Abstract
Animals ubiquitously interact with environmental and symbiotic microbes, and the effects of these interactions on animal physiology are currently the subject of intense interest. Nevertheless, the influence of microbes on nervous system evolution has been largely ignored. We illustrate here how taking microbes into account might enrich our ideas about the evolution of nervous systems. For example, microbes are involved in animals' communicative, defensive, predatory and dispersal behaviours, and have likely influenced the evolution of chemo- and photosensory systems. In addition, we speculate that the need to regulate interactions with microbes at the epithelial surface may have contributed to the evolutionary internalization of the nervous system.
Keywords: holobiont, microbiome, microbiota, symbiosis
1. Introduction
We live in a microbial world. Since their origin, multicellular organisms have been co-evolving with microbes, a collection of organisms including bacteria, archaea, fungi, protozoa and viruses. Microbes colonize the gut and external surface of animals, as well as some reproductive organs. Some animals even have additional, specialized organs that harbour selected groups of microbes. In general, despite the ubiquity of microbes, associations between animals and microbes are not random [1]. Animals do not merely tolerate microbes; we possess complex suites of adaptations to provide beneficial microbes with food, suitable habitats and protection from other microbes.
The contributions of microbes to animal biology—including human biology—are significant [2,3]. Many readers will be aware that current estimates suggest that our bodies contain 10-fold more microbial cells than human cells, and up to 500-fold more microbial genes than human genes [4]. More concretely, the cumulative mass of the human microbiota is 1–2 kg [4], which is sobering considering that the average human brain has a mass of approximately 1.5 kg [5].
Given the relative proportion of microbes and microbial genes in animal bodies as well as the fidelity of these associations across animal generations, researchers have begun to refer to such ensembles as ‘holobionts' and to suggest that holobionts are valid units of selection in animal evolution (e.g. [6,7]). This idea has come to be known as the hologenome model of evolution [7–9]. This change in focus has the potential to upend the way we think about evolutionary change, often defined as a change in gene frequency over time. Brucker & Bordenstein [10], for example, argue that we should be thinking of animal evolution as a change over time in the frequencies of genes in the nucleus, in organelles and in microbial symbionts.
With the recent proliferation of research on animals and their associated microbes, biologists are increasingly learning that microbes play key roles in physiological processes of animals, including in neural function. In this paper, we briefly review three examples of current relationships between microbes and the neurobiology and behaviour of animals: chemosensory detection of microbial products (§2), the role of microbes in foraging and predation (§3), and microbial influences on cognition and social behaviour (§4). In §5, we propose that microbes may have played a key role in the evolution of nervous systems.
2. Chemosensory detection of microbial products
Microbes and microbial products play important roles in animal behaviour. At arguably the simplest level, animals can taste and smell chemical compounds produced by microbes and use this sensory information to avoid pathogenic microbes, as indicators of the presence of food sources or conspecifics, and even as chemical signals.
The nematode Caenorhabditis elegans eats soil bacteria, but it must be discriminating: it has to avoid pathogenic microbes while selecting those that are palatable and beneficial. One pathogen avoided by C. elegans is Serratia marcescens, which, if ingested, produces compounds that kill the nematode and dissolve its eggshells. Nematodes detect Serratia using the paired AWB chemosensory receptor neurons [11]. Of the 302 neurons in C. elegans, 16 paired neurons function as chemosensory receptors; the AWB cells, along with two other pairs of cells, detect cues that mediate avoidance responses [12]. Although the ASJ cells are normally involved in attraction responses and dauer formation [12], they can also mediate avoidance responses induced by bacteria. Specifically, when at high density in low-oxygen environments, Pseudomonas aeruginosa is pathogenic to C. elegans. Under these conditions, the bacterial metabolites phenazine-1-carboxamide and pyochelin are detected by the ASJ neurons, which then produce neuromodulators that alter activity in adjacent neurons. This activity leads the animal to seek higher oxygen environments, away from potential pathogens [13]. Thus, it appears that C. elegans possesses distinct sensory-neural pathways for generating appropriate behavioural responses to specific cues from potential microbial pathogens.
The fruit fly Drosophila melanogaster is strongly repelled by geosmin, a volatile odorant produced by Penicillium fungi and Streptomyces bacteria. These microbes grow on decaying fruit and are lethal to Drosophila [14]. Thus, fruit flies must discriminate fruit that is at the optimal level of ripeness: when fruit is overripe by most human standards, the yeast that grow on it provide food for Drosophila and are highly attractive [15], but fruit that is so overripe that it smells of geosmin must be avoided. Indeed, the odour of geosmin abolishes the normal attraction to vinegar and other food-related odorants; it also inhibits oviposition [14]. Stensmyr et al. [14] have shown that geosmin is detected by a specific class of antennal olfactory receptor neurons, ab4B, that appears to be narrowly tuned to detect only this molecule. These neurons project exclusively to the DA2 glomeruli in the antennal lobe, the first olfactory processing region in insect central nervous systems. The projection neurons from the DA2 glomeruli are also narrowly tuned to geosmin, suggesting the presence of a labelled line for detecting geosmin. Finally, the authors found geosmin-specific olfactory receptor neurons in seven other species of drosophilids, strongly suggesting that microbial products have shaped the evolution of the Drosophila nervous system.
In addition to warning of the presence of dangerous microbes on substrates, microbial products can provide information about the infection status of conspecifics, including potential mates, in a wide diversity of organisms [16–18]. Mice avoid conspecifics that are ill [19], and recent evidence suggests that mice use receptors that normally function as part of the innate immune system to smell compounds characteristic of bacterial infection in conspecifics. Mammalian leucocytes possess specialized receptors for formylated peptides, which are released by bacteria as metabolic by-products. Formylated peptides attract leucocytes to sites of infection (reviewed in [20]). Interestingly, two formyl peptide receptors are expressed in the vomeronasal organ, an accessory olfactory organ, in mice [21,22]. In vitro, the receptors can be activated by formylated peptides and other infection-related molecules [22]. More recent work shows that the vomeronasal organ is essential for mice to distinguish the odours of infected and uninfected conspecifics [23]. Taken together, these results strongly suggest that formyl peptide receptors in the vomeronasal organ warn mice of the presence of infected conspecifics.
Microbial products are not always aversive to animals, as anyone who likes the taste of miso or a ripe cheese, or the smell of wine or baking bread can attest. Stable flies (Stomoxys calcitrans) select oviposition sites based on the presence of particular bacteria, likely detected from a distance by odorant cues [24]. In addition, settlement cues for marine invertebrate larvae, such as barnacles and polychaetes, are often produced by biofilms of microbes either on stable sea floor substrates or on adult conspecifics [25–28]. Given the critical importance of choosing an appropriate habitat—immobile benthic animals that settle far from conspecifics will not reproduce—cues produced by microbes play a critical role in the life history of these animals.
Although the details of the mechanisms whereby marine invertebrate larvae detect settlement cues are still being worked out, studies with drosophilids have recently revealed remarkably precise and robust systems for detecting and responding to volatile odorants produced by microbes. In general, drosophilids are strongly attracted to volatiles produced by yeasts, their main food source—so much so that the Solomon lily (Arum palaestinum) is pollinated by drosophilids attracted to its scent, which contains volatiles normally produced by yeasts rather than by plants [29]. In addition, D. melanogaster is attracted to hydroxycinnamic acids, antioxidant molecules that can prolong life when included in the diet. Although they cannot directly detect the presence of these compounds, flies are acutely sensitive to the presence of the volatile odorants 4-ethylphenol and 4-ethylguaiacol, which are produced by a variety of yeasts as a result of metabolizing hydroxycinnamic acids. The dedicated olfactory receptors used to detect the compounds differ between adults and larvae, but in both cases the animals are attracted to and feed on substrates containing these odorants; adults also preferentially lay eggs on such substrates [30].
