Abstract
We here found that intestinal epithelial Paneth cells secrete FABP4, adipsin and adiponectin in both mice and human. Deletion of Paneth cell results in the decrease of FABP4, adipsin and adiponectin not only in intestinal crypt cells but also in sera, suggesting that they may influence the state of the whole body. We also demonstrate that expression of FABP4, adipsin and adiponectin may be modulated by specific gut microbiota. In germ-free (GF) mice, the expression of FABP4, adipsin and adiponectin were lower or difficult to be detected. Feces transplantation promoted the expression of FABP4, adipsin and adiponectin in gut epithelial Paneth cells. We have found that Lactobacillus NK6 colony, which has the highest similarity with Lactobacillus taiwanensis strain BCRC 17755, may induce the expression of FABP4, adipsin and adiponectin through TRAF2 and TRAF6 ubiquitination mediated NF-κB signaling. Taken together, our findings set up a novel mechanism for FABP4, adipsin and adiponectin through gut microbiota mediating expression in gut Paneth cells.
Metabolism associated factors such as the fatty acid binding protein 4 (FABP4), adipsin and adiponectin play a significant role in the pathogenesis of a cluster of metabolic syndromes such as hypertriglyceridemia, insulin resistance and atherosclerosis1,2,3,4. Adipose tissues4 and adipose associated macrophages5 can secrete these factors to affect the occurrence and development of metabolic diseases. However, it is difficult to explain some observations such that metabolic syndrome can occur in lean individuals6,7 and that some morbidly obese ones are metabolically healthy8,9. These facts imply that adipose tissues may not be a primary origin of metabolic syndrome. Previous studies have suggested that gut epithelial Paneth cells, which are located at the base of the crypts of Lieberkühn, possess the potential to express metabolism associated factors such as adiponectin and adipsin (complement factors D)10,11. We here demonstrate that intestinal epithelial Paneth cells secrete FABP4, adipsin and adiponectin in both mice and human. We also found that the expression of FABP4, adipsin and adiponectin may be regulated by gut bacteria Lactobacillus NK6 colony. Thus, our findings set up a novel regulating axis for FABP4, adipsin and adiponectin through gut microbiota mediating expression in gut Paneth cells.
Results
Expression of FABP4, adipsin and adiponectin in intestinal Paneth cells
Previous studies have implied that gut epithelial Paneth cells may potentially express metabolism associated factors such as adiponectin and adipsin (complement factors D)10,11. Our immunohistochemistry and immunoblot results validated these implications and detected their expression at the similar position in gut tissues (Fig. 1a–d and Supplementary Fig. S1). Meanwhile, at the base of the crypts we also found the expression of FABP4, which was different from FABP2 widely located in gut epithelial tissues (Fig. 1a–d and Supplementary Fig. S1). These metabolism associated factors were also detected in the crypts of jejunum, ileum and colon tissues (Fig. 1e and Supplementary Fig. S2). We also analyzed the perilipin (a specific marker of adipose cells), AEBP1 (adipocyte enhancer-binding protein 1, a specific marker of preadipose cells) and CD11b and FXIIIA (macrophages specific marker)12,13,14 in the isolated crypts cells. No perilipi, AEBP1, CD11b and FXIIIA were detected, indicating that the isolated crypts cells are not contaminated by adipose cells, preadipose cells and macrophages (Fig. 2). Notably, mouse intestinal epithelial cells (MIEC) from mouse embryonic intestinal cells, could also express FABP4, adipsin and adiponectin (Supplementary Fig. S3), implying that Paneth cells were not the only source for FABP4 and adipsin in gut epithelial tissues.
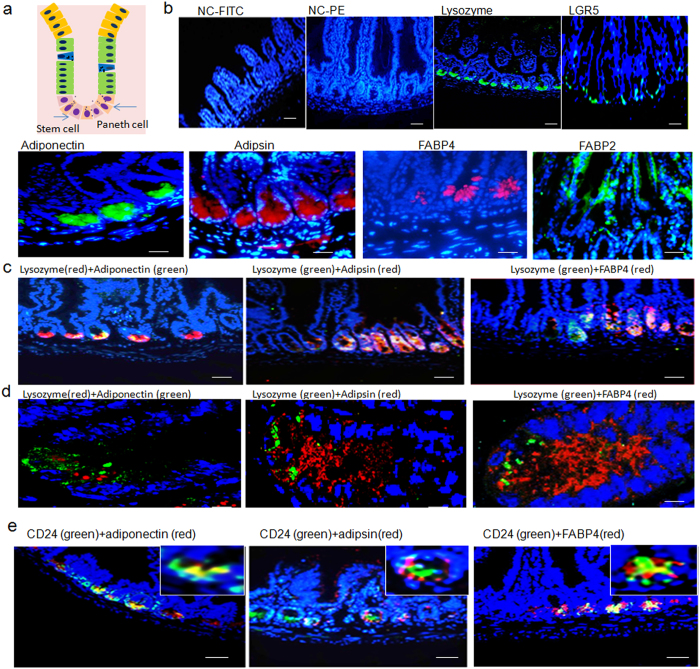
Figure 1. Expression of FABP4, adipsin and adiponectin in intestinal Paneth cells.
