Abstract
High-resolution mass spectrometry maps the cytotoxic T lymphocyte (CTL) proteome and the impact of mammalian target of rapamycin complex 1 (mTORC1) on CTLs. The CTL proteome was dominated by metabolic regulators and granzymes and mTORC1 selectively repressed and promoted expression of subset of CTL proteins (~10%). These included key CTL effector molecules, signaling proteins and a subset of metabolic enzymes. Proteomic data highlighted the potential for mTORC1 negative control of phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3) production in CTL. mTORC1 was shown to repress PtdIns(3,4,5)P3 production and to determine the mTORC2 requirement for activation of the kinase Akt. Unbiased proteomic analysis thus provides a comprehensive understanding of CTL identity and mTORC1 control of CTL function.
Systematic analyses of lymphocyte transcriptomes have yielded important insights about lymphocytes1. However, changes in rates of protein synthesis/turnover create discordances between transcriptomes and proteomes2,3 and there is the need for quantitative proteomics mapping of cellular protein signatures to fully define cell identity4,5. In this context, the serine/threonine kinase mTOR complex 1 (mammalian target of rapamycin complex 1), controls mRNA translation and protein degradation and controls CD8+ cytotoxic T lymphocyte (CTL) differentiation6,7,8. mTORC1 has two known substrates in T cells: p70 S6-Kinase 1 (S6K1) and eIF4E-binding protein 1 (4EBP1), molecules that regulate protein production9. Moreover, one mTORC1 role is to control the translation of mRNAs with 5′-terminal oligopyrimidine (5′-TOP) motifs that encode ribosomal proteins and translation factors to globally enhance cellular protein synthetic capacity10. Understanding mTORC1 function in CTLs thus requires an understanding of how mTORC1 controls proteomes. For example, recent studies showed mTORC1 translational control of the sterol regulatory element-binding proteins (SREBP1 and 2), which mediate expression of sterol biosynthesis enzymes11,12. mTORC1 translational control of the hypoxia-inducible factor 1 (HIF1) transcription factor complex also directs expression of glucose transporters, glycolytic enzymes and cytolytic effector molecules in CTLs13. The relevance of proteomics to understand the impact of mTORC1 in CTLs also stems from the ability of mTORC1 to promote protein degradation. There are thus examples in other cell lineages where mTORC1 regulated phosphorylation of adapter proteins, such as either growth factor receptor-bound protein 10 (GRB10), or insulin receptor substrate (IRS) 1 or 2, modulates the degradation rates of these proteins14,15,16.
A comprehensive analysis of mTORC1 control of T cell proteomes will hence directly inform how mTORC1 controls T cell biology. Accordingly we have used high-resolution mass spectrometry (MS) to map the proteome of CTL and to quantify the regulatory impact of mTORC1 and mTOR inhibition on CTL proteomes. We reveal the CTL proteome diversity and reveal how mTOR inhibitors control T cell function and program T cell signal transduction pathways.
Results
The CTL proteome
High resolution mass spectrometry characterized the proteome of P14 TCR transgenic CTLs (Supplementary Fig. 1), identifying more than 93,000 peptides from 6,800 protein groups in these cells (Fig. 1a). iBAQ intensities, obtained by dividing the summed MS peptide-derived ion extracted ion chromatograms by the theoretically observable numbers of peptides, measure relative protein abundance2,5 and can be transformed into absolute quantification using proteomic ruler methodology17. Copy numbers for proteins from three biological replicates showed strong Pearson correlation coefficients (0.86–0.89), with very few outliers indicating robustness and reproducibility of our MS-based peptide quantitation methods (Fig. 1b).
Figure 1. The cytotoxic T cell proteome.
(a) Scatter plots of estimated protein copy numbers using the proteomic ruler approach show high reproducibility of protein intensities and ~94% of identified proteins are detected in all three biological replicates. R2 = coefficient of determination. (b) CTL proteins ranked by abundance as estimated by mean iBAQ intensities and plotted against the cumulative protein abundance. The proteomic ruler protocol was used to quantify mean protein copy number and relative abundance based on iBAQ intensities17. The 12 most abundant proteins contribute 25% of the CTL proteome; 64 and 249 proteins contribute to 50% and 75% of the CTL proteome. (c) Histogram of log-transformed mean protein copy number quantified using the proteome ruler. Protein expression levels span nearly seven orders of magnitude. Intensity quartiles are depicted in different colors and enriched KEGG pathways (p<0.01, Bonferroni corrected) are displayed above each quartile. The contribution of the most abundant KEGG pathways to the total CTL proteome in terms of molecules or mass is shown in the table. Mean iBAQ values and copy numbers are based on three biological replicates.
Proteomic data revealed protein abundance and specific protein isoforms/orthologues creating an objective description of cell ‘identity’ We ranked CTL proteins by estimated copy number and plotted this against cumulative protein copy number (Fig. 1c). Proteins showed a wide range of expression spanning over seven orders of magnitude. 25 percent of the CTL protein mass comprised 12 proteins; 249 proteins constituted 75% of the total CTL mass; 6562 proteins contributed to the remaining 25% of the CTL. The 20 most abundant CTL proteins included histones and cytoskeleton components vimentin and cofilin (Table 1). They also included translational machinery proteins, ribosomal proteins, initiation and elongation factors. The CTL effector molecule granzyme B and multiple glycolytic enzymes were in the top 20 list (Table 1) and the highest intensity quartile of the CTL proteome was enriched in pathways involved in metabolism and macromolecular biosynthesis (Fig. 1c). As CD8+ T cells differentiate to CTL they switch from metabolizing glucose primarily through oxidative phosphorylation to using the glycolytic pathway18. Proteomic data showed much of the CTL protein mass is dedicated to glycolysis although CTLs retained abundant amounts of the protein machinery for oxidative phosphorylation suggesting that it may be important for them to retain flexibility in terms of their metabolic strategy for glucose metabolism (Fig. 1c).
Table 1.
The 20 most abundant CTL comprising nearly a third of all proteins in CTL are shown. %: percentage of protein relative to total cellular protein pool; cum. %: cumulative relative abundance of all proteins up to that rank.
| rank | protein name | gene name | copy number | % | cum. % |
|---|---|---|---|---|---|
| 1 | Histone H2B | Hist1h2bb | 7.9 × 107 | 5 | 5 |
| 2 | Histone H4 | Hist1h4a | 6.1 × 107 | 4 | 8 |
| 3 | Actin | Actb | 5.0 × 107 | 3 | 11 |
| 4 | Thymosin beta-4 | Tmsb4x | 4.7 × 107 | 3 | 14 |
| 5 | Cofilin-1 | Cfl1 | 3.0 × 107 | 2 | 15 |
| 6 | Histone H2A | Hist1h2ab | 3.0 × 107 | 2 | 17 |
| 7 | Peptidyl-prolyl cis-trans isomerase A | Ppia | 2.8 × 107 | 2 | 19 |
| 8 | Alpha-enolase | Eno1 | 2.5 × 107 | 1 | 20 |
| 9 | Vimentin | Vim | 2.4 × 107 | 1 | 22 |
| 10 | Granzyme B | Gzmb | 2.2 × 107 | 1 | 23 |
| 11 | Profilin-1 | Pfn1 | 2.1 × 107 | 1 | 24 |
| 12 | 60S acidic ribosomal protein P2 | Rplp2 | 2.1 × 107 | 1 | 25 |
| 13 | Histone H3.2 | Hist1h3b | 1.8 × 107 | 1 | 26 |
| 14 | Histone H1.2 | Hist1h1c | 1.7 × 107 | 1 | 27 |
| 15 | Phosphoglycerate kinase 1 | Pgk1 | 1.7 × 107 | 1 | 28 |
| 16 | Elongation factor 1-alpha 1 | Eef1a1 | 1.5 × 107 | 1 | 29 |
| 17 | L-lactate dehydrogenase A chain | Ldha | 1.5 × 107 | 1 | 30 |
| 18 | Eukaryotic translation initiation factor 5A-1 | Eif5a | 1.4 × 107 | 1 | 31 |
| 19 | Fructose-bisphosphate aldolase A | Aldoa | 1.4 × 107 | 1 | 32 |
| 20 | Heat shock cognate 71 kDa protein | Hspa8 | 1.3 × 107 | 1 | 32 |
The proteomic data revealed new insights concerning protein isoform/orthologue expression in T cells. For example, CTLs expressed multiple nutrient transporters (Fig. 2a), but, in terms of abundance, the System L amino acid transporter SLC7A5 and its dimer partner SLC3A2 predominated. This finding explains why SLC7A5 deletion has such a severe impact on CTL function19. Glucose transport is important for CTL20 and previous studies have focused on GLUT1 in T cells21.We showed that GLUT3, which has a higher glucose transport capacity than GLUT122, was expressed at equivalent amounts to GLUT1 in CTLs (Fig. 2a). GLUT1 deletion impacts on T cell function21, but the presence of GLUT3 explains why GLUT1 loss is not catastrophic.
