Abstract
Continued activation of the photocycle of the dim-light receptor rhodopsin leads to accumulation of all-trans-retinal in rod outer segments (ROS). This accumulation can damage the photoreceptor cell. For retinal homeostasis, deactivation processes are initiated in which the release of retinal is delayed. One of the processes involves binding of arrestin to rhodopsin. Here, we investigate the interaction of pre-activated truncated bovine visual arrestin (Tr) with rhodopsin in 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC) micelles by solution NMR techniques and flash photolysis spectroscopy. Our results show that formation of the rhodopsin-arrestin complex markedly influences partitioning in the decay kinetics of rhodopsin that involves the simultaneous formation of meta II state and meta III state from the meta I state. Binding of Tr leads to an increase of meta III state population and consequently to a ~2-fold slower release of all-trans-retinal from rhodopsin.
Keywords: rhodopsin, arrestin splice variant p44, NMR spectroscopy, UV/VIS spectroscopy, retinal regeneration
The mammalian visual cycle is a complex process involving activation, deactivation and regeneration pathways.[1–6] The prototype GPCR rhodopsin plays a pivotal role in this system. The extensive study of this system has provided information about the signaling pathways of other GPCRs making rhodopsin the most crucial member of the largest class of the GPCR protein superfamily.[7–9] One of the breakthroughs in rhodopsin and GPCR research was the elucidation of the crystal structure of bovine rhodopsin[10] which provided a platform for the future structural and functional studies on GPCRs.[7–9]
Rhodopsin adopts a seven transmembrane α-helical structure and carries the retinal chromophore covalently attached. Photo-induced isomerization of the chromophore from its 11-cis to all-trans configuration results in conformational changes in rhodopsin which lead to the formation of several transiently populated intermediate states (Figure 1).[10–16] Early photointermediates are short-lived and convert to a meta I state on a sub-ms range. This meta I state functions as a juncture at which the photocycle bifurcates to form meta II and meta III states.[17] The meta II state interacts with the guanine nucleotide-binding protein (G protein) transducin and triggers the activation signaling cascade, finally leading to hyperpolarization of photoreceptor cells. The meta III state has been reported to function as an energy storage state.[18] The distribution between states populated along the photodecay can be shifted by different conditions including temperature and pH.[19–21] Both meta II and meta III states finally relax to the apoprotein opsin through release of all-trans-retinal.
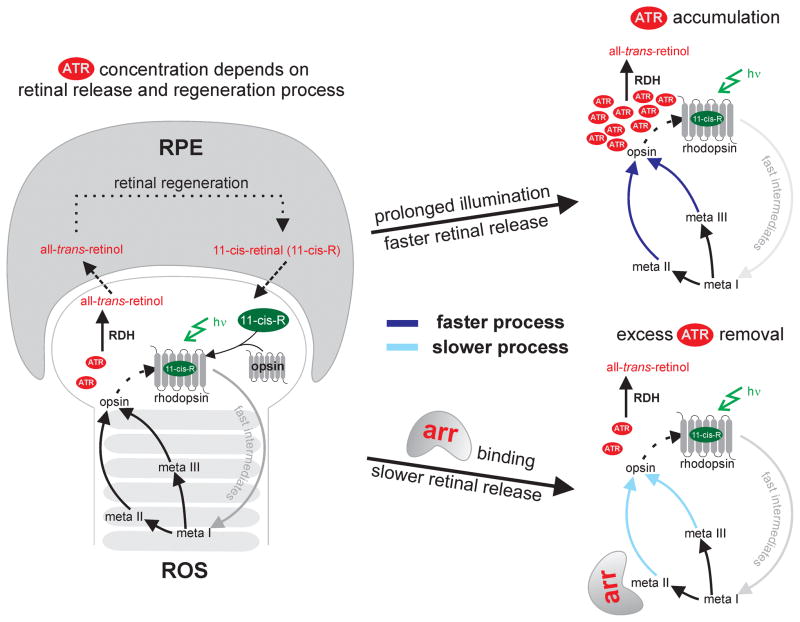
Figure 1.
