Abstract
Using a system of interspecies hybrids, trihybrids, and recombinants with varying proportions of genomes from three distinct Xenopus species, we provide evidence for de novo epigenetic silencing of paternal 45S ribosomal ribonucleic acid (rRNA) genes and their species-dependent expression dominance that escapes transcriptional inactivation after homologous recombination. The same pattern of imprinting is maintained in the offspring from mothers being genetic males (ZZ) sex-reversed to females, indicating that maternal control of ribosomal deoxyribonucleic acid (rDNA) expression is not sex-chromosome linked. Nucleolar dominance (nucleolus underdevelopment) in Xenopus hybrids appears to be associated with a major non-Mendelian reduction in the number of 45S rDNA gene copies rather than a specific pattern of their expression. The loss of rRNA gene copies in F1 hybrids was non-random with respect to the parental species, with the transcriptionally dominant variant preferentially removed from hybrid zygotes. This dramatic disruption in the structure and function of 45S rDNA impacts transcriptome patterns of small nucleolar RNAs and messenger RNAs, with genes from the ribosome and oxidative stress pathways being among the most affected. Unorthodoxies of rDNA inheritance and expression may be interpreted as hallmarks of genetic conflicts between parental genomes, as well as defensive epigenetic mechanisms employed to restore genome integrity.
Keywords: nucleolar dominance, Xenopus, 18S ribosomal RNA (rRNA), interspecies hybrids, genetic conflicts
1. Introduction
Ribosomal DNA (rDNA) encoding rRNA, with its roots descending from the RNA world, is the most ancient part of all known genomes on the Earth, which provides unique insights into the origins of genome architecture, as well as metabolic features of life at the root of the evolutionary tree [1]. To date, however, rRNA-determining sequences and the associated chromatin remain ‘dark matter’ of eukaryotic genomes due to their exceptionally excessive and repetitive nature, hindering efficient characterization and experimentation. The repetitive nature of rDNA was discovered more than three decades ago when it was first isolated from Xenopus and found to contain 400–500 repeat units organized in tandem [2–4]. Each 45S unit consists of 18S, 5.8S, and 28S rRNA genes and spacers oriented mostly in a head-to-tail configuration. Cytogenetic and in situ hybridization data showed that rDNA arrays map either to a single (e.g. Xenopus) or multiple (e.g. mammals) chromosomal pairs and form nucleolar organizer regions (NORs) [3,5,6]. The nucleolus is a prominent, highly autonomous non-membrane nuclear compartment that supports protein synthesis machinery by actively transcribing genes for rRNAs, processing rRNAs, assembling ribosomal subunits, as well as modifying and transporting ribonucleoproteins [7–12]. Although rDNA segregation is completely dependent on the chromosome transmission mechanism, nucleolar autonomy can be perceived as almost a symbiotic mini-life form with its own minigenome, origin of replication [13,14], replication fork block [15,16], and an agglomerate of highly specialized proteins, including 45S rRNA-specific Pol I transcription machinery [17]. As one example, the nucleolus in fission and budding yeast avoids disassembly during mitotic and meiotic division and recruits new protein complexes for the heterochromatization and segregation of rDNA, which results in delayed separation in anaphase relative to other genomic regions [18–22].
In some interspecies hybrids, rRNA genes from only one parental species are transcribed and the number of nucleoli formed per nucleus tends to be halved relative to progenitors, an enigmatic phenomenon known as nucleolar dominance [23]. Unlike X chromosome silencing, which is random with respect to the origin of parental chromosome (except marsupials [24]), nucleolar dominance is progenitor dependent, with rRNA genes from one parental species consistently dominant over the other [25–27]. For example, a hierarchy of dominance has been observed in reciprocal crosses between allotetraploid Brassica species, in which B. nigra dominates over B. rapa and B. oleracea, and B. rapa dominates only over B. oleracea [23]. Similar species-dependent nucleolar dominance has been believed to occur in a palaeo-allotetraploid amphibian system, Xenopus frogs, with parental X. laevis, X. muelleri, and X. borealis having two nucleoli per cell, but their F1 hybrids developing only one nucleolus under X. laevis dominance (figure 1) [25–27]. Nucleolar dominance was mimicked in Xenopus using X. laevis and X. borealis minigenes with intergenic rRNA spacers co-injected into oocytes, resulting in suppression of transcription from X. borealis minigenes [28]. This has led to the hypothesis of enhancer imbalance postulating that either intergenic enhancer repeats alone [28] or enhancer repeats and spacer promoters confer X. laevis dominance [29,30]. However, we have recently re-examined the patterns of 45S rRNA transcription in F1 Xenopus hybrids and found it to be under maternal rather than species control [31]. Here, we dissect nucleolar dominance with respect to rRNA expression, rDNA inheritance, as well as global responses at the transcriptome level in a variety of hybrid recombinants among three Xenopus species. Together, these observations shed light on the genetic basis of nucleolar dominance and its genome-wide impacts.
Figure 1.
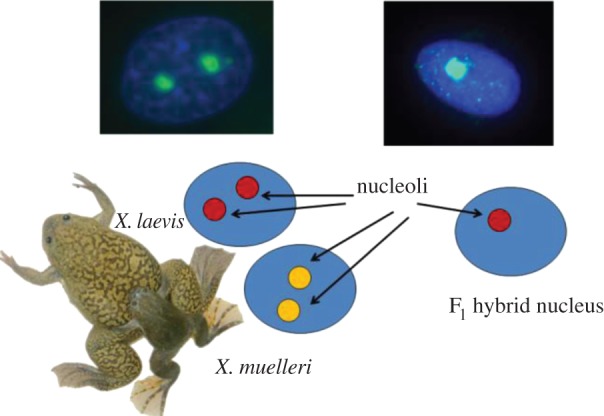
A pictorial representation of nucleolar dominance in Xenopus.
