Abstract
Physics-based force fields are the backbone of molecular dynamics simulations. In recent years, significant progress has been made in the assessment and improvement of commonly-used force fields for describing conformational dynamics of folded proteins. However, the accuracy for the unfolded states remains unclear. The latter is however important for detailed studies of protein folding pathways, conformational transitions involving unfolded states and dynamics of intrinsically disordered proteins. In this work we compare the three commonly-used force fields, AMBER ff99SB-ILDN, CHARMM22/CMAP and CHARMM36, for modeling the natively unfolded fragment peptides, NTL9(1-22) and NTL9(6-17), using explicit-solvent replica-exchange molecular dynamics simulations. All three simulations show that NTL9(6-17) is completely unstructured, while NTL9(1-22) transiently samples various β-hairpin states, reminiscent of the first β-hairpin in the structure of the intact NT9 protein. The radius of gyration of the two peptides is force field independent but likely underestimated due to the current deficiency of additive force fields. Compared to the CHARMM force fields, ff99SB-ILDN gives slightly higher β-sheet propensity and more native-like residual structures for NTL9(1-22), which may be attributed to its known β preference. Surprisingly, only two sequence-local pairs of charged residues make appreciable ionic contacts in the simulations of NTL9(1-22), which are sampled slightly more by the CHARMM force fields. Taken together, these data suggest that the current CHARMM and AMBER force fields are globally in agreement in modeling the unfolded states corresponding to β-sheet in the folded structure, while differing in details such as the native-likeness of the residual structures and interactions.
Keywords: Folding, intrinsically disordered proteins, secondary structure
INTRODUCTION
Physics-based force fields are the backbone of molecular dynamics simulations and hold the key to the de novo predictions of protein structures and folding pathways. Towards this goal, significant effort has been recently made to assess the accuracy of commonly-used force fields leading to significant improvements in recent years.1 For example, the CHARMM36 (C36) force field was recently developed based on additional optimization of the CHARMM22 (C22)/CMAP force field, among other improvements, to more accurately balance the energies of helical and extended states.2 This work was motivated by several benchmark studies that showed a bias of in the C22/CMAP force field toward helical states.3-6 Similarly, AMBER ff99SB was introduced to correct the helical bias that was present in the earlier version ff99.7 Subsequently, a revision (ff99SB-ILDN) was introduced to improve the torsion space sampling of the sidechains of Ile, Leu, Asp and Asn.8
As current protein force fields are parametrized mainly based on the structures and properties of folded states, their accuracy for representing the unfolded states remains unclear. The latter is however important for detailed studies of protein folding pathways, conformational transitions involving unfolded states and dynamics of intrinsically disordered proteins (IDPs). In recent years, several publications have emerged to address this topic. Piana et al. examined several versions of AMBER and a modified C22 force fields in the reversible folding of the miniprotein villin headpiece and found that the folding mechanism and properties of the unfolded state are dependent on the particular force field.9 In studying the folding free-energy landscape of the GB1 hairpin with several AMBER and OPLS force fields, Best and Mittal found that while all force fields gave similar folded fractions at room temperature, the residual secondary structure in the unfolded state differs substantially.6 Furthermore, the overall dimension of the unfolded states is too compact compared to the FRET data.6 The presence of an overly compact unfolded state was also observed in the study of the unfolded state of a cold shock protein CspTm10 using a variant of AMBER force field and the study of the acid-unfolded state of the helix-based protein ACBP using a modified C22 force field.11
The aforementioned work examined the accuracy of commonly-used force fields for modeling the unfolded states of natively folded proteins. In this paper, we will focus on the comparison of the AMBER and CHARMM force fields in describing the conformational dynamics of two natively unfolded peptides, NTL9(1-22) and NTL9(6-17). NTL9(1-22) corresponds to a β-hairpin in the folded protein NTL9 and is a part of the three-stranded β-sheet (1, colored in yellow), while NTL9(6-17) corresponds to the loop region of this β-hairpin, which contains two type II β-turns, between Val9 and Lys12 and between Lys14 and Glu17. The mean charges/mean hydrophobicities of NTL9(1-22) and NTL9(6-17) are 0.23/0.43 and 0.25/0.30, respectively (see Methods and Protocols for details). Thus, both peptides fall into the IDP category according to the classification of Uversky et al.12 However, unlike the typical IDPs with sequences of low complexity and fold only upon binding to targets, NTL9(1-22) is folded in the presence of the remaining sequence of NTL9. The second reason for using NTL9(1-22) to compare force fields is that the unfolded state of the intact NTL9 has been extensively characterized and was suggested to have a non-native, specific electrostatic interaction (between Asp8 and Lys12) based on the combined mutagenesis and pH-dependent stability measurements.13-15 To our best knowledge, the latter represents the only case reported in the literature. In an effort to further characterize the unfolded state of NTL9, Raleigh and coworkers studied several fragment peptides including NTL9(1-22). They found that NTL9(1-22) is a random coil but the pKa of Asp8 is shifted lower by 0.34 units relative to the model value. Although not a proof, the latter is consistent with the proposed specific interaction between Asp8 and Lys12 in the unfolded state of intact NTL9.16
The paper is organized as follows. Following the Methods and Protocols, we will discuss the radius of gyration, secondary structure and backbone dynamics of the two peptides based on the explicit-solvent replica-exchange molecular dynamics simulations using the three force fields, ff99SB-ILDN, C22/CMAP and C36. We then turn to the specific interactions such as backbone hydrogen bonding and sidechain hydrophobic contacts as well as ionic contacts. Finally, we compare the calculated NMR shift deviations from random coil with experiment. This study shows that while all three force fields predict a largely unstructured peptide NTL9(1-22) with transient formation of β hairpin, the degree of native-likeness varies, with ff99SB-ILDN giving the most native-like states although this extent of nativeness is in disagreement with experiment.