Odorants emitted by symbiotic bacteria also play important roles in what used to be called intraspecific communication among animals, although given the importance of microbes in producing some of these odorants, ‘intraspecific’ is not quite the appropriate term. Nevertheless, we here use the word ‘cues' to refer to sensory stimuli that provide information about the animals that emit them, sometimes to the detriment of these animals. ‘Signals' also provide information about the animals that emit them, but signals have been selected over evolutionary time to efficaciously communicate this information between senders and receivers, both of whom benefit from the exchange. To the extent that their contributions to variation in hosts' scents accurately reflect underlying traits, symbiotic microbes can broadcast information about their hosts to other animals through chemical cues and signals [31,32]. Animals rely heavily on chemical signals to communicate their species, group and individual identities, as well as information about their sex and reproductive state [33,34], and odour-producing symbiotic microbes have been broadly hypothesized to contribute to animal chemical signalling [31,32,35–37].
Among insects, symbiotic microbes can produce volatile compounds that are used as host-specific pheromones (reviewed in [36,37]), although some microbes can also convert pheromones into repellents (e.g. [38]). The best-understood example involves the aggregation pheromone of the desert locust (Schistocerca gregaria). Adult desert locusts can be solitary or gregarious, and during the transition to gregariousness, locusts undergo dramatic genetic, hormonal and behavioural changes that render them capable of forming massive swarms that can decimate crops over large distances [39]. After adopting the gregarious morph, locusts aggregate in response to volatiles that are present in the faeces of adult males, but not females or younger animals; the key component of the aggregation pheromone is guaiacol [40,41]. Morphologically distinct sensillae on the antenna respond to sex pheromones, plant odorants and organic acids, and the aggregation pheromone [42]. Notably, the aggregation pheromone inhibits responses in sensillae to plant odorants [42]. Further, the percentage of antennal lobe neurons that respond to guaiacol increases with age and is higher in gregarious than solitary adults [43]. Guaiacol is not produced by locust cells but by gut bacteria, likely as a product of metabolizing vanillic acid derived from lignans in the locusts' diet [44–46]. Surprisingly, the microbe is not unique, as several different microbes, including Pantoea agglomerans, Klebsiella pneumoniae and Enterobacter cloacae, can produce guaiacol within locusts [45]. This example of apparent coevolution between an animal and its symbiotic microbes raises many interesting questions. For example, if guaiacol is produced by gut bacteria, why is it produced by adult males but not females? Do sex hormones play a role? In addition, it would be interesting to determine whether the presence of guaiacol-producing gut bacteria changes with age, and if so whether the bacteria contribute to age-related changes in olfactory sensitivity to guaiacol. Desert locusts have the potential to serve as a powerful model system for understanding the behavioural and physiological processes that contribute to selection of gut microbes as well as their influence on host nervous systems.
Among mammals, preliminary evidence indicates that symbiotic microbes substantially contribute to their hosts' complex signature scents [31,32,37]. For example, many mammals communicate using specialized scent glands, which are known to harbour diverse communities of odour-producing microbes [31]. In original cultivation-based studies of the Indian mongoose (Herpestes auropunctatus), bacteria from scent glands were shown to produce a suite of short-chain fatty acids (SCFAs) typical of scent secretions. In addition, a broad-spectrum antibiotic eliminates both the bacterial communities and scents from these glands, and mongooses respond to both scent secretions and experimental mixtures of SCFAs in a manner suggesting that they communicate information about signallers' individual identities to receivers [47,48]. More recent cultivation-independent studies of spotted hyaenas (Crocuta crocuta) and striped hyaenas (Hyaena hyaena) showed that the bacterial and SCFA profiles of the two hyaena species differ and that the two profiles covary within each hyaena species. Further, among spotted hyaenas, the two profiles are social group-specific, and reflect sex and reproductive state among members of the same social group [49–51]. Similar patterns are evident among mammals that communicate via urine marking as well. For example, microbes associated with the urine marks of male African elephants, Loxodonta africana, are responsible for the production of urinary ketones and alcohols believed to signal male musth status [52,53]. Similarly, microbes in the urine marks of house mice (Mus musculus) appear to function in communicating individual- and genotype-specific information among competitors and prospective mates [54,55].
3. Role of microbes in neural mechanisms of foraging and antipredator behaviour
At an arguably more complex level, microbes play important roles in foraging and antipredator behaviour in a broad spectrum of animals. For example, one of the best known anti-predator defensive molecules is the neurotoxin tetrodotoxin (TTX). TTX blocks the pore of voltage-gated sodium channels, preventing neurotransmission in almost all multicellular animals. TTX is sufficiently toxic that sympatric reef fishes gain protection from predation by visually mimicking pufferfish [56], and in pufferfish and many other animals, TTX is produced by symbiotic bacteria [57]. Although possessing TTX confers obvious advantages to pufferfish, it also requires physiological adaptations beyond providing a suitable habitat for TTX-producing bacteria: all eight genes coding for voltage-gated sodium channels possess mutations that confer TTX resistance [58], and toxic pufferfish possess a novel gene duplication that created a protein involved in the transport and accumulation of TTX [59]. TTX serves not only in defence, as it is used by both blue-ringed octopus (Hapalochlaena maculosa) and planocerid flatworms to envenomate prey [60,61].
Beyond toxicity, symbiotic microbes can function in antipredator defence by providing camouflage. The Hawai'ian bobtail squid (Euprymna scolopes) possesses a ventral light organ that provides counterillumination, preventing the squid from casting a shadow that could attract the attention of predators lurking below. The light organ is colonized by luminescent Vibrio fischeri, and juvenile squid possess complex adaptations for selecting V. fischeri from among the many species of environmental bacteria that can enter the light organ [62,63]. Interestingly, eye-specific genes are expressed during development of the light organ, and the organ expresses phototransduction molecules and produces electrophysiological responses to light [64,65]. The function of light detection in the organ is not yet understood, but it may allow the animal to exclude mutants that fail to luminesce [66] or may enable the squid to match the ambient illumination level. In either case, the relationship with symbiotic Vibrio has clearly led to evolutionary adaptations in the squid nervous system.
In some cases, microbes hijack normal antipredator behaviour, manipulating the nervous systems of animals to produce outcomes that are favourable to the microbe but not the host. For example, the fungal pathogen Ophiocordyceps unilateralis infects the nervous system of ants (Camponotus leonardi), leading the ants to attach themselves to leaves for several days until a fruiting body erupts from the ant's head, scattering fungal spores [67]. Other Ophiocordyceps species infect other species of Camponotus, and the ability to manipulate the ants' behaviour is unique to its naturally occurring pathogen [68,69]. Another well-known example involves infection of rats by the protozoan Toxoplasma gondii, which causes rats to become attracted to cat urine, greatly increasing their risk of predation by the parasites' definitive host [70]. Describing more examples of parasite manipulation of host behaviour is beyond the scope of this review, and the interested reader is directed to recent special issues of The Journal of Experimental Biology, 216(1), Jan. 2013, and Integrative and Comparative Biology, 54(2), July 2014 for more information.