2. Material and methods
See electronic supplementary material, S1.
3. Results
(a). Number of nucleoli and 45S rDNA expression patterns
Unlike parental species that have two nucleoli per nucleus forming from two homologous NORs, F1 hybrid, backcross, and trihybrid frogs tend to develop only one nucleolus per nucleus, regardless of tissue type or developmental stage, and in spite of the presence of both homologous NORs (table 1 and figure 2; electronic supplementary material, figure S1). Based on next generation sequencing (Ion Torrent, Life Technologies) of transcriptomes, as well as allele-specific pyrosequencing and droplet digital polymerase chain reaction (PCR) assays, we demonstrate that F1 hybrids are characterized by the predominantly (91–97%) maternal expression of 45S rDNA in all stages and tissues (table 1; see also [31]). Similarly, all backcrosses with grandmaternal X. laevis, including those from sex-reversed mothers F1(L × M) being genetic males (having ZZ sex chromosomes), resulted in expression patterns consistent with maternal dominance (table 1). However, backcross offspring with grandmaternal X. borealis (i.e. crosses F1(B × L)♀ × L♂ and F1(B × L)♀ × B♂) exhibited X. laevis (grandpaternal) dominance in spite of their mothers having X. borealis expression (table 1). Trihybrids from the cross (L × M)♀ × B♂ had X. laevis (maternal) expression.
Table 1.
The relationship between the number of nucleoli, 45S rRNA gene copies, parental genome, and expression proportions (SR, sex-reversed).
| mother | father | nucleoli/cell | gene copy no. ± s.e. | L rDNA % ± s.e. | M rDNA ± s.e. | B rDNA % ± s.e. | L rRNA % ± s.e. | M rRNA % ± s.e. | B rRNA % ± s.e. |
|---|---|---|---|---|---|---|---|---|---|
| X. laevis (L) | X. laevis (L) | 2 | 761 ± 7 | 100 | 0 | 0 | 100 | 0 | 0 |
| X. muelleri (M) | X. muelleri (M) | 2 | 486 ± 8 | 0 | 100 | 0 | 0 | 100 | 0 |
| X. borealis (B) | X. borealis (B) | 2 | 686 ± 13 | 0 | 0 | 100 | 0 | 0 | 100 |
| X. laevis (L) | X. muelleri (M) | 1 | 335 ± 4 | 20 ± 0.04 | 80 ± 0.04 | 0 | 96 ± 0.01 | 4 ± 0.01 | 0 |
| X. muelleri (M) | X. laevis (L) | 1 | 334 ± 36 | 23 ± 0.10 | 77 ± 0.10 | 0 | 4 ± 0.00 | 96 ± 0.00 | 0 |
| X. laevis (L) | X. borealis (B) | 1 | 418 ± 5 | 10 ± 0.01 | 0 | 90 ± 0.01 | 97 ± 0.19 | 0 | 3 ± 0.68 |
| X. borealis (B) | X. laevis (L) | 1 | 218 ± 8 | 10 ± 0.02 | 0 | 90 ± 0.02 | 9 ± 0.01 | 0 | 91 ± 0.01 |
| F1(L × M) | X. laevis (L) | 1 | 427 ± 0 | 46 ± 2.40 | 54 ± 2.40 | 0 | 98 ± 0.600 | 2 ± 0.600 | 0 |
| F1(L × M) | X. muelleri (M) | 1 | 313 ± 5 | 25 ± 1.07 | 75 ± 1.07 | 0 | 83 ± 0.564 | 17 ± 0.564 | 0 |
| SR F1(L × M) | X. muelleri (M) | 1 | 400 ± 21 | 19 ± 1.80 | 81 ± 1.80 | 0 | 77 ± 2.470 | 23 ± 2.470 | 0 |
| F1(L × B) | X. laevis (L) | 1 | 306 ± 36 | 59 ± 2.40 | 0 | 41 ± 2.400 | 100 ± 0.010 | 0 | 0 ± 0.010 |
| F1(L × B) | X. borealis (B) | 1 | 356 ± 16 | 18 ± 1.40 | 0 | 82 ± 1.400 | 97 ± 0.541 | 0 | 3 ± 0.541 |
| F1(B × L) | X. laevis (L) | 1 | 240 ± 23 | 84 ± 2.60 | 0 | 16 ± 2.600 | 98 ± 0.100 | 0 | 2 ± 0.100 |
| F1(B × L) | X. borealis (B) | 1 | 260 ± 18 | 13 ± 4.40 | 0 | 87 ± 4.400 | 56 ± 0.560 | 0 | 44 ± 0.560 |
| F1(L × M) | X. borealis (B) | 1 | 355 ± 14 | 3 ± 0.70 | 5 ± 0.700 | 92 ± 0.300 | 91 ± 1.800 | 3 ± 1.000 | 6 ± 1.000 |
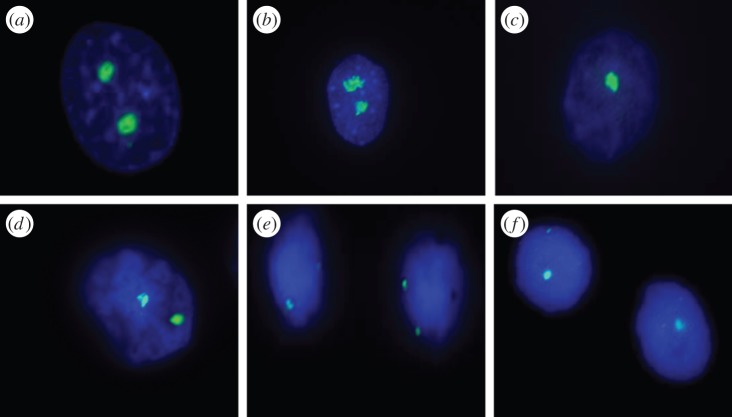
Figure 2.