METHODS AND PROTOCOLS
Structure preparation and equilibration runs
All simulations were performed with the GROMACS package (version 4.517). Proteins were represented by the ff99SB-ILDN,8 C22/CMAP,18,19 or C362 force fields. The CHARMM modified TIP3P model20 was used to represent water. All minimizations and simulations were performed using periodic boundary conditions with the Particle Mesh Ewald method used for treating long-range electrostatics. The bonds containing hydrogen atoms were constrained using the LINCS algorithm21 and a 2-fs time step was used for all MD simulations. A spherical cutoff of 9Å was used for calculations of van der Waals forces. To verify if there is a significant difference with longer cutoffs, the production simulation of NTL9(1-22) was also conducted using a 12Å cutoff and a switching function starting from 10 Å. The results were very similar, confirming that van der Waals interactions beyond 9 Å are not significantly impacting the conformational sampling.
To initiate the simulation of the folded, intact NTL9 protein, the crystal structure (PDB ID: 2HBB, residues 1-51) was used. The missing hydrogen atoms were added and all acidic and lysine residues were in the charged form. The structure was then solvated in a truncated octahedron box filled with TIP3P water.20 The distance between the peptide and the edge of the box was at least 10 Å. Five chloride ions were added to neutralize the system. Energy minimization was performed with the steepest-descent (SD) algorithm until the maximum force was less than 1000 kJ/mol/nm, followed by the 100-ps NVT and 500-ps NPT equilibration runs at 300 K and 1 atm pressure with harmonic restraints of 1000 kJ/mol/nm2 on the heavy atoms of the protein.
Using CHARMM,22 the two fragment peptides, NTL9(1-22) and NTL9(6-17), were built as extended chains with the N-terminus acetylated (ACE) and the C-terminus N-methylamidated (CT3). All acidic and basic (only lysines are present) sidechains were in the charged form. The built structures were solvated in a truncated octahedron water box as described before. Five or three chloride counter-ions were added to neutralize the system, NTL9(1-22) or NTL9(6-17), respectively. Energy minimization was performed with the SD algorithm until the maximum force was less than 1000 kJ/mol/nm. Using GROMACS a 20-ps MD equilibration was performed under constant NVT conditions at 300 K with harmonic restraints of 1000 kJ/mol/nm2 on the heavy atoms of the peptide, followed by a 2-ns unrestraint equilibration step conducted under constant NPT conditions at 400 K and 1 atm pressure. The temperature was controlled by the velocity rescaling algorithm23 with a coupling constant of 0.1 ps, while the pressure was controlled by the Parrinello-Rahman method24 with a coupling constant of 2 ps. To reduce computational cost, a snapshot from the 400 K trajectory was selected, resolvated and equilibrated using 20 ps NVT and 100 ps NPT molecular dynamics at 300 K and 1 atm pressure. The above process was repeated for each force field, resulting in three independent starting structures for the production runs.
Production runs
In the simulation of the folded, intact NTL9, the production run was conducted under constant NPT at 300 K and 1 atm pressure for 20 ns using the three force fields. For the fragment peptides, the temperature replica-exchange protocol was applied with exponentially spaced replicas occupying a temperature range between 280 and 450 K. A total of 55 and 45 replicas were used for NTL9(1-22) and NTL9(6-17), respectively. The replicas adjacent in temperature were allowed to exchange configurations every 2 ps based on the Metropolis criterion. The resulting exchange acceptance ratio was about 10-20% for adjacent replica pairs. The simulation lasted 100 ns for each replica, resulting in an aggregated simulation time of 5.5 and 4.5 μs for NTL9(1-22) and NTL9(6-17), respectively. Unless otherwise indicated, the replica with the temperature closest to 298 K was used for analysis. To verify convergence, the C36 simulation of NTL9(1-22) with the 12Å cutoff for van der Waals interactions was repeated once using a different starting structure. The results were similar, thus confirming that the simulation length of 100 ns per replica is sufficient for converging the properties of interest (see more discussion in Results and Discussion).
Analysis
Unless otherwise stated, the analysis was performed using GROMACS (version 4.517) or VMD (version 1.9.125) based on the last 80 or 18 ns of the production trajectories for the fragment peptides or intact NTL9, respectively. The DSSP program26 was used to identify secondary structures. For the analysis of backbone conformational states, the following definition based on the Ramachandran plot was used: αR: −100° < ϕ < −30° and −67° < ψ < −7°; β: −180° < ϕ < −90° and 50° < ψ < 180° or −180° < ϕ < −90° and −180° < ψ < −120° or 160° < ϕ < 180° and 110° < ψ < 180°; PPII: −90° < ϕ < −20° and 50° < ψ < 180° or −90° < ϕ < −20° and −180° < ψ < −120°; and αL: 30° < ϕ < 90° and −15° < ψ < 80°.
For the analysis of interactions, two sidechains (Cα used for Gly) are defined as in contact if the distance between any pair of heavy atoms is within 4.5 Å. For defining back-bone hydrogen bonds, the cutoff for the donor-acceptor distance is 3.5 Å between heavy atoms and the cutoff for the donor-hydrogen-acceptor angle is 135°. An ionic contact is defined as present if the distance between the amino nitrogen of Lys and the nearest carboxylate oxygen of Asp/Glu is within 5 Å. SPARTA+27 was used to calculate the chemical shifts of Cα protons based on the trajectory snapshots. The values were then averaged over the entire trajectory. The chemical shifts for random coils were taken from Wishart and Sykes.28
To estimate the radius of gyration of a peptide, we used the power-law relation for a self-avoiding chain as follows,29
| (1) |
where = 0.4 is the persistence length, b = 0.38 nm is the distance between two Cα atoms, N is the number of bonds in the chain, and ν is the scaling exponent, which was calculated according to the mean charge Q or mean hydrophobicity H of a sequence.29
| (2) |
| (3) |
where the parameters are a = 0.394, z = 0.09, x0 = 0.114, c = 1.72, and d = 0.9.29 Q is the average net charge per residue in the chain. It is 0.227 for NTL9(1-22) and 0.25 for NTL9(6-17). H is the average hydrophobicity per residue. The hydrophobicity for individual amino acid was taken from Kyte and Doolittle.30 The scale was normalized between 0 and 1. The hydrophobicity of each residue was calculated using a window size of 5 amino acids. Accordingly, H is 0.434 for NTL9(1-22) and 0.299 for NTL9(6-17). Following Hofmann et al.,29 we used polyampholyte theory29,31 to decide which equation to use for the calculation of the scaling exponent. For NTL9(1-22) Equation 2 was used, while for NTL9(6-17) Equation 3 was used.