(a) Schematic representation (right) and staining (left) of the Paneth cells and putative intestinal stem cells. Paneth cells (lysozyme positive) and putative intestinal stem cells (LGR5 positive) localized at the crypt base. Intestinal tissues were sliced and stained using pooled isotypic control (NC) with FITC-labeled (NC-FITC) or PE-labeled (NC-PE) secondary antibodies, anti-lysozyme (lysozyme) and anti-LGR5 (LGR5) antibodies followed by fluorescence-labeled second antibodies. Green indicated lysozyme or LGR5 positive. Nuclei were stained by DAPI (blue). (b) Immunostaining of FABP4, adipsin, adiponectin (adipon) and FABP2 in ileum section from WT mice. Red and green indicated respectively FABP4, adipsin or adiponectin (adipon) and FABP2. Scale bar, 20μm. (c) Double immunostaining of FABP4, adipsin and adiponectin (adipon) with lysozyme in ileum section from WT mice. Scale bar, 40μm. (d) Analysis of confocal microscope of FABP4, adipsin and adiponectin (adipon) with lysozyme in ileum section from WT mice after double staining. Scale bar, 5 μm. (e) Double immuno-staining of adipsin, adiponectin and FABP4 with CD24 in the colon fragments of mice. Colon tissues were sliced and stained using anti-adipsin, anti-adiponectin and anti-FABP4 with anti-CD24 antibodies followed by fluorescence-labeled second antibodies. Green indicated CD24; Red indicated adipsin, adiponectin or FABP4. Nuclei were stained by DAPI (blue). Scale bar, 40 μm.
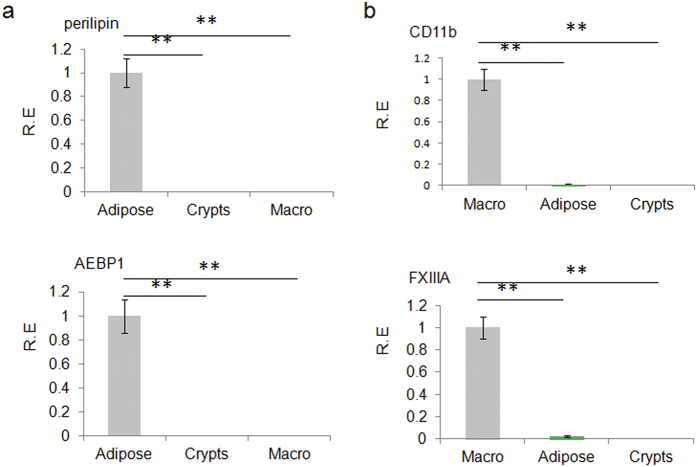
Figure 2. Expression of perilipin, AEBP-1, FXIIIA and CD11b in intestinal crypt cells, adipose tissues and macrophages.
(a) qRT-PCR of perilipin and AEBP-1 in intestinal crypt cells (Crypts), adipose tissues (Adipose) and macrophages (Macro). (b) qRT-PCR of FXIIIA and CD11b in intestinal crypt cells (Crypts), adipose tissues (Adipose tissues) and macrophages (Macro). Intestinal crypt cells were isolated according to the protocol described in the material and methods. Adipose tissues were from mouse fat pat. Macrophages were harvested by peritoneal lavage after 3–4 days following an i.p. injection of 4% sterile thioglycolate medium and then cultured in six-well plates and incubated at 37 °C and 5% CO2. After 6 h, the cells were washed to separate out any nonadhesive cells. R. E., relative expression. *P < 0.05 and **P < 0.01 (t-test, mean ± SD). The data were representative of three independent experiments.
Deletion of Paneth cells impairs the expression of FABP4, adipsin and adiponectin in gut crypt cells
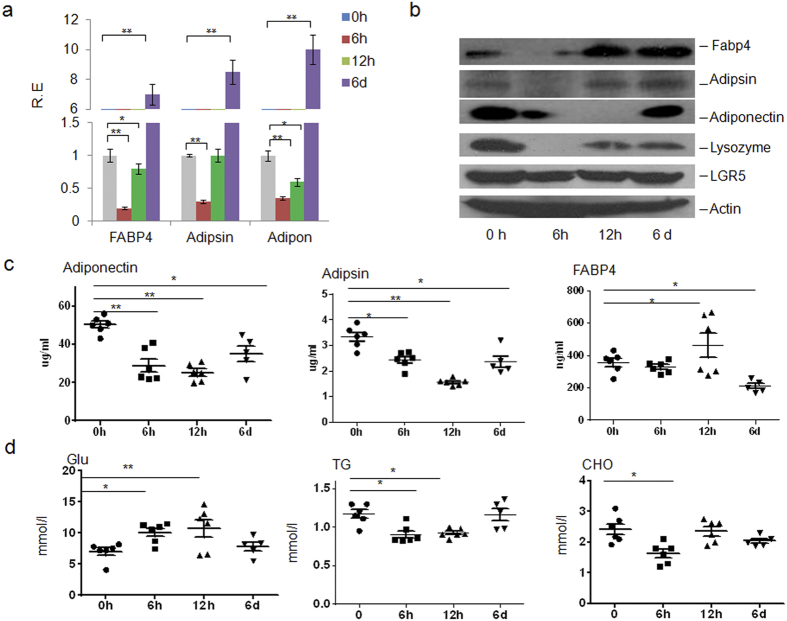
To exactly determine the importance of Paneth cells to produce FABP4, adipsin and adiponectin, we employed zinc depletion experiment with dithizone, which may deplete the secretary granules in Paneth cells15 and damages Paneth cells15,16,17. Consistent with other results15, dithizone treatment eliminated lysozyme staining in Paneth cells, whereas lysozyme staining was still strong in crypt cells of mice treated with control Li2CO3 (Fig. 3a,b and Supplementary Fig. S4). Importantly, this treatment led to the profound decrease of FABP4, adipsin and adiponectin in the isolated crypts (Fig. 3a,b and Supplementary Fig. S4), suggesting that Paneth cells were indeed an important source of these factors. A significant reduction was also detected in serum adipsin and adiponectin (Fig. 3c). Notably, serum levels of triglyceride (TG), glucose (Glu) and cholesterol (Cho) were also profoundly affected by Paneth cell depletion (Fig. 3d). These changes in serum have a great significance because it suggests that the action range of these Paneth cell derived factors are not restricted to intestine, but may be extended to the serum and further, the whole body, which made them able to regulate the systematic metabolism and account for some related diseases. Since metabolism-associated factors such as adiponectin may be considered as negative acute phase protein, we also measured inflammation parameters such as TNF α and IL-6. However, their levels were rather low in sera of both treated and untreated mice (unshown). This may be absence of inflammation in these treated and untreated mice. However, our results clearly show that gut epithelial Paneth cells may be another source of adipsin, adiponectin and FABP4 in addition to adipose tissues, which could largely influence the metabolism of the whole body.