Figure 2. Comparison of the abundance of key CTL proteins.
Histograms of log-transformed mean estimated copy numbers of CTL proteins using the proteomic ruler methodology. Quantification accuracy based on number of detected peptides, fraction of unique/Razor peptides to total peptides and theoretically observable peptides. (a) Key nutrient transporters, (b) CTL effector molecules, (c) transcription factors, (d) tyrosine kinases and phosphatases involved in TCR and IL-2 receptor signaling, (e) IL-2 receptor subunits and associated tyrosine kinases. Protein = protein name, copies = Mean estimated copy number/cell, CV = coefficient of variation, QA = quantification accuracy. Mean copy numbers and CV are based on three biological replicates. Copy numbers for all CTL proteins can be assessed by using the Encyclopedia of Proteome dynamics (http://www.peptracker.com/epd)
We have also used proteomic ruler methodology17 to estimate absolute protein copy numbers to quantify the key cytokine receptors, transcription factors and effector molecules that define CTL identity. Granzymes A and B were present in high copy number in CTLs (4.9 × 106 and 2.2 × 107, respectively, (Fig. 2b)), which explains how CTLs can rapidly kill multiple targets. We observed a wide range in transcription factor copy numbers (Fig. 2c): STAT1, STAT3 and STAT5 copy numbers were high (104–105) compared to T-bet, Foxo1/3, EOMES, STAT4 or STAT6 (103–104). The high copy number of antigen receptor coupled tyrosine kinases Lck and Zap70, compared with the cytokine receptor coupled kinases JAK1/3 and TYK2 was notable (Fig. 2d). Intriguingly, the tyrosine phosphatases CD45 and SHP-1 were expressed at similar abundance to these tyrosine kinases, indicating the importance of negative regulators in intracellular signaling networks. The data also revealed cytokine receptor subunit stoichiometry. For example, interleukin 2 (IL-2) signals to T cells via a high-affinity receptor comprising CD25 (IL-2 receptor alpha chain, IL-2RA), IL-2 receptor beta subunit (IL-2RB or CD122) and the common gamma subunit γc (CD132). CTLs expressed approximately ~100-fold higher copies of CD25 compared to the IL-2RB and γc subunits (Fig. 2e). Earlier studies have reported excesses of CD25 compared to numbers of high-affinity IL-2 receptor complexes on CTL membranes23 and an excess of CD25 over IL-2RB was quantified using flow cytometry24. The present data revealed IL-2RB and γc subunit numbers were limiting at approximately 104 copies of IL-2RB and 2-3×104 γc molecules per cell. IL-2RB and γc subunits bind to JAK1 and JAK3, respectively, and the copy number for these kinases was broadly equivalent to IL-2RB–γc subunit numbers indicating that high affinity IL-2 receptor formation in CTL is limited by availability of IL-2RB and γc and associated tyrosine kinases. These examples all illustrate how understanding protein copy number can afford new insights about cell identity and cellular control mechanisms.
A comparison of the CTL transcriptome and proteome
Systematic analysis of transcriptomes has yielded critical insights about how T cells direct adaptive immune responses25. We assessed whether proteomic data provided additional insights by correlating CTL estimated protein copy numbers with the transcript intensities of corresponding mRNA by using the probe intensities derived from a parallel Affymetrix microarray data set (Fig. 3a). The rather moderate positive correlation between mRNA and protein abundance (Fig. 3a), indicated that post-transcriptional regulatory mechanisms significantly affect the CTL proteome. Examples of discordance between mRNA and protein abundance include that CTLs expressed comparable levels of T-bet and EOMES mRNAs, whereas T-bet protein was much more abundant than EOMES (Fig. 3b). In a second example, the ratio of IL-2 receptor α:β:γ subunits estimated from transcript intensity was 3:1:2, whereas the protein intensity ratio was 92:1:2 (Fig. 3c). There was close correspondence between protein and transcript abundance for some proteins: e.g. ribosomal proteins and granzymes (Fig. 3d, e). Nevertheless the data highlight the importance of direct protein measurements, rather than mRNA measurements as a surrogate, for estimating protein expression.
Figure 3. Comparison of the CTL transcriptome and proteome.
(a) Scatter plots show mean Affymetrix microarray transcript intensities plotted against the corresponding estimated mean copy numbers for CTL proteins. The data show a moderate overall correlation of protein abundance with transcript abundance R2 = coefficient of determination of 0.43. (b, c) Discordance of mean transcript and protein levels for (b) transcription factors T-bet and Eomes and (c) IL-2 receptor subunit α vs β and γc. (d, e) Tight control and correlation of transcript and protein levels for subunits of ribosome protein complexes (d) or granzyme isoforms (e). (b) P-values determined by two-sided, equal variance t-test on transcript intensities or protein copy numbers, respectively. All data are based on three biological replicates.
mTORC1 selectively programs the CTL proteome
CTLs had high mTORC1 activity as judged by high content of phosphorylation of the mTORC1 substrates S6K1 (T389) and 4EBP1 (S37/46, S65) (Fig. 4a). Treatment of CTLs with the mTORC1 inhibitor rapamycin caused dephosphorylation of these two substrates (Fig. 4a). mTOR exists in two protein complexes, mTORC1 and mTORC2, that are defined by their scaffolding/regulator components. CTLs had high activity of mTORC2 as judged by high levels of phosphorylation of the mTORC2 substrate Akt at S473. Rapamycin did not cause dephosphorylation of Akt p-S473 (Fig. 4a). In contrast, an mTOR catalytic inhibitor, KU-0063794, that blocks the activity of both mTORC1 and mTORC2 (ref 26) caused dephosphorylation of Akt p-S473 and p-S6K1 T389 and p-4EBP1 (Fig 4a).
Figure 4. The mTORC1 regulated CTL proteome.
(a) Immunoblot analysis of mTORC1/2 substrates in P14 TCR transgenic CTLs cultured with IL-2/IL-12 ± 48 h treatment with either rapamycin or KU-0063794. (b) Protein synthesis was examined by monitoring 3H-Met incorporation into nascent proteins in CTLs cultured in IL-2/IL-12 and treated with rapamycin for the indicated time. (c) Cellular protein content of CTLs ± 48 h rapamycin. (d, e,) Volcano plots showing fold changes in proteins vs. log-transformed P-values from mass spectrometry analysis of CTLs ± 48 h rapamycin. (d) Total proteins. Known rapamycin sensitive proteins perforin and L-selectin are highlighted. (e) CTL effector molecules. (f) IFN-γ secretion by CTLs ± 48 h rapamycin measured by ELISA. (h) Immunoblot analysis of T-bet in CTLs ± 48 h rapamycin. (h, i) Validation of up-regulated proteins: (h) ELISA of shed CD62L in cell supernatants prepared from CTLs ± 48 h rapamycin. (i) Immunoblot analysis of IRS2 in CTLs ± 48 h rapamycin. (a, g, i): representive immunoblots of at least three biological replicates. (b, c, f, h): individual data points and means are shown. P-values shown determined by (b): one-way ANOVA (Holm-Sidak) vs. DMSO as control on non-normalized data; (c, f, h): two-tailed Student’s t-test. Data based on three (b, f, h) or four (c) biological replicates. *P<0.05, **P<0.01, ***P<0.001. (d, e,): Each data point represents mean fold change of three biological replicates vs. P-value determined by two-tailed, unequal variance t-test; measurements based on three biological replicates.
