Abstract
Sexually dimorphic phenotypes arise from the differential expression of male and female shared genes throughout the genome. Unfortunately, the underlying molecular mechanisms by which dimorphic regulation manifests and evolves are unclear. Recent work suggests that Y-chromosomes may play an important role, given that Drosophila melanogaster Ys were shown to influence the regulation of hundreds of X and autosomal genes. For Y-linked regulatory variation (YRV) to facilitate sexually dimorphic evolution, however, it must exist within populations (where selection operates) and influence male fitness. These criteria have seldom been investigated, leaving the potential for dimorphic evolution via YRV unclear. Interestingly, male and female D. melanogaster differ in immune gene regulation. Furthermore, immune gene regulation appears to be influenced by the Y-chromosome, suggesting it may contribute to dimorphic immune evolution. We address this possibility by introgressing Y-chromosomes from a single wild population into an isogenic background (to create Y-lines) and assessing immune gene regulation and bacterial defence. We found that Y-line males differed in their immune gene regulation and their ability to defend against Serratia marcescens. Moreover, gene expression and bacterial defence were positively genetically correlated. These data indicate that the Y-chromosome has the potential to shape the evolution of sexually dimorphic immunity in this system.
Keywords: Y-linked regulatory variation, gene expression, fitness, sexual dimorphism, Serratia marcescens
1. Introduction
Males and females often exhibit different fitness optima for shared phenotypes, which drives the evolution of sexual dimorphism [1]. However, males and females share a genome, which ultimately constrains the evolution of dimorphism by not allowing sex-specific gene divergence [2]. Despite this constraint, dimorphism does evolve and recent studies have shown that the majority of X and autosomal genes can be differentially regulated between the sexes [3–6]. Unfortunately, the underlying molecular mechanisms by which sexually dimorphic gene regulation manifests and evolves are unclear.
Recent work by Lemos and co-workers [7,8] has shown that the Y-chromosome in Drosophila melanogaster can influence the regulation of hundreds of genes throughout the genome, suggesting that Y-chromosomes may in part facilitate sexually dimorphic evolution. The mechanism underlying this effect appears to be associated with variation in the Y-chromosome's non-coding heterochromatin, which influences the formation of euchromatin boundaries throughout the genome (i.e. areas where tightly packed heterochromatin meets loosely packed euchromatin, which influences gene regulation [7,9,10]). The effect found in Drosophila may be a common phenomenon in most heterogametic systems, as Y-chromosomes tend to have large tracks of heterochromatin [11] and at least one other independently evolved Y-chromosome appears to exhibit a similar effect (e.g. mice; [12–14]). Unfortunately, it is currently unknown if Y-linked regulatory variation (YRV) is widespread among independently evolved Y-chromosomes.
More importantly, it is still unclear if YRV can influence the evolution of autosome or X-chromosome coded traits, as most YRV studies have focused on the molecular mechanisms underlying the phenomenon and not its evolutionary potential [7–9]. In order for YRV to influence the continued evolution of male and female shared traits, it must (i) exist within populations where selection operates and (ii) directly influence male fitness. Regrettably, the original work reporting the YRV phenomenon precluded the detection of within-population variation by using Y-chromosomes captured from disparate geographical regions (i.e. non-coevolving populations; [7,15]). Moreover, no subsequent study has examined if co-evolving Y-chromosomes newly derived from the same population can influence fitness-related traits that are simultaneously expressed by both sexes (e.g. body size, development time, etc.).
Here, we assess the potential for YRV to influence the evolution of immune function in D. melanogaster. Immunity is a central physiological trait that is shared between the sexes and often exhibits dimorphism within numerous animal systems [16]. Sexually dimorphic immunity has likely evolved in these systems owing to differences in pathogen exposure or infection rates between the sexes, or owing to different life-history strategies [17–19]. In D. melanogaster, the regulation of immune-related genes is sensitive to the Y-chromosome [7] and immune gene expression has been shown to be dimorphic [16,20]. However, it is still unknown if contemporary selection can shape YRV. In this study, we use RT-qPCR to determine whether immune-related YRV exists within a wild population of D. melanogaster. We then assess if Y-linked variation influences a male's immune-related fitness. These hitherto untested criteria are essential if the Y-chromosome is to influence the continued evolution of sexually dimorphic immunity.
2. Material and methods
(a). Fly stocks and maintenance
In the autumn of 2010, isofemale lines were established by collecting 40 gravid D. melanogaster females from a single location in Orlando Florida. These lines were maintained via strict single-pair sibling matings to produce genetic homogeneity within each line. Just prior to each assay outlined below, corresponding Y-lines were created for each isofemale line (figure 1). Y-lines consisted of a genetically identical genome across lines, but a unique Y-chromosome within lines. That is to say, all Y-lines had the same X-chromosome and autosomes derived from a common isogenic stock and unique Ys that were derived from the above isofemale lines.