Figure 3. Deletion of Paneth cells impairs the expression of FABP4, adipsin and adiponectin in gut crypt cells.
(a,b) qRT-PCR (a) and immunoblot analyses (b) of intestinal crypts with and without dithizone treatments. The mice were intravenously injected by dithizone and killed at the indicated time (male, n = 6 or indicated number). R. E., relative expression. (c) Serum levels of FABP4, adipsin and adiponectin in mice with (6h, 12h and 6 days after dithizone) and without (0h) dithizone treatments. (d) Serum levels of glucose (Glu), triglyceride (TG) and cholesterol (CHO) in fasting mice with (6h, 12h and 6 days after dithizone) and without (0h) dithizone treatments. *P < 0.05 and **P < 0.01 (t-test, mean ± SD in a; Mann-Whitney U test in c,d). The data were a representative of three independent experiments.
Expression of FABP4, adipsin and adiponectin is modulated by gut microbiota Lacbacillus
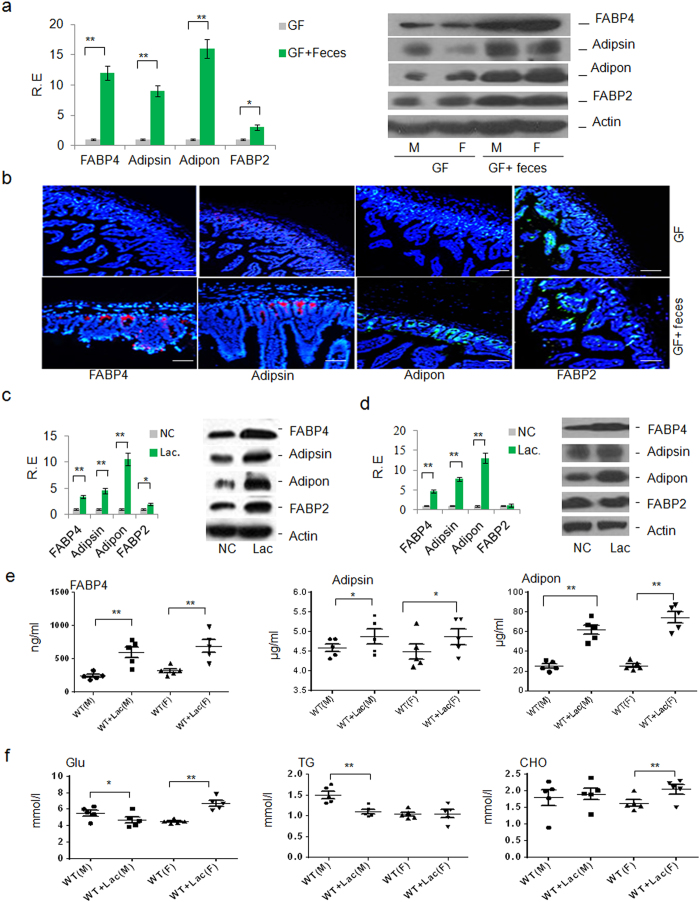
In addition to this finding that gut Paneth cells have an influence on these metabolism associated factors, another important phenomenon is that almost all the metabolic syndromes have a close relationship with the altered gut microbiota. Since both of them could control metabolism in the whole body, it is intriguing for possible regulation of gut microbiota in the expression of FABP4, adipsin and adiponectin in Paneth cells. Paneth cells, which are highly specialized epithelial cells18, directly sense gut microbiota and maintain homeostasis at the intestinal host-microbial interface19. To determine the effects of gut microbiota on the expression of FABP4, adipsin and adiponectin in gut epithelial Paneth cells, we first employed the germ-free (GF) mice, which are often used as a model to explore the effect of gut microbiota on the metabolic diseases. The expression of FABP4, adipsin and adiponectin in GF mice was low (Fig. 4a,b). However, wild-type (WT) mouse feces transplantation could reverse and further promote their expression in gut epithelial Paneth cells (Fig. 4a,b). In addition, the changed expression of FABP4, adiposin and adiponectin also occurred in leptin deficient (Ob/Ob) mice (Supplementary Fig. S5a, b), which have a higher proportion of Firmicutes to Bacteroidetes than lean mice, in high-fat diet mice (Supplementary Fig. S5c, d), which have a different gut microbiota composition and also in pan-antibiotics treated mice (Supplementary Fig. S5e, f). Notably, the expression of FABP4, adipsin and adiponectin was also upregulated by vitamin A and its metabolites 9-cis-retinoid acid (9-cis) and all-trans retinoid acid (ATRA) in gut epithelial crypt cells (Supplementary Fig. S6), in which vitamin A metabolism may be regulated by gut microbiota. All of these evidences support the conclusion that the ability of Paneth cells to express these factors may be modulated by gut microbiota. Thus, it could be seen that gut microbiota influence systemic metabolism at least in part by regulating the expression of FABP4, adipsin and adiponectin in Paneth cells.