Like bobtail squid, cardinalfish (Siphamia versicolor) possess a ventral light organ containing luminescent bacteria (Photobacterium mandapamensis). Instead of using their light organ for antipredator defences, however, cardinalfish use their light organs to attract planktonic prey. Emission of light is under neural control, as the organ can be occluded using a retractable shutter [71]. Like the bobtail squid, cardinalfish can also determine the amount of luminescence in the light organ: the organ abuts the lower jaw, which is largely translucent, and the ventromedial portion of the eyes protrudes into the buccal cavity. Thus it appears that the fish can directly see the luminescence of the light organ [71]. Interestingly, olfactory preference tests suggest that cardinalfish are attracted to chemical cues emitted by P. mandapamensis [72], which colonize the light organ from the external environment. Although symbiotic bacteria are necessary for proper development of the light organ in bobtail squid [73], the bacteria that colonize the light organ of cardinalfish are not necessary for its development [74]. Taken together, these studies indicate that the association with luminescent bacteria has powerfully shaped the nervous system and anatomy of cardinalfish.
In addition to facilitating feeding behaviour, symbiotic microbes may also contribute by influencing dietary decisions. In a recent essay, Alcock et al. [75] postulate that gut microbes may manipulate host feeding behaviour by generating cravings for particular foods that they themselves can readily access as energy sources, or by inducing dysphoria in their hosts until those particular foods are consumed. Gut microbes could accomplish this by affecting host taste receptor expression, vagus nerve activity or neuroendocrine profiles, for example by producing dopamine, serotonin or peptides that influence satiety [75]. Evidence for these hypotheses is to date circumstantial, but elucidating and evaluating the influences of gut microbes on host feeding behaviours should be a research priority moving forward.
4. Influence of microbes on cognition and social behaviour
Recent studies have revealed that microbes contribute to surprisingly complex behaviours, including cognition and social behaviour. Almost all such studies to date have involved laboratory mice and rats, and compare germ-free animals (raised in an environment with no microbes) to both specific pathogen-free and conventionally raised animals.
Interestingly, germ-free mice show reduced exploration of unfamiliar conspecifics and spend less time near conspecifics than do control animals (e.g. [76,77]). In addition, rearing mice in germ-free environments results in an exaggerated physiological response to stress, which can be reversed by restoration of normal microbial communities or even just the bacterium Bifidobacterium infantis [78]. Paradoxically, several studies indicate that germ-free mice show decreased anxiety and increased exploratory behaviour and suggest the existence of a sensitive period after which introducing microbes does not reverse the effects of germ-free rearing (e.g. [79,80]). Gareau et al. [81] have suggested that this discrepancy might be reconciled if the increased exploratory behaviour in germ-free mice is due to deficits in working memory rather than decreased anxiety, leading animals to greater levels of exploration; further, studies using varying measures of anxiety obtain differing results (e.g. [76]).
The neural correlates of altered behaviours in germ-free mice are rarely clear, but their nervous systems are altered in ways that are consistent with changes in anxiety, learning and social behaviour. For example, germ-free mice show widespread changes in gene expression in the amygdala [82]. Such mice also show reduced expression of brain-derived neurotrophic factor in the dentate gyrus and CA1 layer of the hippocampus [79,81], as well as in the amygdala and cingulate cortex [82,83]. Expression of N-methyl-d-aspartate receptor subunits NR1 and NR2A is also reduced in the hippocampus [78], and of NR2B in the central amygdala [79], in germ-free mice. Finally, such mice also show increased turnover of dopamine, noradrenaline and serotonin in the striatum [83]. While such studies are in their infancy, it is clear that the microbiota plays a powerful role in normal neural function as well as in behavioural and neural development in mammals (for recent reviews, see [5,84–86]). Nevertheless, research to date has largely focused on a small number of mammalian species, and broader comparative studies are needed.
5. Could microbes have played a role in internalization of nervous systems?
Given our long history of co-evolving with microbes, as well as the examples described in §§2–4, it seems clear that microbes have played a role in nervous system evolution in at least some specific cases. In addition, we propose that microbes played a role in a major event in nervous system evolution: the internalization of nervous systems in animals derived from ancestors that possessed nerve nets.
The nature of the first neurons is not clear, nor do we agree how many times neurons may have originated [87,88]. Data from demosponges suggest that proto-neurons were sensory cells embedded in the external epithelium [89–91]. These proto-neurons may have signalled to each other or to neighbouring epithelial cells in paracrine fashion, and adaptations for improved signalling fidelity, including neurites and synapses, might have evolved later [92]. The signalling molecules used by these proto-neurons were likely peptides [93]. The molecular components required for the synthesis and release of neuropeptides are present in demosponges, as are homologues of eumetazoan neuropeptide receptors [91]. Thus, even before nerve nets arose, protoneurons, peptide signalling molecules and receptors for these signalling molecules were likely present.
Among animals with nerve nets, peptide neurotransmission may predominate ([94], but see [95]). Many neurobiologists think of neuropeptides as functioning as modulators that activate relatively slow processes; nevertheless, fast, peptide-gated ion channels that are related to acid-sensing ion channels (ASICs) are present in a variety of animals, including cnidarians [96] and molluscs [97,98]. In addition, although neurotransmission in ctenophores is poorly understood, large numbers of fast, peptide-gated ion channels are coded in the genome of Pleurobrachia bachei [99], and the genome of the demosponge Amphimedon queenslandica contains at least one putative ASIC (NCBI reference sequence: XP_011405396.1). Thus, the earliest neural signalling molecules in animals with nerve nets may have been peptides that elicited relative rapid responses from neighbouring neurons.
Although it is not yet clear how many times internal nervous systems arose from animals with nerve nets [93], we propose that microbes may have played an important role in driving the evolution of internalized nervous systems. We can envision three ways in which microbes may have contributed to internalization of the nervous system; these hypotheses are not mutually exclusive.
First, as illustrated in figure 1, nervous systems may have become internalized to optimize signal fidelity. In general, neuropeptides bind to G protein-coupled receptors (GPCRs), evolutionarily ancient receptors that predate the split among plants, fungi and eukaryotes [100,101]. Two of the five major families of GPCRs—the rhodopsin-like and glutamate-like GPCRs—are present in animals without nervous systems, such as Amphimedon and Trichoplax, as well as in ctenophores and cnidarians [102], and could have functioned as the earliest receptors for neural signalling molecules. A basic property of GPCRs is that a single receptor can couple to different intracellular signalling pathways such that different ligands can influence different physiological processes. In other words, GPCRs are inherently promiscuous [103], although in some cases GPCRs have co-evolved with particular ligands to a high degree of specificity (e.g. [104]). Thus, if GPCRs served as receptors for early neuropeptide signalling molecules, these receptors were vulnerable to interference by other molecules.
Figure 1.