Rapamycin treatment decreased CTL protein synthesis over a 24–48 h period and decreased CTL size and protein content (Fig. 4b, c). Quantitative mass spectrometry was then used to assess whether mTORC1 inhibition caused a small reduction of all proteins or targeted a protein subset. Three biological replicates of normal versus rapamycin treated CTL were analyzed at a single time point of 48 hours to assess the sustained effect of mTORC1 inhibition. Rapamycin treatment controlled a small CTL protein subset and decreased expression of 413 proteins and increased expression of 427 proteins (Fig. 4d). Notably, mTORC1 inhibition decreased expression of multiple CTL effector molecules including granzymes, perforin, tumor necrosis factor (TNF) and interferon gamma (IFN-γ) (Fig. 4e). The decrease in IFN-γ production in rapamycin-treated CTLs was confirmed by ELISA (Fig. 4f). Previous studies reported mTORC1 control of IFN-γ by control of expression of the transcription factor T-bet27. We found no difference in T-bet expression between control and rapamycin-treated cells from mass spectrometry or immunoblot analysis (Fig. 4g). Notably, CD62L (L-selectin), an adhesion molecule that controls T cell trafficking into secondary lymphoid tissue, was strongly upregulated in rapamycin-treated CTLs (Fig. 4d): a result validated using ELISA (Fig. 4h). Studies in non-lymphoid cells have reported mTORC1 induced degradation of adapter proteins GRB10 (refs. 14,15) and IRS1/2 (ref. 16). GRB10 or IRS1 were not detected in CTLs but there was an accumulation of IRS2 in rapamycin-treated CTLs, which was validated by immunoblotting (Fig. 4i). In terms of signaling molecules, mTORC1 activity was required to sustain expression of the transcription factor NFIL3 and the PtdIns(3,4,5)P3-phosphatase PTEN.
mTORC1 was also required for the expression of glucose transporters and enzymes that control glycolysis (Fig. 5a), cholesterol biosynthesis enzymes (Fig. 5b), cytosolic aminoacyl-tRNA synthetases and cytosolic ribosomal subunits (Fig. 5c). Conversely, mTORC1 inhibition increased expression of CTL protein subsets including oxidative phosphorylation enzymes (Fig. 5d), mitochondrial aminoacyl-tRNA synthetases and ribosomes (Fig. 5e) and the guanine exchange factor DOCK1. mTORC1 is thus able to both positively and negatively regulate expression of a subset of CTL proteins and its role is selective and cell specific.
Figure 5. mTORC1 regulation of cellular pathways.
(a, b, c) Volcano plots of down regulated KEGG pathways in rapamycin treated CTLs: (a) glycolysis and glucose transporters, (b) terpenoid backbone and steroid biosynthesis with rate limiting enzyme HMGCR highlighted, (c) cytoplasmic subunits of ribosomes and aminoacyl-tRNA biosynthesis. (d, e) Volcano plots of up-regulated KEGG pathways in rapamycin treated CTLs: (d) oxidative phosphorylation, (e) mitochondrial subunits of ribosomes and aminoacyl-tRNA biosynthesis. Each data point represents mean fold change of three biological replicates vs. P-value determined by two-tailed, unequal variance t-test; measurements based on three biological replicates.
mTORC1 controlled transcriptomes versus proteomes
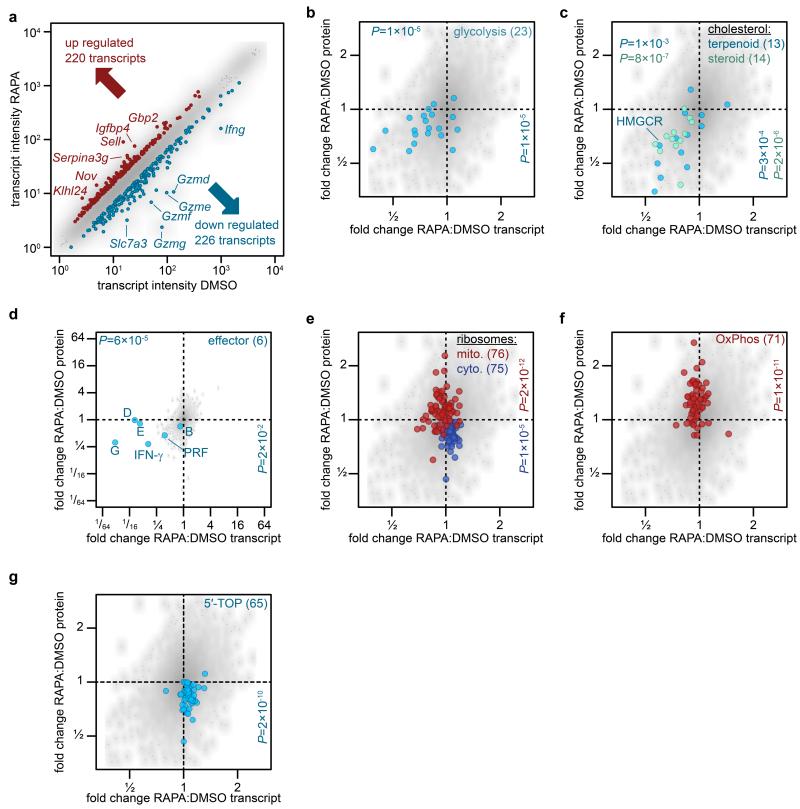
mTORC1 controls expression the transcription factors, SREBP1/2 and HIF-1α12,13. Affymetrix microarray analysis was thus used to assess the full extent of transcriptional changes caused by mTORC1 inhibition. Rapamycin treatment decreased expression of 226 mRNA transcripts and increased of 220 mRNA transcripts in CTLs (Fig. 6a). There was a strong correlation between the effects of mTORC1 inhibition on transcript and protein abundance for glucose transporters, glycolytic enzymes, cholesterol biosynthesis enzymes, granzymes, perforin and IFN-γ (Fig. 6b–d). However, of the 413 proteins down regulated in the rapamycin-treated CTLs, only 95 showed a corresponding change in transcript abundance. Similarly, of the 427 upregulated proteins, only 83 showed increased abundance of mRNA transcripts. mTORC1 inhibition thus regulated expression of cytoplasmic and mitochondrial subunits of ribosomal complexes, oxidative phosphorylation enzymes and proteins encoded by mRNA transcripts carrying a 5′-TOP motif10 at the protein but not the transcript level (Fig. 6e–g). Furthermore, rapamycin treatment of CTL regulated expression of IRS2, DOCK1, and PTEN at the protein but not mRNA level, highlighting the importance of direct proteomic analyses for cell phenotyping.
Figure 6. Comparison of the mTORC1 controlled transcriptome and proteome in CTL.