Figure 1.
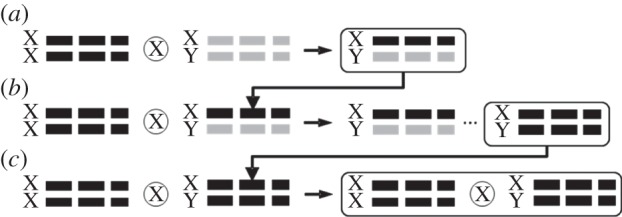
Establishment and maintenance of Y-lines. Here, the Drosophila melanogaster genome is depicted as three sets of autosomes (solid bars) and a pair of sex chromosomes (X and Y). Isogenic 4361 derived chromosomes with recessive markers on each chromosome are depicted in black, while the wild chromosomes from isofemale lines are depicted in grey. Y-lines were established by initially crossing an isogenic 4361 female to a male from each isofemale line (a). The resulting heterozygote male F1 offspring were then backcrossed with 4361 females (b), which produced a range of male genotypes represented by the ellipsis. The F2 male offspring that exhibited all recessive markers (12.5% of the genotypes produced) were then used to establish the Y-lines, as they possessed a common isogenic background, but unique Y-chromosomes. These newly formed Y-line males were then maintained through continual backcrossing with 4361 females (c).
To create the Y-lines, two crosses were conducted. The first cross paired a male from each isofemale line with a female from an isogenic stock (Bloomington Stock no. 4361; made isogenic with 10 generations of single-pair sibling matings prior to experimentation). The isogenic stock contained recessive markers on each chromosome (y[1]; bw[1]; e[4]; ci[1] ey[R]), allowing the detection of homozygous chromosome pairing. The resulting male offspring were all heterozygotes that contained a haploid set of isofemale autosomes and a haploid set of 4361 autosomes, as well as a 4361 X-chromosome (figure 1a). These males were then backcrossed to 4361 to create a range of F2 genotypes. Males that possessed all of the recessive markers were collected to create the Y-lines, as they were genetically identical across lines, but contained a unique Y-chromosome derived from their original isofemale line (figure 1b). Note that the F1 chromosomes are inherited intact, as male Drosophila do not undergo recombination during gametogenesis. The newly created Y-lines were then maintained by mating Y-line males with isogenic 4361 females (figure 1c). All lines were maintained in vials on a cornmeal medium at 25°C 12 L : 12 D photoperiod using Percival incubators (Percival Scientific, Perry, IA).
(b). Y-linked regulatory variation assay for immune-related genes
In the summer of 2011 (14 generations after the establishment of the isofemale lines), 30 isofemale lines were randomly chosen to create corresponding Y-lines and have their immune gene expression assayed. To remain consistent with the original work identifying the YRV effect [7], individuals were not immune-challenged prior to assessing immune gene regulation. From each Y-line, 20 male and 20 female offspring were collected upon adult eclosion and placed into sex-specific vials to ensure virginity. Four days after eclosion, males and females were placed in groups of five into a 1.5 µl microcentrifuge tube containing Trizol and disrupted using a motorized pestle. RNA was extracted according to the Trizol reagent protocol (Invitrogen, Carlsbad, CA), creating four independent RNA samples per sex per Y-line. RNA was then reverse transcribed using the iScript cDNA Synthesis kit (Invitrogen) and the resulting cDNA stored at −80°C. In total, 104 female samples and 111 male samples were used [21] (samples were removed owing to poor extraction results). If immune-related YRV exists within a population, then Y-line males are predicted to differ in their immune gene expression while Y-line females should be the same.
To address our prediction, three immune effector genes associated with the IMD pathway were examined, including attacin-A (CG10146), cecropin A1 (CG1365), and diptericin (CG12763). Attacin-A and diptericin are located on the second chromosome, while cecropin A1 is located on the third chromosome. These genes were chosen because they were shown to be influenced by across-population YRV in a previous study [7]. Gene expression was quantified using SYBR Green Supermix and the MyiQ Single Color Real Time qPCR Detection System (Bio-Rad, Hercules CA). All primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and gene sequences available from Flybase.org. The primer sequences are as follows: attacin-A (left—GGCCTGGATGGACGTGCTAA, right—GTTGGCAAACGGTCCACTCG), cecropin A1 (left—CGTTGGTCAGCACACTCG, right—GACATTGGCGGCTTGTTG), and diptericin (left—AGAGTGCGTCGCCAGTTCCA, right—GGCTGTTGCCATAGGGTCCA). All primers exhibited PCR efficiencies above 95% and no off-target amplifications were detected. Expression estimates were normalized using the housekeeping gene actin5C (CG4027; left—TTGGGAATGGAGGCTTGCGG, right—AGCACGGTGTTGGCATACAGAT; PCR efficiency was 99.3%). For all female samples used in the analysis, actin5c CT-values ranged between 16.3 and 18.4, with a mean and standard error of 17.45 ± 0.12. Male actin5c CT-values ranged between 18.5 and 20.5, with a mean and s.e. of 19.12 ± 0.11. Each 20 µl qPCR reaction was run in triplicate. All technical replicates for a given gene were averaged prior to estimating gene expression. Immune gene expression estimates were generated by calculating the difference between the target gene's cycle threshold (CT) and actin5c's CT within a given sample (i.e. we calculated each immune gene's ΔCT).