Mammalian visual cycle. The concentration of all-trans-retinal (ATR) is dependent on two processes: i) retinal release during the photocycle in the rod outer segment (ROS) and ii) retinal removal by a regeneration pathway in the retinal pigment epithelium (RPE) as schematically shown on the left. Continuous exposure to light leads to the accumulation of ATR (right, top), which initiates rhodopsin deactivation by arrestin (arr). Arrestin binding slows down the retinal release, which gives ample time to retinal dehydrogenase (RDH) to remove excess ATR by converting it to all-trans-retinol (right, bottom).
Prolonged exposure to light can lead to an accumulation of all-trans-retinal in ROS, which can be toxic to the eye due to formation of epoxides via oxidation. Harmful effects of excess retinal include visual dysfunction and retinal damage as found in several diseases like age-related macular degeneration (AMD).[22–24] Previously published data[25,26] suggest that there are two rate limiting steps which maintain the level of all-trans-retinal in ROS: i) the release of free all-trans-retinal that depends on the photodecay kinetics and ii) the conversion of retinal to retinol by the enzyme retinal dehydrogenase (RDH) (Figure 1). Since the rate of enzymatic turnover of RDH is slow compared to meta II decay, retinal can accumulate in ROS.[25,27,28] When retinal accumulates, the eye initiates photocycle deactivation processes to delay the release of retinal.[29]
Visual arrestin, a protein that belongs to the arrestin subfamily, acts as the major facilitator that down-regulates the rhodopsin signaling by tightly binding to rhodopsin and concomitant displacement of G protein.[30,31] A recently published crystal structure of the rhodopsin-arrestin complex has provided insights into the static nature of this interaction.[32] The arrestin binding has been shown to slow down the retinal release due to meta II state stabilization.[33] However the effect of arrestin on meta III photodynamics has not been studied in detail.
Besides full length arrestin, p44 (Arr1–370A), a splice variant of arrestin, [34] is also present in rod cells. Although p44 is not ubiquitously present among all species (for example in mice), it has been suggested to play a role in rhodopsin deactivation.[34–37] The binding mechanism of p44 to rhodopsin is different from that of arrestin due to the absence of the C-terminal tail.[34–37] p44 does not only bind more tightly to phosphorylated rhodopsin, but also binds to the non-phosphorylated form.[36,37] Due to the later interaction, it has previously been argued that phosphorylation is not needed for p44-mediated rhodopsin deactivation.[34,37] However, it is not known how p44 manipulates the rhodopsin decay kinetics especially the meta III dynamics.
In our study, we investigated the effect of p44 on the photodecay kinetics of rhodopsin. For this we employed a truncated mutant (Arr1-378) of visual arrestin (Tr),[38] which is devoid of C-terminal tail. Tr closely resembles p44, including phosphorylation independent deactivation of rhodopsin.[38] We utilized Tr instead of p44 since it is less prone to aggregation. To analyze the interaction, we employed a combined strategy utilizing two biophysical techniques, NMR and flash photolysis.
Our NMR analysis of the dynamics of photo-activated rhodopsin was based on the kinetic behavior of five tryptophan residues present in rhodopsin, used as reporter groups (Figure 2 a). All five tryptophan indole resonances were previously assigned in n-dodecyl-β-D-maltoside (DDM) micelles by utilizing single-point tryptophan to phenylalanine mutants.[17] Since previous studies have shown that arrestin is destabilized in presence of DDM,[39] we utilized DHPC for the interaction studies of rhodopsin and Tr.[34,37] Rhodopsin is expressed from HEK293 cells;[40,41] such expression yields the non-phoshorylated form of the protein.