(a) Plot showing mean microarray probe intensities from RNA isolated from CTLs cultured in IL-2/IL-12 ± 48 h rapamycin. mTORC1 inhibition increased levels of 220 transcripts and decreased 226 transcripts (total of 8198 expressed transcripts). Examples of transcriptional (b-d) and non-transcriptional (e-g) regulation of protein expression by mTORC1. (b) Glycolytic enzymes and glucose transporters (c) terpenoid backbone and steroid biosynthesis pathways (SREBP1/2 targets), (d) cytolytic effector molecules (single letters indicate granzyme isoforms, PRF = Perforin; note different scaling of axes), (e) cytoplasmic and mitochondrial ribosomal subunits, (f) oxidative phosphorylation (OxPhos), (g) proteins encoded by mRNA containing 5′-TOP containing mRNA (as reported10). (b-g) P-values determined by Mann-Whitney U test vs. total transcriptomic (n=5516) or proteomic (n=6641) dataset, respectively. Horizontal values on the top indicate P-values determined from transcriptomic analysis, vertical values at the right for proteomic analysis. Number of transcript-protein pairs for each pathway given in brackets. All data points are based on three biological replicates and represent the mean fold change on transcript or protein level, respectively.
Selective programming of CTL metabolism by mTORC1
The present data show that only a small subclass of metabolic pathways were mTORC1 controlled in CTL, notably steroid biosynthesis and glycolytic pathways. CTLs express at least 72 nutrient transporters and only 6 of these were mTORC1 regulated (Fig. 7a) highlighting the selectivity of mTORC1 control of T cell metabolism. In particular, mTORC1 inhibition did not prevent expression of SLC1A5 (ASCT2) the key glutamine transporter in T cells28 (Fig. 7a) or decrease expression of enzymes that regulate glutamine metabolism (Supplementary Fig. 2). Indeed there was increased expression of some enzymes that control glutaminolytic reactions (e.g. GLUD1), in mTORC1-inhibited CTLs (Fig. 7b). We tested the relevance of these changes by measuring glutaminolysis activity and measured a higher glutaminolytic rate in mTORC1-inhibited CTLs (Fig. 7c).
Figure 7. Selective control of CTL metabolism by mTORC1.
(a) Volcano plot representation of mean fold changes vs. log-transformed P-values of SLC family members in control versus rapamycin treated CTLs. (b, c) Effects of mTORC1 inhibition in CTLs on glutaminolysis: (b) Volcano plot of changes of glutaminolytic proteins depicted in control versus rapamycin treated CTLs. (c) Glutaminolysis rates in CTLs ± 48 h rapamycin as quantified by measuring the formation of 14CO2 derived from radiolabelled L-glutamine. (d) Comparison of the expression of glycolytic (blue) and OxPhos (red) molecules in control versus rapamycin treated CTLs. (e) Pie charts showing the relative contribution of glycolytic pathway to overall CTL mass in control and rapamycin treated CTLs. (f) KEGG pathway analysis of highest intensity quartile in CTLs with enriched pathways (P<0.01). (g, h) Metabolic flux analysis of control versus rapamycin treated CTL: (g) oxygen consumption and (h) extracellular acidification rate of DMSO and rapamycin trated CTL. Oligomycin (oligo.), 2,4-dinitrophenol (DNP), antimycin A (AA) and rotenone (rot.) were added at indicated time points. (i) Glucose uptake levels in control vs. rapamycin treated CTLs. (j) Effects on decreased glucose flux on lactate output by CTLs. (a, b, d): Each data points represents mean fold change of three biological replicates; P-values determined by two-tailed, unequal variance t-test, measurements based on three biological replicates. (c, i, j): individual data points and means are shown. P-values for non-proteomic data (c, i) determined by paired t-test on non-normalized data. *P<0.05. (g, h) Data shown are mean ± SD. Data based on two (g, h), three (a-f, j) or five (i) biological replicates.
Another example of mTORC1 selectivity is that rapamycin caused loss of glycolytic enzymes but increased expression of oxidative phosphorylation enzymes (Fig. 7d). The ability of rapamycin to increase expression of oxidative phosphorylation enzymes is consistent with the ability of rapamycin to promote the development of memory CD8+ T cells7 who are dependent on oxidative phosphorylation rather than glycolysis29. The changes in glycolytic enzymes were statistically significant and systematic albeit not large (Fig. 7d). Inhibition of mTORC1 thus decreased expression of multiple glycolytic enzymes but these enzymes are still abundant. 8% of the proteome of mTORC1 inhibitor-treated CTLs thus consists of glycolytic enzymes (Fig. 7e). We then measured glycolysis and oxygen consumption rates (OCR) of normal and rapamycin-treated CTLs. We assessed the cells in a basal state and after the addition of oligomycin (to block ATP synthesis), DNP (to uncouple ATP synthesis from the electron transport chain, and antimycin A and rotenone (to block electron transport chain complexes) to assess the spare respiratory capacity (SRC) of mitochondria. The baseline data showed that CTLs consumed oxygen (Fig. 7g) and were glycolytic, as judged by their extracellular acidification rate (ECAR) (Fig. 7h). There was no difference between control and mTORC1-inhibited CTLs in baseline oxygen consumption, or spare respiratory capacity (Fig. 7g). However, rapamycin-treated CTLs had decreased rates of extracellular acidification reflecting that these cells have reduced rates of lactate output an indicator for their glycolytic activity (Fig. 7h). They have not however fully ablated lactate production. These metabolic data confirmed the prediction from the proteomic data that CTLs do oxidative phosphorylation and glycolysis and that mTORC1 modulates glycolysis in CTL.
What is the primary cause of the decreased glycolysis in mTORC1 inhibitor treated CTL given that these cells retain abundant expression of glycolytic enzymes. One important factor is that glucose transporters supply the glucose that fuels both oxidative phosphorylation and glycolysis. CTLs expressed 62,000 molecules of GLUT1 and 73,000 molecules of GLUT3 and these decreased to 36,000 and 48,000 molecules, respectively in the mTORC1-inhibitor treated CTLs, which correlated with a 2-fold difference in glucose uptake (Fig. 7i). In this respect, rates of lactate output in CTLs were very sensitive to reduced supply of glucose (Fig. 7j) arguing that reduced glucose supply and not the loss of glycolytic enzymes limits glycolysis in rapamycin-treated CTLs. Decreased glucose uptake in mTORC1-inhibited CTLs explains why there is no detectable increase in oxidative phosphorylation associated with their increased expression of oxidative phosphorylation enzymes.
mTORC1 restrains PI(3)K/Akt signaling in CTL
We consistently saw that PTEN protein was down regulated in rapamycin-treated CTLs and this was validated by immunoblot analysis (Fig. 8a). PTEN dephosphorylates (PtdIns(3,4,5)P3 and its loss raised the possibility that mTORC1 signaling normally restrains cellular accumulation of this lipid. We explored this directly and found that CTLs contained approximately 30,000 molecules of PtdIns(3,4,5)P3/cell (Fig. 8b). Inhibition of Phosphoinositide-3-kinase (PI(3)K) p110δ depletes PtdIns(3,4,5)P3 but rapamycin treatment increases cellular PtdIns(3,4,5)P3 (more than 60,000 molecules PtdIns(3,4,5)P3/CTL after sustained mTORC1 inhibition, (Fig. 8b)).
Figure 8. mTORC1 represses PIP3 production and controls the mTORC2 requirement for activation of AKT.