RESULTS AND DISCUSSION
Overall dimension of the peptides
Explicit-solvent REMD simulations of 100 ns per replica in the temperature range of 280 through 450 K were performed for the two peptides, NTL9(1-22) and NTL9(6-17), using the three force fields, ff99SB-ILDN, C22/CMAP and C36. For brevity, we will refer to C22/CMAP as C22 in the remainder of the paper. For NTL9(1-22) the radius of gyration (Rg) and Cα root-mean-square deviation (RMSD) from the corresponding part in the crystal structure decreased significantly in the first 20 ns and plateaued after 30 ns in all three simulations (Supporting Information). In the remaining 70 ns, the Cα RMSD and the radius of gyration fluctuate around 7 Å and 10 Å, respectively. By contrast, the Cα RMSD and Rg of NTL9(6-17) decreased only slightly in the first 10 ns before plateauing. As a control, we also performed single-temperature MD simulations starting from the crystal structure of the intact NTL9 using all three force fields. The structure remained stable with Cα RMSD fluctuations between 1 and 2 Å within 20 ns for all three force fields (data not shown).
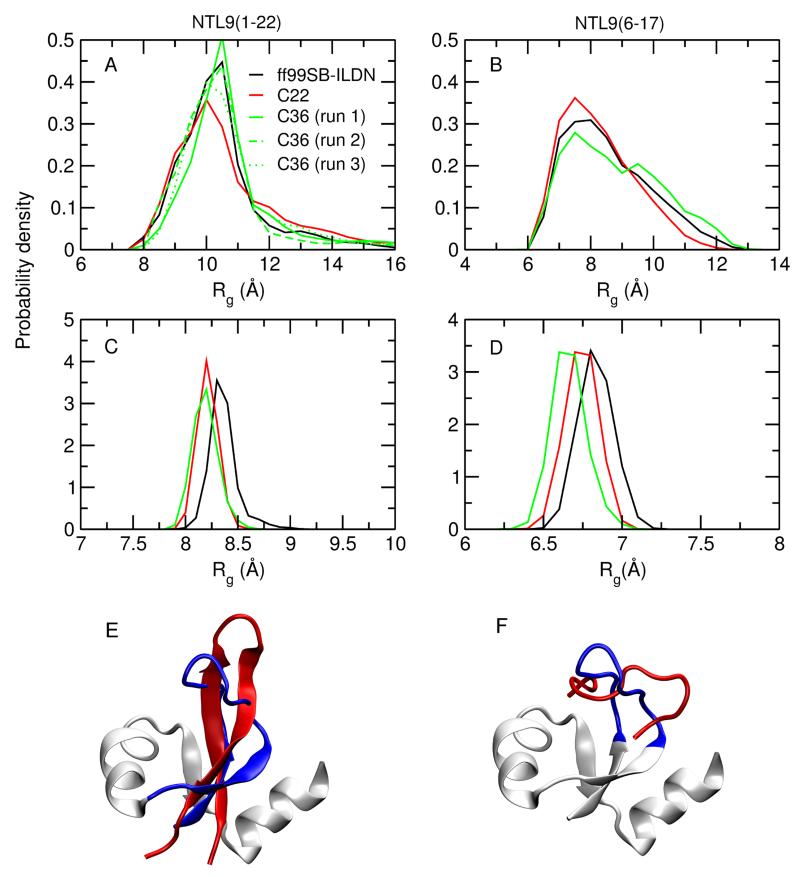
2 shows the probability distribution of Rg for the two peptides as well as their counterparts in the folded structure. Overall, the differences among the three force fields are small. The most probable Rg values for NTL9(1-22) and NTL9(6-17) are about 10.5 and 7.5 Å, respectively. As expected, Rg values for the corresponding segments in the intact protein are much smaller (8.2 and 6.7 Å, respectively) and show a narrower distribution, which reflects the folding and restricted mobility as a result. We consider the above results in light of recent experiments and theoretical studies. Using the scaling law for self-avoiding chains, the radius of gyration of an unfolded protein can be estimated by Rg = ρ0 Nν, where ρ0 is an empirical constant, N is the number of bonds in the chain, and ν is an empirical scaling exponent which has been extensively studied.32 Recently, Hofmann et al. derived empirical relations for calculating the scaling exponents based on single molecular FRET data of several globular proteins and IDPs.29 Following their work (details see Methods and Protocols), we calculated the scaling exponents and radius of gyration for the two fragment peptides (2). The calculated scaling exponents are 0.64 and 0.71 for for NTL9(1-22) and NTL9(6-17), respectively, which fall into the known range for proteins with a mean net charge of 0.2–0.3.29 The estimated Rg is 14.1 Å for NTL9(1-22) and 10.5 Å for NTL9(6-17), which are 3.6 and 3 Å larger than the most probable respective Rg’s obtained from our simulations. Thus, the Rg’s of the two peptides are likely underestimated in our simulations, which is consistent with the current view that additive force fields tend to give overly compact unfolded states.6,10,11,33 Using several versions of AMBER force fields as a testbed, Best and coworkers showed that modestly making the protein-water interactions more favorable is sufficient to recover the correct dimensions of IDPs or unfolded proteins.33 Along this line, Huang and MacKerell demonstrated that more extended states were sampled in simulations of (AAQAA)3 using the Drude polarizable force field, with the polarizabile model also showing a dramatic improvement in the helix-coil cooperativity.34
Secondary structure analysis
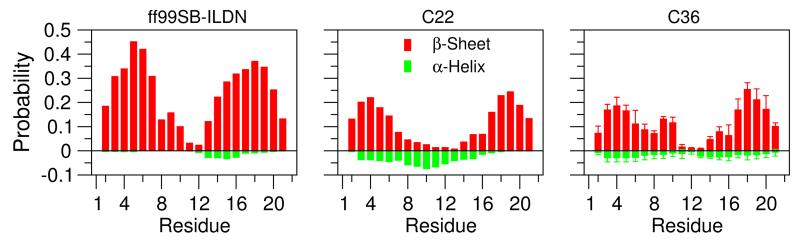
To determine if the two peptides are folded or partially folded, we performed secondary structure analysis using DSSP.26 With all three force fields, no secondary structure was found for NTL9(6-17). However, for NTL9(1-22), significant β-sheet propensity is observed for the N- and C-terminal residues, 2-7 and 15-21 (3), indicating the formation of a β-hairpin. Simulation with ff99SB-ILDN results in higher β propensity for nearly all residues compared to the C22 and C36 simulations, The residue-based β propensity is as high as 45% in the former simulation, while it is below 20% in the latter two. This difference is in agreement with the known β preference of ff99SB.3 The secondary-structure propensity in the C22 and C36 simulations is similar. In the C22 simulation, the β propensity in the N- and C-terminal segments smoothly decreases towards the middle of the peptide, while in the C36 simulations the decrease is not as smooth. The repeated simulations of C36 gave similar results (small error bars in 3), suggesting that the simulations are converged with respect to this property. In contrast to β-sheet, the propensity for α-helix is negligible for all simulations, with the C22 simulation giving slightly higher propensities for α-helix as compared to the other two force fields, in agreement with the known helical bias of C22.3