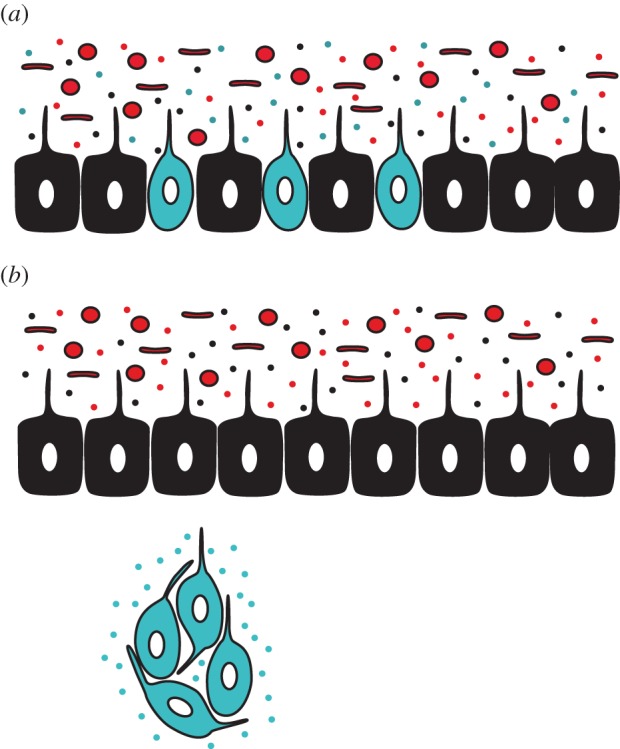
Illustration of a hypothesis concerning the role of microbes in nervous system internalization. Proto-neurons are depicted in blue, epidermal cells in black, and both coccus- and rod-shaped microbes are depicted in red. The smaller blue, black and red dots represent peptides and other small molecules produced by the three classes of cells. (a) The ancestral condition, in which proto-neurons are distributed among epidermal cells. In this state, metabolic products and signalling molecules released by microbes and anti-microbial peptides released by epithelial cells may interfere with interneuronal signalling. (b) In animals with an internal nervous system, neuronal signalling is largely protected from interference; in addition, epithelial cells can regulate the composition of the microbial community and microbes can signal to epithelial cells without interference from neuronal molecules. (Online version in colour.)
Molecules that could create such interference are abundant at the body surface. Epithelial cells produce peptides to regulate microbial communities in and on animals' bodies. Indeed, Hydra use antimicrobial peptides to control species-specific microbiomes [105,106], suggesting that antimicrobial peptides date to at least the last common ancestor of cnidarians and bilaterians and may be widespread across the animal kingdom. Given that the neurons that comprise nerve nets are interspersed among these epithelial cells, the peptides produced by epithelial cells have the potential to interfere with neuronal GPCRs. Further, microbes also produce peptides as metabolic products as well as for use in intraspecific signalling and interspecific competition (for recent reviews, see [107–109]), providing yet another source of potential interference with neural signalling. Finally, many bacterial products are detected by GPCRs (e.g. [110]). As a result, pathogenic bacteria have evolved mechanisms to modulate or block activity of animal GPCRs to avoid detection [111], a process that again could compromise neural signalling. Taken together, these observations suggest that nervous systems may have been internalized to protect neuropeptide receptors from ‘noise’ due to the presence of antimicrobial peptides and microbial products.
Second, a core group of 16 families of peptidases are ubiquitous and as old as life [112], and neuropeptides that are released into the mucus at the epithelial surface are vulnerable to degradation by these enzymes. This creates obvious problems for paracrine signalling, as peptides that are broken down before they reach their target are poor signals. Further, degradation poses an additional problem: when degraded by peptidases, neuropeptides can break down into smaller products that can also activate or antagonize either the parent receptor or receptors for other peptides [113]. Perhaps nervous systems were internalized to avoid problems related to degradation of neuropeptides, which can lead to ineffective signalling and receptor noise.
A brief example may serve to illustrate the kind of cross-talk that we envision among receptors, peptides and peptidases. The nervous systems of vertebrates contain many peptides that possess a C-terminal-RFamide, similar to the invertebrate neuropeptide FMRFamide; such peptides are often referred to as FMRFamide-like peptides (FLPs). FLPs interact with a variety of receptor subtypes [114], and the same receptor can interact with many different FLPs (e.g. [115,116]). For example, frog nervous systems contain members of peptide families that are widely distributed among vertebrates and could be considered FLPs, including gonadotropin inhibiting hormone [117–119]; kisspeptin [120]; met-enkephalin and other opioid peptides [121,122]; prolactin-releasing peptide [123]; and neuropeptide Y, peptide YY and pancreatic polypeptide [124]. The skin of frogs also contains many anti-microbial peptides [125], including several FLPs [126]. Finally, frog skin also contains numerous peptidases [127]. Thus, it seems that unless the nervous system were internal to the epidermis, the multiple FLPs in both neurons and skin cells, multiple FLP receptors on neurons, and multiple peptidases produced by skin cells could all interfere with both neural signalling and epithelial–microbial signalling in frogs.
Third, nervous systems may have become internalized to protect them from invasion by microorganisms. Neural receptors on the surface of an animal are risky, as viruses can gain entry to neurons through receptors; to cite some familiar examples, rabies, herpes and measles viruses all enter neurons through cell-surface receptors [128–130]. Once inside a neuron, viruses can use synapses to rapidly spread from cell to cell, potentially invading large portions of the body [131,132]. Perhaps nervous systems were internalized to protect the organism from infection. Notably, the enteric nervous system of the gastrointestinal tract, which must be in constant communication with the luminal environment [133,134], is protected from gut microbes by a variety of defences, including intestinal epithelial tight junctions, a resistant mucosal barrier and broad and targeted immune responses [134,135].
6. Conclusion
At a time when the influence of microbes is starting to be recognized as fundamental to all aspects of animal biology, it is critical that we consider microbial impacts on the function and evolution of animal nervous systems. All animals necessarily interact with environmental and symbiotic microbes, as well as metabolic products of microbes, and the outcomes of these interactions can be beneficial or detrimental to animals. In this paper, we have described how these interactions have shaped animals' chemosensory and photosensory systems in ways that promote the avoidance of pathogens and predators, the location of important resources including food and suitable mates, and the effective signalling of key information among animal conspecifics. We also introduced a burgeoning line of inquiry regarding the effects of symbiotic microbes on mammalian neurodevelopment, cognition, anxiety and social behaviour, and call for comparative studies on these subjects. Finally, we discussed how beneficial phenotypes conferred by symbiotic microbes to their animal hosts can require concomitant adaptations in animals' nervous systems to accommodate and manage these residents and suggested that a key event in neural evolution—the internalization of the nervous system—may have resulted from selection pressures involving microbes.
Acknowledgements
Thanks to Nicholas Strausfeld and Frank Hirth for organizing the meetings from which this paper emerged. In addition, we thank Emma Coddington, Stephen Thomas and Patric Vaelli for useful discussions of the ideas presented here.
Authors' contributions
H.L.E. and K.R.T. developed the ideas presented here, wrote the article and approved the final version to be published.
Competing interests
We have no competing interests.
Funding
This work was funded by the US National Science Foundation (IOS-1354089 to H.L.E. and subawards from DBI-0939454 to H.L.E. and K.R.T.).