(a) Immunoblot analysis of PTEN expression in CTLs cultured ± 48 h rapamycin. (b) HPLC-MS based analysis of PtdIns(3,4,5)P3 levels in control IL-2/IL-12 maintained CTLs (DM) and CTLs treated with PI(3)K p110δ inhibitor IC-87114 (IC, 1 h) and rapamycin for the indicated times. (c) Immunoblot analysis of Akt T308 and S473 phosphorylation levels in CTLs ± 48 h rapamycin. (d, e) The data show immunoblots of Akt T308 phosphorylation in control and mTORC1 inhibited CTLs treated with (d) IC-87114 or (e) AKTi1/2. (f) Immunoblot analysis of PTEN expression in CTLs cultured ± 48 h rapamycin or KU-0063794. (g) HPLC-MS based analysis of PtdIns(3,4,5)P3 levels in control CTLs and CTLs treated with KU-0063794 for indicated durations. (h) Immunoblot analysis of Akt T308, Akt S473, FOXO1/3A T24/T32 phosphorylation and phosphorylation of the mTORC1 substrates S6K T389 and 4EBP1 S65 in CTLs treated with KU-0063794 for the indicated times. (i) Scatter plot depicting correlation of mean (n=3) fold changes in transcript expression from Affymetrix microarray analysis of control CTLs vs CTLs treated with rapamycin (x-axis) or KU-0063794 (y-axis). Immunoblots are representative of at least three biological replicates. (b, g) Individual data points and means are shown. P-values are determined by one-way ANOVA (Holm-Sidak) vs. DMSO (DM) as control. *P<0.05, **P<0.01. Data based on biological triplicates.
PtdIns(3,4,5)P3 binds to the plextrin homology (PH) domain of Akt allowing the kinase PDK1 to phosphorylate Akt T308, thereby activating the enzyme30. Rapamycin-treated CTLs had more p-T308 as compared to untreated CTLs (Fig. 8c) and this phosphorylation was lost when the rapamycin-treated CTLs were treated with a PI(3)K p110δ inhibitor (Fig. 8d), or with AKTi1/2 (Fig. 8e), which prevents PtdIns(3,4,5)P3 binding the Akt PH-domain31. The increased PtdIns(3,4,5)P3 content in rapamycin-treated CTLs thus increased Akt activity revealing that mTORC1 activity limits Akt function in CTLs.
Many signaling models position mTOR as a positive regulator of Akt. This is because mTORC2 can phosphorylate Akt S473 (ref. 32) creating a docking site for the PDK1 PIF-pocket promoting efficient PDK1 phosphorylation of Akt T308 and activating the enzyme33. In this context, Rictor-deficient T cells that lack mTORC2 reduce S473 and T308 phosphorylation indicating that the docking of phosphorylated Akt S473 to the PDK1 PIF-pocket can control Akt activity in T cells34. However, PDK1 contains a PtdIns(3,4,5)P3 binding PH-domain and PtdIns(3,4,5)P3 mediated co-localization of Akt and PDK1 can occur making Akt activation independent of Akt S473 phosphorylation35.
The mTOR catalytic inhibitor KU-0063794 was as effective as rapamycin in blocking mTORC1 activity and down regulating expression of PTEN (Fig. 8f). KU-0063794 also caused CTLs to accumulate PtdIns(3,4,5)P3 (Fig. 8g). High levels of PtdIns(3,4,5)P3 could switch the balance of whether Akt activity is regulated by the PIF-pocket dependent mechanism for AKT activation versus using a PDK1 PH-domain dependent mechanism. In this respect, the integrity of the PDK1 PH domain is required for optimal Akt activation in CTL36. Therefore, to address the role of mTORC2/PIF-pocket dependent Akt activation in CTLs we examined the effect of KU-0063794 on Akt in CTLs over an 18 h period. KU-0063794 caused a rapid and sustained loss of mTORC1 activity and mTORC2 mediated Akt S473 phosphorylation (Fig. 8h). The impact of KU-0063794 on Akt T308 phosphorylation, however, was biphasic. The rapid de-phosphorylation of Akt S473 in KU-0063794 treated cells was thus initially accompanied by Akt T308 de-phosphorylation and loss of Akt catalytic activity as monitored by the loss of phosphorylation of the Akt substrate: T24 and T32 in Foxo1 and 3A, respectively. Hence treatment of CTLs with an mTOR catalytic inhibitor causes a rapid loss of Akt activity. However, the loss of Akt T308 phosphorylation in KU-0063794-treated CTLs was transient and Akt T308 phosphorylation was restored after approximately 6 h of drug treatment (Fig. 8h). This Akt T308 re-phosphorylation was paralleled by restoration of Akt activity as judged by corresponding re-phosphorylation of Foxo1/3A T24/32. At the 18 h time point, Akt T308 phosphorylation and Akt activity was enhanced compared with the control CTLs despite the absence of any detectable phosphorylation of Akt on S473. Akt activity in CTLs is thus normally dependent on mTORC2-mediated phosphorylation of Akt S473. However, in mTOR inhibitor-treated CTLs there was reprogramming event such that Akt activity became independent of mTOR controlled Akt S473 phosphorylation. The importance of Akt S473 phosphorylation in T cells thus depended on cellular concentrations of PtdIns(3,4,5)P3. If these are high then phosphorylation of Akt S473 was not required for Akt T308 phosphorylation, or catalytic activity (Supplementary Fig. 3).
These result argued that mTORC1 control of PtdIns(3,4,5)P3 dominated mTORC2 control of Akt in CTLs, such that catalytic inhibitors of mTOR did not effectively disrupt Akt activity. In this context, inhibition of Akt in CTLs caused re-expression of Foxo-regulated genes37. However, there was no re-expression of Foxo-regulated genes in KU-0063794-treated CTLs (Table 2) supporting the conclusion that mTOR inhibitors did not disrupt Akt signaling in CTLs. Moreover, we found no difference in the transcriptional changes induced in CTLs by rapamycin inhibition of mTORC1 versus mTOR catalytic inhibition with KU-0063794 (Fig. 8i). A comparison of the effects of rapamycin and KU-0063794 on the CTL proteome using quantitative mass spectrometry also found no major difference between the effects of rapamycin and KU-0063794. (Supplementary Fig. 4) Hence, the mTOR catalytic inhibitor blocked mTORC1 and mTORC2 activity but there was no discernable impact of additional (on or off target) effect of this inhibitor on the T cells compared to loss of mTORC1 activity alone.
Table 2.
Effects of rapamycin vs. KU-0063794 vs. AKTi1/2 inhibition on key Foxo-regulated genes in CTL as determined by microarray analysis. Mean fold changes of microarray transcript intensity of inhibitor treated vs. respective controls are shown. AKTi1/2 data obtained from previous study37.
| Gene | Rapamycin | KU-0063794 | AKTi1/2 |
|---|---|---|---|
| Klf2 | 0.93 | 1.3 | 5.8 |
| Il7r | 0.88 | 0.95 | 2.2 |
| Ccr7 | 1.9 | 1.3 | 3.1 |
| S1pr1 | 0.91 | 0.89 | 2.6 |
All proteomic data presented are available in the Encyclopedia of Proteome Dynamics (http://www.peptracker.com/epd), to maximize accessibility to the scientific community (Supplementary Fig. 5).
Discussion
In this study we have characterized the CTL proteome, mapping the abundance and isoform/orthologue expression of more than 6800 proteins. These proteomic data are available in an online, searchable database, the Encyclopedia of Proteome Dynamics (http://www.peptracker.com/epd). The abundance of some CTL proteins was striking: granzymes comprised collectively 1-2% of the CTL proteome and 9% of the CTL proteome comprised glycolytic enzymes. The threshold of molecules needed for function is often unknown but there is undoubtedly a new perspective to considering the biological relevance of changes in protein expression when protein abundance is factored into the equation. E.g. a 100-fold decrease in granzyme B and perforin would reduce granzyme B to ~105 molecules/CTL and perforin to approximately ~100 copies/CTL; a case in which granzyme B is still abundant whereas perforin seems limiting. In another example, CD25, the IL-2 receptor α chain is expressed ~100-fold excess to the IL-2 receptor β/γc subunits. CD25 levels could decrease 10-fold without impacting on high affinity IL-2 receptor expression. Knowledge of protein copy number is thus invaluable information for a full understanding of cell function. Information about protein isoform expression can also give new ideas about cellular control mechanisms. E.g. CTL express the pyruvate kinase M1 and M2 (PKM1/2) isoforms. However, PKM2 dominates in terms of abundance at >107 molecules/CTL versus <105 molecules/CTL for PKM1. The PKM1/2 isoforms both use phosphoenolpyruvate as a substrate during glycolysis but PKM2 can also function as a protein kinase for STAT3 and MEK538 and is co-activator for HIF1α mediated transcription39. The quantity of PKM2 in CTL (>107/cell) permit this enzyme to have multiple roles as a transcriptional and metabolic regulator.