Figure 4. Expression of FABP4, adipsin and adiponectin is modulated by gut microbiota Lacbacillus.
(a) qRT-PCR and immunoblot analyses of epithelial crypts in ileum fragments of germ-free (GF) mice (n = 10 (male, 5; female, 5)) and feces transplanted GF mice (GF + feces, n = 10 (male (M), 5; female (F), 5)). Actin, a loading control, was detected by anti-β-actin antibody. R. E., relative expression. (b) Immuno-staining of ileum fragments of germ-free (GF) mice and GF mice transplanted with WT mouse feces (GF + feces). Red and green indicated respectively FABP4, adipsin or adiponectin (adipon) and FABP2. Scale bar, 40 μm. The result is a representative of 6 samples. (c) RT-PCR and immunoblot analyses of ileum epithelial crypts in mice after in vivo transplantation with (Lac, N = 10 (male, 5; female, 5)) or without (NC, N = 10 (male, 5; female, 5)) Lacbacillus NK6. Actin, a loading control. R. E., relative expression. (d) qRT-PCR and immunoblot analyses of ileum epithelial crypts after in vitro stimulation with (Lac) or without (NC) Lacbacillus NK6. Actin, a loading control. (e) Serum levels of FABP4, adipsin and adiponectin (adipon) after transplantation with (WT + Lac, N = 10 (male (M), 5; female (F), 5)) or without (WT, N = 10 (male (M), 5; female (F), 5)) Lacbacillus NK6. (f) Serum levels of glucose (Glu), triglyceride (TG) and cholesterol (CHO) in fasting mice after transplantation with (WT + Lac, N = 10 (male (M), 5; female(F), 5)) or without (WT, N = 10 (male(M), 5; female(F), 5)) Lacbacillus NK6. *P < 0.05 and **P < 0.01 (t-test, mean ± SD in a, c, d; Mann-Whitney U test in e, f). The data were representative of three independent experiments.
Several reports have narrowed the range of bacteria associated with metabolic syndrome down to a few kinds of strains. For example, Lactobacillus spp plays a key role in the physiopathology of obesity, type 2 diabetes, and metabolic inflammation20; whereas Lactobacillus rhamnosus and Lactobacillus plantarum were reported to be able to reduce the fat content of mouse adipose tissues21. All of these imply that gut Lactobacillus might be involved in the regulation of metabolism. Indeed, we found that Lactobacillus NK6, which was isolated from WT mice and has the highest similarity with Lactobacillus taiwanensis strain BCRC 17755 (Supplementary Fig. S7–8), could mediate the expression of FABP4, adipsin and adiponectin (Fig. 4c). In vivo transplantation of this strain could affect the levels of FABP4, adipsin and adiponectin in both intestinal tissues (Fig. 4d) and sera (Fig. 4e). Furthermore, the serum levels of TG, Glu and Cho in the transplanted mice also exhibited remarkable changes, corresponding to the whole body effects of these factors, although there exist differences between male and female mice (Fig. 4f). However, as a comparison, expression of FABP4, adipsin and adiponectin were not changed in adipose tissues and macrophages of mice with or without Lactobacillus treatment (Supplementary Fig. S9), indicating that these changes only correlate with the metabolism associated factors secreted by gut Paneth cells. Taken together, our results suggested that gut Lactobacillus NK6 may affect the systematic metabolism through affecting FABP4, adipsin and adiponectin expression in gut epithelial Paneth cells.
Lacbacillus-mediated FABP4, adiposin and adiponectin is through TRAF2/6 ubiquitination mediated NF-κB pathway
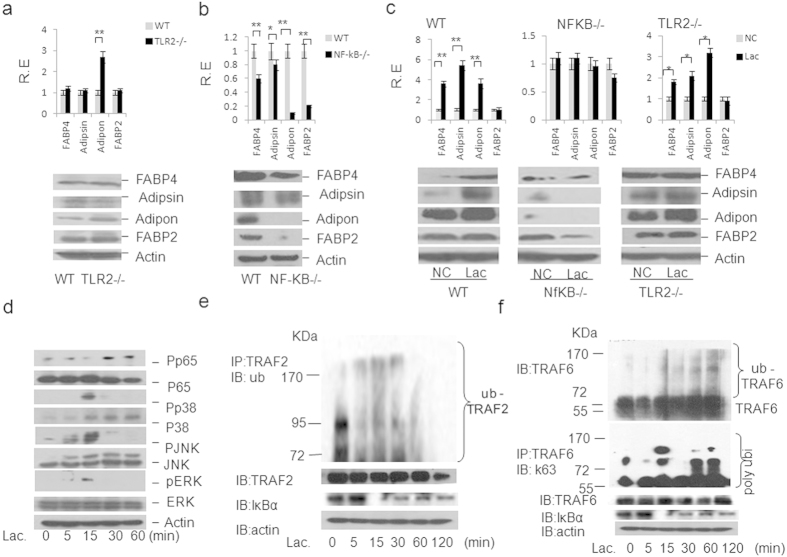
Lactobacillus, as a gram positive bacterium, was also potentially recognized by TLR2 receptor. But beyond our expectation, the level of FABP4, adipsin and adiponectin in the gut epithelial crypts of TLR2 deficient mice was not lower than those in control mice (Fig. 5a, Supplementary Fig. S10). However, NF-κB deficient mice exhibited a remarkably reduced level of FABP4, adipsin and adiponectin in their intestinal epithelial crypt cells (Fig. 5b, Supplementary Fig. S10–11). In vitro experiments also showed that intestinal crypt cells from NF-κB deficient mice had a lower level of FABP4, adipsin and adiponectin as compared to those of WT mice after exposed to heat-inactivated Lactobacillus (Fig. 5c). This reflected that in intestinal crypt cells, the regulation of Lactobacillus on the expression of metabolism associated factors was largely mediated by the downstream signaling molecule NF-κB, but not initiated by TLR2. However, this malfunction of TLR2 was not general, because there were difference in the expression of FABP4, adipsin and adiponectin among jejunum, ileum and colon fragments of both TLR2 and NF-κB deficient mice (Fig. 5c and