(c). Functional immune response assay
In the spring of 2013 (approx. 35 generations after the gene expression assay), 27 isofemale lines were randomly chosen to recreate the Y-lines (Y-lines from the YRV assay were destroyed after that assay). As above, virgin males and females from each newly established Y-line were collected after adult eclosion and maintained separately for 4 days. One male and one female from each Y-line were then randomly chosen and injected with an LD90 of S. marcescens using a Nanoject II (Drummond Scientific, Bromall PA) under light CO2 anesthesia (see below). After injection, each fly was placed in a Drosophila activity monitor (TriKinetics, Waltham MA) for 48 h and their motion recorded every minute to establish an accurate time-to-death (TTD). The TTD assay was replicated 12 separate times (in series) and each Y-line replicate was derived from a new cohort of flies injected with a freshly reared bacterial solution. In total, 212 females and 214 males were assayed across 27 lines. As with the YRV assay, if immune-related Y-linked variation translates into functional immune variation, then Y-line males are predicted to differ in their ability to defend against a live pathogen, while Y-line females should be the same.
Methods for infecting flies with S. marcescens were modified from Apidianakis & Rahme [22]. In short, bacteria were incubated in sterilized LB broth for 18 h at 37°C until log phase. This solution was diluted with sterile broth to an absorbance of 0.4 at 490 nm using a microplate reader (Bio-Rad Model 680, Hercules, CA, USA). One millilitre of the diluted solution was centrifuged at 11 000g for 2 min. The supernatant was discarded and 1 ml of 10 mM MgSO4 wash was added to the remaining pellet and centrifuged at 11 000g for 2 min. Again, the supernatant was discarded and the pellet was re-suspended in 1 ml of MgSO4. Preliminary experiments showed that 90% of flies died within 48 h of being infected with 60 nl of this concentration of MgSO4-suspended bacteria. Negative controls injected with just the bacterial vehicle (MgSO4) showed no mortality, indicating that S. marcesens was the cause of death in these experiments. The S. marcescens used in this study was isolated from wild Drosophila melanogaster and graciously provided by B. Lazzaro.
(d). Statistical analysis
Relative immune gene expression values (ΔCT) were analysed separately for each target gene via a random effects model ANOVA (REML method), with Y-line and replicate representing the independent random effects. The significance of each random effect was determined via a log likelihood ratio test. The sexes were analysed separately, as female expression values serve as a methodological control (all females were isogenic) and we were uninterested in the effect of sex per se or its effect size. All Y-line by replicate interactions were not significant and were therefore not included in the final model. Considering that we examined three genes, we employed a sequential Bonferroni correction within each sex (k = 3) to minimize the potential for Type I error. Repeatability of our qPCR samples was high (0.91, 0.99, 0.96 and 0.85 for actin5C, attacin-A, cecropinA1 and diptericin, respectively). Repeatability was based on the intraclass correlation among our technical well replicates for three randomly chosen PCR plates [23]. To determine whether expression levels of the different genes are correlated across Y-lines, we employed Pearson product moment correlation analysis based on family means, which provides an estimate of the genetic correlation between traits [24].
Time-to-death values were analysed separately for each sex using a proportional hazards model. Flies that survived the 48 h period were censored and included in the analysis. Again, line by replicate interactions were not significant and were therefore not included in the final model. To examine the relationship between immune gene expression (2011 Y-line dataset) and defence against S. marcescens (2013 Y-line dataset), we again employed Pearson correlation analysis based on Y-line means for those Y-lines used in both assays (n = 19 lines). Prior to correlation, gene expression data were transformed from their log-based ΔCT values into the relative proportion of actin5C expression (1/2(ΔCT)).
Last, we estimated the intraclass correlation (ρ) for time-to-death and gene expression using a clonal analysis method [25]. Prior to analysis, the influence of replicate was removed by generating the residuals between replicate and trait (i.e. gene expression or time-to-death). The resulting residuals were then used to estimate Y-linked genetic variation among Y-lines via a one-way ANOVA. In this analysis, the intraclass correlation estimates the proportion of trait variation attributed to the Y-chromosome. Note that the small subsample of wild Y-chromosomes used in this study, coupled with the use of a single isogenic background, may downwardly bias intraclass correlation estimates. By contrast, pooling five isogenic individuals (i.e. clones) into each Y-line replicate sample may decrease our estimate of phenotypic variance, which can upwardly bias the intraclass correlation for the gene expression data [21,26]. Thus, the intraclass correlation estimates may not accurately reflect evolutionary potential in the original wild population. All analyses were conducted using JMPver10.