One key insight herein is that mTORC1 is not a global regulator of protein output in CTL but rather selectively shapes the CTL proteome by controlling expression of a small (<10%) subset of metabolic, effector and adhesion molecules that define CTL identity. mTORC1 repressed and stimulated expression of equal numbers of proteins indicative that it simultaneously controls protein production and degradation. The selectivity of mTORC1 control of CTL proteomes was remarkable as was the finding that there was no considerable difference to CTL of inhibiting mTORC1 versus combined inhibition of mTORC1 and mTORC2. This suggests that mTORC1 has a dominant role in CTL compared with mTORC2. Genetic strategies that selectively delete either mTORC1 or mTORC2 have shown quite different roles for these two complexes in T cells8. In particular loss of mTORC2 complexes as a consequence of Rictor deletion prevents Akt S473 phosphorylation and regulates Akt mediated nuclear exclusion of Foxo transcription factors. We show that mTORC2 does control Akt S473 phosphorylation in T cells. However, we also discovered that mTORC1 repressed the accumulation of PtdIns(3,4,5)P3 in CTL. Specific mTORC1 and mTOR inhibitors thus cause CTL to accumulate very high levels of PtdIns(3,4,5)P3 and reprogram how they regulate Akt such that activation of Akt becomes independent of Akt S473 phosphorylation and mTORC2. These results inform that one cannot predict the biological impact of the combined catalytic inhibition of mTORC1 and mTORC2 from genetic modifications that individually disrupt mTORC1 or mTORC2 complexes. In this context, although previous studies have reported mTORC1 feedback control of Akt in non-lymphoid cells14,15,16, the magnitude of the potentiation of T cell PtdIns(3,4,5)P3-levels by rapamycin, an immunosuppressant, was remarkable and not intuitive because PtdIns(3,4,5)P3 is normally thought of as a positive regulator of T cells. However, constitutive activation of PI(3)K p110δ in humans results in an immunodeficiency syndrome (activated PI(3)Kδ syndrome). Hence, hyper-activation of PtdIns(3,4,5)P3 signaling pathways in T cells is effectively immunosuppressive40,41. The hyper-production of PtdIns(3,4,5)P3 may be part of the mechanism whereby mTORC1 inhibitors suppress T cell immunity. This study thus demonstrates the power of unbiased proteomic analysis to generate a new understanding of mechanisms of drug action.
Previous studies have suggested that mTORC1 phosphorylates and targets for degradation a negative regulator of PI(3)K, GRB1014,15. We show that GRB10 is not expressed in CTL. However, mTORC1 controls the expression of PTEN, another key negative regulator of PI(3)K pathways. In this respect, it has been described that mTORC1 mediated phosphorylation and degradation of the adapter protein IRS2 acts to restrain AKT activity in non-lymphoid cells16. mTORC1 is shown herein to control IRS2 amounts in T cells and to control another key adaptor protein DOCK1. In this respect the increased expression of DOCK1 and IRS2 in rapamycin-treated CTLs indicates that inhibition of mTORC1 would promote the signaling pathways controlled by these molecules. mTOR thus has cell type specific actions and to fully understand its role it will necessitate analysis of its function in different leucocyte populations.
Online Methods
Mice
All mice used were bred and maintained under specific pathogen-free conditions in the Biological Resource Unit at the University of Dundee. The procedures used were approved by the University Ethical Review Committee and authorised by a project licence under the UK Home Office Animals (Scientific Procedures) Act 1986. P14 TCR transgenic mice have been described previously42.
Cell culture
CTLs were generated as prescribed previously36. In brief, lymphocytes isolated from spleens of P14 TCR-transgenic mice (for proteomics and microarray experiments: female mice only, 8–10 weeks old; other experiments: female and male mice, 7-18 weeks old) were activated for 48 h with 100 ng/mL of soluble antigenic peptide gp33-41, 20 ng/mL IL-2 (Proleukin) and 2 ng/mL IL-12 (R&D systems) at 37 °C. Cells were then cultured for another 96 h in 20 ng/mL IL-2 and 2 ng/mL IL-12, resulting in >98% pure CD8+ T cell populations. Where indicated, cells were treated with the following inhibitors: 20 nM rapamycin (EMD Millipore), 1 μM AKTi1/2 (EMD Millipore), 10 μM IC-87114 (synthesized in house) or 1 μM KU-0063794 (Tocris). DMSO was used as a solvent and vehicle control.
Immunoblotting
Cells (20 × 106) were lysed in RIPA buffer (100 mM HEPES, pH 7.4, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM TCEP (Pierce), protease and phosphatase inhibitors (Roche). Lysates were sonicated in a Branson Digital sonicator on ice, centrifuged (4 °C, 16,000 × g for 10 min). Samples were adjusted to 1× LDS sample buffer (life technologies) and 25 mM TCEP was added prior boiling for 10 min. Each lane was loaded with the equivalent of 140,000 CTLs and separated by SDS-PAGE (life technologies NuPAGE precast gels or Bio-Rad Mini-PROTEAN tetra cell system) and transferred to nitrocellulose membranes (Whatman). Blots were probed with the following antibodies: 4EBP1 p-S37/S46 (Cell Signaling Technology (CST) #2855), 4EBP1 - pS65 (CST #9451), 4EBP1 (CST #9452), S6K p-T389 (CST #9239), S6K (CST #9202), Akt p-T308 (CST #4056), Akt p-S473 (CST #4058), SMC1 (Bethyl Laboratories, A300-055A), T-bet (eBioscience 14-5825), IRS2 (CST #4502), PTEN (Santa Cruz sc-7974), Foxo1/3A p-T24/32 (CST #9464), FOXO1 (CST #9454). X-ray films (Konica) were used to monitor chemiluminescence reactions catalysed by HRP-conjugated secondary antibodies. All immunoblots shown are representative of 3 or more biological replicates.
Glucose uptake
Glucose measurements were performed as described previously13. Briefly, 106 CTLs were suspended in 400 μL glucose-free media containing 0.5 μCi/ml 2-deoxy-d-[1-3H]glucose ([3H] 2-DG; GE Healthcare) and incubated for 3 min. Cells were pelleted, washed, and lysed overnight with 200 μL of 1 M NaOH, and the incorporated 3H radioactivity was quantified via liquid scintillation counting. Measurements were performed in technical triplicates per condition.
Lactate measurements
Lactate measurements were performed as described previously13. Briefly, 1 × 106/mL CTLs were cultured for 4 h in RPMI 1640 containing 10% dialyzed fetal calf serum, cells were spun and the supernatants were collected. Lactate concentrations in the supernatants were quantified using an LDH (lactate dehydrogenase)-based enzyme assay, monitoring the emergence of NADH/H+ through increased absorption at 340 nm (the reaction contained 320 mM glycine, 320 mM hydrazine, 2.4 mM NAD+, and 3 U/mL LDH). A standard curve was generated, and the concentration of lactate in the supernatant added to this reaction was calculated.
ELISA
ELISA kits (eBioscience) for CD62L and IFN-γ were used to determine the effects of 5 h rapamycin treatment on the secretion of these proteins. Experiments were performed in biological triplicate according to the manufacturer’s instructions.