Fluorescent imaging of nucleoli and NORs in Xenopus kidney cells. Anti-nucleolin staining: (a) Xenopus laevis, (b) X. borealis, and (c) F1(X. laevis × X. borealis). 18S rDNA FISH assays: (d) X. laevis, (e) X. borealis, and (f) F1(X. laevis × X. borealis).
(b). 45S rDNA copy numbers and parental genomic proportions
F1 hybrids had between 46–55% and 43–70% fewer 45S rRNA genes relative to the mid-parent value in reciprocal crosses that combined X. laevis and X. muelleri, and X. laevis and X. borealis genomes, respectively (table 1). A similar pattern was observed in backcross hybrids that tended to lose 20–27% and 38–49% of rRNA genes relative to the mid-parent value in crosses combining X. laevis and X. muelleri, and X. laevis and X. borealis genomes, respectively. Assuming Mendelian ratio expectations, X. laevis rRNA genes were 20–42% underrepresented in F1 hybrid zygotes (table 1).
(c). Expression profiling of messenger RNAs and small nucleolar RNAs in Xenopus laevis, Xenopus muelleri, and their hybrid F1
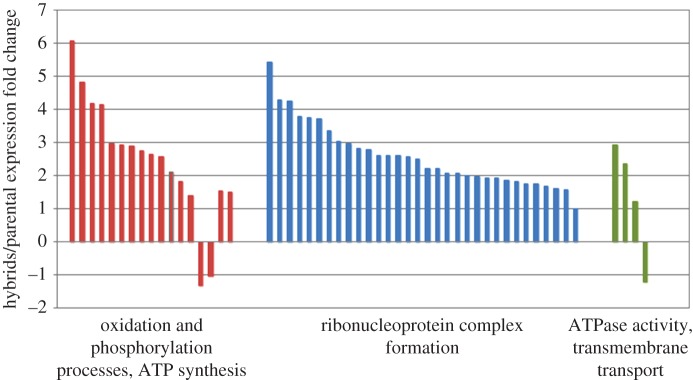
We used Illumina (HiSeq) RNA-sequencing of X. laevis, X. muelleri, and their hybrid F1 to assess transcriptome-wide changes associated with observed rDNA and rRNA unorthodoxies (electronic supplementary material, figure S2). The largest fraction (18–31%) of the 116 and 279 differentially expressed genes (false discovery rate (FDR p < 0.05) in, respectively, X. laevis × X. muelleri, and X. muelleri × X. laevis hybrids relative to parental species belonged to Gene Ontology (GO) categories that were directly related to ribosome structure, rRNA binding, and the polysome (electronic supplementary material, tables S1–S3). Ribosome-related categories were the only significantly over-represented GO terms at the FDR p < 0.05 level. Ribosomal protein genes (e.g. rplp1, rplp2, rplp3, rplp11, rps3a-a, rps15, and rpl28-a) were among the most overexpressed (fivefold to 107-fold) genes in hybrids, presumably due to overcompensation for rDNA depletion and rDNA-related incompatibilities. However, ATP5A1, a nuclear gene encoding a subunit of mitochondrial ATP-synthase, was the most significantly anomalously expressed gene (FDR p < 2.3 × 10−21) in X. laevis × X. muelleri hybrids (electronic supplementary material, table S1). Similar to ribonucleoprotein complex genes, ATP5A1 and COX7A2, genes from the oxidative phosphorylation pathway, were both highly upregulated in hybrid genomes. A number of other genes associated with oxidative stress were also upregulated in hybrids, including three ATPases, ALDH1L1, AASS, and LOX (figure 3; electronic supplementary material, table S1). Overall, very few genes had maternally biased transcript frequencies (adam10, cwf19l2, myrf, and pacsin3; however, one gene, ppp1r14c, was paternally biased).
Figure 3.
Relative changes in expression in functional groups over-represented among genes with significantly differential expression (FDR p < 0.05). Each bar represents a single gene.
Based on SOLiD (ABI) sequencing of small RNAs, we found that two small nucleolar RNAs (snoRNAs), SNORD22 and SNORD96, were consistently differentially expressed in F1 hybrids relative to their parental X. laevis and X. muelleri. Three additional snoRNAs (SNORD49, snoR38, and SNORA73) were downregulated; with only one upregulated in F1 hybrids (SNORD29) relative to parental X. laevis (electronic supplementary material, table S4). At least two of the snoRNAs are derived from introns of the U22 host gene whose primary role seems to be transcriptional production of snoRNA [32]. No snoRNAs were subject to parental imprinting with the possible exception of snosnR60_Z15, expression of which was consistent with paternal (or X. muelleri) imprinting.
Together, these results indicate that nucleolar dominance is not only associated with a distinct rDNA expression pattern, but also with rDNA hereditary instability, having profound impacts on the rest of transcriptome. As discussed below, Xenopus provide a unique model system to investigate the relationship between these phenomena in animals.
4. Discussion
(a). Number of nucleoli and 45S rDNA expression patterns
In agreement with earlier studies [25–27], we show that X. laevis, X. muelleri, and X. borealis typically develop two nucleoli per nucleus, whereas F1 hybrids among the species tend to suppress one of the nucleoli, a pattern interpreted as nucleolar dominance (figures 1 and 2; and electronic supplementary material, figure S1). However, the early reports have incorrectly linked this phenomenon to species-specific genome dominance, claiming that only X. laevis 45S rRNA gene copies are transcribed, regardless of the cross direction. We have established that 45S rRNA (but not 5S rRNA) in F1 hybrids is under genomic imprinting, and maternal copies are predominantly expressed in all stages and tissues [31]. Interestingly, parental genome imprinting was believed to have evolved in response to parental conflicts involving embryonic growth; and therefore are absent from amphibians, animals devoid of intimate parent–offspring interactions during the embryo development [33]. We thus ask if this dramatic epigenetic phenomenon, likely affecting more than 50% of all transcriptional output, is a transient feature limited to F1 hybrids or a more general, although overlooked, trait of Xenopus chromatin. To address the question, we backcrossed F1 hybrid females (F1 hybrid males are sterile [34,35]) to all three parental species. Similar to F1 hybrids, backcross hybrids also develop only one nucleolus (electronic supplementary material, figure S1).