Backbone torsion sampling
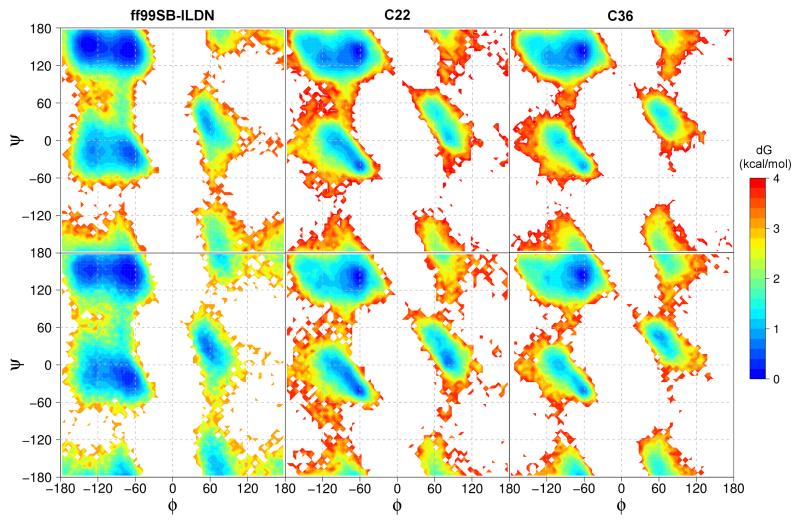
To understand the formation of secondary structure in NTL9(1-22) and the lack thereof in the short fragment NTL9(6-17), we examine the backbone torsion sampling using Ramachandran plots of (ϕ, ψ) dihedral angles. With all three force fields, the most preferred regions of sampling for both peptides β are and PPII, followed by αR and αL (4), confirming that both peptides mainly sample the extended states. Let us consider NTL9(1-22) first (4, top panel). A major difference between ff99SB-ILDN and C22/C36 is the relative weight and shape of the β and PPII regions. The β region is more extensive and has a higher probability than PPII in the simulation with ff99SB-ILDN, while it is smaller and has a lower probability than PPII in the simulations with C22 and C36. The preference for β is in agreement with the DSSP analysis (3), and consistent with the known β-preference of ff99SB. Another difference lies in the shape of the αR region, which is narrow in the simulations with C22 and C36, a feature that has been shown to yield improved helix-coil cooperativity in C36.35 Furthermore, the barrier between β/PP-II and αR is higher with C22 and C36, a property that may impact helix-coil transition rates. We note that some helix cooperativity has been demonstrated by the OPLS-AA/L force field for polyalanine36 but not for (AAQAA)3.37 The ABSINTH implicit-solvent model showed some cooperativity though the helical transition temperature is overestimated.38
Similar analysis was performed on NTL9(6-17) (4, bottom panel). Only small differences are seen between the (ϕ, ψ) maps of NTL9(1-22) and NTL9(6-17). The β region is sampled slightly less while the αL region is sampled slightly more in the shorter fragment. Similar differences between the ff99SB-ILDN and CHARMM simulations are seen. To further explore the difference between the ff99SB-ILDN and CHARMM force fields, we examine the temperature dependence of the (ϕ, ψ) free energy map (Supporting Information). An obvious difference is in the αR region, which remains rarely sampled as temperature increases in the simulations with CHARMM force field, but becomes significantly populated in the simulation with ff99SB-ILDN. This may be related to the higher barrier between the extended and helical states in the CHARMM force fields, a property which may impact helix-coil cooperativity.
To further quantify the backbone torsion sampling, 1 lists the populations of β, PPII, αR and αL states. Interestingly, the β population is higher than PPII for both peptides using ff99SB-ILDN. By contrast, the reverse trend holds using the CHARMM force fields. This confirms the aforementioned β preference with ff99SB. It is also consistent with the slightly better agreement of the C36 results with NMR data as compared to ff99SB for (Ala)5.2
Backbone hydrogen bonding in NTL9(1-22)
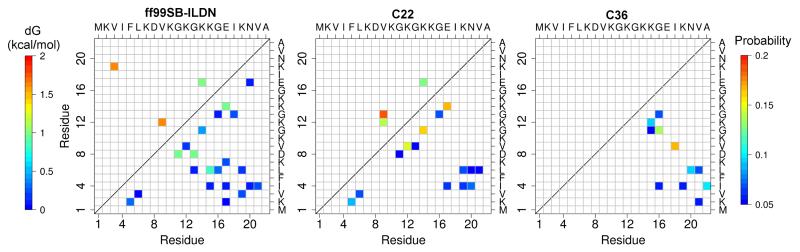
Analysis was undertaken on backbone hydrogen bonds in the simulations of NTL9(1-22). We first consider the native hydrogen bonds, defined as those sampled for at least 10% of the time (occupancy) by all three force fields in the simulation of the intact protein. To highlight the stability of these native contacts in the fragment simulations, we calculated the probabilities relative to those in the simulation of the intact protein with the same force field. The relative probabilities were then converted to relative free energies (ΔG, 5, upper triangle). Considering an occupancy cutoff of 5% for the fragment simulations, ff99SB-ILDN and C22 sample three native hydrogen bonds, while C36 does not sample any. Among the native contacts, two are identical, sequence-local native hydrogen bonds, Val9-Lys12 and Lys14-Glu17, the latter of which is the most stable in the fragment simulations and corresponds to the native β-turn. ff99SB-ILDN and C22 also sample two sequence-distant native hydrogen bonds, Val3-Lys19 and Val9-Gly13, respectively.