Seahorse metabolic flux analyser
A XF24 cell culture microplate was coated with 50 μL/well of a 22.4 μg/mL Cell-Tak, 0.1 M NaHCO3, pH 8.0 solution for 30 min. The plate was washed twice with sterile H2O and air dried at 22 °C overnight. A XF24 cartridge was equilibrated with 1 mL Seahorse calibrant solution and equilibrated at 37 °C overnight. Unbuffered RPMI media without FBS was prepared as per manufacturer’s instruction and sterile filtered. 150,000 CTL/well were used in the experiments. The manufacturer’s recommended settings for an oxygen consumption rate of 200-500 pmol/min were used (3 minutes mixing, 2 minutes waiting, 3 minutes measuring). A mitochondrial stress test was performed, sequentially using oligomycin, DNP and rotenone/antimycin to affect the ATP synthase and electron transport chain. Three measurements were taken for the baseline OCR and ECAR and each inhibitor treatment giving a total of 12 measurements. The average for each data point was calculated from at least 3 wells and 2 biological replicates were performed.
Glutaminolysis assay
Glutaminolytic rates were measured as described previously43. 1 × 106 cells per data point were harvested and resuspended in 1 mL fresh glutamine free media containing appropriate cytokines and required drugs. The cells were transferred into 7 mL vials with a PCR tube containing 50 μL 1 M KOH glued to the inner side wall to collect produced CO2. 50 μL of a 20% [U-14C]-glutamine (equivalent to 0.5 μCi [U-14C] glutamine) were added to each sample and the vial closed with a screw cap with rubber septum. Samples were then incubated for 1 h at 37 °C and the assay was stopped by injecting 100 μL 5M HCl through the septum with a Hamilton syringe. The vials were kept at 22 °C overnight to trap the released CO2. The KOH solution in the PCR inside the glass vials was then transferred to scintillation vials, 3 mL of Optiphase HiSafe 3 was added and the samples were analysed in a scintillation counter. All measurements were performed in technical triplicates.
Mass spectrometry measurements of inositol lipids
Mass spectrometry was used to measure inositol lipid concentrations essentially as previously described44, using a QTRAP 4000 (AB Sciex) mass spectrometer and employing the lipid extraction and derivitization method described for cultured cells, with the modification that 1 ng C17:0/C16:0 PtdIns(3,4,5)P3 internal standard was added to primary extracts and that final samples were dried in a speedivac concentrator rather than under N2. Measurements were conducted, in triplicate per experiment, on 1 × 106 cells per sample. The HPLC-MS peak area of the ISD was used as a reference to calculate absolute numbers of PtdIns(3,4,5)P3 per cell in each sample by using the respective sample peak areas.
Sample lysis and in-solution digest for mass spectrometry (label-free quantification)
25 × 106 CTLs treated with either DMSO or rapamycin were harvested in a 50 mL falcon tube and washed three times in cold HBSS and transferred into a 2.0 mL protein LoBind Eppendorf tube. Cells were lysed in 0.5 mL urea lysis buffer (8 M urea, 100 mM Tris pH 8.0, protease and phosphatase inhibitors) and vigorously mixed for 15 min at 22 °C. The samples were then sonicated with a Branson digital sonicator before vigorously mixed for another 15 min. The protein concentration was determined by BCA assay as per manufacturer’s instructions before DTT at a working concentration of 10 mM was added. Lysates were incubated at 30 °C for 30 min. Iodoacetamide was added to a working concentration of 50 mM and lysates were incubated in the dark at 22 °C for 45 min. Lysates were diluted with digest buffer to 4 M urea. LysC was added to the samples in a 50:1 (protein:LysC) ratio and the samples were then incubated at 30 °C overnight. The samples were then split in half. One half was diluted to 0.8 M urea with digest buffer and Trypsin was added in a 50:1 ratio. The other half was kept a LysC fraction. The samples were then incubated at 30 °C for a further 8 h. Samples were adjusted to 1% TFA prior to desalting. C18 Sep-Pak cartridges were washed twice with 1 mL elution buffer and equilibrated twice with 1 mL wash buffer before the acidified peptide samples were loaded. The flow through was loaded again to ensure maximal peptide binding. The peptide loaded cartridges were washed three times with 1 mL washing buffer. Peptides were eluted into 2 mL Eppendorf Protein LoBind tubes by 2 subsequent elutions with 600 μL elution buffer each. The eluted samples were reduced to dryness in a vacuum concentrator.
Strong anion exchange chromatography
Samples were separated via hSAX chromatography as described previously3. Samples were resuspended in 210 μL SAX sample buffer (10 mM sodium borate, pH 9.3, 20% acetonitrile) and the pH was readjusted to pH 9.3 with 1 M NaOH if necessary. Samples were injected and peptides separated by a Dionex Ultimate 3000 HPLC system equipped with an AS24 strong anion exchange column. The following buffers were used for the separation of peptides: 10 mM sodium borate, pH 9.3 (Buffer A) and 10 mM sodium borate, pH 9.3, 500 mM NaCl (Buffer B). An exponential elution gradient starting with Buffer A was used to separate the peptides into 12 fractions of 750 μL which were desalted prior to analysis via LC-MS/MS.
Sample lysis, size exclusion chromatography and in-solution digest for mass spectrometry (SILAC-based quantification)
CTLs were cultured in SILAC media as described previously45. In brief, 50 × 106 CTLs grown in ‘light’ SILAC media and treated with either rapamycin or KU-0063794 were mixed with 50 × 106 CTLs grown in ‘heavy’ SILAC treated with DMSO and washed twice with ice cold HBSS. Cells were lysed and fractionated into five different subcellular fractions (cytoplasmic, membrane, soluble nuclear, chromatin-bound nuclear and cytoskeletal fractions) using a Subcellular Fractionation Kit for Cultured Cells (Pierce) following the manufacturer’s instructions for a 200 μL packed cell volume. Protein contents for each fraction were measured by BCA assay. 300 μg of each subcellular fraction were used for the chloroform-methanol precipitation. Samples were adjusted to a final concentration of 2% SDS, 10 mM TCEP and 20 mM NEM in 1 mL and heated to 65 °C for 10 min to denature proteins. Samples were precipitated using a chloroform-methanol method and air dried. The precipitated cytoplasmic, membrane, nuclear and chromatin bound nuclear fraction were resuspended in 60 μL SEC sample buffer and separated by a mAbPacSEC column (Dionex) using a 0.2% SDS, 100 mM NaCl, 10 mM sodium phosphate buffer, pH 6.0. The flow rate was 0.2 mL min−1 and 8 fractions of 200 μL were collected into Protein LoBind 1 mL 96-deep well plates (Eppendorf). TEAB was added to the SEC fractions to a final concentration of 0.1 M. Trypsin was added in a 50:1 (protein:trypsin) ratio. The unseparated cytoskeletal fraction was diluted with digest buffer to a urea concentration of 1 M. Trypsin was added in a 50:1 (protein:trypsin) ratio. All samples were incubated at 37 °C overnight. Detergents were removed using detergent removal 96-well spin plates (Pierce). The detergent free flow through and the cytoskeletal fraction were then kept for desalting as described above and further sample processing.
Liquid chromatography mass spectrometry analysis (LC-MS/MS)
Samples from desalting were resuspended in 5% formic acid and 1 μg of peptides was used for analysis. A Dionex RSLCnano HPLC was used for the peptide chromatography. A 5 mm PepMap-C18 pre-column with an inner diameter of 0.3 mm was used and a 75 μm × 50 cm PepMap-C18 column was used for the subsequent chromatography. The mobile phase consisted of 2% acetonitrile + 0.1% formic acid (solvent A) and 80% acetonitrile + 0.1% formic acid (solvent B). A constant flow rate of 300 nL/min was used and the linear gradient increased from 5% to 35% solvent B over a runtime of 156 minutes. The eluted peptides were injected into a Velos Orbitrap mass spectrometer (Thermo) through a nanoelectrospray emitter. A typical ‘Top15’ acquisition method was used. The primary mass spectrometry scan (MS1) was performed at a resolution of 60,000. The aforementioned top 15 most abundant m/z signals from the MS1 scan were selected for subjected for collision-induced dissociation and MS2 analysis in the Orbitrap mass analyzer at a resolution of 17,500.