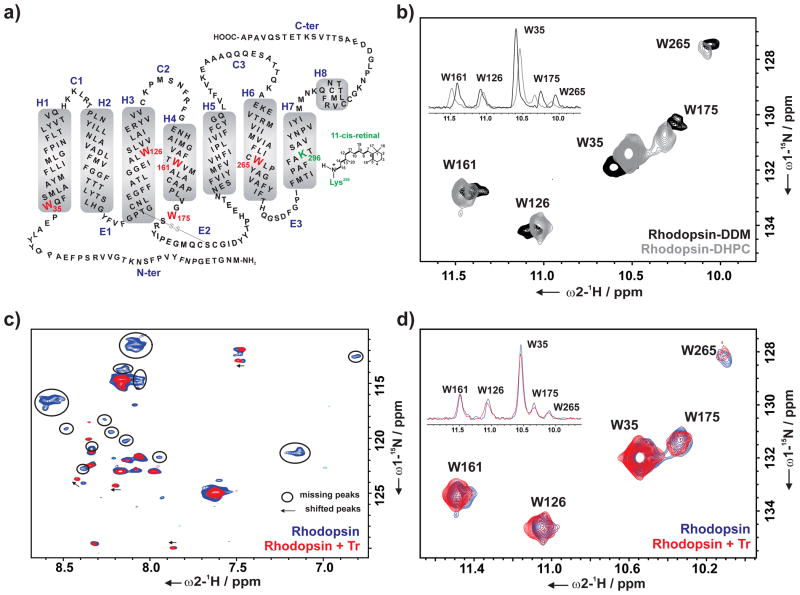
Figure 2.
a) Schematic representation of the secondary structure of bovine rhodopsin with the five tryptophan residues (W35, W126, W161, W175, and W265) used as reporter groups in the NMR experiments. b) Overlay of the indole region of 2D 1H, 15N SOFAST-HMQC spectra of α,ε-15N-tryptophan-labeled rhodopsin in DDM (black) and DHPC (gray) micelle. Overlay of 2D 1H, 15N SOFAST-HMQC spectra of α,ε-15N-tryptophan-labeled rhodopsin without (blue) and with Tr (red) (rhodopsin : Tr – 1 : 2) showing c) backbone region, d) indole region of the spectra. Overlay of 1D projection of the corresponding cross peaks of the indole region of 2D spectra is shown in inset (b and d). All spectra were recorded at 800 MHz (T = 298 K) in the following buffer: DDM spectrum – 20 mM sodium phosphate buffer pH 7.4; DHPC spectra – 25 mM Tris pH 7.5, 100 mM NaCl, 0.1 μM EDTA, 10% D2O and 1 mM 3-(trimethylsilyl)-2,2′,3,3′-tetradeuteropropionic acid (TSP-d4).
Initially, the difference in the ground state structure of DDM- and DHPC-solubilized rhodopsin was characterized using 2D NMR spectra, followed by the investigation of the effect of different detergents on the photodecay of rhodopsin. The chemical shift perturbations (CSP) for the tryptophan indole resonances in DHPC micelles remain almost identical to those in DDM (Figure 2 b). Hence, the assignments for each of these tryptophan resonances could be easily transferred from the previous assignment in DDM. The similarity of the CSPs also indicates that the overall fold of rhodopsin is not significantly altered upon the change of the micelle condition from DDM to DHPC.