Now we turn to all backbone hydrogen bonds formed in the simulations of NTL9(1-22) (5, lower triangle). Overall, the probabilities are very low (below 17%) with all three force fields, consistent with the transient nature of the β-hairpin and the lack of αR conformations. Considering an occupancy cutoff of 5%, the total number of hydrogen bonds are 21, 14 and 11 with ff99SB-ILDN, C22 and C36, respectively. Not surprisingly, most of the non-native hydrogen bonds are different in the three simulations. The sampling of more hydrogen bonds by ff99SB-ILDN, including those both local and distant in sequence, is consistent with the overall higher β-sheet content with this force field (3). Increasing the cutoff to 10%, the number of non-native contacts drops down significantly (light blue or green data points in 5). Among these non-native backbone hydrogen bonds Leu6–Lys15 and Asp8–Gly13 are sampled by ff99SB-ILDN, are only one amino acid out of the registry of the native β-sheet. By contrast, C36 samples two pairs of hydrogen bonds, Val9–Ile18 and Gly11–Gly16, which are five amino acids out of the registry of the native β-sheet. These data are consistent with the enhanced β-sheet propensity of ff99SB-ILDN, leading to sampling of more native hydrogen bonds present in residues 1–22 of the intact NTL9 protein.
Hydrophobic sidechain contacts in NTL9(1-22)
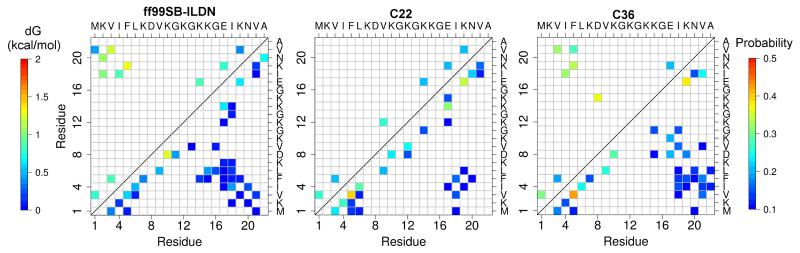
In addition to hydrogen bonds, analysis was also undertaken on hydrophobic contacts as compared to those in the intact NTL9. A sidechain contact is considered native if it is sampled for more than 20% of the time by all three force fields in simulations of the intact NTL9. Excluding the direct neighbors, a total of 21 native contacts are found. Among them, sequence-distant hydrophobic contacts are formed between residues on the opposite strands of the β-hairpin, i.e., between Met1, Val3, Phe5 and Val21, and between Ile4, Leu6 and Ile18. The sampling of native sidechain contacts was next analyzed in simulations of the fragment peptide NTL9(1-22) (6, upper triangle). With an occupancy cut-off of 10%, more native contacts are observed with ff99SB-ILDN (11 contacts) than with C22 and C36 (6 and 8, respectively) (6, lower triangle). Interestingly, native contacts with more than 10 amino acids apart are sampled by ff99SB-ILDN and C36 (with similar relative free energies) but not by C22.
Sampling of non-native sidechain contacts follows the same trend (6, lower triangle). The number of non-native contacts decreases in the order of ff99SB-ILDN (28), C36 (25) and C22 (21). These data suggest that the sidechain contacts are most abundant and native-like with ff99SB-ILDN, while the native-likeness is about the same with C22 and C36. The former observation is consistent with the differences found for the backbone hydrogen bonds and with the β tendency in general of the ff99SB-ILDN force field.
Ionic interactions in NTL9(1-22)
NTL9(1-22) contains 1 Asp, 1 Glu and 7 Lys. Four pairs of them, Asp8–Lys14, Asp8–Lys15, Glu17–Lys14, and Glu17–Lys19, spent a significant fraction of time making ionic contacts in the simulation of the intact NTL9. An ionic contact is defined with a cutoff of 5 Å between the amino nitrogen of Lys and the closest carboxylate oxygen of Asp or Glu. Compared to ff99SB-ILDN, the ionic contacts Asp8–Lys14 and Glu17–Lys19 are slightly more favored in the simulations with C22 and C36 (Supporting Information).
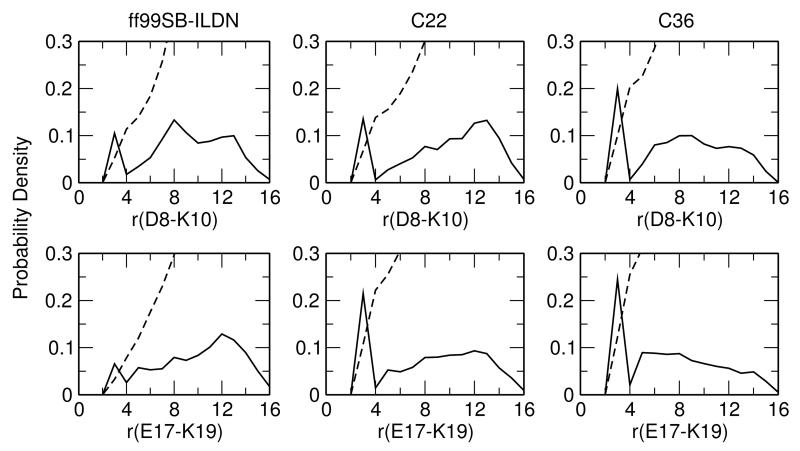
In simulations of the fragment NTL9(1-22), all charged pairs of sidechains make occasional ionic contacts. However, only one native interaction, Glu17–Lys19, shows a significant probability for ionic contact, with values of 7.9%, 22.2% and 25.5% with ff99SB-ILDN, C22 and C36, respectively (see dashed integration lines in 7, bottom). We examine Asp8, as it was indicated by both experiments and simulations to form a non-native contact with Lys12 in the unfolded state of the intact NTL9.15,16,39 In simulations of NTL9(1-22), Asp8 does not form any ionic contact with Lys12. However, it interacts with Lys10, with an ionic-contact probability of 11.3%, 13.9% and 20.3% with ff99SB-ILDN, C22 and C36, respectively (see dashed in 7, top). Both interactions, Glu17-Lys19 (native) and Asp8-Lys10 (non-native) are sequence local and slightly increase in probability in the order of ff99SB-ILDN, C22 and C36. These data suggest that, in contrast to the hydrophobic contacts, the electrostatic interactions in the fragment (1-22) are sequence local and slightly more favorable with the CHARMM force fields than ff99SB-ILDN.