3. Results
If within-population YRV exists for immune-related genes, we predicted that Y-line males would exhibit differences in gene expression while Y-line females would not. We found that Y-line males did indeed exhibit differences in their gene expression for two of the three immune genes: attacin-A and cecropin-A1 (table 1). Furthermore, the proportion of gene expression variation attributed to the Y-chromosome was ρ = 0.18 for attacin-A (p = 0.0150) and ρ = 0.20 (p = 0.0072) for cecropin-A1. Y-line females showed no among-line variation for any of the immune genes. Male expression levels for all three genes were positively correlated (all r > 0.79 and all p < 0.0001) and Y-lines exhibited consistent expression across all three genes (e.g. consistently high or low; figure 2). This is not surprising considering that these genes share the same regulatory pathway. These data indicate that immune-related YRV exists within the examined population.
Table 1.
Evidence for within-population Y-chromosome regulatory variation (YRV) for immune-related genes. As expected, Y-line males exhibited significant variation in immune gene expression for attacin-A and cecropin-A1, while female Y-lines were not associated with expression variation. Significance of the random effects models was determined via log likelihood ratio tests (degrees of freedom = 1). Bolded values remain significant after a sequential Bonferroni test (k = 3).
| source | var. comp. | χ2 | p-value |
|---|---|---|---|
| females | |||
| attacin | |||
| line | 0.08 | 0.17 | <0.6766 |
| replicate | 0.07 | 0.68 | <0.4102 |
| residual | 3.04 | ||
| cecropin | |||
| line | 0.16 | 0.62 | <0.4296 |
| replicate | −0.05 | 0.55 | <0.4575 |
| residual | 3.29 | ||
| diptericin | |||
| line | −0.10 | 0.04 | <0.8415 |
| replicate | 0.64 | 4.24 | <0.0395 |
| residual | 8.41 | ||
| males | |||
| attacin | |||
| line | 0.51 | 6.25 | <0.0124 |
| replicate | 0.27 | 9.44 | <0.0021 |
| residual | 2.43 | ||
| cecropin | |||
| line | 0.43 | 7.10 | <0.0077 |
| replicate | 0.32 | 20.7 | <0.0001 |
| residual | 1.58 | ||
| diptericin | |||
| line | 0.43 | 0.90 | <0.3428 |
| replicate | 0.48 | 5.39 | <0.0202 |
| residual | 6.55 |
Figure 2.
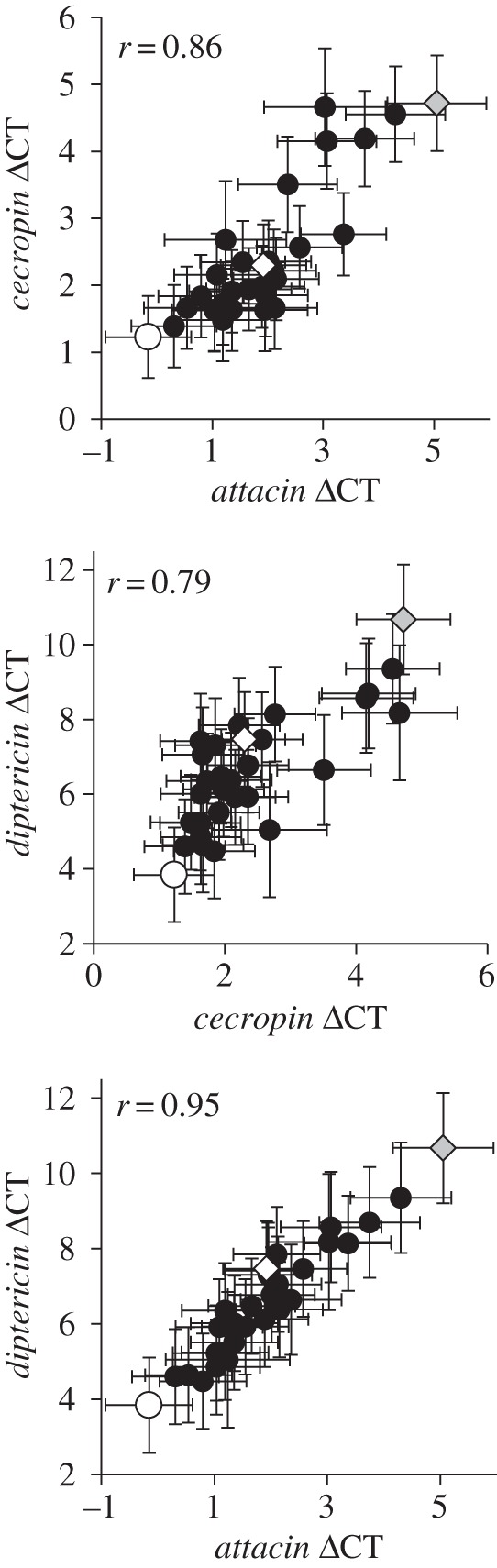
Correlation between immune gene expression levels across male Y-line means. All three immune genes were positively correlated (all p < 0.0001) and Y-lines exhibited consistency in their expression across the genes. For clarity, the Y-lines with the highest, lowest and middle levels of attacin-A expression have been identified on all three graphs (open circle, grey diamond and open diamond, respectively). Error bars represent standard errors of Y-line means.