Supplementary Fig. S11). Further studies showed that isolated Lactobacillus NK6 could extensively activate the gut cell signal pathways, including NF-κB and MAPK pathways in intestinal crypts (Fig. 5d). TRAF2 and TRAF6 in gut epithelial crypt cells were ubiquitinated, especially TRAF6 K63 ubiquitination after exposed to Lactobacillus NK6 (Fig. 5e,f). Thus, gut microbiota Lactobacillus NK6 triggered FABP4, adipsin and adiponectin is through TRAF2 and TRAF6 ubiquitination mediated NF-κB signaling pathways. In human, some clinical evidences have implied the effects of adipsin, adiponectin and FABP4 from gut epithelial Paneth cells on colitis associated metabolic syndrome. Statistically, the number of Paneth cells were found dramatically increased in the caecum, ascending, transverse and descending colons in patients with ulcerative colitis (UC) and Crohn’s disease (CD)22. Our results further exhibited a stronger staining of lysozyme and LGR-5 in the gut epithelial crypt of patients with colitis (Supplementary Fig. S12a) too. Meanwhile, stronger staining of FABP4, adipsin and adiponectin in the epithelial crypts of ulcerative colitis patients could be detected (Supplementary Fig. S12b). Thus, increased FABP4, adipsin and adiponectin in colitic epithelial Paneth cells may be involved in the occurrence and development of metabolic syndrome.
Figure 5. Lacbacillus-mediated FABP4, adiposin and adiponectin is through TRAF2/6 ubiquitination mediated NF-κB pathway.
(a) qRT-PCR (upper) and immunoblot (lower) of FABP4, adipsin, adiponectin and FABP2 in the jejunum epithelial crypt cells of WT and TLR2 deficient (−/−) mice (n = 6). R. E., relative expression. (b) qRT-PCR (upper) and immunoblot (lower) of FABP4, adipsin, adiponectin and FABP2 in the ileum epithelial crypt cells of WT and NF-κB deficient (−/−) mice (n = 6). R. E., relative expression. (c) qRT-PCR and immunoblot of FABP4, adipsin, adiponectin and FABP2 in the ileum epithelial crypts of WT, TLR2 and NF-κB deficient (−/−) mice with (Lac) or without (NC) Lacbacillus NK6 stimulation. R. E., relative expression. (d) Phosphorylation analyses of p65, p38, JNK, and ERK in the intestinal crypts after exposed to Lacbacillus NK6. Murine intestinal crypts were stimulated with Lacbacillus NK6 and lysed at the indicated time. Phosphor-p65, p38, -ERK and -JNK were detected using anti-phosphor-p-65, p38, -ERK or –JNK antibodies. Actin, a loading control, was detected by anti-β-actin antibody. (e) Analyses of TRAF2 ubiquitination in intestinal crypt cells after exposed to Lacbacillus NK6. Immunoblot analysis of ubiqitinated TRAF2 and TRAF2 immunoprecipitated from intestinal crypt cells stimulated with Lacbacillus NK6 at the indicated time (upper blots), and TRAF2, IκBα and actin (below blot) in the same cells without immunoprecipitation. IP, immunoprecipitation; IB, immunoblot assay. (f) Analyses of TRAF6 ubiquitination in intestinal crypt cells after exposed to Lacbacillus NK6. Immunoblot analysis of K63-linked ubiquitination (K63-Ub) (middle blot) of endogenouse TRAF6 immunoprecipitated from intestinal crypts stimulated with Lacbacillus NK6, and immunoblot analysis of Ub-TRAF6 (top blot), TRAF6, IkBα and actin (below blots) in the same cells without immunoprecipitation. PolyUbi., polyubiquitination; IP, immunoprecipitation; IB, immunoblot assay. *P < 0.05 and **P < 0.01 (t-test in a, b, c, mean ± SD). The data were at least a representative of three independent experiments.
Discussion
Paneth cells may produce FABP4, adipsin and adiponectin, which can regulate multiple physiological functions and are implicated in the pathogenesis of clinical entities, such as the metabolic syndrome in autocrine, paracrine and endocrine manners. Therefore, these cells may be considered as a modulator of systemic metabolism. The regulation of FABP4, adipsin and adiponectin in gut epithelial Paneth cells by gut microbiota Lactobacillus may offer a clue to disclose the association between gut microbiota and metabolic syndrome. Our studies suggest a new mechanism for systemic metabolism from gut microbiota, FABP4, adipsin and adiponectin in gut epithelial Paneth cells to metabolic syndrome. Because of the less bias of intestinal crypt cells distribution than that of adipose tissues between obese and lean people, our findings explain the disjunction between metabolism malfunction and obesity level in some cases. Finally, the observed abnormal state of Paneth cells and the metabolism associated factors in some metabolism related diseases also stimulates an important consideration regarding therapeutic strategies for these diseases.