We were surprised to observe that in offspring from a backcross of F1(X. borealis ♀ × X. laevis ♂) ♀ × X. laevis the expression pattern completely shifted to X. laevis dominance (table 1), despite the fact that their F1 mother had X. borealis expression. Even in a backcross between both parents having X. borealis expression, i.e. F1(X. borealis ♀ × X. laevis ♂) ♀ × X. borealis ♂, offspring showed a slight bias towards X. laevis expression (56%) that originated from grandpaternal genomic contribution. Unlike paternal copies, grandpaternal X. laevis copies appear to escape silencing because they segregate through maternal chromosomes. Since all other backcrosses also predominantly expressed X. laevis copies regardless of the paternal species, we conclude that (i) although paternal genes are silenced, (ii) once maternal NOR contains genes from X. laevis, these genes transcriptionally dominate over genes from the other species, even if X. laevis is in the minority (table 1), and (iii) nucleolar dominance, or the number of expressed nucleoli, is uncoupled from gene expression patterns and genomic imprinting.
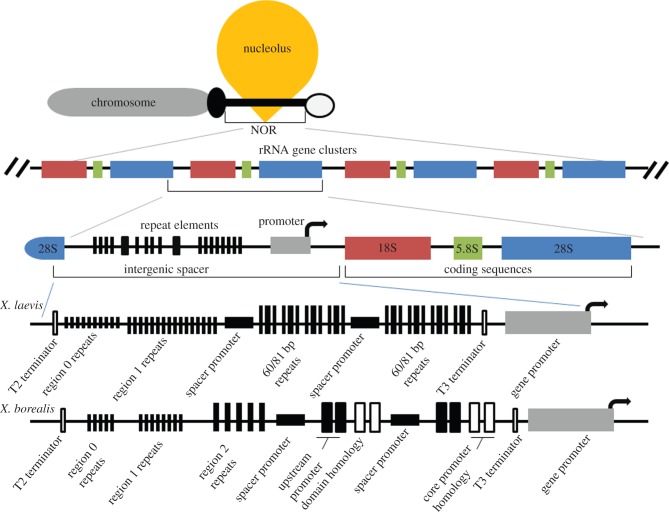
The expression patterns are consistent with de novo silencing of 45S rDNA from the sperm pronucleus within the maternal cytoplasm, which may be an ancient epigenetic feature of amphibian chromatin. The Xenopus sperm nucleus is rapidly reprogrammed by the egg cytoplasm following fertilization to form the paternal pronucleus, and both egg and sperm pronuclei start very intense DNA replication within 20 min of fertilization [36,37]. The process takes place in an environment extremely enriched for maternal rRNAs, associated small RNAs, and proteins, as amphibian oocytes undergo rDNA amplification leading to 500–2 500 extrachromosomal nucleoli containing at least 2 million rRNA gene copies [38]. This transcriptional repression of paternal genes does not prevent meiotic cross-overs between homologous clusters in the F1 germline, consistent with the fact that recombination has been long known to play a critical role in the stabilization of 45S rDNA repeats [39]. Once recombined into maternal chromosomes, X. laevis 45S rDNA alleles transcriptionally outcompete X. muelleri and X. borealis alleles in backcross hybrid genomes. Although 18S, 5.8S, and 28S rDNA sequences are well conserved among taxa, intergenic spacer regions (IGSs) are highly variable among Xenopus species [40]. Unlike X. laevis, which has about 10 60 or 81 bp repeats between the gene promoter and the nearest IGS promoter, X. borealis and X. muelleri have only two of these elements in the analogous location (figure 4) [41,42]. These repeats contain 42 bp enhancer elements in all three species, but because X. laevis has more than five times more of them, its alleles transcriptionally dominate in experiments with minigene constructs [28–30] or when combined in backcross (but not F1) hybrids, as demonstrated in this study.
Figure 4.
NOR of Xenopus. An annotated close-up of the X. laevis and X. borealis enhancer regions are shown on the lowest portion (after Caudy & Pikaard [30]).
This species-dependent expression from a maternal chromosome is a confounding factor in backcrosses, as only two alleles are available for the analysis, and it is difficult to determine if the recombinants from backcrosses in fact undergo de novo silencing of paternal genes. To introduce a third allele as a marker for the paternal chromosome, we produced trihybrids by crossing an X. laevis × X. muelleri F1 female with an X. borealis male. Despite the fact that 45S rRNA gene clusters consisted mainly of paternal X. borealis alleles (more than 90%, table 1) in such trihybrids, they still predominantly expressed maternal X. laevis alleles (90%), similar to F1 and backcross hybrids, and consistent with the hypothesis of paternal gene inactivation. However, we still lack direct evidence for the silencing of paternal rDNA in backcrosses or trihybrids, and alternative explanations need to be taken into consideration. For example, additional smaller clusters of 45S rRNA genes on other chromosomes may go undetected, creating a more complex rDNA landscape shifting expression from one direction to another. Indeed, Xenopus are pseudo-polyploids with very complex genomes [43], and even though they have lost 50–75% of all duplicated genes [44], remnants of 45S rRNA clusters may exist in addition to the main NOR.