To investigate the influence of Tr on the dark state of rhodopsin, we performed 2D 1H, 15N correlation experiments. The interaction with Tr induces both, CSPs and the disappearance of a subset of NMR signals in the amide region of the spectra arising from residues in the flexible C-terminal part of rhodopsin (Figure 2 c).[42–46] The five indole sidechain tryptophan NMR resonances (Figure 2 d) are, however, not altered, indicating that low-affinity transient binding of Tr to rhodopsin in the dark does not lead to a major structural rearrangement in the transmembrane helical part.[37,39]
Our previous results on the kinetic analysis of rhodopsin in DDM micelles showed complex photodecay kinetics with two different rate constants detected for different tryptophan reporter groups and a third substantialy slower decay constant due to protein aggregation.[17] In DDM, W126, W161, and W265 formed a first group of residues with a common lifetime of 8 min (t1). The second process had a lifetime of 34 min (t2) which could be observed for W35 and W175. We proposed that these two rate constants represent decay kinetics via a direct route involving meta II and via a branched route populating an alternative pathway meta III, respectively.[17]
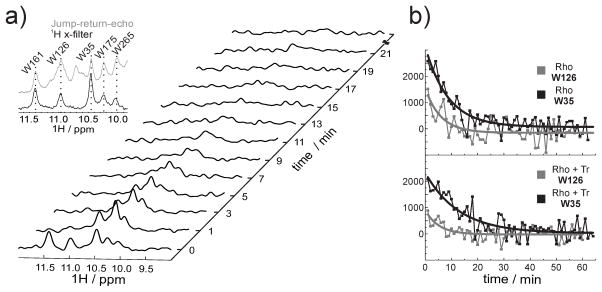
Time-resolved NMR experiments of the rhodopsin kinetics in a DHPC micelle (Figure 3 a) revealed faster decay of the five tryptophan resonances in comparison to the DDM micelle. A lower signal-to-noise ratio and the significantly faster decay in DHPC made the analysis of the NMR-derived kinetic data more difficult. Thus, we focused on the kinetic analysis of the two most intense indole signals, W126 and W35 (Figure 3 b). W126 showed a decay time of 5 min, while W35 showed a decay time of 7 min (Table 1). Similar to our previous kinetic analysis in DDM, the presence of two lifetimes in our current data suggest that the kinetic partitioning of the photodecay, seen in DDM[17] and even in native disk,[21] was preserved in DHPC.
Figure 3.
NMR kinetics. a) A series of 1D 1H NMR spectra of α,ε-15N-tryptophan-labeled rhodopsin recorded at different time intervals after illumination. The indole region of the spectrum is shown. The five resonances visible in the dark state correspond to the five tryptophan residues present in rhodopsin. Subsequent to illumination, 1D 1H spectra were recorded with a temporal resolution of one minute. Inset: overlay of spectrum of the jump-return-echo (gray) and 1H x-filter (black) NMR experiment of rhodopsin samples. The dashed lines connect the corresponding resonances of the five tryptophan residues. b) Extracted signal intensities from the series of 1D 1H NMR spectra of α,ε-15N-tryptophan-labeled rhodopsin in presence and absence of Tr. A mono-exponential fit was applied for the signal intensities of W35 (black curve) and W126 (gray curve).
Table 1.
The rate constants and lifetimes of the tryptophan resonances of rhodopsin (150μM) in absence (−) and presence (+) of Tr
| lifetime/min
|
||
|---|---|---|
| −Tr (0 μM) | +Tr (300 μM) | |
| W35 | 6.92 ± 0.38 | 9.58 ± 1.94 |
| W126 | 5.03 ± 0.40 | 5.23 ± 2.05 |
Upon addition of Tr, all five tryptophan indole resonances were broadened, indicating the formation of a Tr-rhodopsin complex. After illumination, Tr binding does not change the lifetime of the W126 resonance but it increased the lifetime of the W35 resonance by a factor of 1.6 from 7 min to 9.6 min. This indicates that Tr induced a change in decay kinetics of rhodopsin photointermediates.