Methods
Reagents
Anti-mouse FABP4 (ab13979, Abcam), adipsin (M-120, Santa cruz), adiponectin (19F1, Abcam), FABP2 (9A9B7B3, Milipore), LGR5 (EPR3065Y, Abcam), lysozyme (EPR2994, Abcam), CD24 (Mi/69, Abcom), lysozyme (W-20, Santa), p38(D13E1,Cell Signaling), JNK(sc-137020, Santa), ERK(197G2,Cell Signaling), STAT3 (124H6, Cell Signaling), phosphorylated-STAT3 (EP2147Y,Abcam), phosphorylated-p38 (pp38) (D3F9,Cell Signaling), phosphorylated -JNK (pJNK)(sc-81502,Santa), phosphorylated- ERK (pERK) (D13.14.4E,Cell Signaling), phosphorylated -IκBα (pIkBα) (14D4,Cell Signaling), IκBα (L35A5,Cell Signaling), p-65 (sc8008, Santa),phosphorylated-p-65 (sc-52401,Santa), ubiquitin (YT4793,Immunoway), K63-Ub (HWA4C4,Enzo), K48-Ub (D9D5,Cell Signaling), TRAF2 (EPR6048, Epitomics), TRAF6 (sc-8408,Santa), β-actin (sc-47778,Santa) antibodies, IFKine green conjugated anti-goat IgG (Abbkine), IFKine red conjugated anti-rabbit IgG (Abbkine),
IFKine red conjugated anti-mouse IgG (Abbkine), fluorescein-conjugated anti-mouse IgG (ZSGB-BIO), fluorescein-conjugated anti-rat IgG (ZSGB-BIO), rhodamine-conjugated anti-rabbit IgG (ZSGB-BIO), and DAPI staining reagent (D9542,Sigma) were purchased. Vitamin-A, 9-cis-retinoid acid (9-cis), all-trans retinoid acid (ATRA) were from Sigma and solved in sunflower seed oil. Dithizone were from Sigma. The concentration of antibodies used in this study was based on the company instruction.
Mice and human tissues samples
4–6-week-old male or female C57BL/6 and BALB/c mice were from Beijing Animal Center. TLR2 deficient (−/−) mice and Ob/Ob mice were from Nanjing Animal Center (Nanjing, China). NF-κB deficient (−/−) mice were offered by Prof. Zhexong Lian in Chinese Technique University (Anhui, China). These mice were bred and maintained under specific pathogen-free conditions at the animal facility of Nankai University (Tianjin, China). Germ-free (GF) mice were generated by the Fourth Military Medical University, bred and maintained at the animal facility of the Fourth Military Medical University. Experiments were carried out using age and gender matched groups. All procedures were conducted according to the Institutional Animal Care and Use Committee of the Model Animal Research Center. Animal experiments were approved by the Institute’s Animal Ethics Committee of Nankai University. The experiments for human tissue samples were approved by the Institute’s Ethics Committee of Nankai University and conducted according to the Institutional Use Committee of the Human Tissue Samples. The colon tissues from healthy individuals (gut disease-free) and patients with Crohn’s disease were offered by Tianjin People Hospital. These experiments were approved by all subjects.
Experimental mice
For microbiota transplantation mice, caecal contents were pooled from five WT mice. Each caecal content (150 mg) was sampled in an anaerobic chamber and suspended in 1 mL PBS (phosphate buffered saline) and intragastrically administered (0.1 mL per mouse) immediately to sterilely-packed 6–7 week old germ-free mice three times per week for 4 weeks. Lacbacillus NK colony was selected and cultured in Lactobacillus selected medium (Barebio., China) and then suspended in PBS and intragastrically administered (1 × 109 CFU/mouse) immediately to sterilely-packed 6–7 week old mice three time per week for four weeks.
For antibiotics-treated mice, 6- to 8-week-old mice were treated with ampicillin (A, 1 g/L, Sigma), vancomycine (V, 0.5g/L), neomycin sulfate (N, 1 g/L), and metronidazole (M, 1g/L) in drinking water for 4 weeks23 via the drinking water. Water containing antibiotic was exchanged every three days. To confirm the elimination of bacteria, stool was collected from antibiotic-treated and untreated mice and cultured in anaerobic and aerobic condition. The bacteria were counted under microscope.
For the deletion of Paneth cells, mice were treated with dithizone (100 mg/kg, i.v.)24; whereas control mice were treated with Li2CO3 vehicle. Dithizone is dissolved as 10 mg/mL of final concentration in the saturated Li2CO3 (1 g/100 mL). Gut tissues and sera were collected respectively in 0 hrs, 6 hrs, 12 hrs and 6 days for further experiments.
For high-fat-diet mouse model, 6–8 week old mice were fed using high-fat diet (lipid, 18%, yolk powder 10%, cane sugar (10%), cholesterol 1%, bile salts (0.2%) and other basic substance (60.8%) for 3 months.
In vitro stimulation
For in vitro stimulation, gut epithelial crypts were stimulated using bacteria (crypt cells: bacteria = 1: 100) or 9-cis-retinoid acid (9-cis, 1μM), all-trans retinoid acid (ATRA, 2μM) for 12 hrs, then lysed for immunoblot of FABP2, FABP4, adipsin and adiponectin. To isolate crypts, samples were transferred to 5 mM EDTA in PBS (pH 8), followed by three 1 min shakings by hand, a 15-min incubation at 4 °C, and passage through 70-μm filters (BD Falcon) to collect the flowthrough. Fraction containing intact and isolated crypts were collected by centrifugation at 75 g for 5 min. at 4oC and washed with PBS. No adipocytes and macrophages were detected in the collected intestinal crypts.
To identify gut specific Lacbacillus colonies, which could mediate the expression of FABP4, adipsin and adiponectin in gut epithelial crypt cells, isolated bacteria colonies from the culture under strictly anaerobic conditions were killed by heating and then used as stimulator to induce the expression of FABP4, adipsin and adiponectin in the gut epithelial crypts. Finally, 18 bacteria colonies with or without stimulation ability were selected and sequenced.