References
- 1.Hacquard S, et al. 2015. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. ( 10.1016/j.chom.2015.04.009) [DOI] [PubMed] [Google Scholar]
- 2.Huttenhower C, et al. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. ( 10.1038/nature11234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsythe P, Kunze WA. 2013. Voices from within: gut microbes and the CNS. Cell. Mol. Life Sci. 70, 55–69. ( 10.1007/s00018-012-1028-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stilling RM, Bordenstein SR, Dinan TG, Cryan JF. 2014. Friends with social benefits: host–microbe interactions as a driver of brain evolution and development? Front. Cell Infect. Microbiol. 4, 147 ( 10.3389/fcimb.2014.00147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114, 185–190. ( 10.1016/j.zool.2011.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. ( 10.1111/j.1574-6976.2008.00123.x) [DOI] [PubMed] [Google Scholar]
- 8.Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226 ( 10.1371/journal.pbio.1002226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg E, Zilber-Rosenberg I. 2013. The hologenome concept: human, animal, and plant microbiota. Berlin, Germany: Springer. [Google Scholar]
- 10.Brucker RM, Bordenstein SR. 2013. The capacious hologenome. Zoology 116, 260–261. ( 10.1016/j.zool.2013.08.003) [DOI] [PubMed] [Google Scholar]
- 11.Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. 2007. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 104, 2295–2300. ( 10.1073/pnas.0610281104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart AC, Chao MY. 2010. From odors to behaviors in Caenorhabditis elegans. In The neurobiology of olfaction (ed. Menini A.), pp. 1–33. Boca Raton, FL: CRC Press. [PubMed] [Google Scholar]
- 13.Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. 2014. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280. ( 10.1016/j.cell.2014.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stensmyr MC, et al. 2012. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. ( 10.1016/j.cell.2012.09.046) [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Park KC, Baker TC. 2003. Identification of odors from overripe mango that attract vinegar flies, Drosophila melanogaster. J. Chem. Ecol. 29, 899–909. ( 10.1023/A:1022931816351) [DOI] [PubMed] [Google Scholar]
- 16.Beltran-Bech S, Richard FJ. 2014. Impact of infection on mate choice. Anim. Behav. 90, 159–170. ( 10.1016/j.anbehav.2014.01.026) [DOI] [Google Scholar]
- 17.Penn D, Potts WK. 1998. Chemical signals and parasite-mediated sexual selection. Trends Ecol. Evol. 13, 391–396. ( 10.1016/S0169-5347(98)01473-6) [DOI] [PubMed] [Google Scholar]
- 18.Zala SM, Potts WK, Penn DJ. 2004. Scent-marking displays provide honest signals of health and infection. Behav. Ecol. 15, 338–344. ( 10.1093/beheco/arh022) [DOI] [Google Scholar]
- 19.Arakawa H, Cruz S, Deak T. 2011. From models to mechanisms: odorant communication as a key determinant of social behavior in rodents during illness-associated states. Neurosci. Biobehav. Rev. 35, 1916–1928. ( 10.1016/j.neubiorev.2011.03.007) [DOI] [PubMed] [Google Scholar]
- 20.Bloes DA, Kretschmer D, Peschel A. 2015. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Microbiol. 13, 95–104. ( 10.1038/nrmicro3390) [DOI] [PubMed] [Google Scholar]
- 21.Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, Siltberg-Liberles J, Liberles DA, Buck LB. 2009. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proc. Natl Acad. Sci. USA 106, 9842–9847. ( 10.1073/pnas.0904464106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. 2009. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature 459, 574–577. ( 10.1038/nature08029) [DOI] [PubMed] [Google Scholar]
- 23.Boillat M, Challet L, Rossier D, Kan C, Carleton A, Rodriguez I. 2015. The vomeronasal system mediates sick conspecific avoidance. Curr. Biol. 25, 251–255. ( 10.1016/j.cub.2014.11.061) [DOI] [PubMed] [Google Scholar]
- 24.Albuquerque TA, Zurek L. 2014. Temporal changes in the bacterial community of animal feces and their correlation with stable fly oviposition, larval development, and adult fitness. Front. Microbiol. 5, 590 ( 10.3389/fmicb.2014.00590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clare AS. 2011. Toward a characterization of the chemical cue to barnacle gregariousness. In Chemical communication in crustaceans (eds Breithaupt T, Thiel M), pp. 431–450. New York, NY: Springer. [Google Scholar]
- 26.De Gregoris TB, Khandeparker L, Anil AC, Mesbahi E, Burgess JG, Clare AS. 2012. Characterisation of the bacteria associated with barnacle, Balanus amphitrite, shell and their role in gregarious settlement of cypris larvae. J. Exp. Mar. Biol. Ecol. 413, 7–12. ( 10.1016/j.jembe.2011.11.014) [DOI] [Google Scholar]
- 27.Hadfield MG. 2011. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann. Rev. Mar. Sci. 3, 453–470. ( 10.1146/annurev-marine-120709-142753) [DOI] [PubMed] [Google Scholar]
- 28.Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F. 2012. The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3, 292 ( 10.3389/fmicb.2012.00292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stökl J, Strutz A, Dafni A, Svatos A, Doubsky J, Knaden M, Sachse S, Hansson BS, Stensmyr MC. 2010. A deceptive pollination system targeting drosophilids through olfactory mimicry of yeast. Curr. Biol. 20, 1846–1852. ( 10.1016/j.cub.2010.09.033) [DOI] [PubMed] [Google Scholar]
- 30.Dweck HK, Ebrahim SA, Farhan A, Hansson BS, Stensmyr MC. 2015. Olfactory proxy detection of dietary antioxidants in Drosophila. Curr. Biol. 25, 455–466. ( 10.1016/j.cub.2014.11.062) [DOI] [PubMed] [Google Scholar]
- 31.Archie EA, Theis KR. 2011. Animal behaviour meets microbial ecology. Anim. Behav. 82, 425–436. ( 10.1016/j.anbehav.2011.05.029) [DOI] [Google Scholar]
- 32.Ezenwa VO, Williams AE. 2014. Microbes and animal olfactory communication: where do we go from here? Bioessays 36, 847–854. ( 10.1002/bies.201400016) [DOI] [PubMed] [Google Scholar]
- 33.Bradbury JW, Vehrencamp SL. 2011. Principles of animal communication, 2nd edn Sunderland, MA: Sinauer Associates. [Google Scholar]
- 34.Wyatt TD. 2014. Pheromones and animal behavior, 419 New York, NY: Cambridge University Press. [Google Scholar]
- 35.Albone ES. 1984. Mammalian semiochemistry. New York, NY: John Wiley. [Google Scholar]
- 36.Davis TS, Crippen TL, Hofstetter RW, Tomberlin JK. 2013. Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol. 39, 840–859. ( 10.1007/s10886-013-0306-z) [DOI] [PubMed] [Google Scholar]
- 37.Wyatt TD. 2010. Pheromones and signature mixtures: defining species-wide signals and variable cues for identity in both invertebrates and vertebrates. J. Comp. Physiol. A 196, 685–700. ( 10.1007/s00359-010-0564-y) [DOI] [PubMed] [Google Scholar]
- 38.Davis TS. 2015. The ecology of yeasts in the bark beetle holobiont: a century of research revisited. Microb. Ecol. 69, 723–732. ( 10.1007/s00248-014-0479-1) [DOI] [PubMed] [Google Scholar]