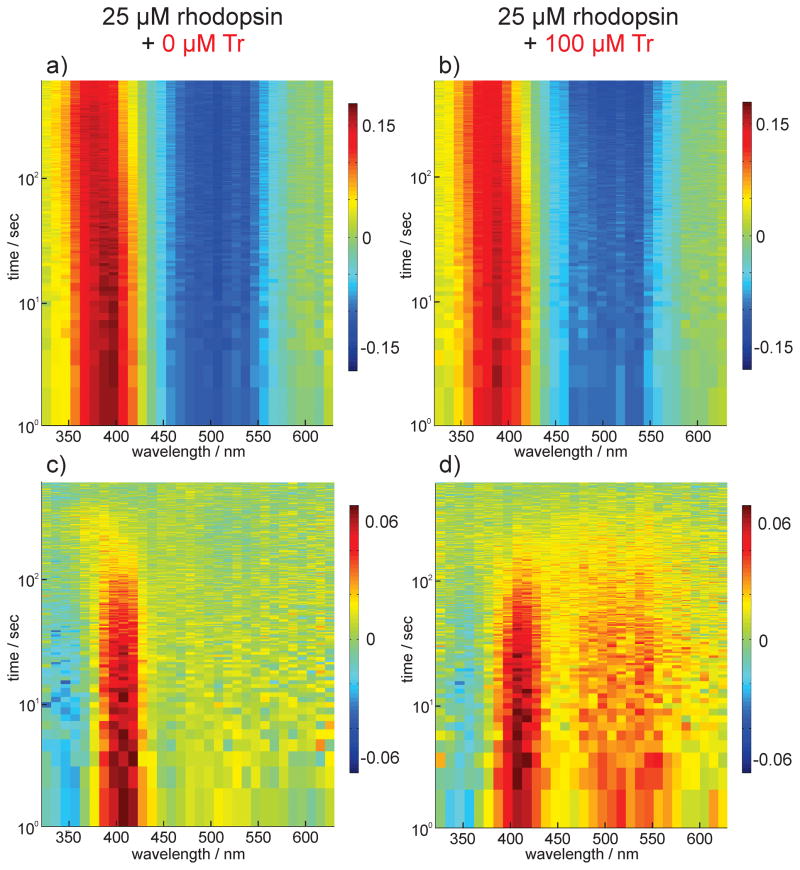
In order to investigate this complex photo dynamics in more detail, we performed broadband flash photolysis. We employed a multichannel detection system to achieve spectrally resolved transient absorption data (see Supplementary Information (SI)). We used the reported absorption maxima of the relevant intermediates of rhodopsin determined for hypotonically extracted rod outer segment disk membranes from retina[47] as reference for the assignment of the spectral signatures in our transient absorption data. The transient spectra of DHPC-solubilized rhodopsin in absence (Figure 4 a) or presence of Tr (Figure 4 b) are mainly determined by ground state bleaching (GSB) at ~ 500 nm and the formation of free all-trans-retinal at ~ 380 nm. Double difference spectra (Figure 4 c, Figure 4 d) are calculated by subtracting the absorption change recorded at the maximum delay time from the transient absorption data. They depicted a positive signature at 400 nm reporting on meta II decay and a spectrally broad positive change in absorption at 460–560 nm representing meta III and potentially meta I. With Tr, the meta III signature was clearly more pronounced.
Figure 4.
Transient absorption spectra of rhodopsin a) without and b) with Tr. Red is positive and blue is negative change in absorption. Double difference spectra of a) and b) calculated by subtracting the absorption change recorded at the maximum delay time from the transient absorption data are depicted in c) and d) respectively.
To visualize and quantify differences in the kinetics of these intermediates with and without Tr, a global lifetime analysis (GLA)[48,49] was performed on the transient absorption data I (λ, t) (see SI). The GLA provided the lifetimes of meta II (1–2 min) and meta III (6–7 min) decay (Figure S3) and also suggested an increased meta III formation in presence of Tr. The GLA lifetimes are comparable to the two lifetimes observed in the NMR kinetic analysis (Table 1).
To further corroborate the GLA results, we additionally performed a lifetime density analysis (LDA).[49] With this method the pre-exponential amplitudes of the sum of a large number (typically ~100) of exponential functions with fixed lifetimes (τ) were determined,
where t is time, I(λ, t) is the normalized decay function and Φ(λ, τ) is the spectral density function. The kinetic information contained in the time-resolved data is then presented in the form of a 2D lifetime distribution map. In contrast to GLA, the LDA is model independent and thereby also extracts non-exponential or stretched exponential kinetics.