As with the immune gene assay, if Y-line males but not females exhibit variation in their ability to defend against a pathogen, then it would suggest that YRV influences immune-related fitness. Accordingly, we found that only Y-line males differed in their ability to defend against the gram-negative bacteria S. marcescens (table 2). The proportion of variation in time-to-death explained by the Y-chromosome was ρ = 0.05 (p = 0.0583). Moreover, we found a significant positive correlation between immune gene expression and defence against S. marcescens across male Y-lines for attacin-A, cecropin-A1 and diptericin (r = 0.64, p = 0.0029; r = 0.60, p = 0.0071; r = 0.57, p = 0.0109, respectively). We found no significant relationship across female Y-lines (all p > 0.2703; figure 3). These data suggest that Y-chromosomes that induce high baseline levels of immune gene expression in males also induce a greater defence against S. marcescens infection. Importantly, the lack of a female correlation does not imply that variation in female immune gene expression is unassociated with immune defence. Rather, it indicates that female Y-lines did not differ significantly in immune gene expression, which resulted in no significant relationship between expression and survival.
Table 2.
Evidence for within-population Y-chromosome variation for defence against a bacterial pathogen. As expected, Y-line males exhibit variation in their time-to-death after infection with Serratia marcescens, while Y-line females exhibited no variation.
| source | d.f. | χ2 | p-value |
|---|---|---|---|
| female | |||
| model | 37 | 59.46 | <0.0110 |
| line | 26 | 31.00 | <0.2281 |
| rep | 11 | 31.24 | <0.0010 |
| male | |||
| model | 37 | 74.00 | <0.0003 |
| line | 26 | 41.95 | <0.0248 |
| rep | 11 | 45.59 | <0.0001 |
Figure 3.
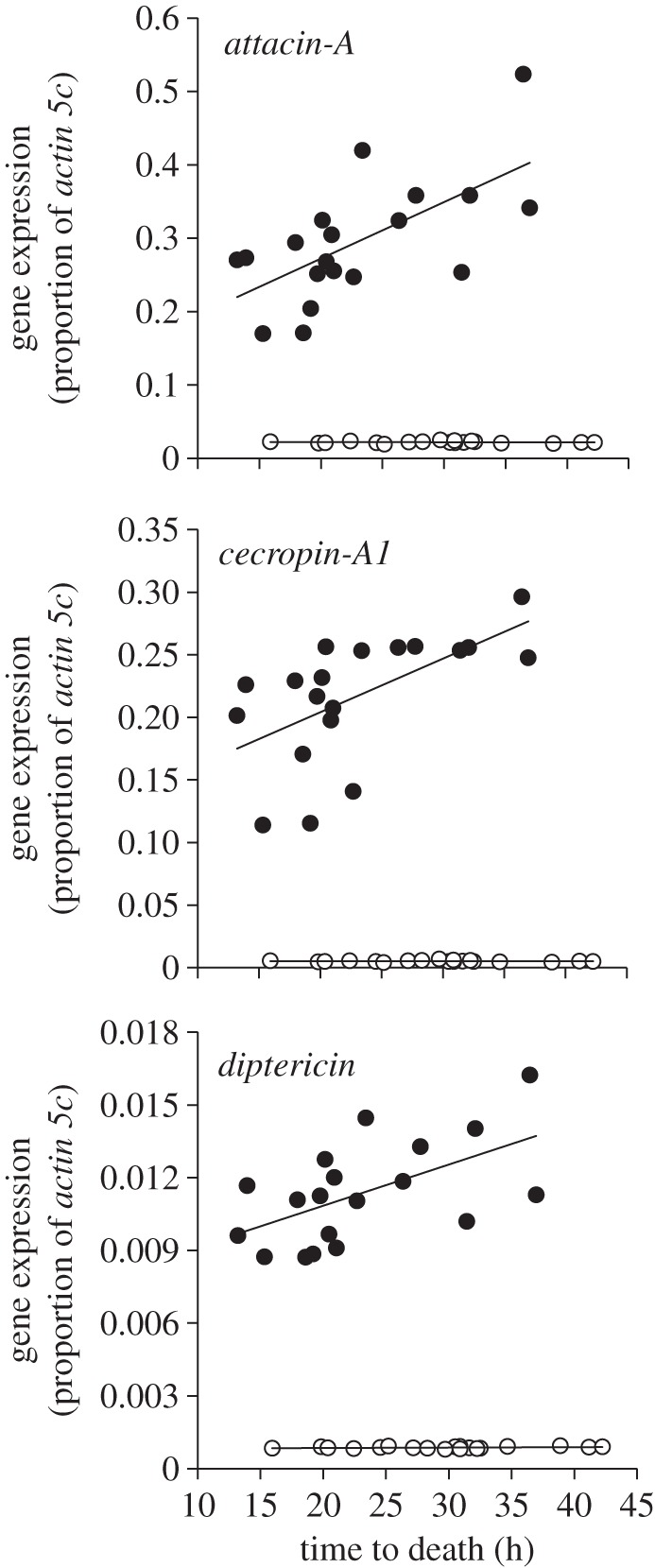
Male and female immune gene expression as a function of pathogen resistance for all three immune-related genes. As baseline expression increased for each immune gene, resistance against S. marcescens increased for males (closed circles), but not for females (open circles). Immune gene expression is relative to actin5C and expression for all three genes is male-biased. Although female expression levels are low relative to the housekeeping gene (on average 2% of actin5C, making them appear close to zero), expression levels remain robust. Each data point represents the least-squared Y-line means for the models presented in tables 1 and 2.
4. Discussion
For the YRV phenomenon to have an impact on the continued evolution of immune gene regulation, immune-related YRV must exist within populations and have a functional influence on a male's ability to defend against pathogens. Here we show that Y-chromosomes sampled from a single wild population affect immune-related fitness variation when introgressed into an isogenic laboratory background. Specifically, we show that Y-line males differed significantly in their immune gene expression (table 1) and functional immune response to a bacterial pathogen (table 2). By contrast, Y-line females did not exhibit variation in either assay. Furthermore, immune gene expression level and ability to defend against a pathogen were genetically correlated in all three genes in males but not females (figure 3). This correlation