Female Xenopus frogs have heterogametic sex chromosomes (ZW), whereas males are homogametic (ZZ). We asked a question whether maternal control of the imprinting pattern extends to a cross in which the mother is a genetic male (ZZ) sex-reversed to phenotypic female to resolve if the system is dependent upon sex chromosomes, which are prone to genetic conflicts [45]. To address this question, we used a sex-reversed X. laevis × X. muelleri F1 hybrid male that was a fertile female [35] and backcrossed it to a X. muelleri male. Offspring from such a cross are all male, and their expression pattern of 45S rDNA was predominantly X. laevis (77%), consistent with maternal control of expression observed in other crosses. However, in crosses with sex-reversed mothers, maternal effects are sex-chromosome independent.
(b). 45S rDNA copy numbers and parental genomic proportions
We hypothesized that nucleolus underdevelopment (nucleolar dominance) results from disruptions in hybrid genomes and hereditary instability of rRNA genes, a hypothesis that could unambiguously be tested in frogs because NOR has been relatively well described in this system [3,4,40]. To test this hypothesis, we quantified gene copy variation and discovered that F1 hybrids tended to lose nearly half of all 45S rRNA genes relative to the mid-parent value in reciprocal crosses that combine X. muelleri, and X. laevis and X. laevis and X. borealis genomes, respectively. This dramatic rDNA volume reduction corresponds well with the number of nucleoli being halved in F1 and backcross hybrids relative to parental species.
Interestingly, X. laevis rRNA genes were preferentially excluded from all F1 hybrid zygotes, leading to a consistent 20–42% deficit in this allele compared to Mendelian ratio expectations, although this segregation disadvantage of X. laevis alleles ceased after recombination (table 1). Longer IGSs give X. laevis a transcriptional advantage, but presumably at the cost of compromised stability during replication, recombination, or repair. Ribosomal DNA transcription and replication are not completely independent, as overexpression of the chromatin remodelling complex NoRC not only silences rDNA transcription, reduces the size and the number of nucleoli, but also impairs cell proliferation and resets replication timing from early to late, as observed in mice [46]. In yeast, rDNA recombination and replication are dependent on transcription from a non-coding bidirectional promoter (E-pro) within the rDNA spacer, which stimulates the dissociation of cohesin, a DNA-binding protein complex suppressing sister-chromatid-based changes in the rDNA copy number [39]. Histone deacetylase Sir2 controls both E-pro transcription [39] and replication timing [47]. Uniparental loss of 45S rDNA was also reported in the allotetraploid grass Zingeria trichopoda [48], allotetraploid Tragopogon (Asteraceae) [49], and allopolyploid yeast Pichia sorbitophila [50]. All these observations from hybrid (allopolyploid) genomes suggest that parent-of-origin effects may be a pervasive feature of rDNA chromatin.
Hybrid genomes from interspecies crosses tend to be disrupted by genetic incompatibilities among genes having their own distinct evolutionary histories, brought together in a new epigenomic environment (Bateson–Dobzhansky–Muller effect) [51]. This instability helps uncover the hidden world of selfish genetic elements and understand how illusive the unity of the organism is, being continuously challenged by genomic conflicts [45]. Indeed, rDNA shows many characteristics of ‘selfish’ genetic elements, capable of violating Mendelian rules of fair segregation, and spreading in genomes without contributing to organismal fitness. First, rDNA seems to be highly redundant, as many of the rRNA genes remain heterochromatinized and are never transcribed. In chickens, for example, a mutant genotype with about 160 out of the typical complement of 290 45S rRNA genes (56%) still supports normal development [52]. The phenotypic effect of a new mutation in one of the multiple copies is thus expected to be negligible, and biased gene conversion may play a major role in the fate of new mutations [45]. Second, rDNA is subject to very rapid copy number evolution between species [53–55] and within species [56–58]. It can extrachromosomally circularize [59,60], attract transposable elements [58,61] as well as invade other selfish elements, such as B chromosomes [62–64] and germline-limited DNA [65]. Finally, rRNA gene clusters are recombinational hotspots in cancer [66], and abnormalities in the nucleolar morphology of cancer cells attracted the attention of tumour pathologists as early as the nineteenth century [67,68].
(c). Expression profiling of messenger RNAs and small nucleolar RNAs in Xenopus laevis, Xenopus muelleri, and their hybrid F1
Since snoRNAs are critical for the processing of 5.8S, 18S, and 28S rRNAs [69], a question arises as to how such a large disruption of rRNA expression due to nucleolar dominance affects snoRNA profiles. Other studies have found that some snoRNAs could indeed be subject to parental imprinting, at least in mammals [70]. Although we observed significant alterations in snoRNA expression, genome imprinting does not affect this system of small RNAs. It appears that imprinting the sperm pronucleus does not extend beyond Pol I transcription, as Pol III-transcribed 5S rDNA was not uniparentally silenced [31], nor were genes under Pol II transcriptional control, among which we found very few with maternally biased transcript frequencies. In addition to affecting the pattern of snoRNAs expression, nucleolar dominance and the de novo genome imprinting of 45S rDNA had a significant impact on mRNA levels, especially those from genes related to ribosome structure, rRNA binding, the polysome, and oxidative stress. Mitochondrial-encoded proteins (maternal) and nuclear-encoded proteins (maternal and paternal), therefore, must work in concert when forming functional complexes for efficient oxidative phosphorylation and energy production. Maternal–paternal cytonuclear incompatibilities may thus lead to oxidative shock and severe hybrid dysfunctions [71]. Also, direct interactions between ribosomes and mitochondria via ribosome-sensing receptors on the outer mitochondrial membrane are central to co-translational and post-translational protein transport [72], which may provide a novel link between mitochondrial and ribosome pathways, consistent with the observed patterns of transcriptional disruption.