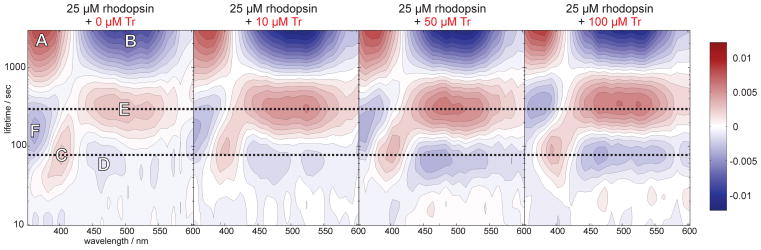
The lifetime density maps (LDM) (Figure 5) consisted of four prominent distributions as indicated by the letters C – F describing the photodecay dynamics (Table 2, Table S1). The spectral signature of A (free retinal absorption) and B (ground state bleach) depicted the latest spectrum of the transient absorption data. These two signatures are long-lived nondecaying components which do not describe any photodecay dynamics. Similar to the GLA results, we observed a distribution for a fast lifetime describing meta II decay (C) and meta III formation (D). The distribution for the slower lifetime corresponds to meta III decay (E). We assigned signature F to the formation of free retinal. The temporal position of F indicated that the retinal release proceeds via both meta II and meta III decay in absence of Tr. In presence of Tr, the free retinal formation (F) was significantly delayed which is shown by the strong shift in the lifetime of signature F from 2.5 min to 5.3 min. Moreover the amplitudes signifying the formation (D) and the decay (E) of meta III were more pronounced. From these observations, we concluded that the Tr-rhodopsin interaction slowed down the photodecay and thereby inhibited retinal release via meta II. As a consequence, the meta II – meta I equilibrium shifted towards meta I which led to the accumulation of meta III. Subsequently, the retinal release is delayed by a factor of 2, since meta III has a longer decay lifetime than meta II. A similar effect was observed in the NMR kinetic data in which the decay lifetime of the W35 resonance has been prolonged upon addition of Tr (Table 1). This effect can be attributed to a change in the protein dynamics influenced by the retinal release process.
Figure 5.
Lifetime density analysis of transient absorption data. LDMs of rhodopsin samples with increasing Tr concentration. The reading of the LDMs is as for DAS with positive (red) amplitudes accounting for decay of absorption and negative (blue) amplitudes accounting for rise of absorption. The lifetimes and the distribution maxima of the signatures C-F are shown in table 1.
Table 2.
The lifetimes, the distribution maxima and the amplitudes of the signatures C-F of the LDMs of rhodopsin (25 μM) in absence (−) and presence (+) of Tr.
| lifetime/min | wavelength/nm | amplitude/a.u. | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| −Tr[5] | +Tr[6] | −Tr | +Tr | −Tr | +Tr | |
| C[1] | 1.3 | 1.5 | 400 | 390 | 0.003 | 0,004 |
| D[2] | 1.2 | 1.3 | 465 | 465 | −0,001 | −0,003 |
| E[3] | 5.0 | 5.3 | 460–540 | 460–540 | 0,003 | 0,007 |
| F[4] | 2.5 | 5.3 | 365 | 375 | −0,004 | −0,004 |
meta II decay
meta III formation
meta III decay
free retinal formation
Tr concentration - 0 μM
Tr concentration - 100 μM
Our findings indicate that meta III decay may have a physiological relevance on the regulation of the free all-trans-retinal concentration in ROS. We suggest that besides the previously discussed rate limiting factor for retinal regeneration within the photocycle of rhodopsin, namely the release of retinal from meta II, also the meta III decay can be considered as an additional important factor for homeostasis of free all-trans-retinal concentration in ROS essential for the functionality of the eye at varying light intensities. The role of meta III envisioned as an energy storage state, as previously proposed by Bartl and Vogel[50] who stated that under ‘bright light and thus high bleaching conditions, a delayed release of all-trans retinal during meta II decay by formation of a meta III storage might be physiologically advantageous’, is fully consistent with our analysis. Here we thus provide evidence for the function of meta III as a physiologically relevant energy storage state.