Gut Lacbacillus colonies
For isolation of gut microbiota, the caecal contents from WT mice were serially diluted with PBS and seeded onto Lacbacillus selected culture plates. After culture under aerobic conditions or strictly anaerobic conditions at 37 °C for 24 hrs, individual colonies were picked up and cultured for an additional 2 or 4 days at 37 °C in Lacbacillus selected culture medium. The isolated colonies were collected into stock medium (10% glycerol) and stored at −80 °C. The sequences of the 16S rRNA of the isolated colonies were obtained by cycle sequencing and then were aligned with the 16S rRNA database of GeneBank using BLAST. Each inquiry gave 100 most similar sequence results including different bacterial genera. For each genus, the one bacterial strain with the highest Max Score was selected and its sequence was downloaded. Next, all obtained sequences were aligned by MUSLE and then neighbor-joining method with a bootstrap of 1000 replicates was used to construct the phylogenetic tree.
Histological and immunostaining
For histological and immunostaining analysis, previously reported protocol25 was used in this study. Briefly, tissues were fixed in 10% formalin-buffered saline and embedded in paraffin, 5-μm-thick sections were prepared from embedded tissue and fixed in acetone (−20 °C) for 10 min. After rehydration in PBS for 5 min and further washing in PBS, tissue sections were blocked with 1% (w/v) BSA and 0.2% (w/v) milk powder in PBS. The primary antibody was added and incubated overnight at 4 °C. After PBS washing (three times, 5 min each), tissue was detected with DAB kit or fluorescence labeled second antibody. Notably, intestinal epithelial Paneth cells could be stained using anti-lysozyme antibody; whereas putative intestinal stem cells, which were localized at the crypt base, were stained by anti-LGR5 antibodies. Nuclei were stained by DAPI.
For immune-staining of intestinal epithelial cells, intestinal epithelial cells (Tongwei company, Shanghai, China) were seeded onto coverslips in 24 well dishes and were grown for 24 hrs, and then the cells were stained with different antibody and second fluorescence-conjugated antibody and then counterstained for nucleic acid with DAPI. Nuclei were stained with DAPI (blue). Cells were analyzed with a Zeiss LSM 710 laser-scanning confocal microscope.
RT-PCR and qRT-PCR
RT-PCR and qRT-PCR were performed using previously reported protocol25. Briefly, semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qRT-PCR) were performed. Total RNA was extracted from the cells, tissues and organs using TRIzol reagent (Invitrogen Corp). First-strand cDNA was generated from the total RNA using oligo-dT primers and reverse transcriptase (Invitrogen Corp). The PCR products were visualized on 1.0% (wt/vol) agarose gel. Real-time PCR was conducted using QuantiTect SYBR Green PCR Master Mix (Qiagen) and specific primers in an ABI Prism 7000 analyzer (Applied Biosystems). GAPDH mRNA expression was detected using each experimental sample as an endogenous control. The fold changes were calculated using the ∆∆Ct method according to the manufacturer’s instructions (Applied Biosystems). All the reactions were run in triplicate. Following primers were used: adiponectin, 5′TGTTGGAATGACAGGAGCTG and 5′CGAATGGGTACATTGGGAAC; adipsin, 5′GCTATCCCAGAATGCCTCGTT and 5′GGTTCCACTTCTTTGTCCTCGTAT; FABP4, 5′ATGATCATCAGCGTAAATGG and 5′GCCTTTCATAACACATTCCA; FABP2, 5′TTGCTGTCCGAGAGGTTTCT and 5′GCTTTGACAAGGCTGGAGAC; Adipocyte enhancer-binding protein 1 (AEBP-1), 5′CACGTTCCTCGCGCCCTTTC and 5′GTTGCACCATGAATCAGGTC; perilipin1(PLIN1), 5′GGGACCTGTGAGTGCTTCC and 5′GTATTGAAGAGCCGGGATCTTTT; FXIIIA, 5′CAGAGAGACTACCAGAGCACC and 5′GCATTGGAGTTATTGGGCGG; CD11b, 5′CTCCATGCATTGACCTCCCC and 5′GGCATTGGTCACAGGCAAGA; GAPDH, 5′TCAACGGCACAGTCAAGG and 5′TACTCAGCACCGGCCTCA.
Immunoprecipitation and immunoblot
Immunoprecipitation and immunoblot were performed according to the previously reported protocol25. Briefly, the cells were lysed with cell-lysis buffer (Cell Signaling Technology), which was supplemented with a protease inhibitor ‘cocktail’ (Calbiochem). The protein concentrations of the extracts were measured using a bicinchoninic acid assay (Pierce). Immunoprecipitation (IP) was performed essentially the same as described by the manufacturer (Thermo Scientific, USA). Cell lysates were preabsorbed with protein A/G agarose beads at 4 °C for 1 h. After centrifuging, the supernatants were incubated with antibody overnight at 4oC followed by incubation with protein A/G agarose beads (Santa Cruz biotechnology) for 2 hr at 4 °C. For the immunoblot, hybridizations with primary antibodies were conducted for 1 h at room temperature in blocking buffer. The protein-antibody complexes were detected using peroxidase-conjugated secondary antibodies (Boehringer Mannheim) and enhanced chemiluminescence (Amersham).
Serum FABP4, adipsine and adiponecin
Commercial kits for FABP4 (CY-8077, CycLex), adipsine (KA3822, Abnova) and adiponectin (A05187, bertinpharma) were used to quantitate mouse serum FABP4, adipsin and adiponecin. The absorbance on a microplate reader (Labsystems Dragon Wellscan MK2) were detected at a wavelength of 450 nm. The serum levels of FABP4, adipsin and adiponecin were quantified from two to three titrations using standard curves.