NMR chemical shifts of NTL9(1-22)
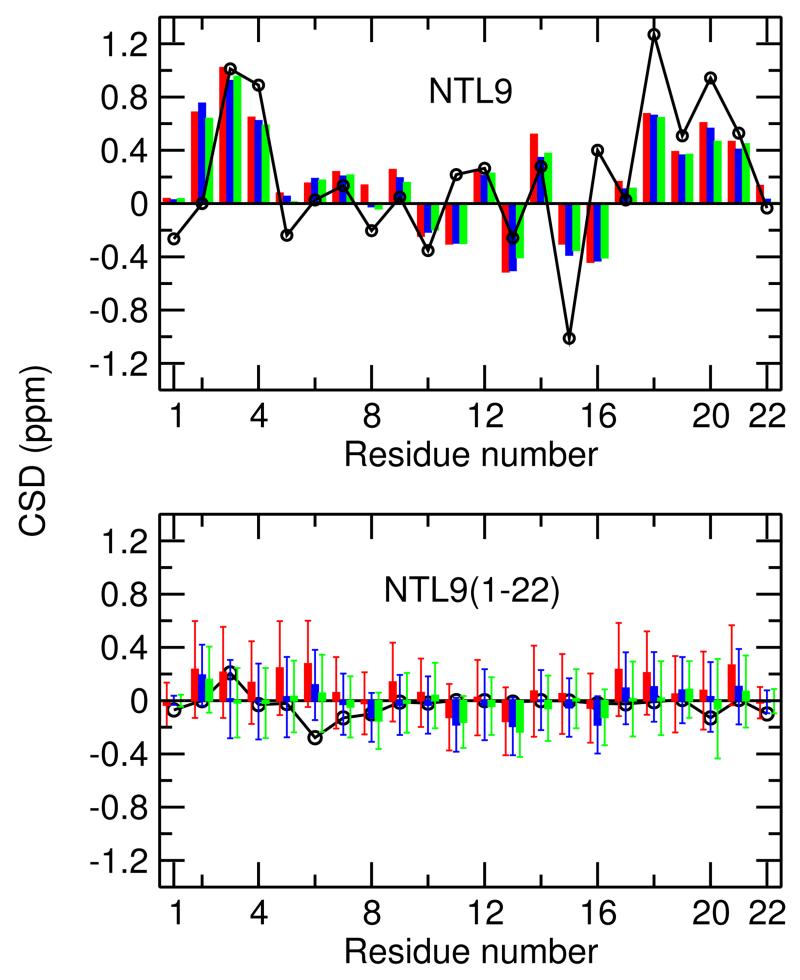
CD and NMR experiments suggest that NTL9(1-22) is mainly unstructured.16 However, our simulations, especially with the ff99SB-ILDN force field, predicted an appreciable amount of residue-based propensity for β-sheet formation. Thus, we were investigated if the simulations would predict β states as the macroscopic (ensemble average) behavior of the peptide, which would be in disagreement with experiment. To test this, we calculated the trajectory averaged chemical shift deviations (CSD) from the random-coil values for NTL9(1-22) as well as the intact NTL9 using the program SPARTA+.27 The latter was used as a control to gauge the calculation errors. As shown in 8 top, calculated CSD’s for sequence 1–22 of the intact NTL9 are generally in good agreement with experiment. However, differences in magnitude exist, especially for residues 15, 16, 18, and 20, for which experimental CSD’s are larger. The former two residues are in a hydrogen-bonded turn while the latter two are on the β-strand. In contrast, the calculated CSD’s for residues 6, 9, 13 are larger by 0.2–0.3 ppm in magnitude as compared to experiment. Residue 2 shows the largest difference. The calculated CSD is as large as 0.8 ppm, while the experimental value is nearly zero. These residues are either in a unstructured region or involved in transient β-bridges. Together, these data suggest that, compared to experiment with Wishart and Skyes’s estimates for random-coil states,28 the calculated CSD’s tend to be somewhat larger in magnitude for unstructured regions and somewhat smaller in magnitude for hydrogen-bonded β-strand regions. The larger calculated values for the unstructured regions is likely due to limited sampling of the configurations for these regions.
Analysis next involves the comparison of calculated and experimental CSD’s for the fragment peptide NTL9(1-22) (8, bottom). The calculated CSD’s are all within about 0.3 ppm of the experimental estimates in all three simulations. The absolute values in the middle region are smaller than the rest, as the corresponding residues are always unstructured and not affected by the transient sampling of β-hairpin states. The absolute CSD’s from the ff99SB-ILDN simulation are slightly larger overall, which is consistent with the higher β-sheet propensity as discussed above. Compared to the calculation, the measured absolute CSD’s are smaller and mostly approaching zero, except for residues 3, 6, 20 and 22, where the measured CSD’s are slightly larger than the calculated values. Considering the overestimation of CSD’s for the unstructured regions in the simulations of the intact NTL9 and the β preference of ff99SB-ILDN, we suggest that the differences for the fragment peptide can be partially attributed to the limited sampling in the replica-exchange simulations and to the overstabilization of the minimally stable β region notably by the ff99SB-ILDN force field. Thus, the CSD data from the simulations is consistent with the experiment16 supporting a model in which NTL9(1-22) is largely unstructured but transiently samples β-hairpin states. The small propensity of the latter suggests that stable folding of the β-hairpin requires stabilization by tertiary contacts.