Nucleolar architecture and rDNA transcription respond to cellular stresses such as UV irradiation, viral infection, and temperature shock [73]. Given the transcriptome signatures, it might be tempting to speculate that maternal rDNA imprinting could be an epigenetic defensive response to mitonuclear incompatibilities and oxidative stress. However, this explanation would be inconsistent with the observed pattern of rDNA expression in backcrosses (i.e. X. laevis dominance), as in one backcrossing direction maternal cytoplasm matches the paternal genome and is expected to reduce mitonuclear conflict, whereas in the reciprocal direction, maternal cytoplasm and paternal genome mismatch and are thus predicted to increase mitonuclear incompatibilities relative to F1 hybrids.
5. Conclusion
Earlier investigations of nucleolar dominance were mainly focused on the mechanistic aspects of rDNA expression control. By extending our explorations to rDNA inheritance patterns, as well as their consequences on global changes in transcriptomes, including snoRNAs, we showed that nucleolar dominance results in a major disruption at both genomic and transcriptional levels. We also discovered maternal control over rDNA expression, which can be an ancient epigenetic mechanism or its relic to keep genomic parasites and genetic conflicts in check. Highly repeatable rDNA not only harbours multiple mobile elements [74] but itself has all the major features of selfish DNA [45], and its stability poses a constant metabolic challenge to cells.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Zalman Vaksman for his help with some of the crosses and cell cultures.
Ethics
All animal work meets the legal requirements relating to animal welfare in the United States of America and the Commonwealth of Virginia (IACUC protocol no. 14–061 (VBI)).
Data accessibility
Sequence data are available via NCBI's SRA, accession no. SRP064976.
Authors' contributions
K.M. participated in the design of the study, conducted research and participated in the analysis of the data and drafting the manuscript. S.M. conducted research and participated in the analysis of the data. Y.B.K., J.H.O., and L.K. participated in the analysis of sequence data. G.S. was responsible for the animal husbandry. H.R.G. helped draft the manuscript. P.M. conceived of the study, coordinated the study, and helped draft the manuscript.
Competing interests
We have no competing interests.
Funding
An internal fund from the Medical Informatics and Systems Division within the Virginia Bioinformatics Institute was used to support the project, and Mobility Plus Program supported S.M.
References
- 1.Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2, a003483 ( 10.1101/cshperspect.a003483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace H, Birnstiel ML. 1966. Ribosomal cistrons and the nucleolar organizer. Biochim. Biophys. Acta 114, 296–310. ( 10.1016/0005-2787(66)90311-X) [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Wensink PC, Jordan E. 1972. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 63, 57–73. ( 10.1016/0022-2836(72)90521-9) [DOI] [PubMed] [Google Scholar]
- 4.Wensink PC, Brown DD. 1971. Denaturation map of the ribosomal DNA of Xenopus laevis. J. Mol. Biol. 60, 235–247. ( 10.1016/0022-2836(71)90290-7) [DOI] [PubMed] [Google Scholar]
- 5.Henderson AS, Warburton D, Atwood KC. 1972. Location of ribosomal DNA in the human chromosome complement. Proc. Natl Acad. Sci. USA 69, 3394–3398. ( 10.1073/pnas.69.11.3394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson AS, Eicher EM, Yu MT, Atwood KC. 1974. The chromosomal location of ribosomal DNA in the mouse. Chromosoma 49, 155–160. ( 10.1007/BF00348887) [DOI] [PubMed] [Google Scholar]
- 7.Carmo-Fonseca M, Mendes-Soares L, Campos I. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107–E112. ( 10.1038/35014078) [DOI] [PubMed] [Google Scholar]
- 8.Olson MO, Dundr M, Szebeni A. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196. ( 10.1016/S0962-8924(00)01738-4) [DOI] [PubMed] [Google Scholar]
- 9.Olson MO, Hingorani K, Szebeni A. 2002. Conventional and nonconventional roles of the nucleolus. Int. Rev. Cytol. 219, 199–266. ( 10.1016/S0074-7696(02)19014-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung AK, Lamond AI. 2003. The dynamics of the nucleolus. Crit. Rev. Eukaryot. Gene Expr. 13, 39–54. ( 10.1615/CritRevEukaryotGeneExpr.v13.i1.40) [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Verdun D. 2006. Nucleolus: from structure to dynamics. Histochem. Cell Biol. 125, 127–137. ( 10.1007/s00418-005-0046-4) [DOI] [PubMed] [Google Scholar]
- 12.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. ( 10.1038/nrm2184) [DOI] [PubMed] [Google Scholar]
- 13.Walmsley RM, Johnston LH, Williamson DH, Oliver SG. 1984. Replicon size of yeast ribosomal DNA. Mol. Gen. Genet. 195, 260–266. ( 10.1007/BF00332757) [DOI] [PubMed] [Google Scholar]
- 14.Muller M, Lucchini R, Sogo JM. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5, 767–777. ( 10.1016/S1097-2765(00)80317-2) [DOI] [PubMed] [Google Scholar]
- 15.Mais C, McStay B, Scheer U. 2002. On the formation of amplified nucleoli during early Xenopus oogenesis. J. Struct. Biol. 140, 214–226. ( 10.1016/S1047-8477(02)00526-9) [DOI] [PubMed] [Google Scholar]
- 16.Wiesendanger B, Lucchini R, Koller T, Sogo JM. 1994. Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res. 22, 5038–5046. ( 10.1093/nar/22.23.5038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung AK, Andersen JS, Mann M, Lamond AI. 2003. Bioinformatic analysis of the nucleolus. Biochem. J. 376, 553–569. ( 10.1042/BJ20031169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strunnikov AV. 2005. A case of selfish nucleolar segregation. Cell Cycle 4, 113–117. ( 10.4161/cc.4.1.1488) [DOI] [PubMed] [Google Scholar]