- 39.Wang XH, Kang L. 2014. Molecular mechanisms of phase change in locusts. Ann. Rev. Entomol. 59, 225–244. ( 10.1146/annurev-ento-011613-162019) [DOI] [PubMed] [Google Scholar]
- 40.Obeng-Ofori D, Torto B, Njagi PG, Hassanali A, Amiani H. 1994. Fecal volatiles as part of the aggregation pheromone complex of the desert locust, Schistocerca gregaria (Forskal) (Orthoptera: Acrididae). J. Chem. Ecol. 20, 2077–2087. ( 10.1007/BF02066244) [DOI] [PubMed] [Google Scholar]
- 41.Torto B, Obeng-Ofori D, Njagi PG, Hassanali A, Amiani H. 1994. Aggregation pheromone system of adult gregarious desert locust Schistocerca gregaria (Forskal). J. Chem. Ecol. 20, 1749–1762. ( 10.1007/BF02059896) [DOI] [PubMed] [Google Scholar]
- 42.Ochieng SA, Hansson BS. 1999. Responses of olfactory receptor neurones to behaviourally important odours in gregarious and solitarious desert locust, Schistocerca gregaria. Physiol. Entomol. 24, 28–36. ( 10.1046/j.1365-3032.1999.00107.x) [DOI] [Google Scholar]
- 43.Anton S, Ignell R, Hansson BS. 2002. Developmental changes in the structure and function of the central olfactory system in gregarious and solitary desert locusts. Microsc. Res. Tech. 56, 281–291. ( 10.1002/jemt.10032) [DOI] [PubMed] [Google Scholar]
- 44.Dillon RJ, Vennard CT, Charnley AK. 2000. Exploitation of gut bacteria in the locust. Nature 403, 851 ( 10.1038/35002669) [DOI] [PubMed] [Google Scholar]
- 45.Dillon RJ, Vennard CT, Charnley AK. 2002. A note: gut bacteria produce components of a locust cohesion pheromone. J. Appl. Microbiol. 92, 759–763. ( 10.1046/j.1365-2672.2002.01581.x) [DOI] [PubMed] [Google Scholar]
- 46.Dillon R, Charnley K. 2002. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res. Microbiol. 153, 503–509. ( 10.1016/S0923-2508(02)01361-X) [DOI] [PubMed] [Google Scholar]
- 47.Gorman ML. 1976. A mechanism for individual recognition by odour in Herpestes auropunctatus (Carnivora: Viverridae). Anim. Behav. 24, 141–145. ( 10.1016/S0003-3472(76)80107-8) [DOI] [Google Scholar]
- 48.Gorman ML, Nedwell DB, Smith RM. 1974. An analysis of the contents of the anal scent pockets of Herpestes auropunctatus (Carnivora: Viverridae). J. Zool. 172, 389–399. ( 10.1111/j.1469-7998.1974.tb04115.x) [DOI] [Google Scholar]
- 49.Burgener N, East ML, Hofer H, Dehnhard M. 2008. Do spotted hyena scent marks code for clan membership? In Chemical signals in vertebrates 11 (eds Hurst JL, Beynon RJ, Roberts SC, Wyatt TD), pp. 169–178. New York, NY: Springer. [Google Scholar]
- 50.Theis KR, Schmidt TM, Holekamp KE. 2012. Evidence for a bacterial mechanism for group-specific social odors among hyenas. Sci. Rep. 2, 615 ( 10.1038/srep00615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theis KR, Venkataraman A, Dycus JA, Koonter KD, Schmitt-Matzen EN, Wagner AP, Holekamp KE, Schmidt TM. 2013. Symbiotic bacteria appear to mediate hyena social odors. Proc Natl Acad Sci USA 110, 19 832–19 837. ( 10.1073/pnas.1306477110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodwin TE, et al. 2012. Chemical signals of elephant musth: temporal aspects of microbially-mediated modifications. J Chem Ecol 38, 81–87. ( 10.1007/s10886-011-0056-8) [DOI] [PubMed] [Google Scholar]
- 53.Goodwin TE, et al. In press The role of bacteria in chemical signals of elephant musth: proximate causes and biochemical pathways. In Chemical signals in vertebrates (eds Schulte BA, Ferkin MH, Goodwin TE). New York, NY: Springer. [Google Scholar]
- 54.Lanyon CV, Rushton SP, O'Donnell AG, Goodfellow M, Ward AC, Petrie M, Jensen SP, Gosling LM, Penn DJ. 2007. Murine scent mark microbial communities are genetically determined. FEMS Microbiol. Ecol. 59, 576–583. ( 10.1111/j.1574-6941.2006.00252.x) [DOI] [PubMed] [Google Scholar]
- 55.Zomer S, et al. 2009. Consensus multivariate methods in gas chromatography mass spectrometry and denaturing gradient gel electrophoresis: MHC-congenic and other strains of mice can be classified according to the profiles of volatiles and microflora in their scent-marks. Analyst 134, 114–123. ( 10.1039/b807061j) [DOI] [PubMed] [Google Scholar]
- 56.Caley MJ, Schluter D. 2003. Predators favour mimicry in a tropical reef fish. Proc. R. Soc. Lond. B 270, 667–672. ( 10.1098/rspb.2002.2263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chau R, Kalaitzis JA, Neilan BA. 2011. On the origins and biosynthesis of tetrodotoxin. Aquat. Toxicol. 104, 61–72. ( 10.1016/j.aquatox.2011.04.001) [DOI] [PubMed] [Google Scholar]
- 58.Jost MC, Hillis DM, Lu Y, Kyle JW, Fozzard HA, Zakon HH. 2008. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 25, 1016–1024. ( 10.1093/molbev/msn025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashiguchi Y, Lee JM, Shiraishi M, Komatsu S, Miki S, Shimasaki Y, Mochioka N, Kusakabe T, Oshima Y. 2015. Characterization and evolutionary analysis of tributyltin-binding protein and pufferfish saxitoxin and tetrodotoxin-binding protein genes in toxic and nontoxic pufferfishes. J. Evol. Biol. 28, 1103–1118. ( 10.1111/jeb.12634) [DOI] [PubMed] [Google Scholar]
- 60.Ritson-Williams R, Yotsu-Yamashita M, Paul VJ. 2006. Ecological functions of tetrodotoxin in a deadly polyclad flatworm. Proc. Natl Acad. Sci. USA 103, 3176–3179. ( 10.1073/pnas.0506093103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yotsu-Yamashita M, Mebs D, Flachsenberger W. 2007. Distribution of tetrodotoxin in the body of the blue-ringed octopus (Hapalochlaena maculosa). Toxicon 49, 410–412. ( 10.1016/j.toxicon.2006.10.008) [DOI] [PubMed] [Google Scholar]
- 62.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid–Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. ( 10.1038/nrmicro957) [DOI] [PubMed] [Google Scholar]
- 63.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl Acad. Sci. USA 97, 10 231–10 235. ( 10.1073/pnas.97.18.10231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. 2014. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech. Dev. 131, 111–126. ( 10.1016/j.mod.2013.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ. 2009. Evidence for light perception in a bioluminescent organ. Proc. Natl Acad. Sci. USA 106, 9836–9841. ( 10.1073/pnas.0904571106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Visick KL, McFall-Ngai MJ. 2000. An exclusive contract: specificity in the Vibrio fischeri–Euprymna scolopes partnership. J. Bacteriol. 182, 1779–1787. ( 10.1128/JB.182.7.1779-1787.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andersen SB, Gerritsma S, Yusah KM, Mayntz D, Hywel-Jones NL, Billen J, Boomsma JJ, Hughes DP. 2009. The life of a dead ant: the expression of an adaptive extended phenotype. Am. Nat. 174, 424–433. ( 10.1086/603640) [DOI] [PubMed] [Google Scholar]
- 68.de Bekker C, Merrow M, Hughes DP. 2014. From behavior to mechanisms: an integrative approach to the manipulation by a parasitic fungus (Ophiocordyceps unilateralis s.l.) of its host ants (Camponotus spp.). Integr. Comp. Biol. 54, 166–176. ( 10.1093/icb/icu063) [DOI] [PubMed] [Google Scholar]