CONCLUSIONS
Explicit-solvent replica-exchange molecular dynamics simulations were carried out to investigate the conformational sampling of two natively unfolded fragment peptides from the NTL9 protein with three force fields, ff99SB-ILDN, C22 and C36. All three simulations reveal that NTL9(6-17) is almost completely unstructured, while NTL9(1-22) transiently samples β-hairpin states with various non-native registries. The latter is in agreement with the NMR chemical shifts which suggest a largely random-coil model.16 The radius of gyration of the two peptides is similar with the three force fields and likely smaller than experiment, which supports the previous findings that additive force fields give overly compact unfolded states for proteins.33,34 In agreement with the known β preference of ff99SB,3 we found that ff99SB-ILDN most frequently samples the β region of the back-bone torsion space and as a result gives higher β-sheet propensity for NTL9(1-22) than C22 and C36. The differences between C22 and C36 are small, consistent with the lack of sampling of αR states for these peptides, the region which C36 was designed to sample less. Some differences occur in the PPII and αL regions, which are sampled slightly more by C36. Expectedly, both the backbone hydrogen bonds and hydrophobic sidechain contacts are most abundant and native-like with ff99SB-ILDN, consistent with its known preference for β states. Surprisingly, only two ionic contacts, both sequence local, are sampled, with the CHARMM force fields giving slightly stronger interactions than ff99SB-ILDN. Notably, Asp8 and Lys12 do not interact as was proposed for the unfolded state of the intact NTL9.13-15,39 Taken together, these data suggest that the three force fields predict similar global properties for the unfolded states, while differing in details such as the native-likeness of the residual structures and interactions.
Supplementary Material
Figure 1.
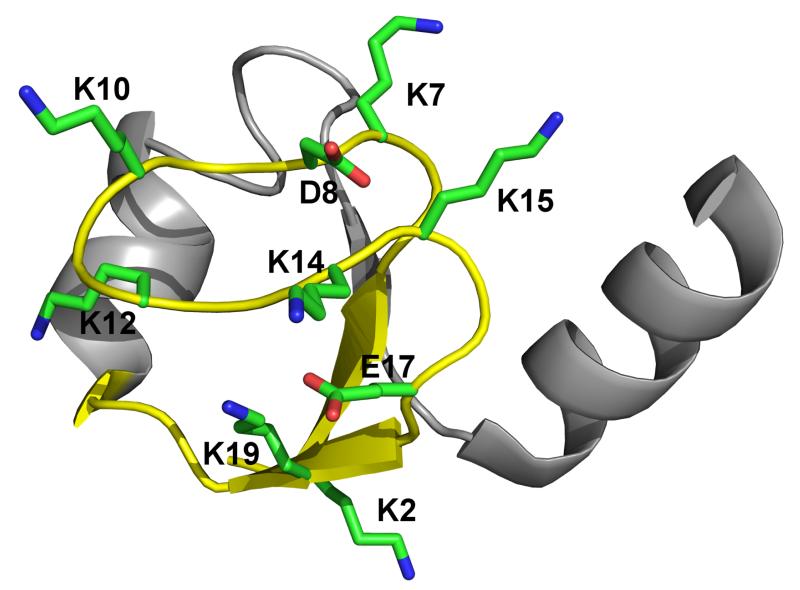
Crystal structure of NTL9 (pdb ID: 2HBB). The fragment NTL9(1-22) (sequence MKVIFLKDVKGKGKKGEIKNVA), encompassing two β-strands connected by a loop NTL9 (6-17) (sequence LKDVKGKGKKGE), is colored yellow, with acidic and basic sidechains shown in stick representation.
Figure 2.
Radius of gyration of the two fragment peptides and the corresponding regions in the intact protein. (A and B) Distributions of radius of gyration for NTL9(1-22) (A) and NTL9(6-17) (B). (C and D) Distributions of radius of gyration for the segments (1-22) (C) and (6-17) (D) in the intact NTL9. Results with ff99SB-ILDN, C22 and C36 are colored in black, red and green, respectively. The three runs with C36 are indicated as solid, dash and dotted lines. (E and F) Snapshots for NTL9(1-22) (E) and NTL9(6-17) (F) in red. The corresponding segments in NTL9 are shown in blue. The remainder of the NTL9 structure is shown in gray.
Figure 3.
Residue-based probability of β-sheet and α-helix for NTL9(1-22). For clarity, the probabilities of α-helix are shown in negative values.
Figure 4.
Distribution of backbone (ϕ, ψ) angles. Top panel: simulations of NTL9(1-22). Bottom panel: simulations of NTL9(6-17).
Figure 5.
Maps of backbone hydrogen bonds from the simulations of NTL9(1-22). Upper triangle. Relative free energies of the native backbone hydrogen bonds in reference to the simulation of the intact NTL9. Lower triangle. Probabilities of native and non-native backbone hydrogen bonds. See Methods for definition of hydrogen bonds. 5% was used as a cutoff to show the contacts, as it is close to the standard deviation of the contact probabilities for all three runs with C36.
Figure 6.
Maps of sidechain contacts in the simulations of NTL9(1-22). Upper triangle. Relative free energies of the native sidechain contacts in reference to the simulation of the intact NTL9. Lower triangle. Probabilities of native and non-native sidechain contacts. Contacts between direct neighbors and those with probabilities below 10% are hidden. See Methods for definition of sidechain contacts. 10% was used as a cutoff to show the contacts, as it is close to the standard deviation of the contact probabilities for all three runs with C36.
Figure 7.
Probability distribution of the distance between Asp/Glu and Lys in the simulations of NTL9(1-22). Distance is defined as that between the amino nitrogen atom of Lys and the closest carboxylate oxygen atom of Asp8/Glu17. Dashed lines represent integration of the probability density. The data for the C36 simulation is the average of the three runs.
Figure 8.
1Hα chemical shift deviations (CSD) from the random-coil values. Top. The intact NTL9. Only the first 22 residues are shown. Bottom. NTL9(1-22). Red, blue and green bars represent calculated CSD’s from the simulations with ff99SB-ILDN, C22 and C36, respectively. Experimental data are in black.16 The error bars represent the standard deviations. See Methods for details of calculation.