- 19.Sullivan M, Higuchi T, Katis VL, Uhlmann F. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117, 471–482. ( 10.1016/S0092-8674(04)00415-5) [DOI] [PubMed] [Google Scholar]
- 20.D'Amours D, Stegmeier F, Amon A. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117, 455–469. ( 10.1016/S0092-8674(04)00413-1) [DOI] [PubMed] [Google Scholar]
- 21.Wang BD, Yong-Gonzalez V, Strunnikov AV. 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 3, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira G, Schiebel E. 2004. Cdc14 phosphatase resolves the rDNA segregation delay. Nat. Cell Biol. 6, 473–475. ( 10.1038/ncb0604-473) [DOI] [PubMed] [Google Scholar]
- 23.Chen ZJ, Pikaard CS. 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl Acad. Sci. USA 94, 3442–3447. ( 10.1073/pnas.94.7.3442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graves JA. 1996. Mammals that break the rules: genetics of marsupials and monotremes. Annu. Rev. Genet. 30, 233–260. ( 10.1146/annurev.genet.30.1.233) [DOI] [PubMed] [Google Scholar]
- 25.Blackler AW, Gecking CA. 1972. Transmission of sex cells of one species through the body of a second species in the genus Xenopus. II. Interspecific matings. Dev. Biol. 27, 385–394. ( 10.1016/0012-1606(72)90177-7) [DOI] [PubMed] [Google Scholar]
- 26.Cassidy DM, Blackler AW. 1974. Repression of nucleolar organizer activity in an interspecific hybrid of the genus Xenopus. Dev. Biol. 41, 84–96. ( 10.1016/0012-1606(74)90285-1) [DOI] [PubMed] [Google Scholar]
- 27.Honjo T, Reeder RH. 1973. Preferential transcription of Xenopus laevis ribosomal RNA in interspecies hybrids between Xenopus laevis and Xenopus mulleri. J. Mol. Biol. 80, 217–228. ( 10.1016/0022-2836(73)90168-X) [DOI] [PubMed] [Google Scholar]
- 28.Reeder RH, Roan JG, Dunaway M. 1983. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell 35, 449–456. ( 10.1016/0092-8674(83)90178-2) [DOI] [PubMed] [Google Scholar]
- 29.Mougey EB, Pape LK, Sollner-Webb B. 1996. Virtually the entire Xenopus laevis rDNA multikilobase intergenic spacer serves to stimulate polymerase I transcription. J. Biol. Chem. 271, 27 138–27 145. ( 10.1074/jbc.271.43.27138) [DOI] [PubMed] [Google Scholar]
- 30.Caudy AA, Pikaard CS. 2002. Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J. Biol. Chem. 277, 31 577–31 584. ( 10.1074/jbc.M202737200) [DOI] [PubMed] [Google Scholar]
- 31.Michalak P. 2014. Evidence for maternal imprinting of 45S ribosomal RNA genes in Xenopus hybrids. Dev. Genes Evol. 224, 125–128. ( 10.1007/s00427-014-0464-1) [DOI] [PubMed] [Google Scholar]
- 32.Tycowski KT, Shu MD, Steitz JA. 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379, 464–466. ( 10.1038/379464a0) [DOI] [PubMed] [Google Scholar]
- 33.Pardo-Manuel de Villena F, de la Casa-Esperon E, Sapienza C. 2000. Natural selection and the function of genome imprinting: beyond the silenced minority. Trends Genet. 16, 573–579. ( 10.1016/S0168-9525(00)02134-X) [DOI] [PubMed] [Google Scholar]
- 34.Malone JH, Chrzanowski TH, Michalak P. 2007. Sterility and gene expression in hybrid males of Xenopus laevis and X. muelleri. PLoS ONE 2, e781 ( 10.1371/journal.pone.0000781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malone JH, Michalak P. 2008. Physiological sex predicts hybrid sterility regardless of genotype. Science 319, 59 ( 10.1126/science.1148231) [DOI] [PubMed] [Google Scholar]
- 36.Poccia D. 1986. Remodeling of nucleoproteins during gametogenesis, fertilization, and early development. Int. Rev. Cytol. 105, 1–65. ( 10.1016/S0074-7696(08)61061-X) [DOI] [PubMed] [Google Scholar]
- 37.Gurdon J. 2009. Nuclear reprogramming in eggs. Nat. Med. 15, 1141–1144. ( 10.1038/nm1009-1141) [DOI] [PubMed] [Google Scholar]
- 38.Thiebaud CH. 1979. Quantitative determination of amplified rDNA and its distribution during oogenesis in Xenopus laevis. Chromosoma 73, 37–44. ( 10.1007/BF00294842) [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Ganley AR. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309, 1581–1584. ( 10.1126/science.1116102) [DOI] [PubMed] [Google Scholar]
- 40.Furlong JC, Maden BE. 1983. Patterns of major divergence between the internal transcribed spacers of ribosomal DNA in Xenopus borealis and Xenopus laevis, and of minimal divergence within ribosomal coding regions. EMBO J. 2, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bach R, Allet B, Crippa M. 1981. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 9, 5311–5330. ( 10.1093/nar/9.20.5311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labhart P, Reeder RH. 1987. DNA sequences for typical ribosomal gene spacers from Xenopus laevis and Xenopus borealis. Nucleic Acids Res. 15, 3623–3624. ( 10.1093/nar/15.8.3623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uno Y, Nishida C, Takagi C, Ueno N, Matsuda Y. 2013. Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Heredity 111, 430–436. ( 10.1038/hdy.2013.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. 2007. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 5, 31 ( 10.1186/1741-7007-5-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burt A, Trivers R. 2006. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Belknap Press of Harvard University Press. [Google Scholar]
- 46.Li J, Santoro R, Koberna K, Grummt I. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 24, 120–127. ( 10.1038/sj.emboj.7600492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida K, et al. 2014. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol. Cell 54, 691–697. ( 10.1016/j.molcel.2014.04.032) [DOI] [PubMed] [Google Scholar]
