 10.1093/hmg/8.5.839. [DOI] [PubMed] [Google Scholar]
- Vamos E, Voros K, Vecsei L, Klivenyi P. Neuroprotective effects of L-carnitine in a transgenic animal model of Huntington’s disease. Biomed Pharmacother. 2010;64:282–286. doi: 10.1016/j.biopha.2009.06.020. [DOI] [PubMed] [Google Scholar]
- van Dellen A, Blakemore C, Deacon R, York D, Hannan AJ. Delaying the onset of Huntington’s in mice. Nature. 2000;404:721–722. doi: 10.1038/35008142. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Murphy Z, Slow EJ, Leavitt BR, Hayden MR. Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Human Molecular Genetics. 2005a;14:3823–3835. doi: 10.1093/hmg/ddi407. [DOI] [PubMed] [Google Scholar]
- van Raamsdonk JM, Pearson J, Rogers DA, Lu G, Barakauskas VE, Barr AM, Honer WG, Hayden MR, Leavitt BR. Ethyl-EPA treatment improves motor dysfunction, but not neurodegeneration in the YAC128 mouse model of Huntington disease. Experimental Neurology. 2005b;196:266–272. doi: 10.1016/j.expneurol.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Pearson J, Slow EJ, Hossain SM, Leavitt BR, Hayden MR. Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington’s disease. J Neurosci. 2005c;25:4169–4180. doi: 10.1523/JNEUROSCI.0590-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsavayai SC, Dallérac GM, Milnerwood AJ, Cummings DM, Rezaie P, Murphy KPSJ, Hirst MC. Progressive CAG expansion in the brain of a novel R6/1-89Q mouse model of Huntington’s disease with delayed phenotypic onset. Brain Res Bull. 2007;72:98–102. doi: 10.1016/j.brainresbull.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Walker AG, Miller BR, Fritsch JN, Barton SJ, Rebec GV. Altered information processing in the prefrontal cortex of Huntington’s disease mouse models. J Neurosci. 2008;28:8973–8982. doi: 10.1523/JNEUROSCI.2804-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen X, Li Y, Tang T-S, Bezprozvanny I. Tetrabenazine is neuroprotective in Huntington’s disease mice. Mol Neurodegener. 2010;5:18. doi: 10.1186/1750-1326-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks RA, Piccini P, Harding AE, Brooks DJ. Striatal D1 and D2 dopamine receptor loss in asymptomatic mutation carriers of Huntington’s disease. Ann Neurol. 1996;40:49–54. doi: 10.1002/ana.410400110. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Human Molecular Genetics. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Gutekunst C-A, Vrbanac V, Lebel L-A, Schilling G, Hersch S, Friedlander RM, Gusella JF, Vonsattel J-P, Borchelt DR, MacDonald ME. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Human Molecular Genetics. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Lebel L-A, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Human Molecular Genetics. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Persichetti F, McNeil SM, Mysore JS, Mysore SS, MacDonald ME, Myers RH, Gusella JF, Wexler NS, Group U-VCR. Factors associated with HD CAG repeat instability in Huntington disease. J Med Genet. 2007;44:695–701. doi: 10.1136/jmg.2007.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, White JK, Gutekunst CA, Vrbanac V, Weaver M, Li XJ, Li SH, Yi H, Vonsattel JP, Gusella JF, et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Human Molecular Genetics. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- Woodman B, Butler R, Landles C, Lupton MK, Tse J, Hockly E, Moffitt H, Sathasivam K, Bates GP. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res Bull. 2007;72:83–97. doi: 10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Yamada M, Sato T, Tsuji S, Takahashi H. CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:71–86. doi: 10.1007/s00401-007-0287-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yu Z-X, Li S-H, Evans J, Pillarisetti A, Li H, Li X-J. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. Journal of Neuroscience. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádori D, Geisz A, Vámos E, Vécsei L, Klivényi P. Valproate ameliorates the survival and the motor performance in a transgenic mouse model of Huntington’s disease. Pharmacol Biochem Behav. 2009;94:148–153. doi: 10.1016/j.pbb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Peng Q, Li Q, Jahanshad N, Hou Z, Jiang M, Masuda N, Langbehn D, Miller M, Mori S, et al. Longitudinal Characterization of Brain Atrophy of a Huntington Disease Mouse Model by Automated Morphological Analyses of Magnetic Resonance Images. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker B, Luthi-Carter R, Kama JA, Dunah AW, Stern EA, Fox JH, Standaert DG, Young AB, Augood SJ. Transcriptional dysregulation in striatal projection- and interneurons in a mouse model of Huntington’s disease: neuronal selectivity and potential neuroprotective role of HAP1. Human Molecular Genetics. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]