Data analysis for mass spectrometry data
The data were processed, searched, and quantified using the MaxQuant software package version 1.5.0.0 as described previously46, using the default settings and employing the mouse Uniprot database from April 2014 and the contaminants database supplied by MaxQuant. The following settings were used: two miscleavages were allowed; fixed modification was carbamidomethylation on cysteine; enzyme specificities were Trypsin and/or LysC were applicable; variable modifications included in the analysis were methionine oxidation, deamidation of glutamine or asparagine, N-terminal pyroglutamic acid formation, and protein N-terminal acetylation. Default MaxQuant settings included a mass tolerance of 7 ppm for precursor ions, and a tolerance of 0.5 Da for fragment ions. A reverse database was used to apply a maximum false positive rate of 1% for both peptide and protein identification. This cut-off was applied to individual spectra and whole proteins in the MaxQuant output. The match between runs feature was activated with an allowed time window of 2 minutes. All proteins were quantified on the basis of unique and Razor peptides with the requantification feature enabled. iBAQ intensities were calculated by dividing the summed peptide intensities for each protein by the number of theoretically observable peptides. The MaxLFQ algorithm47 was used to assess fold changes between control and rapamycin treated CTLs. Estimated abundances und fold changes were calculated separately for LysC-only or LysC/Trypsin double-digested samples. Abundances or fold changes stated for each biological replicate are the mean of log2-transformed LysC-only and double-digested samples. Further down-stream analysis was performed using Microsoft Excel, Perseus 1.5.1.6 (developed by the Matthias Mann lab), SigmaPlot 12.5 and the language R (version 3.1.3, using R Studio 0.98.11.03). An initial pilot proteomics experiment was performed to determine fold-changes of known rapamycin sensitive proteins perforin13 and L-selectin6 and a total of three biological replicates was required to determine these changes with a P-value of ≤ 0.05 (two-tailed, unequal variance t-test). The same two-tailed, unequal variance t-test without further adjustment was used to calculate significance of all fold changes in proteomic experiments. For SILAC based proteomics, SILAC ratios obtained for each subcellular fraction were weighted by the contribution of the respective subcellular fraction to the overall cellular protein content and by the contribution of reported SILAC ratio counts for each subcellular fraction ratios to the number of total SILAC ratio counts for each experiment. Log-normalized SILAC ratios were then used to determine statistical significance (p ≤ 0.01 as determined two-tailed, unequal variance t-test). Pathway analyses were performed using the Database for Annotation, Visualisation and Integrated Discovery (DAVID) bioinformatics tools based on Kyoto Encyclopedia of Genes and Genomes (KEGG)48. A plugin for Perseus was used to calculate protein copy numbers using the proteomic ruler17: Relative protein abundance for each protein group was calculated by dividing the iBAQ protein intensity for the respective protein group by the sum of iBAQ intensities of all protein groups in each dataset. Total histone numbers in a diploid murine cell (~2.2 × 108) were calculated from the size of the mouse genome and assigned to the summed relative abundance of all histones with the control CTLs (~13%) as inferred by the relative protein intensities and subsequently used to estimate copy numbers for proteins within the dataset. Protein groups were assigned “high” (min 8 peptides detected, min 75% all peptides unique+razor, min 3 observable peptides per 100 amino acid), “medium” (min. 3 peptides detected, min 50% all peptides unique+razor, min 2 observable peptides per 100 amino acid) or “low” (all other) quantification accuracy for both LysC and LysC-Trypsin double-digest derived peptides and averaged, resulting in five classification from “high”/“high” to “low”/“low”.
Affymetrix GeneChip mouse genome array analysis
CTLs were treated either DMSO (control) or rapamycin or KU-0063794 for 48 h in triplicate as described above. RNA was extracted with the RNeasy RNA purification minikit (QIAGEN) according to the manufacturer’s specifications. Microarray analysis was carried out by the Finnish DNA Microarray Centre at the Centre for Biotechnology, Turku, Finland via 430_2.0 mouse expression arrays (Affymetrix) and the manufacturer’s recommended protocol. Affymetrix Expression Console v1.1 (Affymetrix) was used for normalization of data. Normalization with Microarray Suite 5 (MAS5) was used for the selection of probes present in at least one sample, and robust multi-array averaging was used for normalization of data. Statistically significant differences in gene expression were identified with Multiple Experiment Viewer v4.349 by performing a Significance Analysis of Microarrays (SAM) algorithm, with the 90th-percentile false-discovery rate set to 5%. Transcript data was matched to proteomics data by matching the gene symbol of the Affymetrix probes to the corresponding gene symbols reported by the Uniprot FASTA-headers.
Statistical Methods
All statistical tests not involved in the analysis of the raw proteomic and microarray data were performed using SigmaPlot 12.5 (Systat Software) for Windows or Prism V6 (Graphpad Software) for Mac. A Shapiro-Wilk test for normality was performed to determine suitable tests for parametric or non-parametric populations. F-tests were performed to determine equal variance of populations. All utilized tests were two-tailed and are stated in the respective figure legends. Samples were considered biological replicates if CTLs were generated from separate spleens.
Supplementary Material
ACKNOWLEDGEMENTS
We thank present and past colleagues in the Cantrell Group for advice and discussion, particularly C. Feijoo-Carnero for help with microarray analysis. We thank T. Ly for help with peptide fractionation using hSAX. We thank D. Lamont and team of the mass spectrometry facility at the College of Life Sciences, University Dundee and the Finnish DNA microarray Centre at the Centre for Biotechnology (Turku, Finland). Supported by the Wellcome Trust (093713/Z/10/A to J.L.H., 073980/Z/03/Z and 105024/Z/14/Z to A.I.L., 065975/Z/01/A and 097418/Z/11/Z to D.A.C.). J.L.H. was supported by a Wellcome Trust PhD Studentship. D.A.C. and A.I.L. are Wellcome Trust Principal Research Fellows.
Footnotes
Accession numbers.
All of the label-free .raw MS and MaxQuant search output data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD002928 .
The microarray data are available in the Gene Expression omnibus (GEO) database (http:/www.ncbi.nlm.nih.gov/geo) under the accession number GSE70925.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Heng TSP, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 2.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Ly T, et al. A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. eLife. 2014;3:e01630. doi: 10.7554/eLife.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larance M, Lamond AI. Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Bio. 2015;16:269–280. doi: 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- 5.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollizzi KN, et al. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 10.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington LS, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell. Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiśniewski JR, Hein MY, Cox J, Mann M. A ‘proteomic ruler’ for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics: MCP. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson IA, et al. The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol. Endocrinol. Metab. 2008;295:E242–53. doi: 10.1152/ajpendo.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith KA, Cantrell DA. Interleukin 2 regulates its own receptors. Proc. Natl. Acad. Sci. U.S.A. 1985;82:864–868. doi: 10.1073/pnas.82.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinerman O, et al. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Mol. Syst. Biol. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-Martínez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem. J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao RR, Li Q, Bupp MRG, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakaya M, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Windt GJW, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 31.Barnett SF, et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 33.Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 2001;20:4380–4390. doi: 10.1093/emboj/20.16.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Najafov A, Shpiro N, Alessi DR. Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors. Biochem. J. 2012;448:285–295. doi: 10.1042/BJ20121287. [DOI] [PubMed] [Google Scholar]
- 36.Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol. Cell. Biol. 2009;29:5952–5962. doi: 10.1128/MCB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macintyre AN, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang P, Li Z, Fu R, Wu H, Li Z. Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell. Signal. 2014;26:1853–1862. doi: 10.1016/j.cellsig.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Palsson-McDermott EM, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angulo I, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat. Immunol. 2014;15:88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark J, et al. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat. Immunol. 2011;12:352–361. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.