shows that baseline immune gene expression is sexually dimorphic, with males exhibiting higher values than females for all three genes, a trend that has previously been documented by our laboratory for IMD-related genes [20]. It also shows that Y-lines with the highest levels of expression were associated with the strongest pathogen defence (figure 3). Although the proportion of variation in time-to-death explained by the Y-chromosome appears small (ρ = 0.05), small amounts of genetic variation can have profound implications on evolutionary time scales. Furthermore, this value is likely underestimated owing to our use of only a small subset of wild population Y-chromosomes and a single isogenic background. These results suggest that the YRV phenomenon has the potential to influence the adaptive evolution of immune function in D. melanogaster. Future work should examine more genes and other types of fitness assays in order to obtain a robust understanding of how YRV influences male immunity.
Although these data suggest that YRV can influence immune system evolution, the question of how it influences immune system evolution remains to be answered. Specifically, the type of genetic variation on the Y-chromosome can influence immune evolution in different ways. If the Y-linked genetic variation detected in this study is entirely additive, then selection can efficiently shape adaptive sexual dimorphism. If the variation is a mix of additive and epistatic variance, then adaptive evolution could be greatly hindered, depending on the proportion of the Y-linked variation that is epistatic. Epistasis can hinder adaptation by (i) making fitness landscapes rugged and (ii) reducing overall trait heritability, both of which reduce the efficiency of selection. In general, ruggedness is driven by local sign epistasis within the landscape, which can also cause a population to become marooned on a sub-optimal fitness peak and impede further evolution [27,28]. The reduction in heritability stems from the fact that all types of epistasis increase phenotypic variance (the denominator of the heritability estimate; [29]) and can also decrease additive genetic variance (the numerator of the heritability estimate; [30]).
Importantly, if Y-linked variation is entirely epistatic, then dimorphic evolution via the Y-chromosome cannot proceed. Moreover, it could hinder monomorphic selection (i.e. when males and females share the same fitness optimum), considering that alleles must spend 50% of their time in a male background (assuming a 1 : 1 sex ratio) where epistasis alters their phenotypic effects and fitness values in a non-additive manner. The extent to which monomorphic selection is hindered would depend on the magnitude of the YRV effect relative to the rest of the genome. Interestingly, one of the few studies to have estimated Y-linked epistatic contributions on a trait did so for male mating success (a male-specific trait) and found that at least 40% of the variation in male fitness was linked to the Y-chromosome and this variation was entirely epistatic [31]. If Y-linked epistasis similarly influences the regulation of a male–female shared trait (i.e. not sex-specific) experiencing monomorphic selection, then it may be a common mechanism constraining adaptive evolution throughout the genome.