In conclusion, this study not only provides detailed insight into the rhodopsin deactivation and the retinal regeneration process, but also pushes our time-resolved NMR and flash photolysis techniques to the limits. The data analysis conducted in this study is based on single-shot experiments, due to the non-cyclic photodynamics of bovine rhodopsin. It required high NMR fields and advanced data analysis techniques (GLA and LDA). By two different spectroscopic techniques, the role of arrestin in stabilizing the meta III storage in the bifurcated photodecay of visual rhodopsin could be unraveled, at high temporal and amino acid resolution.
Supplementary Material
Footnotes
The work was supported by the DFG grant SFB807. H.S. and J.W. are members of the DFG-funded cluster of excellence: macromolecular complexes. BMRZ is supported by the state of Hesse. C.R.S. is supported by NIH grant U54 GM094608.
Contributor Information
Prof. Dr. Josef Wachtveitl, Email: wveitl@theochem.uni-frankfurt.de.
Prof. Dr. Harald Schwalbe, Email: schwalbe@nmr.uni-frankfurt.de.
References
- 1.Ridge KD, Palczewski K. J Biol Chem. 2007;282:9297–9301. doi: 10.1074/jbc.R600032200. [DOI] [PubMed] [Google Scholar]
- 2.Parker RO, Crouch RK. Exp Eye Res. 2010;91:788–792. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang PH, Kono M, Koutalos Y, Ablonczy Z, Crouch RK. Prog Retin Eye Res. 2013;32:48–63. doi: 10.1016/j.preteyeres.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylor DA, Burns ME. Eye (Lond) 1998;12:521–525. doi: 10.1038/eye.1998.140. [DOI] [PubMed] [Google Scholar]
- 5.Zhou XE, Melcher K, Xu HE. Acta Pharmacol Sin. 2012;33:291–299. doi: 10.1038/aps.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arshavsky VY, Lamb TD, Pugh EN. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 7.Deupi X. Biochim Biophys Acta - Bioenerg. 2014;1837:674–682. doi: 10.1016/j.bbabio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf S, Grünewald S. PLoS One. 2015;10:e0123533. doi: 10.1371/journal.pone.0123533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 11.Nakamichi H, Okada T. Angew Chem Int Ed Engl. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]; Angew Chemie. 2006;118:4376–4379. [Google Scholar]
- 12.Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 13.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe H-W, Hofmann KP, Ernst OP. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 14.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc Natl Acad Sci. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham TD, Farrens DL. J Biol Chem. 1999;274:1683–1690. doi: 10.1074/jbc.274.3.1683. [DOI] [PubMed] [Google Scholar]
- 16.Nakamichi H, Okada T. Proc Natl Acad Sci U S A. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stehle J, Silvers R, Werner K, Chatterjee D, Gande S, Scholz F, Dutta A, Wachtveitl J, Klein-Seetharaman J, Schwalbe H. Angew Chem Int Ed Engl. 2014;53:2078–2084. doi: 10.1002/anie.201309581. [DOI] [PubMed] [Google Scholar]; Angew Chemie. 2014;126:2110–2116. [Google Scholar]
- 18.Ritter E, Elgeti M, Bartl FJ. Photochem Photobiol. 2008;84:911–920. doi: 10.1111/j.1751-1097.2008.00324.x. [DOI] [PubMed] [Google Scholar]
- 19.Matthews RG, Hubbard R, Brown PK, Wald G. J Gen Physiol. 1963;47:215–240. doi: 10.1085/jgp.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann K, Ritter E, Bartl FJ, Hofmann KP, Heck M. J Biol Chem. 2004;279:48112–48119. doi: 10.1074/jbc.M406856200. [DOI] [PubMed] [Google Scholar]
- 21.Vogel R, Siebert F, Zhang XY, Fan G, Sheves M. Biochemistry. 2004;43:9457–9466. doi: 10.1021/bi049337u. [DOI] [PubMed] [Google Scholar]