Serum Glu, CHO and TG
Serum glucose (Glu), cholesterin (Cho) and triglyceride (TG) were analyzed using blood biochemical Analyzer (GRT-3006).
Data analysis
The statistical significance of the comparisons between the two groups was determined using a student’s t-test. The statistical significance of the comparisons between the multiple groups was determined using an ANOVA test. Serum FABP4, adipsin and adiponectin and serum glucose (Glu), cholesterin (Cho) and triglyceride (TG) were analyzed by a Mann-Whitney U test. A 95% confidence interval was considered significant and was defined as P < 0.05. *p < 0.05, **p < 0.01, ***p < 0.001.
Additional Information
How to cite this article: Su, X. et al. Expression of FABP4, adipsin and adiponectin in Paneth cells is modulated by gut Lactobacillus. Sci. Rep. 5, 18588; doi: 10.1038/srep18588 (2015).
Supplementary Material
Acknowledgments
This research was supported by NSFC grants 31470876, 91029736, 91442111 and ISF-NSFC program 31461143010; a Ministry of Science and Technology grant (863 program, 2008AA02Z129); and the National Key Scientific Program (2011CB964902). The Program for Changjiang Scholars and Innovative Research team in University (No. IRT13023) and State Key Laboratory of Medicinal Chemical Biology.
Footnotes
Author Contributions R.Y. designed the research and wrote the paper; H.Y., Y.H., B.Z., E.W., H.Y. and Y. Z. conducted in vivo experiments and immunoassay, participated in the study design and performed the statistical analysis; X.S., Y.H., H.Y., E.W. and H.Y. carried out in vitro assay; Y.C. offered assistances for animal experiments; Z.Z., Y.L. and F.L. read and gave some suggestions for manuscript. All authors read and approved the final manuscript.
References
- Ghoshal K. & Bhattacharyya M. Adiponectin: Probe of the molecular paradigm associating diabetes and obesity. World J Diabetes 6, 151–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M., Saitoh S., Shimamoto K. & Miura T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clin Med Insights Cardiol 8, 23–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo J. C. et al. Adipsin is an adipokine that improves beta cell function in diabetes. Cell 158, 41–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T. et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 7, 941–946 (2001). [DOI] [PubMed] [Google Scholar]
- Lesniewski L. A. et al. Bone marrow-specific Cap gene deletion protects against high-fat diet-induced insulin resistance. Nat Med 13, 455–462 (2007). [DOI] [PubMed] [Google Scholar]
- Weiss R., Bremer A. A. & Lustig R. H. What is metabolic syndrome, and why are children getting it? Ann N Y Acad Sci 1281, 123–140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie A. G. et al. The genes influencing adiponectin levels also influence risk factors for metabolic syndrome and type 2 diabetes. Hum Biol 79, 191–200 (2007). [DOI] [PubMed] [Google Scholar]
- Chan J. M., Rimm E. B., Colditz G. A., Stampfer M. J. & Willett W. C. Obesity, fat distribution, and weight gain as risk factors for clinical diabetes in men. Diabetes Care 17, 961–969 (1994). [DOI] [PubMed] [Google Scholar]
- McLaughlin T. et al. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann Intern Med 139, 802–809 (2003). [DOI] [PubMed] [Google Scholar]
- Cadwell K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searfoss G. H. et al. Adipsin, a biomarker of gastrointestinal toxicity mediated by a functional gamma-secretase inhibitor. J Biol Chem 278, 46107–46116 (2003). [DOI] [PubMed] [Google Scholar]
- Meijer K. et al. Human primary adipocytes exhibit immune cell function: adipocytes prime inflammation independent of macrophages. PLoS One 6, e17154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashty M. et al. Characterization of coagulation factor synthesis in nine human primary cell types. Sci Rep 2, 787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi B. et al. Lipid droplets hypertrophy: a crucial determining factor in insulin regulation by adipocytes. Sci Rep 5, 8816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada M., Takahashi K., Sawada S. & Midorikawa O. Selective killing of Paneth cells by intravenous administration of dithizone in rats. Int J Exp Pathol 72, 407–421 (1991). [PMC free article] [PubMed] [Google Scholar]
- Sherman M. P., Bennett S. H., Hwang F. F., Sherman J. & Bevins C. L. Paneth cells and antibacterial host defense in neonatal small intestine. Infect Immun 73, 6143–6146 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshchenko V. A. [Damage to Paneth cells in rats following administration of dithizone and 8-(arensulfonylamino)-quinolines]. Biull Eksp Biol Med 83, 494–496 (1977). [PubMed] [Google Scholar]
- Bevins C. L. & Salzman N. H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9, 356–368 (2011). [DOI] [PubMed] [Google Scholar]
- Vaishnava S., Behrendt C. L., Ismail A. S., Eckmann L. & Hooper L. V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA 105, 20858–20863 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M. et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA 108 Suppl 1, 4623–4630 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y. et al. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta 1761, 736–744 (2006). [DOI] [PubMed] [Google Scholar]
- Tanaka M. et al. Spatial distribution and histogenesis of colorectal Paneth cell metaplasia in idiopathic inflammatory bowel disease. J Gastroenterol Hepatol 16, 1353–1359 (2001). [DOI] [PubMed] [Google Scholar]
- Fagarasan S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002). [DOI] [PubMed] [Google Scholar]
- Park S. W. et al. Paneth cell-mediated multiorgan dysfunction after acute kidney injury. J Immunol 189, 5421–5433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. et al. LRRC19 expressed in the kidney induces TRAF2/6-mediated signals to prevent infection by uropathogenic bacteria. Nat Commun 5, 4434 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.