It is important to note that the maintenance of Y-chromosome additive variation within populations may be quite difficult, requiring either frequency-dependent selection or strong interactions with other genetic elements [32]. Unfortunately, the original work reporting the YRV phenomenon precluded the detection of within-population variation by using Y-chromosomes captured from disparate geographical regions (i.e. non-coevolving populations; [7,15]). Recent work in D. simulans did find evidence for within-population YRV associated with sex ratio distortion [33]. However, the D. simulans population used in the previous study had been maintained in the laboratory as isofemale lines for approximately 200 generations prior to investigation. Thus, the within-population YRV reported may have evolved under laboratory isolation and not under wild conditions (owing to the likely occurrence of independent evolution among D. simulans isofemale line genetic elements), making the determination of whether YRV persists within populations difficult.
In our study, the assessment of immune-related YRV was conducted in the absence of infection, and therefore represents a ‘baseline’ investment in immune gene products. Some Y-chromosomes exhibited a higher baseline investment than others (figure 2), which may induce a greater energetic cost on the bearer when no pathogen is present (i.e. a higher immune function maintenance cost). If the probability of infection is temporally or spatially variable, then a chromosome by environment interaction could help to maintain Y-chromosome variation within a population. This is an interesting result that deserves further investigation.
The original work describing the YRV phenomenon [7] noted several immune genes under the influence of the Y-chromosome, including attacin-A, cecropin A1, and diptericin (the three genes studied here). In contrast to the previous work, we found no effect of the Y-chromosome on diptericin (table 1). The reason underlying this discrepancy may be the use of different Y-chromosomes, which influenced the formation of the euchromatin–heterochromatin boundaries differently [7]. For instance, the Orlando population may lack a Y-chromosome that has a large influence on diptericin, or such a Y-chromosome was not captured during the initial phase of the study. These possibilities raise interesting questions. For instance, which immune genes are most sensitive to the YRV effect, and do sensitive genes share similarities in function (i.e. recognition proteins versus signal transduction proteins)? An assay of YRV-sensitive genes may provide insight into which immune traits and/or pathways are readily being shaped by YRV.
There are hundreds of heterochromatin–euchromatin boundaries in the Drosophila genome [34], each of which may be susceptible to the Y-effect. Furthermore, YRV is not limited to immune-related genes, but may affect any area of organismal physiology. The more fitness-related traits found to be influenced by YRV, the broader its evolutionary implications. Additionally, this phenomenon has only been studied in detail in a few Drosophila species and these species all share the same ancestral Y-chromosome. We still do not know if the within-population fitness consequences of YRV are simply a unique characteristic of this ancestral Drosophila Y, a characteristic of all Y-chromosomes, or a common characteristic of all heterogametic systems, including ZW systems. Work in mice does suggest that murid Y-chromosomes also influence the expression of autosomal genes, hinting that the YRV phenomenon may be widespread; however, these studies examined Y-chromosomes drawn from independent populations thereby limiting any conclusions about the ubiquity of within-population YRV [12–14]. Thus, a widespread examination of Y-chromosomes drawn from natural populations across numerous evolutionarily unique Y-chromosome systems could greatly increase our basic understanding of genome evolution.
Acknowledgements
We thank Brian Lazzaro for donating the strain of Serratia marcescens.
Ethics
All research methods adhered to state and federal guidelines for the treatment of invertebrate animals.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.2kr90
Authors' contributions
This project was conceived by K.M.F., designed and written by I.C.K. and K.M.F. The data were collected and analysed by I.C.K.
Competing interests
We declare no competing interests.
Funding
This work was supported in part by a National Science Foundation grant to K.M.F. (IOS-0722123).