- 69.de Bekker C, Quevillon LE, Smith PB, Fleming KR, Ghosh D, Patterson AD, Hughes DP. 2014. Species-specific ant brain manipulation by a specialized fungal parasite. BMC Evol Biol. 14, 166 ( 10.1186/s12862-014-0166-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vyas A, Kim SK, Giacomini N, Boothroyd JC, Sapolsky RM. 2007. Behavioral changes induced by Toxoplasma infection of rodents are highly specific to aversion of cat odors. Proc. Natl Acad. Sci. USA 104, 6442–6447. ( 10.1073/pnas.0608310104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunlap PV, Nakamura M. 2011. Functional morphology of the luminescence system of Siphamia versicolor (Perciformes: Apogonidae), a bacterially luminous coral reef fish. J. Morphol. 272, 897–909. ( 10.1002/jmor.10956) [DOI] [PubMed] [Google Scholar]
- 72.Gould AL, Harii S, Dunlap PV. 2015. Cues from the reef: olfactory preferences of a symbiotically luminous cardinalfish. Coral Reefs 34, 673–677. ( 10.1007/s00338-015-1278-y) [DOI] [Google Scholar]
- 73.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120, 1719–1729. [DOI] [PubMed] [Google Scholar]
- 74.Dunlap PV, Gould AL, Wittenrich ML, Nakamura M. 2012. Symbiosis initiation in the bacterially luminous sea urchin cardinalfish Siphamia versicolor. J. Fish. Biol. 81, 1340–1356. ( 10.1111/j.1095-8649.2012.03415.x) [DOI] [PubMed] [Google Scholar]
- 75.Alcock J, Maley CC, Aktipis CA. 2014. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 36, 940–949. ( 10.1002/bies.201400071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crumeyrolle-Arias M, Jaglin M, Bruneau A, Vancassel S, Cardona A, Dauge V, Naudon L, Rabot S. 2014. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology 42, 207–217. ( 10.1016/j.psyneuen.2014.01.014) [DOI] [PubMed] [Google Scholar]
- 77.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. 2014. Microbiota is essential for social development in the mouse. Mol. Psychiatry 19, 146–148. ( 10.1038/mp.2013.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 558, 263–275. ( 10.1113/jphysiol.2004.063388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neufeld KA, Kang N, Bienenstock J, Foster JA. 2011. Effects of intestinal microbiota on anxiety-like behavior. Commun. Integr. Biol. 4, 492–494. ( 10.4161/cib.4.4.15702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neufeld KM, Kang N, Bienenstock J, Foster JA. 2011. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 23, 255–264. ( 10.1111/j.1365-2982.2010.01620.x) [DOI] [PubMed] [Google Scholar]
- 81.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, Macqueen G, Sherman PM. 2011. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 60, 307–317. ( 10.1136/gut.2009.202515) [DOI] [PubMed] [Google Scholar]
- 82.Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, Dinan TG, Cryan JF. 2015. Microbes and neurodevelopment—absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav. Immun. 50, 209–220. ( 10.1016/j.bbi.2015.07.009) [DOI] [PubMed] [Google Scholar]
- 83.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl Acad. Sci. USA 108, 3047–3052. ( 10.1073/pnas.1010529108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen-Blevins CR, Sela DA, Hinde K. 2015. Milk bioactives may manipulate microbes to mediate parent-offspring conflict. Evol. Med. Public Health 2015, 106–121. ( 10.1093/emph/eov007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Montiel-Castro AJ, Gonzalez-Cervantes RM, Bravo-Ruiseco G, Pacheco-Lopez G. 2013. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 7, 70 ( 10.3389/fnint.2013.00070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sampson TR, Mazmanian SK. 2015. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17, 565–576. ( 10.1016/j.chom.2015.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moroz LL, Kohn AB. 2016. Independent origins of neurons and synapses: insights from ctenophores. Phil. Trans. R. Soc. B 371, 20150041 ( 10.1098/rstb.2015.0041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ryan JF, Chiodin M. 2015. Where is my mind? How sponges and placozoans may have lost neural cell types. Phil. Trans. R. Soc. B 370, 20150059 ( 10.1098/rstb.2015.0059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS. 2007. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2, e506 ( 10.1371/journal.pone.0000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. 2008. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 18, 1156–1161. ( 10.1016/j.cub.2008.06.074) [DOI] [PubMed] [Google Scholar]
- 91.Srivastava M, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. ( 10.1038/nature09201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jekely G. 2011. Origin and early evolution of neural circuits for the control of ciliary locomotion. Proc. R. Soc. B 278, 914–922. ( 10.1098/rspb.2010.2027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moroz LL. 2009. On the independent origins of complex brains and neurons. Brain Behav. Evol. 74, 177–190. ( 10.1159/000258665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grimmelikhuijzen CJ, Hauser F. 2012. Mini-review: the evolution of neuropeptide signaling. Regul. Pept. 177, S6–S9. ( 10.1016/j.regpep.2012.05.001) [DOI] [PubMed] [Google Scholar]
- 95.Kass-Simon G, Pierobon P. 2007. Cnidarian chemical neurotransmission, an updated overview. Comp. Biochem. Physiol. A 146, 9–25. ( 10.1016/j.cbpa.2006.09.008) [DOI] [PubMed] [Google Scholar]
- 96.Durrnagel S, Kuhn A, Tsiairis CD, Williamson M, Kalbacher H, Grimmelikhuijzen CJ, Holstein TW, Grunder S. 2010. Three homologous subunits form a high affinity peptide-gated ion channel in Hydra. J. Biol. Chem. 285, 11 958–11 965. ( 10.1074/jbc.M109.059998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cottrell GA, Green KA, Davies NW. 1990. The neuropeptide Phe-Met-Arg-Phe-NH2 (FMRFamide) can activate a ligand-gated ion channel in Helix neurones. Pflugers Arch. 416, 612–614. ( 10.1007/BF00382698) [DOI] [PubMed] [Google Scholar]
- 98.Lingueglia E, Champigny G, Lazdunski M, Barbry P. 1995. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature 378, 730–733. ( 10.1038/378730a0) [DOI] [PubMed] [Google Scholar]
- 99.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoyle CHV. 2011. Evolution of neuronal signalling: transmitters and receptors. Auton. Neurosci. Bas. Clin. 165, 28–53. ( 10.1016/j.autneu.2010.05.007) [DOI] [PubMed] [Google Scholar]
- 101.Stäubert C, Le Duc D, Schoneberg T. 2014. Examining the dynamic evolution of G protein-coupled receptors. In G protein-coupled receptor genetics: research and methods in the post-genomic era (ed. Stevens CW.), pp. 23–43. New York, NY: Humana Press. [Google Scholar]
- 102.Krishnan A, Schioth HB. 2015. The role of G protein-coupled receptors in the early evolution of neurotransmission and the nervous system. J. Exp. Biol. 218, 562–571. ( 10.1242/jeb.110312) [DOI] [PubMed] [Google Scholar]