Table 1. Backbone conformational states of NTL9(1-22) and NTL9(6-17).
| Conformation | ff99SB-ILDN % |
C22 % |
C36 % |
|---|---|---|---|
| NTL9(1-22) | |||
| α R | 11.3 (0.7) | 12.4 (0.4) | 9.0 (1.1) |
| β | 41.0 (1.8) | 32.3 (0.8) | 30.4 (0.7) |
| PPII | 24.7 (0.7) | 39.0 (0.2) | 43.0 (2.0) |
| α L | 4.6 (0.3) | 4.3 (0.1) | 7.8 (1.9) |
| NTL9(6-17) | |||
| α R | 15.2 (3.2) | 14.7 (4.4) | 10.2 (5.2) |
| β | 31.3 (3.2) | 24.2 (3.8) | 20.8 (4.1) |
| PPII | 25.1 (5.4) | 41.4 (6.2) | 44.3 (7.6) |
| α L | 6.3 (3.0) | 6.0 (3.7) | 11.3 (6.6) |
A percentage population for a conformational state (see Methods for definition) was calculated for each non-Gly residue based on the last 80-ns data. The populations for all residues were then avaraged and standard deviations were taken (shown in parentheses). Note: CD spectrum indicates that NTL9(1-22) is unstructured.16
Table 2. Calculation of the radius of gyration for NTL9(1-22) and NTL9(6-17).
| Peptide | Q | H | ν | Rg |
|---|---|---|---|---|
| NTL9(1-22) | 0.23 | 0.43 | 0.64 | 14.1 |
| NTL9(6-17) | 0.25 | 0.30 | 0.71 | 10.5 |
Q, H, ν and Rg represent the calculated mean charge, mean hydrophobicity, scaling exponent and radius gyration, respectively. See Methods and Protocols for calculation details.
Acknowledgement
Financial support is provided by National Science Foundation (MCB1305560 to JS) and National Institutes of Health (GM098818 to JS and GM051501 and GM072558 to ADM).
References
- (1).Piana S, Klepeis JL, Shaw DE. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr. Opin. Struct. Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- (2).Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Best RB, Buchete N-V, Hummer G. Are current molecular dynamics force fields too helical? Biophys. J. 2008;95:L07–L09. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Freddolino PL, Park S, Roux B, Schulten K. Force field bias in protein folding simulations. Biophys. J. 2009;96:3772–3780. doi: 10.1016/j.bpj.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Patapati KK, Glykos NM. Three force fields’ views of the 310 Helix. Biophys. J. 2011;101:1766–1771. doi: 10.1016/j.bpj.2011.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Best RB, Mittal J. Free-energy landscape of the GB1 hairpin in all-atom explicit solvent simulations with different force fields: similarities and differences. Proteins. 2011;79:1318–1328. doi: 10.1002/prot.22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Nettels D, Müller-Späath S, Küster F, Hofmann H, Haenni D, Rüegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Bestd RB, Schuler B. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. USA. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lindorff-Larsen K, Trbovic N, Maragakis P, Piana S, Shaw DE. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J. Am. Chem. Soc. 2012;134:3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- (12).Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- (13).Kuhlman B, Luisi DL, Young P, Raleigh DP. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 1999;38:4896–4903. doi: 10.1021/bi982931h. [DOI] [PubMed] [Google Scholar]
- (14).Cho J-H, Sato S, Raleigh DP. Thermodynamics and kinetics of non-native interactions in protein folding: a single point mutant significantly stabilizes the N-terminal domain of L9 by modulating non-native interactions in the denatured state. J. Mol. Biol. 2004;338:827–837. doi: 10.1016/j.jmb.2004.02.073. [DOI] [PubMed] [Google Scholar]
- (15).Cho J-H, Raleigh DP. Mutational analysis demonstrates that specific electrostatic interactions can play a key role in the denatured state ensemble of proteins. J. Mol. Biol. 2005;353:174–185. doi: 10.1016/j.jmb.2005.08.019. [DOI] [PubMed] [Google Scholar]
- (16).Horng J, Moroz V, Rigotti DJ, Fairman R, Raleigh DP. Characterization of large peptide fragments derived from the N-Terminal domain of the ribosomal protein L9: definition of the minimum folding motif and characterization of local electrostatic interactions. Biochemistry. 2002;41:13360–13369. doi: 10.1021/bi026410c. [DOI] [PubMed] [Google Scholar]
- (17).Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).MacKerell AD, Jr., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- (19).Mackerell AD, Jr., Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- (20).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- (21).Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- (22).Brooks BR, et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity-rescaling. J. Chem. Phys. 2007;127:14101–14107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- (24).Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- (25).Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Grapics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (26).Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- (27).Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methos Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- (29).Hofmanna H, Sorannoa A, Borgiaa A, Gastb K, Nettelsa D, Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kyte J, Doolittle RF. A simple method for displaying the hydrophathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- (31).Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition. 4 Comparison of theretical results with experimental data. Macromolecules. 1992;25:1996–2003. [Google Scholar]
- (33).Best RB, Zheng W, Mittal J. Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association. J. Chem. Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Huang J, Alexander D, Mackerell J. Induction of peptide bond dipoles drives cooperative helix formation in the (AAQAA)3 peptide. Biophys. J. 2014;107:991–997. doi: 10.1016/j.bpj.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Best RB, Mittal J, Feig M, Mackerell AD. Inclusion of Many-Body Effects in the Additive CHARMM Protein CMAP Potential Results in Enhanced Cooperativity of α-Helix and β-Hairpin Formation. Biophys. J. 2012;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gnanakaran S, García AE. Helix-coil transition of alanine peptides in water: force field dependence on the folded and unfolded structures. Proteins. 2005;59:773–782. doi: 10.1002/prot.20439. [DOI] [PubMed] [Google Scholar]
- (37).Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic validation of protein force fields against experimental data. PLoS ONE. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Vitalis A, Pappu RV. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Shen JK. Uncovering specific electrostatic interactions in the denatured states of proteins. Biophys. J. 2010;99:924–932. doi: 10.1016/j.bpj.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.