References
- 1.Andersson M. 1994. Sexual selection. Princeton, NJ: Princeton University Press. [Google Scholar]
- 2.Griffin RM, Dean R, Grace JL, Ryden P, Friberg U. 2013. The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol. Biol. Evol. 30, 2168–2176. ( 10.1093/molbev/mst121) [DOI] [PubMed] [Google Scholar]
- 3.Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl Acad. Sci. USA 100, 9894–9899. ( 10.1073/pnas.1630690100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742–1745. ( 10.1126/science.1085881) [DOI] [PubMed] [Google Scholar]
- 5.Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 98, 218–223. ( 10.1073/pnas.98.1.218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Snyder M. 2005. Sexual dimorphism in mammalian gene expression. Trends Genet. 21, 298–305. ( 10.1016/j.tig.2005.03.005) [DOI] [PubMed] [Google Scholar]
- 7.Lemos B, Branco AT, Hartl DL. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl Acad. Sci. USA 107, 15 826–15 831. ( 10.1073/pnas.1010383107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang PP, Hartl DL, Lemos B. 2010. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics 186, U109–U221. ( 10.1534/genetics.110.118109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. 2011. Ribosomal DNA deletions modulate genome-wide gene expression: ‘rDNA-sensitive’ genes and natural variation. PLoS Genet. 7, e1001376 ( 10.1371/journal.pgen.1001376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francisco FO, Lemos B. 2014. How do y-chromosomes modulate genome-wide epigenetic States: genome folding, chromatin sinks, and gene expression. J. Genomics 2, 94–103. ( 10.7150/jgen.8043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124. ( 10.1038/nrg3366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spach KM, et al. 2009. Cutting edge: the Y chromosome controls the age-dependent experimental allergic encephalomyelitis sexual dimorphism in SJL/J mice. J. Immunol. 182, 1789–1793. ( 10.4049/jimmunol.0803200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teuschert C, Spach KM, Blake M, Bunn JY, McElvany B, Noubade R, Blankenhorn EP. 2006. Evidence that the Y chromosome influences autoimmune disease in male and female mice. Proc. Natl Acad. Sci. USA 103, 8024–8029. ( 10.1073/pnas.0600536103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case LK, et al. 2013. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 23, 1474–1485. ( 10.1101/gr.156703.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemos B, Araripe LO, Hartl DL. 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319, 91–93. ( 10.1126/science.1148861) [DOI] [PubMed] [Google Scholar]
- 16.Rolff J. 2002. Bateman's principle and immunity. Proc. R. Soc. Lond. B 269, 867–872. ( 10.1098/rspb.2002.1959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuk M, Thornhill R, Ligon JD, Johnson K. 1990. Parasites and mate choice in red jungle fowl. Am. Zool. 30, 235–244. ( 10.1093/icb/30.2.235) [DOI] [PubMed] [Google Scholar]
- 18.Zuk M, McKean KA. 1996. Sex differences in parasite infections: patterns and processes. Int. J. Parasit. 26, 1009–1023. ( 10.1016/S0020-7519(96)00086-0) [DOI] [PubMed] [Google Scholar]
- 19.Poulin R. 1996. Sexual inequalities in helminth infections: a cost of being a male? Am. Nat. 147, 287–295. ( 10.1086/285851) [DOI] [Google Scholar]
- 20.Winterhalter WE, Fedorka KM. 2009. Sex-specific variation in the emphasis, inducibility and timing of the post-mating immune response in Drosophila melanogaster. Proc. R. Soc. B 276, 1109–1117. ( 10.1098/rspb.2008.1559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krebs RA, Loeschcke V. 1997. Estimating heritability in a threshold trait: heat-shock tolerance in Drosophila buzzatii. Heredity 79, 252–259. ( 10.1038/hdy.1997.152) [DOI] [PubMed] [Google Scholar]
- 22.Apidianakis Y, Rahme LG. 2009. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 4, 1285–1294. ( 10.1038/nprot.2009.124) [DOI] [PubMed] [Google Scholar]
- 23.Sokal RR, Rohlf FJ. 2012. Biometry, 4th edn New York, NY: W. H. Freeman and Company. [Google Scholar]
- 24.Via S. 1984. The quantitative genetics of polyphagy in an insect herbivore. II. Genetic correlations in larval performance within and among host plants . Evolution 38, 896–905. ( 10.2307/2408399) [DOI] [PubMed] [Google Scholar]
- 25.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates Inc. [Google Scholar]
- 26.Krebs RA, Feder ME, Lee J. 1998. Heritability of expression of the 70KD heat-shock protein in Drosophila melanogaster and its relevance to the evolution of thermotolerance. Evolution 52, 841–847. ( 10.2307/2411278) [DOI] [PubMed] [Google Scholar]
- 27.Kvitek DJ, Sherlock G. 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7, e1002056 ( 10.1371/journal.pgen.1002056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poelwijk FJ, Tanase-Nicola S, Kiviet DJ, Tans SJ. 2011. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 272, 141–144. ( 10.1016/j.jtbi.2010.12.015) [DOI] [PubMed] [Google Scholar]
- 29.Roff D. 1997. Evolutionary quantitative genetics. New York, NY: Chapman and Hall. [Google Scholar]
- 30.Cheverud JM, Routman EJ. 1995. Epistasis and its contribution to genetic variance-components. Genetics 139, 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chippindale AK, Rice WR. 2001. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 98, 5677–5682. ( 10.1073/pnas.101456898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark AG. 1987. Natural selection and Y-linked polymorphism. Genetics 115, 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branco AT, Tao Y, Hartl DL, Lemos B. 2013. Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans. Heredity 111, 8–15. ( 10.1038/hdy.2013.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kharchenko PV, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485. ( 10.1038/nature09725) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.2kr90