- 22.Radu RA, Mata NL, Bagla A, Travis GH. Proc Natl Acad Sci U S A. 2004;101:5928–5933. doi: 10.1073/pnas.0308302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sparrow JR, Fishkin N, Zhou J, Cai B, Jang YP, Krane S, Itagaki Y, Nakanishi K. Vision Res. 2003;43:2983–2990. doi: 10.1016/s0042-6989(03)00475-9. [DOI] [PubMed] [Google Scholar]
- 24.Wenzel A, Grimm C, Samardzija M, Remé CE. Prog Retin Eye Res. 2005;24:275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Tsina E, Cornwall MC, Crouch RK, Vijayaraghavan S, Koutalos Y. Biophys J. 2005;88:2278–2287. doi: 10.1529/biophysj.104.054254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer ME, Smith WC, Farrens DL. J Biol Chem. 2005;280:6861–6871. doi: 10.1074/jbc.M411341200. [DOI] [PubMed] [Google Scholar]
- 27.Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC. Biochemistry. 1999;38:12012–12019. doi: 10.1021/bi990504d. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann KP, Pulvermüller A, Buczyłko J, Van Hooser P, Palczewski K. J Biol Chem. 1992;267:15701–15706. [PubMed] [Google Scholar]
- 29.Sommer ME, Hofmann KP, Heck M. Nat Commun. 2012;3:1–10. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 31.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Büldt G. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 32.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer ME, Farrens DL. Vision Res. 2006;46:4532–4546. doi: 10.1016/j.visres.2006.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith WC, Milam AH, Dugger D, Arendt A, Hargrave PA, Palczewski K. J Biol Chem. 1994;269:15407–15410. [PubMed] [Google Scholar]
- 35.Granzin J, Cousin A, Weirauch M, Schlesinger R, Büldt G, Batra-Safferling R. J Mol Biol. 2012;416:611–618. doi: 10.1016/j.jmb.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe H-W, Sommer ME. Nature. 2013;497:142–146. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- 37.Schröder K, Pulvermüller A, Hofmann KP. J Biol Chem. 2002;277:43987–43996. doi: 10.1074/jbc.M206211200. [DOI] [PubMed] [Google Scholar]
- 38.Gurevich VV. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang T, Chen Q, Cho M-K, Vishnivetskiy SA, Iverson TM, Gurevich VV, Sanders CR. Proc Natl Acad Sci U S A. 2013;110:942–947. doi: 10.1073/pnas.1215176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves PJ, Kim J-M, Khorana HG. Proc Natl Acad Sci U S A. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langen R, Cai K, Altenbach C, Gobind Khorana H, Hubbell WL. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 43.Werner K, Lehner I, Dhiman HK, Richter C, Glaubitz C, Schwalbe H, Klein-Seetharaman J, Khorana HG. J Biomol NMR. 2007;37:303–312. doi: 10.1007/s10858-007-9143-0. [DOI] [PubMed] [Google Scholar]
- 44.Werner K, Richter C, Klein-Seetharaman J, Schwalbe H. J Biomol NMR. 2008;40:49–53. doi: 10.1007/s10858-007-9205-3. [DOI] [PubMed] [Google Scholar]
- 45.Getmanova E, Patel AB, Klein-Seetharaman J, Loewen MC, Reeves PJ, Friedman N, Sheves M, Smith SO, Khorana HG. Biochemistry. 2004;43:1126–1133. doi: 10.1021/bi030120u. [DOI] [PubMed] [Google Scholar]
- 46.Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung J, Schwalbe H, Wright PE, Khorana HG. Proc Natl Acad Sci U S A. 2002;99:3452–3457. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst OP, Bartl FJ. Chembiochem. 2002;3:968–974. doi: 10.1002/1439-7633(20021004)3:10<968::AID-CBIC968>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Van Stokkum IHM, Larsen DS, Van Grondelle R. Biochim Biophys Acta - Bioenerg. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Slavov C, Hartmann H, Wachtveitl J. Anal Chem. 2015;87:2328–2336. doi: 10.1021/ac504348h. [DOI] [PubMed] [Google Scholar]
- 50.Bartl FJ, Vogel R. Phys Chem Chem Phys. 2007;9:1648–1658. doi: 10.1039/b616365c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.