- 48.Kotseruba V, Gernand D, Meister A, Houben A. 2003. Uniparental loss of ribosomal DNA in the allotetraploid grass Zingeria trichopoda (2n=8). Genome 46, 156–163. ( 10.1139/g02-104) [DOI] [PubMed] [Google Scholar]
- 49.Malinska H, Tate JA, Matyasek R, Leitch AR, Soltis DE, Soltis PS, Kovarik A. 2010. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol. Biol. 10, 291 ( 10.1186/1471-2148-10-291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Louis VL, et al. 2012. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda) 2, 299–311. ( 10.1534/g3.111.000745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 52.Delany ME, Taylor RL Jr, Bloom SE. 1995. Teratogenic development in chicken embryos associated with a major deletion in the rRNA gene cluster. Dev. Growth Differ. 37, 403–412. ( 10.1046/j.1440-169X.1995.t01-3-00007.x) [DOI] [PubMed] [Google Scholar]
- 53.Maleszka R, Clark-Walker GD. 1993. Yeasts have a four-fold variation in ribosomal DNA copy number. Yeast 9, 53–58. ( 10.1002/yea.320090107) [DOI] [PubMed] [Google Scholar]
- 54.Keller I, Chintauan-Marquier IC, Veltsos P, Nichols RA. 2006. Ribosomal DNA in the grasshopper Podisma pedestris: escape from concerted evolution. Genetics 174, 863–874. ( 10.1534/genetics.106.061341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar A, Rai KS. 1990. Chromosomal localization and copy number of 18S+28S ribosomal RNA genes in evolutionarily diverse mosquitoes (Diptera, Culicidae). Hereditas 113, 277–289. ( 10.1111/j.1601-5223.1990.tb00094.x) [DOI] [PubMed] [Google Scholar]
- 56.Delany ME, Muscarella DE, Bloom SE. 1991. Formation of nucleolar polymorphisms in trisomic chickens and subsequent microevolution of rRNA gene clusters in diploids. J. Hered. 82, 213–220. [DOI] [PubMed] [Google Scholar]
- 57.Stults DM, Killen MW, Pierce HH, Pierce AJ. 2008. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 18, 13–18. ( 10.1101/gr.6858507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eagle SH, Crease TJ. 2012. Copy number variation of ribosomal DNA and Pokey transposons in natural populations of Daphnia. Mob. DNA 3, 4 ( 10.1186/1759-8753-3-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng JC, Karpen GH. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9, 25–35. ( 10.1038/ncb1514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Cavalier-Smith T, Green BR. 2001. A family of selfish minicircular chromosomes with jumbled chloroplast gene fragments from a dinoflagellate. Mol. Biol. Evol. 18, 1558–1565. ( 10.1093/oxfordjournals.molbev.a003942) [DOI] [PubMed] [Google Scholar]
- 61.Eickbush MT, Eickbush TH. 2011. Retrotransposition of R2 elements in somatic nuclei during the early development of Drosophila. Mob. DNA 2, 11 ( 10.1186/1759-8753-2-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maluszynska J, Schweizer D. 1989. Ribosomal RNA genes in B chromosomes of Crepis capillaris detected by non-radioactive in situ hybridization. Heredity 62(Pt 1), 59–65. ( 10.1038/hdy.1989.8) [DOI] [PubMed] [Google Scholar]
- 63.Donald TM, Leach CR, Clough A, Timmis JN. 1995. Ribosomal RNA genes and the B chromosome of Brachycome dichromosomatica. Heredity 74(Pt 5), 556–561. ( 10.1038/hdy.1995.77) [DOI] [PubMed] [Google Scholar]
- 64.Ruiz-Estevez M, Lopez-Leon MD, Cabrero J, Camacho JP. 2012. B-chromosome ribosomal DNA is functional in the grasshopper Eyprepocnemis plorans. PLoS ONE 7, e36600 ( 10.1371/journal.pone.0036600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Etter A, Bernard V, Kenzelmann M, Tobler H, Muller F. 1994. Ribosomal heterogeneity from chromatin diminution in Ascaris lumbricoides. Science 265, 954–956. ( 10.1126/science.8052853) [DOI] [PubMed] [Google Scholar]
- 66.Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. 2009. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 69, 9096–9104. ( 10.1158/0008-5472.CAN-09-2680) [DOI] [PubMed] [Google Scholar]
- 67.Maggi LB Jr, Weber JD. 2005. Nucleolar adaptation in human cancer. Cancer Invest. 23, 599–608. ( 10.1080/07357900500283085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montanaro L, Trere D, Derenzini M. 2008. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 173, 301–310. ( 10.2353/ajpath.2008.070752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maxwell ES, Fournier MJ. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64, 897–934. ( 10.1146/annurev.bi.64.070195.004341) [DOI] [PubMed] [Google Scholar]
- 70.Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP. 2002. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 11, 1527–1538. ( 10.1093/hmg/11.13.1527) [DOI] [PubMed] [Google Scholar]
- 71.Burton RS, Barreto FS. 2012. A disproportionate role for mtDNA in Dobzhansky–Muller incompatibilities? Mol. Ecol. 21, 4942–4957. ( 10.1111/mec.12006) [DOI] [PubMed] [Google Scholar]
- 72.MacKenzie JA, Payne RM. 2004. Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J. Biol. Chem. 279, 9803–9810. ( 10.1074/jbc.M307167200) [DOI] [PubMed] [Google Scholar]
- 73.Stimpson KM, Sullivan LL, Kuo ME, Sullivan BA. 2014. Nucleolar organization, ribosomal DNA array stability, and acrocentric chromosome integrity are linked to telomere function. PLoS ONE 9, e92432 ( 10.1371/journal.pone.0092432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou J, Eickbush TH. 2009. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genet. 5, e1000386 ( 10.1371/journal.pgen.1000386) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available via NCBI's SRA, accession no. SRP064976.