- 103.Kenakin T. 1995. Agonist-receptor efficacy. I: mechanisms of efficacy and receptor promiscuity. Trends Pharmacol. Sci. 16, 188–192. ( 10.1016/S0165-6147(00)89020-3) [DOI] [PubMed] [Google Scholar]
- 104.Moyle WR, Campbell RK, Myers RV, Bernard MP, Han Y, Wang X. 1994. Co-evolution of ligand–receptor pairs. Nature 368, 251–255. ( 10.1038/368251a0) [DOI] [PubMed] [Google Scholar]
- 105.Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, Fraune S. 2013. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl Acad. Sci. USA 110, E3730–E3738. ( 10.1073/pnas.1304960110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fraune S, Bosch TC. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA 104, 13 146–13 151. ( 10.1073/pnas.0703375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Douglas AE. 2014. The molecular basis of bacterial–insect symbiosis. J. Mol. Biol. 426, 3830–3837. ( 10.1016/j.jmb.2014.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kouzuma A, Kato S, Watanabe K. 2015. Microbial interspecies interactions: recent findings in syntrophic consortia. Front. Microbiol. 6, 477 ( 10.3389/fmict.2015.00477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandrini S, Aldriwesh M, Alruways M, Freestone P. 2015. Microbial endocrinology: host–bacteria communication within the gut microbiome. J. Endocrinol. 225, R21–R34. ( 10.1530/joe-14-0615) [DOI] [PubMed] [Google Scholar]
- 110.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. 2015. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849. ( 10.3390/nu7042839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bestebroer J, de Haas CJC, van Strijp JAG. 2010. How microorganisms avoid phagocyte attraction. FEMS Microbiol. Rev. 34, 395–414. ( 10.1111/j.1574-6976.2009.00202.x) [DOI] [PubMed] [Google Scholar]
- 112.Page MJ, Di Cera E. 2008. Evolution of peptidase diversity. J. Biol. Chem. 283, 30 010–30 014. ( 10.1074/jbc.M804650200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hallberg M. 2015. Neuropeptides: metabolism to bioactive fragments and the pharmacology of their receptors. Med. Res. Rev. 35, 464–519. ( 10.1002/med.21323) [DOI] [PubMed] [Google Scholar]
- 114.Roszer T, Banfalvi G. 2012. FMRFamide-related peptides: anti-opiate transmitters acting in apoptosis. Peptides 34, 177–185. ( 10.1016/j.peptides.2011.04.011) [DOI] [PubMed] [Google Scholar]
- 115.Walker SE, Stell WK. 1986. Gonadotropin-releasing-hormone (GnRF), molluscan cardioexcitatory peptide (FMRFamide), enkephalin and related neuropeptides affect goldfish retinal ganglion-cell activity. Brain Res. 384, 262–273. ( 10.1016/0006-8993(86)91162-5) [DOI] [PubMed] [Google Scholar]
- 116.Mousley A, Marks NJ, Halton DW, Geary TG, Thompson DP, Maule AG. 2004. Arthropod FMRFamide-related peptides modulate muscle activity in helminths. Int. J. Parasitol. 34, 755–768. ( 10.1016/j.ijpara.2004.02.005) [DOI] [PubMed] [Google Scholar]
- 117.Chartrel N et al. . 2002. Isolation, characterization, and distribution of a novel neuropeptide, Rana RFamide (R-RFa), in the brain of the European green frog Rana esculenta. J. Comp. Neurol. 448, 111–127. ( 10.1002/cne.10253) [DOI] [PubMed] [Google Scholar]
- 118.Kanetoh T, Sugikawa T, Sasaki I, Muneoka Y, Minakata H, Takabatake I, Fujimoto M. 2003. Identification of a novel frog RFamide and its effect on the latency of the tail-flick response of the newt. Comp. Biochem. Physiol. C 134, 259–266. ( 10.1016/s1532-0456(02)00277-6) [DOI] [PubMed] [Google Scholar]
- 119.Ukena K, Ubuka T, Tsutsui K. 2003. Distribution of a novel avian gonadotropin-inhibitory hormone in the quail brain. Cell Tiss. Res. 312, 73–79. ( 10.1007/s00441-003-0700-x) [DOI] [PubMed] [Google Scholar]
- 120.Lee YR, et al. 2009. Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology 150, 2837–2846. ( 10.1210/en.2008-1679) [DOI] [PubMed] [Google Scholar]
- 121.Askin A, Camlica Y, Comelekoglu U. 2007. Opioid peptides as possible neuromodulators in the frog peripheral nerve system. Neuropeptides 41, 73–81. ( 10.1016/j.npep.2006.12.002) [DOI] [PubMed] [Google Scholar]
- 122.Bojnik E, Magyar A, Toth G, Bajusz S, Borsodi A, Benyhe S. 2009. Binding studies of novel, non-mammalian enkephalins, structures predicted from frog and lungfish brain cDNA sequences. Neuroscience 158, 867–874. ( 10.1016/j.neuroscience.2008.09.056) [DOI] [PubMed] [Google Scholar]
- 123.Sakamoto T et al. . 2006. Molecular cloning and functional characterization of a prolactin-releasing peptide homolog from Xenopus laevis. Peptides 27, 3347–3351. ( 10.1016/j.peptides.2006.08.003) [DOI] [PubMed] [Google Scholar]
- 124.Sundstrom G. et al. . 2012. Characterization of the neuropeptide Y system in the frog Silurana tropicalis (Pipidae): three peptides and six receptor subtypes. Gen. Comp. Endocrinol. 177, 322–331. ( 10.1016/j.ygcen.2012.04.027) [DOI] [PubMed] [Google Scholar]
- 125.Phoenix D, Dennison S, Harris F. 2013. Antimicrobial peptides. Weinheim, Germany: Wiley-VCH. [Google Scholar]
- 126.Wang L, Smyth A, Johnsen AH, Zhou M, Chen T, Walker B, Shaw C. 2009. FMRFamide-related peptides (FaRPs): A new family of peptides from amphibian defensive skin secretions. Biochem. Biophys. Res. Commun. 383, 314–319. ( 10.1016/j.bbrc.2009.04.002) [DOI] [PubMed] [Google Scholar]
- 127.Liberio MD, Bastos IMD, Pires OR, Fontes W, Santana JM, Castro MS. 2014. The crude skin secretion of the pepper frog Leptodactylus labyrinthicus is rich in metallo and serine peptidases. PLoS ONE 9, e96893 ( 10.1371/journal.pone.0096893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. 2010. The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8, 51–61. ( 10.1038/nrmicro2260) [DOI] [PubMed] [Google Scholar]
- 129.Kramer T, Enquist LW. 2013. Directional spread of alphaherpesviruses in the nervous system. Viruses 5, 678–707. ( 10.3390/v5020678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fukuhara H, Chen S, Takeda S, Maenaka K. 2013. Entry mechanism of morbillivirus family. Yakugaku Zasshi 133, 549–559. (doi:DN/JST.JSTAGE/yakushi/13-00001-5) [DOI] [PubMed] [Google Scholar]
- 131.Mothes W, Sherer NM, Jin J, Zhong P. 2010. Virus cell-to-cell transmission. J. Virol. 84, 8360–8368. ( 10.1128/JVI.00443-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sattentau Q. 2008. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 6, 815–826. ( 10.1038/nrmicro1972) [DOI] [PubMed] [Google Scholar]
- 133.Carabotti M, Scirocco A, Maselli MA, Severi C. 2015. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 28, 203–209. [PMC free article] [PubMed] [Google Scholar]
- 134.Mayer EA, Tillisch K, Gupta A. 2015. Gut/brain axis and the microbiota. J. Clin. Invest. 125, 926–938. ( 10.1172/JCI76304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336, 1268–1273. ( 10.1126/science.1223490) [DOI] [PMC free article] [PubMed] [Google Scholar]


